Abstract
With increasing number of novel bacteria being isolated from the human gut ecosystem, there is a greater need to study their role in the gut ecosystem and their effect on the host health. In the present study, we carried out in silico genome-wide analysis of two novel Megasphaera sp. isolates NM10 (DSM25563) and BL7 (DSM25562), isolated from feces of two healthy individuals and validated the key features by in vitro studies. The analysis revealed the general metabolic potential, adaptive features and the potential effects of these isolates on the host. The comparative genome analysis of the two human gut isolates NM10 and BL7 with ruminal isolate Megasphaera elsdenii (DSM20460) highlighted the differential adaptive features for their survival in human gut. The key findings include features like bile resistance, presence of various sensory and regulatory systems, stress response systems, membrane transporters and resistance to antibiotics. Comparison of the “glycobiome” based on the genomes of the ruminal isolate with the human gut isolates NM10 and BL revealed the presence of diverse and unique sets of Carbohydrate-Active enzymes (CAZymes) amongst these isolates, with a higher collection of CAZymes in the human gut isolates. This could be attributed to the difference in host diet and thereby the environment, consequently suggesting host specific adaptation in these isolates. In silico analysis of metabolic potential predicted the ability of these isolates to produce important metabolites like short chain fatty acids (butyrate, acetate, formate, and caproate), vitamins and essential amino acids, which was further validated by in vitro experiments. The ability of these isolates to produce important metabolites advocates for a potential healthy influence on the host. Further in vivo studies including transcriptomic and proteomic analysis will be required for better understanding the role and impact of these Megasphaera sp. isolates NM10 and BL7 on the human host.
Introduction
The human gut microbiome is a complex ecological niche and the interaction of this microbiome with its host is an important factor contributing towards the health status of the host [1]–[3]. Studies based on 16S rRNA gene amplicon sequencing, using next generation sequencing technologies have successfully established the relationship of the human gut microbiome with the health and disease conditions of the host [3], [4]. The representation of various bacteria through 16S rRNA gene does give insights into ‘who are present?’ but the question ‘who does what?’ remains obscure. Recent efforts have been directed towards exploring the gene content of the human microbiome using shotgun metagenomics. These studies have helped in unraveling the complex gene repertoire, which exists within the human gut. Genes coding for central metabolic pathways, production of amino acids, biosynthesis of vitamins and cofactors, degradation of xenobiotic compounds, etc., are reported to be the major genes in this complex gene repertoire [5]. In addition, efforts have been made to sequence genomes of all available isolates of human origin, which are expected to be between 1000 and 1,150 bacterial species [6]. Genomic studies give an opportunity to unravel the underlying genetic potential of the bacteria to encode a given protein and help in assigning putative adaptive features as well as functional role for a particular bacterium in an ecosystem.
The human gut microbiota is dominated by phylum Firmicutes and Bacteroidetes [3], [4]. Genus Megasphaera is a member of the phylum Firmicutes, it belongs to the class Negativicutes and comprises of Gram-negative coccoid shaped obligate anaerobic bacteria. This genus till date includes 5 validly published species (M. cerevisiae, M. elsdenii, M. micronuciformis, M. paucivorans and M. sueciensis) that have been isolated from various sources such as rumen, spoiled beer, human clinical specimens [7]–[9]. Studies on Megasphaera sp. from the rumen have suggested that it is an important member of the rumen microbiome, having beneficial effects on the host [10]. On the other hand, there are no studies reporting the role of Megasphaera sp. in the human gut.
As a part of our larger culturomics study on the Indian gut microbiota, we have isolated two potential novel bacteria belonging to the genus Megasphaera (isolate NM10 and BL7). In the present study, we carried out genome sequencing of these isolates in order to identify the adaptive features and to determine their gene repertoire. These isolates were the closest phylogenetic neighbors of Megasphaera elsdenii DSM20460, which was previously isolated from rumen. Comparative genome analysis of the genomes of the human gut isolates and the publicly available genome of ruminal isolate revealed the differential adaptive features of Megasphaera sp. NM10 and BL7 that are crucial for the survival in the human gut [11]. In addition, the in silico genome wide analysis and in vitro experiments revealed metabolic traits that suggest a potential beneficial effect of Megasphaera sp. on the human health.
Results and Discussion
Isolates Used in the Study
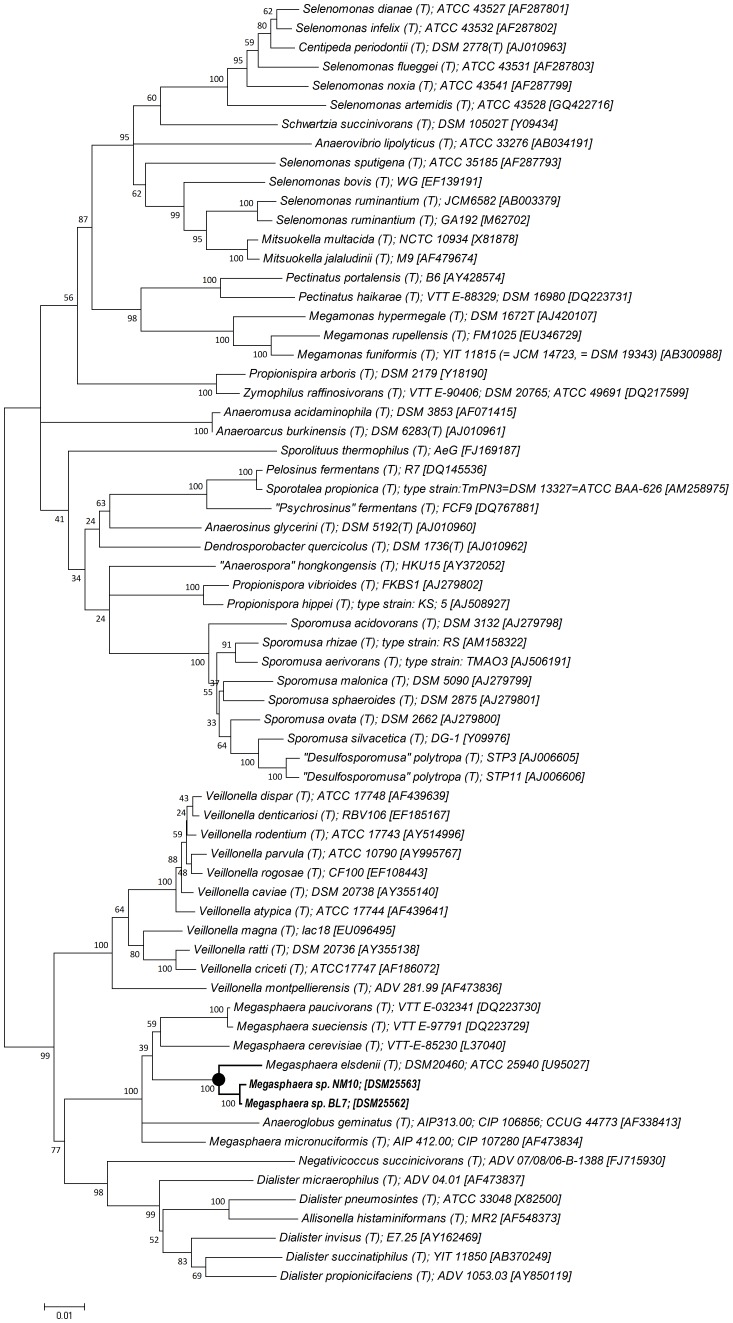
The two isolates of Megasphaera sp. NM10, BL7 were isolated from the feces of two healthy Indian individuals. The strain BL7 was reported in our previous study and the strain NM10 was isolated as a part of our larger culturomics study on gut microbiota of the Indian individuals [12]. Institutional ethical clearance (NCCS, Pune, India) and informed consent was obtained from the individuals before the sampling. Megasphaera sp. isolates NM10 and BL7 are deposited with DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen, Germany) under accession numbers DSM25563 and DSM25562 respectively. The 16S rRNA gene phylogeny revealed that these strains belong to the family Veillonellaceae and showhigh similarity with Megasphaera hominis and M. elsdenii. However, M. hominis is not included in the list of validly published species (http://www.bacterio.net) and is not in the list of prokaryotic names with standing nomenclature. Therefore, we considered the closest validly published type strain, that is, Megasphaera elsdenii (DSM20460) (Figure 1). The 16S rRNA gene sequences of Megasphaera sp. NM10 and BL7 are deposited at GenBank under accession numbers HM990965 and HM990964 respectively. Recently, Megasphaera massiliensis, a novel species belonging to genus Megasphaera was proposed and its genome sequence has been described [13]. The 16S rRNA gene sequences of Megasphaera sp. NM10 and BL7 were 96% similar to 16S rRNA sequence of M. massiliensis, suggesting that these human gut isolates belong to a different species. Hence, the genome of M. massiliensis was not considered for comparative genome analysis. The polyphasic taxonomy suggested that the isolates Megasphaera sp. NM10 and BL7 represent a novel species belonging to genera Megasphaera (data not shown).
Figure 1. Phylogenetic tree of family Veillonellaceae based on 16S rRNA gene.
The phylogenetic tree was constructed in MEGA4 using neighbor-joining method. The bootstrap values (expressed as percentages of 1000 replications) are shown at branch points. The scale bar represents genetic distance (1 substitution per 100 nucleotides). Isolates in present study are in bold.
General Features of Megasphaera sp. NM10 and Megasphaera sp. BL7 Genomes
The draft genomes of Megasphaera sp. NM10 and BL7 are deposited at GenBank under accession numbers APHY00000000 and APHX00000000 respectively. The draft genome of Megasphaera sp. NM10 was 2,615,280 bp and that of Megasphaera sp. BL7 was 2,656,480 bp with a G+C content of approximately 54.3% for both NM10 and BL7. The sequencing coverage obtained for the genomes of NM10 was 74X and for BL7 was 83X. The total consensus length of draft genomes obtained for both of the isolates in this study is larger than the genome sequence available for the type strain M. elsdenii DSM20460 (∼2.47 Mb). The functional annotation of sequence data was performed using RAST server that uses a subsystem-based approach [14]. The number of subsystems identified for NM10 were 297 and the predicted number of open reading frames (ORF’s) were 2436. For BL7, the number of subsystems identified were 293 and the predicted number of open reading frames (ORF’s) were 2432. In addition to these features, phage elements were detected in both the NM10 and BL7 genome sequence assembly. The publicly available genome of M. elsdenii DSM20460 (accession number HE576794) was also reanalyzed using RAST, for having consistency in comparison [11]. The subsystems features predicted by RAST server based on the genome sequences of NM10, BL7 and M. elsdenii DSM20460 are represented in the Table 1.
Table 1. The predicted sub-system features in the genomes of Megasphaera sp. NM10, BL7 and M. elsdenii DSM20460.
| Subsystem Feature | Megasphaerasp. NM10 | Megasphaerasp. BL7 | M. elsdeniiDSM20460 |
| Cofactors, Vitamins, Prosthetic Groups, Pigments | 123 | 123 | 141 |
| Cell Wall and Capsule | 103 | 102 | 108 |
| Virulence, Disease and Defense | 44 | 49 | 45 |
| Potassium metabolism | 15 | 15 | 15 |
| Phages, Prophages, Transposable elements, Plasmids | 14 | 12 | 5 |
| Membrane Transport | 44 | 44 | 35 |
| RNA Metabolism | 110 | 107 | 115 |
| Nucleosides and Nucleotides | 71 | 107 | 75 |
| Protein Metabolism | 162 | 134 | 136 |
| Cell Division and Cell Cycle | 21 | 21 | 21 |
| Regulation and Cell signaling | 8 | 8 | 8 |
| Secondary Metabolism | 6 | 0 | 6 |
| DNA Metabolism | 84 | 79 | 94 |
| Regulons | 3 | 2 | 3 |
| Fatty Acids, Lipids, and Isoprenoids | 43 | 43 | 46 |
| Nitrogen Metabolism | 13 | 14 | 16 |
| Dormancy and Sporulation | 2 | 2 | 2 |
| Respiration | 46 | 46 | 47 |
| Stress Response | 44 | 44 | 43 |
| Metabolism of Aromatic Compounds | 4 | 4 | 4 |
| Amino Acids and Derivatives | 273 | 271 | 273 |
| Phosphorus Metabolism | 33 | 32 | 33 |
| Carbohydrates | 255 | 219 | 201 |
| Miscellaneous | |||
| Niacin-Choline transport and metabolism | 4 | 4 | 3 |
| Phosphoglycerate mutase protein family | 3 | 3 | 2 |
| Muconate lactonizing enzyme family | 1 | 1 | 1 |
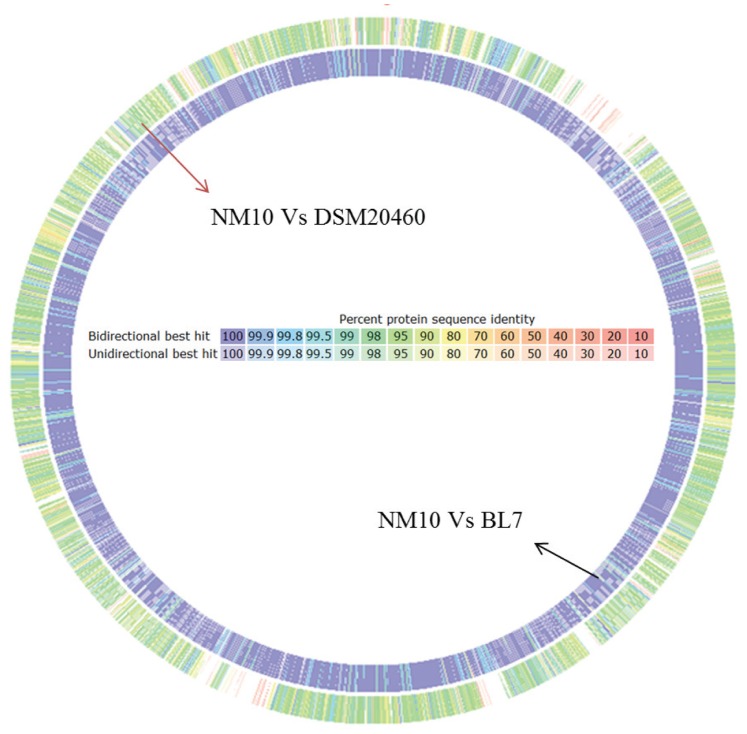
The comparison of protein sequences of all predicted ORF’s in the genomes of NM10, BL7 and M. elsdenii DSM20460 showed that the genomes of human gut isolates NM10 and BL7 are highly similar to each other, with around 2200 proteins sharing more than 99.5% similarity between them (Figure 2 and Table S1). Whereas M. elsdenii genome had low sequence similarity to the genomes of the human gut isolates, with only 252 proteins having more than 99% similarity to the human gut isolates, suggesting that the genomes of human gut isolates differ from ruminal isolate DSM20460. In addition, more than 400 proteins encoded by the genomes of the human gut isolates were not detected in the ruminal isolate (Table S1).
Figure 2. Comparison of protein sequences of Megasphaera sp. NM10, BL7 and M. elsdenii DSM20460.
The color code indicates the percent similarity between the predicted protein sequences.
General Metabolic Potential of Megasphaera sp. NM10 and BL7
The draft metabolic model for the three Megasphaera sp. obtained from Model SEED suggested the potential of these isolates to produce various primary and secondary metabolites. The predicted genes by RAST for NM10, BL7 and DSM20460 are enlisted in Table S1. The Model SEED predicts the presence of a pathway based on the presence of a protein or a single step in the pathway. The genomes under study were unclosed drafts; this would lead to imprecise estimation of the metabolic capabilities of these bacteria. To avoid this inflated estimate of the biological pathways and the metabolic capabilities of the bacteria, we carried out MinPath (Minimal set of Pathways) analysis. The MinPath analysis uses protein family predictions for biological pathway reconstructions; this yields a more conservative and more faithful, estimation of the biological pathways for a query dataset [15]. This assisted in avoiding overestimation of the functional capabilities of the isolates and to have a more conservative and concrete prediction of the metabolic capabilities based on draft genomes. The metabolic functions predicted for all three isolates by MinPath analysis are given in Tables S2, S3 and S4.
Central metabolism
The human gut metagenomic and metatranscriptomic studies have revealed that the genes for central metabolic pathways like carbohydrate metabolism, amino acid metabolism, nucleotide metabolism, etc., are abundantly expressed in the human microbiome [5], [16]–[18]. The general metabolic features of Megasphaera sp. NM10 and BL7 were highly similar to each other and most of the predicted features in the subsystem are shared by these isolates.
Carbohydrate metabolism
The genomes of NM10 and BL7 coded for enzymes essential for carrying out glycolysis and gluconeogenesis. Out of the total 42 SEED families for glycolysis and gluconeogenesis, 11 were present in NM10 and 10 were identified in BL7. The genomes also had genes coding for enzymes of Tricarboxylic acid cycle, Pentose phosphate pathway and Entner-Doudoroff pathway. Out of the 56 SEED families for the Entner-Doudoroff pathway, 14 were represented in NM10 and BL7. Both the isolates were capable of utilizing fructose and had genes coding for 1-phosphofructokinase (EC 2.7.1.56), a PTS system (EC 2.7.1.69), fructose-specific IIA component, fructose-specific IIB component, and fructose-specific IIC component. These isolates were also capable of mixed acid fermentation. Butyryl-CoA dehydrogenase (EC 1.3.99.2) was present in both the genomes; this enzyme is involved in the Acetyl-CoA fermentation to produce butyrate. NM10 and BL7 genomes had higher number of genes involved in carbohydrate metabolism as compared to the ruminal isolate DSM20460 (255 and 219 compared to 201) (Table 1), this may be attributed to the difference in host diet.
Amino acid and nucleotide metabolism
Amino acids are broadly classified as non-essential and essential amino acids. The latter group of amino acids are the ones that the human body cannot synthesize and consequently depends largely on food and the microbiome as its source [19]. The genomes of NM10 and BL7 had 273 and 271 subsystem counts associated with subsystem feature for amino acid metabolism. The MinPath analysis predicted the capability of these isolates for the biosynthesis of essential amino acids like histidine, lysine, methionine, threonine and tryptophan. The synthesis of lysine was predicted via the diaminopimelate (DAP) pathway which leads to the production of lysine from aspartate [20]. The important feature of this pathway is the intermediate diaminopimelate, which is important for peptidoglycan synthesis. In addition to the essential amino acids, the genomes of isolates NM10 and BL7 encode for the biosynthetic pathways for the synthesis of non-essential amino acids such as glycine (biosynthesis from L-threonine by threonine aldolase), arginine (via the arginine biosynthesis extended), glutamine, glutamate, aspartate, asparagine, cysteine (produced via super pathway of cysteine biosynthesis), proline (using glutamate as substrate to obtain proline) and homoserine. The Model Seed predicted the ability for synthesis of all three branched-chain amino acid (leucine, isoleucine and valine) but, the MinPath analysis suggested that these isolates were incapable for the same. The presence of membrane transporters for branched chain amino acids in the genomes of these isolates would aid in acquisition of these branched-chain amino acids.
In case of nucleotide synthesis, the genomes of the isolates NM10 and BL7 had genes involved in De Novo purine and pyrimidine synthesis pathways. The enzymes required to generate 5-phosphoribosyl-1-pyrophosphate i.e. ADP-ribose pyrophosphatase (EC 3.6.1.13), Ribose-phosphate pyrophosphokinase (EC 2.7.6.1) were present in the genomes of NM10 and BL7.
Adaptive Features of Megasphaera sp. NM10 and BL7 for Survival in Human Gut
Mammalian gut is one of the most densely populated ecosystems in which the microbial populations are governed by a dynamic process of selection and competition [21], [22]. Survival in such a challenging ecosystem necessitates the bacteria to constantly adapt and evolve. In the human gut these different adaptive features include presence of carbohydrate degrading genes, resistance to stress conditions, sensing the surroundings, membrane transporters, etc.
‘Glycobiome’
The entire gene repertoire involved in the breakdown of carbohydrates is termed as the ‘glycobiome’ [23]. Studies on the human gut metagenome has revealed the presence of an extensive glycobiome harbored by the gut microbial community [24], [25]. This feature has been studied in most of the bacteria associated with the human gut. The most studied bacterium with the largest reported glycobiome is Bacteroides thetaiotaomicron. This bacterium encodes more than 170 glycosylhydrolases [22]. Similarly, other gut symbionts add up to the glycobiome potential of the gut microbiome.
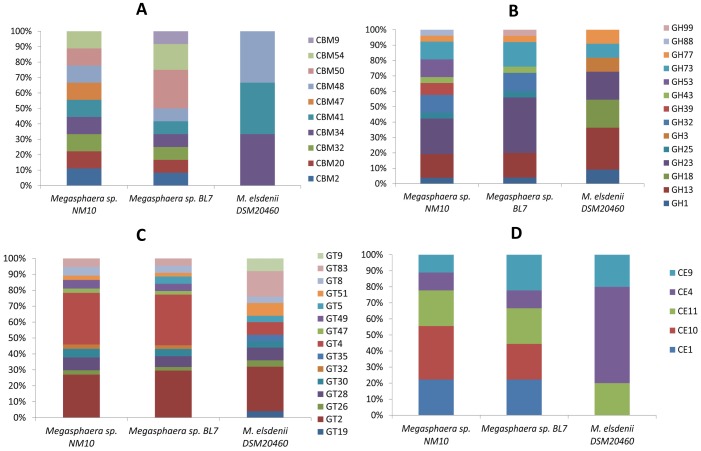
Comparisons of the glycobiome of the Megasphaera sp. NM10 and BL7 of the human fecal origin with that of M. elsdenii DSM20460 of the ruminal origin, showed the presence of a higher repertoire of complex carbohydrate utilizing genes in the human gut isolates (Figure 3). The Polysaccharide Lyase Family (PLs) was not detected in all the three Megasphaera sp. genomes. M. elsdenii is not a primary metabolizer in the rumen, but rather is involved in utilizing the end products of fermentation especially; lactic acid [26]. Megasphaera sp. might play a similar role in the human gut.
Figure 3. The distribution of different CAZyme families in genome of Megasphaera sp. NM10, BL7 and M. elsdenii (DSM20460).
A) Distribution of Carbohydrate-Binding Modules (CBMs). B) Distribution of Glycoside Hydrolases (GHs). C) Distribution of Glycosyl Transferases (GTs), D) Distribution of Carbohydrate Esterases (CEs).
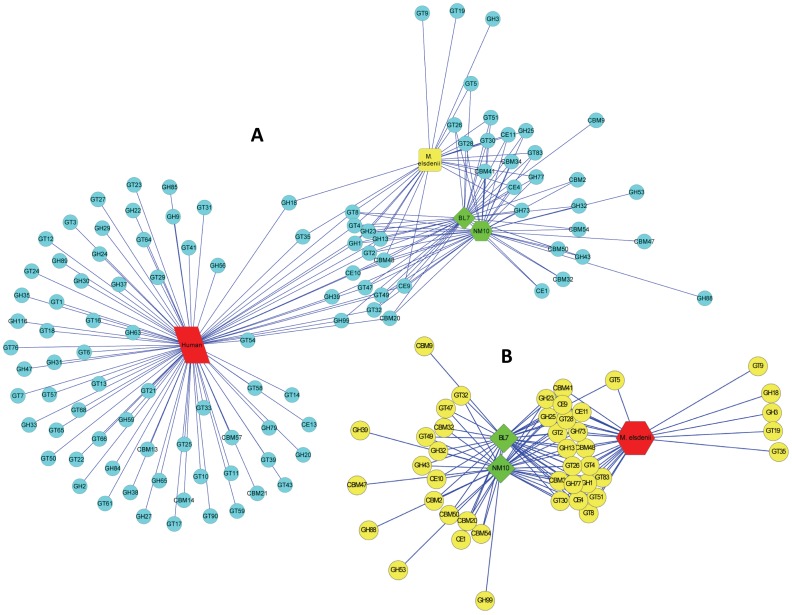
The Carbohydrate-Active EnZymes (CAZymes) of Megasphaera sp. NM10 and BL7 were compared to the CAZymes that are encoded by the human genome (Figure 4A). The human genome encodes 29 glycosyl hydrolases families. However, only a few of these enzymes are involved in the digestion of carbohydrates present in the daily diet. On comparing the glycobiome of the human genome and the ruminal isolate with the human gut isolates, it was observed that the human gut isolates encode enzymes that help fill the enzymatic lacunae required to degrade carbohydrates present in the human diet. These enzymes belong the glycosyl hydrolase (GH) families GH25, GH32, GH43, GH53, GH73 and GH77 (GH88 was detected only in NM10) thereby adding to the potential of the gut microbiome for degrading various carbohydrates. This observation suggests that Megasphaera sp. NM10 and BL7 play a role in utilization of carbohydrates that the host is incapable of degrading. A study by Cantarel et al in 2012, suggested that the gastrointestinal tract has the highest abundance of CAZymes among all the body sites [24]. The CAZy family GH53 (involved in plant cell wall degradation), is among the six over-represented families in the digestive tract. GH53 was present in the genome of isolate NM10. The human gut isolates also carried the genes that encode amino acid sequence having carbohydrate-binding activity. These are classified as carbohydrate-binding module (CBM). The genomes of NM10 and BL7 encoded glycosyl hydrolase GH13 associated with CBM48 and CBM41, which help in binding to starch. In addition, the Megasphaera sp. NM10 and BL7 genomes also encoded amylomaltase that is associated with CBM34, which is a starch binding domain.
Figure 4. The Glycobiome network of Megasphaera sp.
A) The glycobiome network of Human (red), Megasphaera sp. NM10, BL7 (green) and M. elsdenii DSM20460 (yellow). B) The glycobiome network of Megasphaera sp. NM10, BL7 (green) and M. elsdenii DSM20460 (red). CBMs- Carbohydrate-Binding Modules, GHs- Glycoside Hydrolases, GTs- Glycosyl Transferases, CEs- Carbohydrate Esterases. The nodes represent the CAZyme superfamilies and the edges are connecting the nodes based on the presence or absence of respective superfamilies in the organism.
The glycobiome network (represented in Figure 4B) indicated that only a few CAZymes are shared between the rumen and the human gut isolates. The ruminal isolate genome encoded glycosyl hydrolases, GH18 (a conserved domain protein) and GH3 (beta-glucosidase-related glycosidases), glycosyltransferases GT19 (lipid-A-disaccharide synthase), GT9 (lipopolysaccharide heptosyltransferase I) and GT35 (phosphorylase). However, eighteen CAZyme families were observed to be present only in the human gut isolates. These included various glycosyl hydrolases such as GH43, GH32, GH99, GH53, GH88, and GH39 and glycosyltransferases like GT32, GT47 and GT49. This could be attributed to adaptation to the host specific diet. The human diet is diverse as compared to the ruminals. Diet of humans contains different sources of carbohydrates ranging from simple sugars to complex polysaccharides. Consequently, to adapt to these diverse sources of carbohydrates the bacteria in the human gut need to have a higher repertoire of genes involved in carbohydrate degradation. The unique set of CAZymes present in human gut isolates and the diverse CAZyme repertoire compared to ruminal isolate signify a probable host specific evolution and adaptation of the genomes of the isolates of Megasphaera sp. isolated from the human gut.
Bile resistance
One of the selective pressures in the gut is the presence of bile. The bacteria that are sensitive to bile are not capable of survival in the gut. The bile tolerance test suggested that all the three isolates in this study can tolerate 0.3% bile. Bile salt hydrolase (bsh) is the enzyme responsible for bile resistance in many bacteria [27]. The bile salt hydrolase activity was not detected in the human gut isolates as well as M. elsdenii in the in vitro assay, Figure S1. On analysis of the genome of all the three Megasphaera sp., the gene encoding bile salt hydrolase (bsh) was not detected [27]. This is consistent with the absence of bile salt hydrolase activity, in the in vitro analysis, among these isolates. However, bile acid: sodium symporter and bile acid transporters were identified in the genomes of both the human gut isolates. Additionally, several MDR pumps conferring bile resistance along with resistance to drugs were detected in the genomes of NM10 and BL7. These include MDR efflux pump CmeABC which is essential for bile resistance in Campylobacter [28], [29]. The acrAB has been reported to confer bile resistance in Salmonella typhi [30], while TolC efflux pump is reported to confer bile resistance in Vibrio cholerae [31]. Presence of all these efflux pumps, explains the observed bile resistance detected in these isolates.
Oxidative stress
Presence of oxygen and reactive oxygen species is a major stress for obligate anaerobic organisms. The genomes of the isolates NM10 and BL7 had several mechanisms for protection against oxidative stress. These include the presence of glutathione peroxidase and glutathione-dependent enzyme systems like lactoylglutathione lyase (EC 4.4.1.5) and hydroxyacylglutathione hydrolase (EC 3.1.2.6) (also called glyoxalase I and glyoxalase II), these enzymes detoxify oxidative stress and are important for bacterial survival [32], [33]. The Ferritin-like Dps protein, transcriptional regulator Rex and peroxide stress regulator PerR belonging to FUR family are known to regulate oxidative stress response in bacteria, all these genes were detected in the genomes of the isolates NM10 and BL7 [34]–[36]. Rubrerythrin was found to be one of the abundant proteins in human gut metaproteomic studies [5]. Rubrerythrin and superoxide reductase (EC 1.15.1.2) system are shown to be involved in the protection against oxidative stress [37]. These systems were present in the genomes of the isolates NM10 and BL7. Thus, the isolates NM10 and BL7 have various oxidative stress management systems that aid in the survival of these isolates in the gut environment.
Stress due to antibiotics
In addition to the internal selective pressures, external factors such as antibiotics pose a major challenge for the survival of bacteria in the human gut. This is mainly because of the constant selective pressure due to consumption of antibiotics during the treatment of infections. Studies have demonstrated that the human gut microbiota can act as a reservoir of antibiotic resistance [38], [39]. Megasphaera sp. NM10 and BL7 harbor multidrug resistance efflux pumps and genes that confer resistance to specific antibiotics. The genome of NM10 encodes for genes that confer resistance to beta-lactams, quinolones, fosmidomycin, polymyxins, macrolides and vancomycin. BL7 has genes for resistance to vancomycin, quinolones, macrolides and metallo-betalactamases. These genes would give adaptive advantage for these isolates during antibiotic treatment of the host.
Horizontal gene transfer (HGT) is considered as one of the major factors contributing to increased antibiotic resistance in the human gut commensals. Antibiotic resistance genes are associated with mobile elements such as plasmids and transposons, and are frequently transferred between gut commensals [40], [41]. The presence of plasmid conjugal transfer proteins, mobile element proteins and transposes in the genomes of isolates NM10 and BL7 suggest that these genes are acquired by human gut isolates for adaptation and survival in human gut. All these genes were not detected in the genome of M. elsdenii DSM20460 (Table S1).
Sensing the surroundings
Sensing the environmental metabolites is an important factor in the survival of the bacteria in any environment, as it is associated with various responses such as uptake of the available nutrients [42]. Both NM10 and BL7 genomes had an elaborate repertoire of genes encoding proteins associated with sensory response and transcriptional regulation. Sigma factors viz. sigma factor –54 and sigma factor –70, that are involved in sensing the environmental clues, were detected in both the isolates [42]. In addition to sigma factors, genes encoding substrate specific sensory proteins were identified in the genomes of Megasphaera sp. NM10 and BL7. Various two-component systems were identified in the genomes of NM10 and BL7. The two-component regulatory systems play a crucial role as regulators of various environmental signaling transduction pathways. These two-component systems have a histidine kinase sensor containing histidine kinase and phosphoacceptor domains that is localized in the membrane [43]. PhoR-PhoB two-component regulatory system associated with a high affinity phosphate transporter was detected in the Megasphaera genomes. Both NM10 and BL7 had a two-component sensor regulator linked to the carbon starvation protein-A, belonging to sensory box/GGDEF family protein and several histidine kinases. A hybrid two component regulatory system which is an AraC (arabinose specific helix-turn-helix domain) i.e. arabinose operon control protein; was present in both NM10 and BL7. This hybrid two component system with DNA binding domain was first identified in B. thetaiotaomicron, which consisted of 32 of these novel hybrid histidine kinases with a DNA-binding domain [22]. The hybrid two-component regulatory system present in the isolates NM10 and BL7 is for rhamnose utilization, highlighting the fact that these isolates have an ability to sense the presence of a carbon and energy source in the gut environment. In addition, the genomes of NM10 and BL7 have a methyl-accepting chemotaxis protein. Methyl-accepting chemotaxis proteins are a class of sensory receptors mediating chemotaxis to diverse extracellular and intracellular signals [44].
The presence of various sensory and regulatory systems suggests that the isolates NM10 and BL7 have the features required to sense the available nutrients in the environment and subsequently regulate a wide array of genes for the uptake and utilization of these nutrients. This would help Megasphaera sp. to overcome one of the major challenges in the complex gut environment i.e., the ability to distinguish between varied environmental chemical molecules.
Membrane transporters
Membrane transporters are important for the survival of bacteria in any environment. These membrane transporters facilitate the exchange of nutrients and metabolites with the surrounding environment. ATP-binding cassette transporters (ABC-transporter) are the most prevalent membrane transporters found in all organisms, involved in the transport of various substrates across cell membrane [45]. ABC-transporters for several substrates were detected in the genomes of isolates NM10 and BL7, these included transporters for important substrates like sugars, phosphate, iron, zinc, sulphate, nickel, molybdenum and important metabolites like amino acids, vitamin B12, spermidine-putricine and formate/nitrate (Table S1). The presence of D-serine/D-alanine/glycine, glutamine ABC transporter, methionine ABC transporter permease protein, serine transporter, transporters for aromatic, branched chain amino acid and several other amino acid transporters was detected in both the isolates. The MinPath analysis predicted the inability of the human gut isolates to synthesize serine and alanine. The presence of transporters for serine and alanine would facilitate acquisition of serine and alanine by these isolates from the environment and thereby help in the survival of these isolates.
Oligopeptide transport systems are important systems that facilitate uptake of oligopeptides from the environment. The presence of opp operon (OppABCDF) for oligopeptide transporter, suggests the ability of NM10 and BL7 to compete in the gut environment with an ability to use various sources of amino acids.
Another family of transporters, the major facilitator superfamily (MFS) involved in solute transport, was detected in both the isolates [46]. These include nitrate/nitrite transporter, Na+/H+ antiporter NhaA type, formate efflux pump and sugar efflux pump. Drug/metabolite efflux pumps of MATE family and RND type were detected, these efflux pumps along with magnesium and cobalt efflux protein CorC and Co/Zn/Cd efflux system may be responsible for protection against antibiotics and heavy metals.
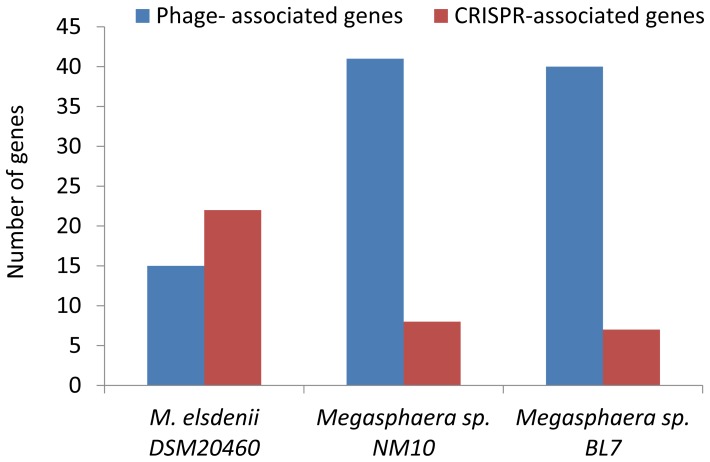
Phage associated genes
Along with the bacterial population, the human gut harbors viruses. In order to survive in such ecosystem, bacteria have evolved defense mechanisms that help them in counter acting the infection by viruses [47]. Two of these mechanisms involve prokaryotic restriction–modification (R/M) systems and clustered regularly interspaced short palindromic repeats (CRISPRs), which give protection against foreign DNA like phages and plasmids [48], [49]. CRISPRs are short, direct repeating sequences (typically 30–40 nucleotide long) that separate variable sequences of similar size. These act as a database of fragments derived from phage and plasmid genomes and provide protection to the bacteria [49]. The genomic analysis revealed the presence of different CRISPR families in all the three studied Megasphaera sp. genomes. The highest number was detected in the ruminal isolate; on the contrary relatively fewer CRISPRs were detected in the human gut isolates (Figure 5 and Table S1). Consequently, the number of phage-associated genes in the ruminal isolate was comparatively lower to the human gut isolates, suggesting a higher resilience to phage attack in ruminal isolate. Metagenomic study of the human gut microbiome showed that approximately 5% of the genes in the gut metagenome were phage associated genes [16]. This study speculated a probable role of pro-phages in the evolution of gut microbiome. In order to adapt to the human gut environment, carrying mobile DNA such as plasmids (invariably carrying antibiotic resistance genes) and phages seem to be important for adaptation to stress. The presence of the CRISPR elements in the human gut isolates suggests the ability of these bacteria to sustain attack from specific bacteriophages, while allowing exchange of some mobile DNA; this is an important adaptation for survival in the gut ecosystem. The human gut isolates harbor some type II and III restriction modification systems, which were absent in the genome of ruminal isolate (Table S1), suggesting presence of an alternative mechanism for the defense against phages.
Figure 5. Distribution of CRISPRs and phage-associated genes in the genomes of Megasphaera sp. NM10, BL7 and M. elsdenii DSM20460.
Potential Beneficial Effects on the Host
The beneficial effect of the gut microbes on the host is by and large through the metabolites utilized and/or produced through various metabolic pathways. These range from fermentation end products (mostly short chain fatty acids), vitamins and co-factors to numerous other bio-molecules [50]. In the present study, we identified the metabolic capabilities as defined by the genomic data for production of such important metabolites and validated by detecting the in vitro production.
Short Chain Fatty Acids (SCFA’s) production
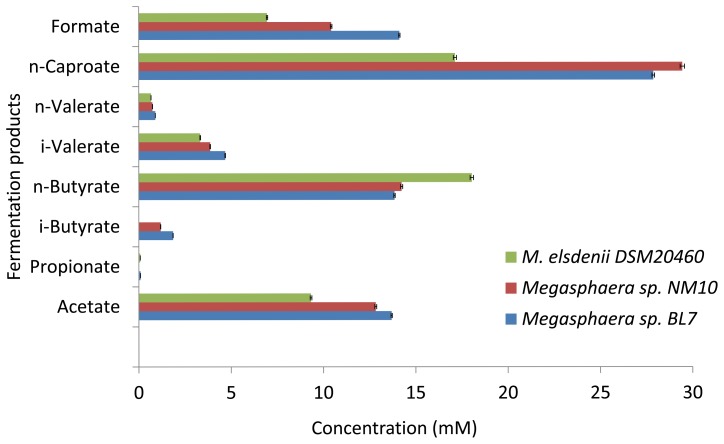
The SCFA’s are products of degradation of the dietary fibers by bacteria. Most prominent SCFA’s produced in human gut are acetate, butyrate, propionate and valerate [50]. The In silico metabolic analysis predicted the ability of the human gut isolates NM10 and BL7 to synthesize SCFA’s like butyrate, formate, acetate and valerate. Analysis of the fermentation products of these bacteria confirmed the ability of these isolates to produce acetate, butyrate, valerate and formate (Figure 6 and 7). The glucose fermentation yielded butyrate, formate, acetate, valerate and caproate; while negligible amounts of propionate were produced (Figure 6). Caproate was the major product of glucose fermentation. Human gut isolates produced ∼3.5 mg/L caproate, which is significantly higher than ruminal isolate (∼1.98 mg/L). Caproate is an industrially important product and serves as a precursor for cholesterol synthesis and synthesis of hormone progesterone [51].
Figure 6. Fermentation products of glucose utilization by Megasphaera sp. NM10, BL7 and M. elsdenii DSM20460.
The error bar represents standard deviation of three technical repliactes.
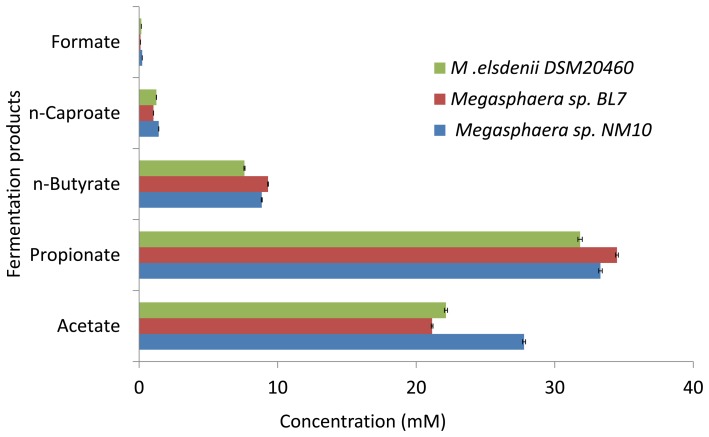
Figure 7. Fermentation products of lactate utilization by Megasphaera sp. NM10, BL7 and M. elsdenii DSM20460.
The error bar represents standard deviation of three technical repliactes.
Lactate is produced by microbes colonizing human gut as the end product of carbohydrate fermentation. Lactate accumulation is observed in short bowel syndrome and ulcerative colitis, this accumulation can be serious, causing neurotoxicity and cardiac arythmia [52], [53]. A study in porcine cecal digesta has shown that a co-culture of lactic acid producing bacteria, Lactobacillus acidophilus and M. elsdenii stimulates butyrate production [54]. M. elsdenii is known to produce SCFA’s by utilizing lactate and reduce acidosis in ruminals [55], [56]. Utilization of lactate by Megasphaera sp. may serve a similar function (reducing lactate toxicity and producing important metabolites like SCFA’s) in human gut. In order to validate this, SCFA’s production by NM10, BL7 and M. elsdenii by utilizing lactate was checked. In contrast to glucose fermentation, propionate was the major product of lactate fermentation followed by acetate and butyrate (Figure 7). Negligible amount of formate was detected on lactate utilization, while low amount of caproate was produced.
Many of these SCFA’s have a positive effect on the human health. SCFA’s stimulate water and sodium absorption in the epithelial cells. SCFA’s play an important role in the proliferation, differentiation and regulation of gene expression in the epithelial cells. Butyrate and acetate are used as an energy source by the colonic epithelium and the muscles [50], [57], [58]. In addition, acetate, propionate and to some extent butyrate, act as ligands for signaling molecules. Propionate and acetate are transported to liver through blood stream and used for gluconeogenesis [50]. The production of these SCFA’s by NM10 and BL7 suggests that these isolates may have a potential positive effect on the host health. Caproate has a protective effect on the host, it is reported to reduce colonization of pathogens in the gut [59]. Formate and caproate production would give competitive advantage to these isolates in human gut as formate and caproate are reported to have antibacterial activity against both Gram-negative and Gram-positive organisms [60].
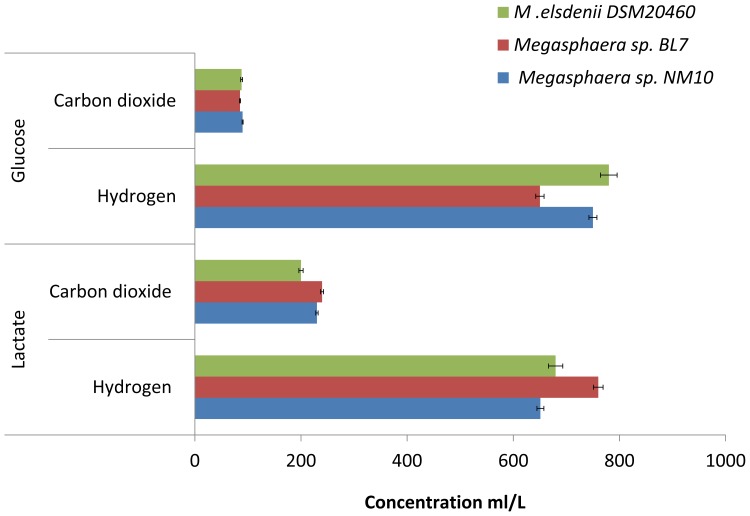
Hydrogen and carbon dioxide are produced as a byproduct of fermentation by both NM10 and BL7 using both glucose and lactate as substrates (Figure 8). Hydrogen and carbon dioxide produced during fermentation are utilized by methanogens, while hydrogen is utilized by acetogens and sulphate-reducing bacteria as a substrate for metabolism [61]–[63]. This makes Megasphaera sp. an important part of the food chain in the human gut environment.
Figure 8. Gas production by Megasphaera sp. NM10, BL7 and M. elsdenii DSM20460 using glucose and lactate as substrates.
The error bar represents standard deviation of three technical repliactes.
Production of vitamins
The microbes serve as a complementary source of some vitamins and a primary source for other vitamins such as biotin, pyridoxine and its derivatives which, the humans are not capable of synthesizing [64]. RAST annotation of the genomes identified genes coding enzymes for production of various vitamins such as biotin, thiamine, folate, pyridoxine, niacin, riboflavin. The MinPath analysis assigned functional annotations for biosynthesis of five different vitamins viz. biotin, thiamine, folate, B12 and pyridoxine. In case of riboflavin, the gene encoding riboflavin synthase was detected. The ribAH operon for riboflavin synthesis was present in both the isolates. This suggests that these isolates are capable of converting riboflavin into flavin mononucleotide via riboflavin kinase and flavin mononucleotide to riboflavin via a putative phosphotyrosine protein phosphatase. Entire pathway for vitamin B12 production was identified in the genomes of the isolates NM10 and BL7. The production of vitamin B12 by these isolates was validated by the in vitro assay. The isolate BL7 produced 3.83 ng vitamin B12/g of cell mass, while NM10 produced 6.52 ng vitamin B12/g of cell mass respectively. Thus, Megasphaera sp. can potentially provide vitamins to the host.
Conclusion
In the present study, we sequenced genomes of two potential novel Megasphaera sp. (NM10 and BL7) that were isolated from stools of two healthy Indian individuals. Previously, we have discussed the importance of gut microbiome studies in the Indian population [65]. The significance of developing population specific indigenous probiotics by exploring novel bacteria from the human gut has also been highlighted. Comparative genome analysis of two potential novel isolates of the Megasphaera sp. (NM10 and BL7) and Megasphaera elsdenii (DSM20460), in the current study, demonstrates the differences in genomic capabilities of these bacteria. Previous study on the genomes of Bacteroides species suggested that the variation between genomes may help the bacterium to evolve for a specific niche in the gut environment [22]. The genomic differences observed in this study may be attributed to the niche specialization and adaptation of the human gut isolates for survival in the gut environment. The observed difference between the genomes of the human gut isolates and the ruminal isolate, and similarities amongst the human gut isolates; demonstrates the evolutionary delineation of genomes towards adaptation to the human gut ecosystem. The human gut isolates are characterized by the presence of an enriched set of CAZymes compared to their ruminal counterpart. Additionally, the presence of various stress related genes, sensory systems, membrane transporters and resistance to antimicrobials provide an evidence for their adaptation in order to survive in the human gut and interact with the intestinal microbial community. The metabolic features of the human gut isolates suggest that these isolates play an important role in the complex gut environment, and in part, do add to the overall metabolic functions of the human gut microbiome. The human gut isolates have an ability to produce essential amino acids, vitamins and utilize lactic acid to produce SCFA’s. Overall, the study highlights the crucial adaptive features of Megasphaera sp. NM10 and BL7 for survival in the human gut and their potential for having a positive effect on the host health. The knowledge about the genome sequence of the two potential novel bacteria in this study is an addition to existing knowledge about the metabolic and genomic capabilities of the bacterial species from the human gut. This would help in further understanding the role of different bacterial species found in the human gut environment. This study also provides a basis for future in vivo studies, using these isolates for better understanding the host-microbe interaction and confirming the predicted beneficial effect of these isolates on the host.
Materials and Methods
Genomic DNA Extraction, 16S rRNA Sequencing and Genome Sequencing
The cultures were grown in Peptone Yeast Glucose (PYG) medium under anaerobic conditions at 37°C. The DNA was extracted from freshly grown cultures using standard Phenol:Chloroform method [66]. Additionally, RNase treatment was given to obtain RNA free DNA from these isolates. The 16S rRNA gene sequencing was carried out as described earlier [12]. Phylogenetic analyses was carried out using MEGA, version 4 [67], and the phylogenetic tree was constructed using neighbor-joining method with Kimura 2 parameter with 1000 replicates [68]. For whole shot-gun genome sequencing (WGS) the purified genomic DNA was quantified using nanodrop ND-1000 spectrophotometer (JH Bio innovations, Hyderabad India). One µg of pure genomic DNA was fragmented into approximately 250 to 300 bp fragments. End-repaired fragmented DNA was used for adaptor ligation. Ligated DNA fragments were Size selected using E-Gel® SizeSelect™ 2% Agarose Gel (Invitrogen, USA) to prepare gDNA library. Size selected library was quantified using Agilent 2100 Bioanalyzer high-sensitivity chip (Agilent Technologies, Germany). The template dilution factor (TDF) was calculated and an ePCR with 26 pM DNA was done. The ISPs (Ion Sphere Particles) were purified and enriched before sequencing. Two 316 chips (one for each genome) were used for sequencing on Ion torrent PGM™ following the manufacturer’s protocol for 200 bp chemistry.
Assembly and annotation
The reads obtained in FASTAQ format were quality checked and assembled de novo using Mira assembler v3.0, this program relies on the overlap-layout-consensus approach, where each read is represented as a node and each detected overlap as an arch between the appropriate nodes [69]. Gene prediction and annotation were done using the RAST [70] server and the NCBI Prokaryotic Genomes Automatic Annotation Pipeline (PGAAP) (http://www.ncbi.nlm.nih.gov/genomes/static/Pipeline.html). RAST server is a rapidly growing, manually curated library of subsystems and annotation is based on protein families derived from the subsystems (FIGfams), RAST uses tRNAscan-SE for predicting tRNA genes [14], [71], [72]. The MultiGenomeCompare tool of the SEED was used for genome comparison where Megasphaera sp NM10 was used as the reference and was compared with BL7 and M. eldesnii DSM20460 [14]. This system computes a bidirectional BLAST comparison of each genome to the reference genome. The hits are given as bi- or uni- (bi stands for bidirectional best hit and uni for uni-directional.
Investigating biochemical potential and network based representation of glycobiome
The draft metabolic model was constructed using Model SEED version 1.0, which is a web-based resource for high-throughput generation, optimization and analysis of genome-scale metabolic models (theSEED.org). In addition, for a more faithful representation of metabolic capabilities of bacteria under study, MinPath (Bayesian method) analysis for prediction of metabolic capabilities was performed [15]. The glycobiome for Megasphaera eldesnii DSM20460 and Human was obtained from CAZy database (http://www.cazy.org/). Whereas, glycobiome of Megasphaera sp NM10 and Megasphaera sp BL7 was identified using CAZymes Analysis Toolkit (CAT) web-based server [73]. The comparative network of the glycobiome was constructed using Cytoscape v2.8 [74].
In vitro Validation
Analysis of fermentation products
The isolates were grown in Peptone Yeast Glucose (PYG) and Peptone Yeast Lactic acid (PYL) broth for analysis of metabolic end products of glucose and lactate fermentation respectively. The metabolic end products were prepared as described by Holdeman et al. [75]. Short-chain fatty acids produced by the strains were extracted from 48 hrs broth cultures as previously described [76]. Volatile fatty acids were analyzed by Chemito 8610 GC equipped with Flame Ionization Detector, with oven temperature 150°C, injector temperature 170°C, detector temperature 190°C), column used was Chromosorb W (HP) (1.83 m×3.2 mm. SS) packed with 10% FFAP and 2% H3PO4. The carrier gas used was N2 at the flow rate of 30 ml min [77].
Gas production was determined in Peptone Yeast extract (PY), PYL and PYG broth using GC equipped with TCD [77]. H2 and CO2 were analyzed by Perkin Elmer GC (oven temperature 40°C, injector temperature 70°C, detector temperature 100°C ) using Porapak Q column, carrier gas used was Argon (Ar) at the flow rate of 40 ml min−1. In all the GC analyses, data analysis was done using IRIS 32 and total chrome navigations software. All experiments were carried out in triplicate.
Tolerance to bile
Brain Heart infusion yeast extract (BHI-YE) broth containing (per litre): 10 g BHI (Oxoid Ltd., England) supplemented with 10 g yeast extract, 10 ml hemin (Sigma-Aldrich, USA) solution (0.1%), 0.5 g cysteine HCl was used as the growth medium with pH 7 and N2 gas in headspace. Oxgall was added to this medium to get 3 concentrations of bile 0.2%, 0.3%, 0.4% (w/v), respectively. The BHI-YE broth was inoculated with 10% (v/v) of 48 hrs old culture and incubated at 37°C for 72 hrs. Bacterial growth was monitored every 24 hrs interval by measuring absorbance at 600 nm. Tolerance level of the strains was evaluated in terms of time required for increase in absorbance by 0.3 units (U) with respect to growth of the strains in the broth with and without oxgall [78].
Bile Salt Hydrolase (BSH) activity
BSH activity of the cultures was evaluated using the procedure described earlier [79]. Forty eight hrs old cultures were spot inoculated on BHI-YE medium plates supplemented with (per litre): 0.3 g oxgall (Sigma-Aldrich, USA) and 0.37 g CaCl2, 0.5 g cysteine HCl, 20 g Bacto Agar (Difco, USA). The diameters of precipitation zones were measured after incubation at 37°C for 72 hrs. BHI-YE agar plates without supplementation were used as controls. The strains which displayed the precipitation zone were considered positive for BSH activity.
Screening for vitamin B12 production
The fermentation was carried out in 100 ml Vitamin B12 production medium under anaerobic conditions at 37°C for 5 days [80]. Cyanide method was used for vitamin B12 extraction from bacterial cells [81]. The analysis of cell extract samples was conducted using HPLC system equipped with RP-C18 column 250×4.6 mm, 5 µm (LiChroCART, Germany) with an UV detector at 340 nm, 75% of 0.25 M NaH2PO4, pH 3.5, and 25% of methanol was used as the mobile phase with a flow of 1 ml min−1 at 40°C. Cyanocobalamin (Sigma, USA) was used as standard.
Supporting Information
The plate representing absence of bile salt hydrolase (BSH) activity in the isolates Megasphaera sp. NM10, BL7 and M. elsdenii. The white zone of precipitation represents a positive result. A) Represents the absence of BSH activity in isolate NM10 and BL7, while presence of activity in the other isolates from the study. A) Represents the absence of BSH activity in isolate NM10, BL7 and M. elsdenii, E. coli is used a negative control.
(TIF)
Comparison and functions of all predicted protein sequences in Megasphaera sp. NM10, BL7 and M. elsdenii DSM20460 using MultiGenomeCompare tool.
(XLS)
MinPath analysis results for Megasphaera sp. NM10. Legend: The MinPath results are interpreted as follows. Naive 1 or 0: the pathway is reconstructed, or not, by the naive mapping approach; MinPath 1 or 0: 1-the pathway is kept, 0- removed by MinPath; fam0: the total number of families involved in the corresponding pathway; fam-found: the total number of involved families that are annotated; name: the description of the corresponding subsystem.
(XLS)
MinPath analysis results for Megasphaera sp. BL7. Legend: The MinPath results are interpreted as follows. Naive 1 or 0: the pathway is reconstructed, or not, by the naive mapping approach; MinPath 1 or 0: 1-the pathway is kept, 0- removed by MinPath; fam0: the total number of families involved in the corresponding pathway; fam-found: the total number of involved families that are annotated; name: the description of the corresponding subsystem.
(XLS)
MinPath analysis results for Megasphaera elsdenii DSM20460. Legend: The MinPath results are interpreted as follows. Naive 1 or 0: the pathway is reconstructed, or not, by the naive mapping approach; MinPath 1 or 0: 1-the pathway is kept, 0- removed by MinPath; fam0: the total number of families involved in the corresponding pathway; fam-found: the total number of involved families that are annotated; name: the description of the corresponding subsystem.
(XLS)
Acknowledgments
We wish to thank Mr Dhiraj Dhotre for his assistance and healthy discussions related to bioinformatic analysis. We thank Ms Ankita Vaishampayan for the critical review of the manuscript.
Funding Statement
The work was funded by the Department of Biotechnology (DBT), Government of India; Microbial Culture Collection Project. Nachiket Marathe is grateful to Council of Scientific and Industrial Research (CSIR), New Delhi, for fellowship. The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Clemente JC, Ursell LK, Parfrey LW, Knight R (2012) The impact of the gut microbiota on human health: an integrative view. Cell 148: 1258–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Flint HJ, Karen P, Louis P, Duncan SH (2012) The role of the gut microbiota in nutrition and health. Nature Reviews Gastroenterology & Hepatology 9: 577–589. [DOI] [PubMed] [Google Scholar]
- 3. Turnbaugh PJ, Gordon JI (2009) The core gut microbiome, energy balance and obesity. The Journal of physiology 587: 4153–4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Samuel BS, Shaito A, Motoike T, Rey FE, Backhed F, et al. (2008) Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proceedings of the National Academy of Sciences of the United States of America 105: 16767–16772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Verberkmoes NC, Russell AL, Shah M, Godzik A, Rosenquist M, et al. (2009) Shotgun metaproteomics of the human distal gut microbiota. The ISME journal 3: 179–189. [DOI] [PubMed] [Google Scholar]
- 6. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, et al. (2010) A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Juvonen R, Suihko M-L (2006) Megasphaera paucivorans sp. nov., Megasphaera sueciensis sp. nov. and Pectinatus haikarae sp. nov., isolated from brewery samples, and emended description of the genus Pectinatus. International journal of systematic and evolutionary microbiology 56: 695–702. [DOI] [PubMed] [Google Scholar]
- 8. Marchandin H, Jumas-Bilak E, Gay B, Teyssier C, Jean-Pierre H (2003) Phylogenetic analysis of some Sporomusa sub-branch members isolated from human clinical specimens: description of Megasphaera micronuciformis sp. nov. International Journal of Systematic and Evolutionary Microbiology 53(2): 547–553. [DOI] [PubMed] [Google Scholar]
- 9. Rogosa M (1971) Transfer of Peptostreptococcus elsdenii Gutierrez et al. to a new genus, Megasphaera [M. elsdenii (Gutierrez et al.) comb. nov.]. International Journal of Systematic and Evolutionary Microbiology 21(2): 187–189. [Google Scholar]
- 10. Klieve AV, Hennessy D, Ouwerkerk D, Forster RJ, Mackie RI, et al. (2003) Establishing populations of Megasphaera elsdenii YE 34 and Butyrivibrio fibrisolvens YE 44 in the rumen of cattle fed high grain diets. Journal of Applied Microbiology 95: 621–630. [DOI] [PubMed] [Google Scholar]
- 11. Marx H, Graf AB, Tatto NE, Thallinger GG, Mattanovich D, et al. (2011) Genome Sequence of the Ruminal Bacterium Megasphaera elsdenii Genome Sequence of the Ruminal Bacterium Megasphaera elsdenii . Journal of Bacteriology 193(19): 5578–5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marathe N, Shetty S, Lanjekar V, Ranade D, Shouche Y (2012) Changes in human gut flora with age: an Indian familial study. BMC microbiology 12: 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Padhmanabhan R, Lagier JC, Makaya Dangui NP, Michelle C, Couderc C, et al. (2013) Non-contiguous finished genome sequence and description of Megasphaera massiliae. Standards in Genomic Sciences 8(3): 525–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang H, et al. (2005) The Subsystems Approach to Genome Annotation and its Use in the Project to Annotate 1000 Genomes. Nucleic Acids Research 33: 5691–5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ye Y, Doak TG (2009) A Parsimony Approach to Biological Pathway Reconstruction/Inference for Genomes and Metagenomes. PLoS Computational Biology 5: 1–8 doi:10.1371/journal.pcbi.1000465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, et al. (2010) A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pérez-Cobas AE, Gosalbes MJ, Friedrichs A, Knecht H, Artacho A, et al.. (2012) Gut microbiota disturbance during antibiotic therapy: a multi-omic approach. Gut: 1–11. [DOI] [PMC free article] [PubMed]
- 18. Booijink CC, Boekhorst J, Zoetendal EG, Smidt H, Kleerebezem M, et al. (2010) Metatranscriptome analysis of the human fecal microbiota reveals subject-specific expression profiles, with genes encoding proteins involved in carbohydrate metabolism being dominantly expressed. Applied and Environmental Microbiology 76: 5533–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, et al. (2012) Host-gut microbiota metabolic interactions. Science (New York, NY) 336: 1262–1267. [DOI] [PubMed] [Google Scholar]
- 20. Sun G, Huang J (2011) Horizontally acquired DAP pathway as a unit of self-regulation. Journal of evolutionary biology 24: 587–595. [DOI] [PubMed] [Google Scholar]
- 21. Xu J, Gordon JI (2003) Honor thy symbionts. Proceedings of the National Academy of Sciences of the United States of America 100: 10452–10459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xu J, Mahowald MA, Ley RE, Lozupone CA, Hamady M, et al. (2007) Evolution of symbiotic bacteria in the distal human intestine. PLoS biology 5: e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kelly WJ, Leahy SC, Altermann E, Yeoman CJ, Dunne JC, et al. (2010) The glycobiome of the rumen bacterium Butyrivibrio proteoclasticus B316(T) highlights adaptation to a polysaccharide-rich environment. PloS one 5: e11942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cantarel BL, Lombard V, Henrissat B (2012) Complex carbohydrate utilization by the healthy human microbiome. PloS one 7: e28742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Flint HJ, Bayer E, Rincon MT, Lamed R, White B (2008) Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nature reviews Microbiology 6: 121–131. [DOI] [PubMed] [Google Scholar]
- 26. Aikman PC, Henning PH, Humphries DJ, Horn CH (2011) Rumen pH and fermentation characteristics in dairy cows supplemented with Megasphaera elsdenii NCIMB 41125 in early lactation. Journal of dairy science 94: 2840–2849. [DOI] [PubMed] [Google Scholar]
- 27.Begley M, Hill C, Gahan CGM (2006) Bile Salt Hydrolase Activity in Probiotics Bile Salt Hydrolase Activity in Probiotics. Applied and Environmental microbiology 72(3), 1729–1738. [DOI] [PMC free article] [PubMed]
- 28. Lin J, Martinez A (2006) Effect of efflux pump inhibitors on bile resistance and in vivo colonization of Campylobacter jejuni . The Journal of antimicrobial chemotherapy 58: 966–972. [DOI] [PubMed] [Google Scholar]
- 29. Lin J, Cagliero C, Guo B, Barton Y-W, Maurel M-C, et al. (2005) Bile salts modulate expression of the CmeABC multidrug efflux pump in Campylobacter jejuni . Journal of bacteriology 187: 7417–7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Prouty AM, Brodsky IE, Falkow S, Gunn JS (2004) Bile-salt-mediated induction of antimicrobial and bile resistance in Salmonella typhimurium . Microbiology 150: 775–783. [DOI] [PubMed] [Google Scholar]
- 31. Bina JE, Mekalanos JJ (2001) Vibrio cholerae tolC Is Required for Bile Resistance and Colonization. Infection And Immunity 69(7): 4681–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Murata K, Tani K, Kato J, Chibata I (1980) Excretion of Glutathione by Methylglyoxal-resistant Escherichia coli. . Journal of General Microbiology 120(2): 545–547. [DOI] [PubMed] [Google Scholar]
- 33. Li Y, Hugenholtz J, Abee T, Molenaar D (2003) Glutathione Protects Lactococcus lactis against Oxidative Stress. Applied and Environmental Microbiology 69(10): 5739–5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Halsey TA, Gravdahl DJ, Fang FC, Libby SJ (2004) The Ferritin-Like Dps Protein Is Required for Salmonella enterica Serovar Typhimurium Oxidative Stress Resistance and Virulence. Infection And Immunity 72: 1155–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Verneuil N, Rincé A, Sanguinetti M, Posteraro B, Fadda G, et al. (2005) Contribution of a PerR-like regulator to the oxidative-stress response and virulence of Enterococcus faecalis . Microbiology (Reading, England) 151: 3997–4004. [DOI] [PubMed] [Google Scholar]
- 36.Bitoun JP, Liao S, Yao X, Xie GG, Wen ZT (2012) The Redox-Sensing Regulator Rex Modulates Central Carbon Metabolism, Stress Tolerance Response and Biofilm Formation by Streptococcus mutans. PloS one 7. doi:10.1371/journal.pone.0044766. [DOI] [PMC free article] [PubMed]
- 37. Lumppio HL, Shenvi N V, Summers AO, Voordouw G, Kurtz DM (2001) Rubrerythrin and Rubredoxin Oxidoreductase in Desulfovibrio vulgaris: a Novel Oxidative Stress Protection System. Journal of Bacteriology 183(1): 101–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sommer MOA, Church GM, Dantas G (2009) Functional characterization of the antibiotic resistance reservoir in the human microflora. Science 325(5944): 1128–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. De Vries LE, Vallès Y, Agersø Y, Vaishampayan P, García-Montaner A, et al. (2011) The gut as reservoir of antibiotic resistance: microbial diversity of tetracycline resistance in mother and infant. PloS one 6: e21644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Blake DP, Hillman K, Fenlon DR, Low JC (2003) Transfer of antibiotic resistance between commensal and pathogenic members of the Enterobacteriaceae under ileal conditions. Journal of Applied Microbiology 95: 428–436. [DOI] [PubMed] [Google Scholar]
- 41. Salyers AA, Gupta A, Wang Y (2004) Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends in Microbiology 12(9): 412–6. [DOI] [PubMed] [Google Scholar]
- 42. Kazmierczak MJ, Wiedmann M, Boor KJ (2005) Alternative Sigma Factors and Their Roles in Bacterial Virulence. Microbiology and Molecular Biology Reviews 69(4): 527–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Martens EC, Lowe EC, Chiang H, Pudlo N a, Wu M, et al. (2011) Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS biology 9: e1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Alexander RP, Zhulin IB (2007) Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proceedings of the National Academy of Sciences of the United States of America 104: 2885–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Davidson AL, Dassa E, Orelle C, Chen J (2008) Structure, Function, and Evolution of Bacterial ATP-Binding Cassette Systems. Microbiology and Molecular Biology Reviews 72(2): 317–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pao SS, Paulsen IT, Saier MH (1998) Major Facilitator Superfamily. Microbiology and Molecular Biology Reviews 62(1): 1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stern A, Mick E, Tirosh I, Sagy O, Sorek R (2012) CRISPR targeting reveals a reservoir of common phages associated with the human gut microbiome: Genome Research. 22(10): 1985–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kobayashi I (2001) Behavior of restriction – modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Research 29: 3742–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang Q, Rho M, Tang H, Doak TG, Ye Y (2013) CRISPR-Cas systems target a diverse collection of invasive mobile genetic elements in human microbiomes. Genome Biology 14(4): R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hijova E, Chmelarova A (2007) Short chain fatty acids and colonic health. Bratislavské lekárske listy 108(8): 354–8. [PubMed] [Google Scholar]
- 51. Kenealy WR, Cao Y, Weimer PJ (1995) Production of caproic acid by cocultures of ruminal cellulolytic bacteria and Clostridium kluyveri grown on cellulose and ethanol. Applied microbiology and biotechnology 44: 507–513. [DOI] [PubMed] [Google Scholar]
- 52. Kaneko T, Bando Y, Kurihara H, Satomi K, Nonoyama K, et al. (1997) Fecal microflora in a patient with short-bowel syndrome and identification of dominant lactobacilli. Journal of clinical microbiology 35: 3181–3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Duncan SH, Louis P, Flint HJ (2004) Lactate-Utilizing Bacteria, Isolated from Human Feces, That Produce Butyrate as a Major Fermentation Product. Applied and Environmental Microbiology 70(10): 5810–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tsukahara T, Hashizume K, Koyama H, Ushida K (2006) Stimulation of butyrate production through the metabolic interaction among lactic acid bacteria, Lactobacillus acidophilus, and lactic acid-utilizing bacteria, Megasphaera elsdenii, in porcine cecal digesta. Animal Science Journal 77: 454–461. [Google Scholar]
- 55. Chiquette J (2009) The Role of Probiotics in Promoting Dairy Production. Advances in Dairy Technology 21: 143–157. [Google Scholar]
- 56. Marounek M, Fliegrova K, Bartos S (1989) Metabolism and some characteristics of ruminal strains of Megasphaera elsdenii . Applied andEnvironmental Microbiology 55: 1570–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Topping DL, Clifton PM (2001) Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiological reviews 81: 1031–1064. [DOI] [PubMed] [Google Scholar]
- 58. Scheppach W (1994) Effects of short chain fatty acids on gut morphology and function. Gut 35: S35–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Immerseel F V, Buck J De, Boyen F, Bohez L, Volf J, et al. (2004) Medium-Chain Fatty Acids Decrease Colonization and Invasion through hilA Suppression Shortly after Infection of Chickens with Salmonella enterica Serovar Enteritidis. Applied and Environmental Microbiology 70(6): 3582–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Huang CB, Alimova Y, Myers TM, Ebersole JL (2011) Short- and medium-chain fatty acids exhibit antimicrobial activity for oral microorganisms. Archives of oral biology 56: 650–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rey FE, Faith JJ, Bain J, Muehlbauer MJ, Stevens RD, et al. (2010) Dissecting the in vivo metabolic potential of two human gut acetogens. The Journal of biological chemistry 285: 22082–22090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Samuel BS, Hansen EE, Manchester JK, Coutinho PM, Henrissat B, et al. (2007) Genomic and metabolic adaptations of Methanobrevibacter smithii to the human gut. Proceedings of the National Academy of Sciences of the United States of America 104: 10643–10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gibson GR, Macfarlane GT, Cummings JH (1993) Sulphate reducing bacteria and hydrogen metabolism in the human. Gut 34(4): 437–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI (2005) Host-bacterial mutualism in the human intestine. Science 307: 1915–1920. [DOI] [PubMed] [Google Scholar]
- 65. Shetty SA, Marathe NP, Shouche YS (2013) Opportunities and challenges for gut microbiome studies in the Indian population. Microbiome 1(1): 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sambrook J, Russell D (2000) Molecular Cloning - A Laboratory Manual, volume 1. 3rdedition. New York: CSHL press. 1.32–1.3.
- 67. Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular biology and evolution 24: 1596–1599. [DOI] [PubMed] [Google Scholar]
- 68. Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular biology and evolution 4: 406–425. [DOI] [PubMed] [Google Scholar]
- 69. Chevreux B, Wetter T, Suhai S (1999) Genome sequence assembly using trace signals and additional sequence information. Comput. Sci. Biol. 99: 45–56. [Google Scholar]
- 70. Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, et al. (2008) The RAST Server: rapid annotations using subsystems technology. BMC genomics 9: 75 doi:10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Meyer F, Overbeek R, Rodriguez A (2009) FIGfams: yet another set of protein families. Nucleic acids research 37: 6643–6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Schattner P, Brooks AN, Lowe TM (2005) The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic acids research 33: W686–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Park BH, Karpinets TV, Syed MH, Leuze MR, Uberbacher EC (2010) CAZymes Analysis Toolkit (CAT ): Web service for searching and analyzing carbohydrate-active enzymes in a newly sequenced organism using CAZy database. Glycobiology 20(12): 1574–1584. [DOI] [PubMed] [Google Scholar]
- 74. Smoot ME, Ono K, Ruscheinski J, Wang P, Ideker T (2011) Cytoscape 2. 8: new features for data integration and network visualization. Bioinformatics 27(3): 431–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Holdeman LV, Elizabeth P, Cato, Moore WEC (1997) Anaerobe Laboratory Manual.4th edition. Blacksburg, Virginia: Virginia Polytechnic Institute and State University Press. 1–156.
- 76.Jousimies-Somer HR, Summanen P, Citron DM, Baron EJ, Wexler H M, et al.. (2002). Wadsworth – KTL Anaerobic Bacteriology Manual, 6th edn. Belmont, CA: Star Publishing.
- 77. Dighe AS, Shouche YS, Ranade DR (1998) Selenomonas lipolytica sp. nov., an obligately anaerobic bacterium possessing lipolytic activity. International journal of systematic and evolutionary microbiology 48 783–91: 783–791. [DOI] [PubMed] [Google Scholar]
- 78. Liong MT, Shah NP (2005) Acid and bile tolerance and cholesterol removal ability of lactobacilli strains. Journal of Dairy Science 88: 55–66. [DOI] [PubMed] [Google Scholar]
- 79. Pereira DIA, Mccartney AL, Gibson GR, Pereira DIA, Mccartney AL, et al. (2003) An In Vitro Study of the Probiotic Potential of a Bile-Salt-Hydrolyzing Lactobacillus fermentum Strain, and Determination of Its Cholesterol-Lowering Properties. Applied and Environmental Microbiology 69(8): 4743–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Madhu AN, Giribhattanavar P, Narayan MS, Prapulla SG (2010) Probiotic lactic acid bacterium from kanjika as a potential source of vitamin B 12: evidence from LC-MS, immunological and microbiological techniques. Biotechnology Letters 32: 503–506. [DOI] [PubMed] [Google Scholar]
- 81. Quesada-Chanto A, Schmid-Meyer AC, Schroeder AG, Carvalho-Jonas M, Blanco I, et al. (1998) Effect of oxygen supply on biomass, organic acids and vitamin B12 production by Propionibacterium shermanii . World Journal of Microbiology & Biotechnology 14: 843–846. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The plate representing absence of bile salt hydrolase (BSH) activity in the isolates Megasphaera sp. NM10, BL7 and M. elsdenii. The white zone of precipitation represents a positive result. A) Represents the absence of BSH activity in isolate NM10 and BL7, while presence of activity in the other isolates from the study. A) Represents the absence of BSH activity in isolate NM10, BL7 and M. elsdenii, E. coli is used a negative control.
(TIF)
Comparison and functions of all predicted protein sequences in Megasphaera sp. NM10, BL7 and M. elsdenii DSM20460 using MultiGenomeCompare tool.
(XLS)
MinPath analysis results for Megasphaera sp. NM10. Legend: The MinPath results are interpreted as follows. Naive 1 or 0: the pathway is reconstructed, or not, by the naive mapping approach; MinPath 1 or 0: 1-the pathway is kept, 0- removed by MinPath; fam0: the total number of families involved in the corresponding pathway; fam-found: the total number of involved families that are annotated; name: the description of the corresponding subsystem.
(XLS)
MinPath analysis results for Megasphaera sp. BL7. Legend: The MinPath results are interpreted as follows. Naive 1 or 0: the pathway is reconstructed, or not, by the naive mapping approach; MinPath 1 or 0: 1-the pathway is kept, 0- removed by MinPath; fam0: the total number of families involved in the corresponding pathway; fam-found: the total number of involved families that are annotated; name: the description of the corresponding subsystem.
(XLS)
MinPath analysis results for Megasphaera elsdenii DSM20460. Legend: The MinPath results are interpreted as follows. Naive 1 or 0: the pathway is reconstructed, or not, by the naive mapping approach; MinPath 1 or 0: 1-the pathway is kept, 0- removed by MinPath; fam0: the total number of families involved in the corresponding pathway; fam-found: the total number of involved families that are annotated; name: the description of the corresponding subsystem.
(XLS)