Abstract
Reconstruction of host-pathogen protein interaction networks is of great significance to reveal the underlying microbic pathogenesis. However, the current experimentally-derived networks are generally small and should be augmented by computational methods for less-biased biological inference. From the point of view of computational modelling, data scarcity, data unavailability and negative data sampling are the three major problems for host-pathogen protein interaction networks reconstruction. In this work, we are motivated to address the three concerns and propose a probability weighted ensemble transfer learning model for HIV-human protein interaction prediction (PWEN-TLM), where support vector machine (SVM) is adopted as the individual classifier of the ensemble model. In the model, data scarcity and data unavailability are tackled by homolog knowledge transfer. The importance of homolog knowledge is measured by the ROC-AUC metric of the individual classifiers, whose outputs are probability weighted to yield the final decision. In addition, we further validate the assumption that only the homolog knowledge is sufficient to train a satisfactory model for host-pathogen protein interaction prediction. Thus the model is more robust against data unavailability with less demanding data constraint. As regards with negative data construction, experiments show that exclusiveness of subcellular co-localized proteins is unbiased and more reliable than random sampling. Last, we conduct analysis of overlapped predictions between our model and the existing models, and apply the model to novel host-pathogen PPIs recognition for further biological research.
Introduction
Accurate mapping of protein interactome is essential to reveal protein functions, biological processes, signal transduction pathways. In recent years, although high throughput experimental techniques have drastically accumulated much knowledge about protein-protein interactions (PPI), the derived PPI networks are far incomplete and noisy [1], [2]. As a good complement to the labour-intensive biological experiments, computational methods can accelerate the reconstruction of PPI networks at low cost [3].
At present most of the existing computational methods are developed for intra-species PPI networks reconstruction, e.g. yeast PPI network [3], Arabidopsis thaliana PPI network [4], human PPI network [5], etc. As compared to intra-species PPI networks reconstruction, inter-species host-pathogen PPI networks (the interacting partners are from two different species) reconstruction is faced up with more challenges in that the scale of the host-pathogen PPI networks is generally rather small. Small PPI network results in data scarcity that would easily lead to poor generalization ability of computational model. Data integration is an effective method to compensate for data scarcity. By simultaneously leveraging a catalog of biological feature information, data integration can greatly increase the information abundance for sufficient model training. Tastan et al. [6] applied Random Forest to integrate the feature information of binding motif, gene expression profile, gene ontology, sequence similarity, post-translational modification, tissue distribution and PPI network topology for HIV-human protein interaction prediction. Based on the work, Qi et al. [7] further proposed a semi-supervised multi-task learning method to exploit the weakly labelled data. Dyer et al. [8] combined protein domain profile, sequence k-mer and PPI network properties for HIV-human protein interaction prediction. For another pathogenetic microbe Plasmodium falciparum, Dyer et al. [9] combined protein domain profile, gene expression, gene ontology and gene co-expression to predict and validate the host-pathogen protein interactions. Wuchty S [10] combined sequence k-mer, interlog, gene ontology and signal transduction pathways to predict and validate the protein interactions between Plasmodium falciparum and Homo sapiens. In the latter two models, the validation information (gene co-expression, signal transduction pathways, gene ontology) was used to manually filter the predicted PPIs. It has been claimed that gene ontology (GO) is one of the strongest indicators for host-pathogen PPI prediction [6] and intra-species PPI prediction [3], [4], [11], [12], [13], [14], [15], [16], [17] among the catalog of feature information. The work [14] explained the reasons why GO feature outperformed the other feature information based on the observations: (1) proteins localized in identical cellular compartments are more likely to interact than are proteins that reside in spatially distant compartments; (2) proteins that participate in similar biological processes or perform similar molecular functions are likely to interact. Hence the three aspects of gene ontology (cellular compartments, biological processes and molecular functions) are informative to indicate PPI.
Although data integration can simultaneously exploit multiple aspects of biological knowledge, the difficulty in availability of some feature information such as gene co-expression poses a great challenge on host-pathogen PPI networks reconstruction [9]. Once the feature information is unavailable for the proteins to be predicted, the data integration methods [4], [6], [7], [11], [16] would fail to work. Even for those methods that exploit only one type of non-sequence feature information (e.g. gene ontology) [14], data integration would also fail to work because the information required for prediction (e.g. GO annotations) may be potentially not available. For the reasons, data integration model should deliberately take into account the case of data unavailability and provide effective solutions to information substitution. Less demanding data constraint helps the model gain wide applicability. Like the other feature information, structural similarity, is also a strong indicator of protein-protein interaction. Doolittle et al. [18] exploited the information of protein structural similarity to predict host-pathogen PPI. However, the potential unavailability of the spatial structural information would likewise restrict the model application. As compared to the costly feature information such as structural information, gene ontology, gene co-expression and metabolic pathways, etc., obtaining protein sequence information is less expensive, thus the computational model based on protein sequence only has the least data constraint nearly without the problem of data unavailability. Unfortunately, the work [19] argued that protein sequence alone was not sufficient to train a satisfactory model for PPI prediction.
HIV-human PPI prediction can be viewed as a problem of 2-class classification that needs both positive data and negative data to define the decision function. Positive data contains the information of interaction and negative data contains the information of non-interaction. Unfortunately, there are far few experimentally derived negative data available to computational modelling for host-pathogen PPI networks reconstruction. At present negative data construction is a hard-tackling problem and the common method is random sampling. Random sampling is simple but has the demerits of model uncertainty and potential inclusion of interacting protein pairs. The work [20] proposed one-class Biclustering method to mine association rules from the positive data for HIV-human PPI prediction. Biclustering need not construct the negative data, so that the computational modelling is much simplified. However, the model that does not learn the non-interacting patterns would run the risk of high rate of false positive.
In this work, we are motivated to address the concerns of data scarcity, data unavailability and negative data sampling for HIV-human PPI prediction. To reduce data dependency, we choose gene ontology as the only feature information for host-pathogen PPI prediction. Unlike the existing GO-based PPI prediction models [3], [4], [6], [7], [11], [12], [13], [14], [15], [16], [17], we attempt to exploit the homolog GO information (GO annotations from the homologs) to compensate for data scarcity and data unavailability. We deliberately investigate the assumption that only the homolog GO information is sufficient to train a satisfactory model for HIV-human PPI prediction. If the assumption is validated, effective information substitution could make the model more robust against data unavailability with less demanding data constraint. To validate the assumption, we conduct three experimental settings, namely the Optimistic case, the Moderate case and the Pessimistic case. The Optimistic case assumes that both the target GO information (GO annotations from the protein itself) and the homolog GO information are available for model training and model evaluation. Good performance can indicate that data scarcity is properly tackled to a certain degree. The Moderate case assumes that the target GO information of the test data is not available and the Pessimistic case assumes that the target GO information of the training data and the test data is not available. If any of the two cases achieves good performance, it can be convincingly concluded that data unavailability is well tackled. As regards with negative data sampling, we further conduct two experimental settings for each case, one is random sampling and the other is exclusiveness of subcellular co-localized proteins. All the tasks are implemented by our proposed probability weighted ensemble transfer learning model (PWEN-TLM). The target GO information and the homolog GO information are used to train individual support vector machine (SVM) and are assigned different weights according to their contributions to the model performance. The merit is that the weights could depress the potential noise from the homolog GO information. To investigate the importance of molecular functions, cellular compartments and biological processes (three aspects of gene ontology) to HIV-human PPI prediction, the three aspects of the target GO information and the homolog GO information are used to train three individual classifiers respectively, thus there are totally 6 individual classifiers. The ensemble classifier yields the final decision in the form of probability by linearly weighting the probability outputs of the individual classifiers. For critical model performance estimation, we conduct cross validation, independent test and novel PPI detection on the benchmark HIV-human PPI dataset [21].
Methods
Transfer Learning
Transfer learning is a hot research topic in machine learning community. As compared to traditional supervised learning, transfer learning aims at leveraging useful information from auxiliary data. In most cases, the auxiliary data and the target data show different distributions or heterogeneous representations [22]. Especially in bioinformatics field, the biological data from different laboratories are usually subjected to different distributions, heterogeneous representations and noise levels [23]. Thus it is necessary for us to develop sophisticated transfer learning models to exploit useful information from the auxiliary data for the target domain learning. The work [24], [25], [26] proposed several non-parametric multiple kernel learning based transfer learning models (GO-TLM, MK-TLM and MLMK-TLM) to reduce the risk of negative knowledge transfer. In this work, we propose a probability weighted ensemble learning model (PWEN-TLM) to transfer the homolog GO information to enrich or substitute for the target GO information. As compared to multiple kernel learning based transfer learning models, the ensemble based transfer learning method can take full advantages of SVM (support vector machine) sparseness to reduce the computational complexity. The details are described in the section Probability weighted ensemble learning.
GO Feature Construction
The homologs are extracted from SwissProt 57.3 database [27] using PSI-BLast
[28]. Here we adopt the default parameters setting (e.g. default E-value = 10) to enlarge the GO term coverage. The GO terms are extracted from the latest GOA database [29] (114 Release, as of 28 November, 2012). For each protein i, we separate the target set of GO terms (denoted as 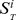 ) from the homolog set of GO terms (denoted as
) from the homolog set of GO terms (denoted as 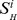 ), and further divide
), and further divide 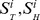 into three subsets corresponding to the three aspects of gene ontology, denoted as
into three subsets corresponding to the three aspects of gene ontology, denoted as 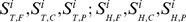 , respectively. Here T denotes the target protein, H denotes the homolog protein, F denotes molecular functions, C denotes cellular components and P denotes biological processes. It is noted that the term target here is used to denote the protein itself (comparative to homolog), it does not refer to the virus-targeted protein. Let capital I denote the set of proteins, then the total set of GO terms can be defined as follows:
, respectively. Here T denotes the target protein, H denotes the homolog protein, F denotes molecular functions, C denotes cellular components and P denotes biological processes. It is noted that the term target here is used to denote the protein itself (comparative to homolog), it does not refer to the virus-targeted protein. Let capital I denote the set of proteins, then the total set of GO terms can be defined as follows:
| (1) |
Based on the denotations, we can formally define the feature vector for each PPI pair (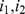 ) as follows:
) as follows:
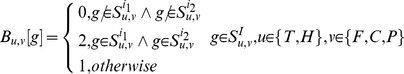 |
(2) |
where 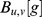 denotes the component g of PPI feature vector
denotes the component g of PPI feature vector 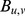 (each PPI pair follows the same feature representation, so we use
(each PPI pair follows the same feature representation, so we use 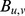 instead of
instead of 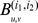 as the general definition). Formula (2) means that if the interacting protein pair shares the same GO term g, then the corresponding component in the feature vector B is set 2; if neither protein in the protein pair (
as the general definition). Formula (2) means that if the interacting protein pair shares the same GO term g, then the corresponding component in the feature vector B is set 2; if neither protein in the protein pair (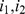 ) possesses GO term g, then the value is set 0; otherwise the value is set 1. From the formula, we can see that the above definition is symmetrical, i.e., (
) possesses GO term g, then the value is set 0; otherwise the value is set 1. From the formula, we can see that the above definition is symmetrical, i.e., (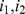 ) and (
) and (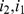 ) have identical feature representation, thus the order of the proteins in each protein pair does not change the feature representation.
) have identical feature representation, thus the order of the proteins in each protein pair does not change the feature representation.
Probability Weighted Ensemble Learning
Sparseness is one of the graceful characteristics of SVM, which means that the parameters are optimized on a small working set instead of the whole training set [30]. Kuhn-Tucker Theorem states that only the training examples that lie on the surface of the optimal hypersphere have their corresponding Lagrange parameters non-zero, and the corresponding Lagrange parameters are all zero for the remaining examples. The training examples with non-zero Lagrange parameters are referred to as support vectors. Only the support vectors are informative to support the optimal hypersphere and the other data can be discarded. Assuming there are 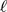 training data, the working set that helps define the final decision function generally contains rather small number of data points
training data, the working set that helps define the final decision function generally contains rather small number of data points 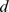 ,
, 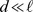 , that’s, we only need to compute the kernel matrix on the working set (
, that’s, we only need to compute the kernel matrix on the working set (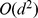 ) instead of the whole training dataset (
) instead of the whole training dataset (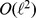 ), thus the runtime complexity and the space complexity are greatly reduced. In our method, the six independent individual SVMs (denoted as
), thus the runtime complexity and the space complexity are greatly reduced. In our method, the six independent individual SVMs (denoted as 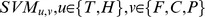 ) trained by the six feature vectors (
) trained by the six feature vectors (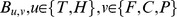 ) have time complexity
) have time complexity 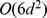 , much smaller than the multiple kernel learning method
, much smaller than the multiple kernel learning method 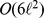 .
.
Traditional two-class labels {−1, +1} are not convenient to reveal the confidence level of the prediction. Probability output is a good alternative to the classical 2-class output and is especially applicable to vote-weighted ensemble learning for the final decisions combination. Platt [31] proposed a method to yield posterior class probability output for binary SVM as defined below:
| (3) |
where the coefficient A and B can be derived from data by cross validation, and f(x) is the decision value of binary SVM. The final decision function of the ensemble classifier is defined as follows:
| (4) |
where  denotes the test protein,
denotes the test protein, 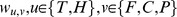 denotes the weight of the individual classifier
denotes the weight of the individual classifier 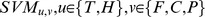 and
and 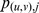 denotes the probability that the individual classifier
denotes the probability that the individual classifier 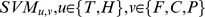 assigns protein
assigns protein  to the jth class. ROC curve [32] is a frequently-used statistical tool to illustrate the predictive performance of 2-class classification. In this work, we use AUC score (area under the ROC curve) to measure the individual SVM weight
to the jth class. ROC curve [32] is a frequently-used statistical tool to illustrate the predictive performance of 2-class classification. In this work, we use AUC score (area under the ROC curve) to measure the individual SVM weight 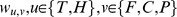 :
:
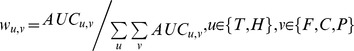 |
(5) |
where 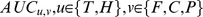 can be derived by 2-fold cross validation on the training set. The individual
can be derived by 2-fold cross validation on the training set. The individual 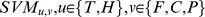 adopts Gaussian kernel defined as follows:
adopts Gaussian kernel defined as follows:
| (6) |
where  denotes 2-norm of a vector, and the hyperparameter
denotes 2-norm of a vector, and the hyperparameter  controls the flexibility of the kernel.
controls the flexibility of the kernel.
Model Evaluation and Model Selection
We design three experimental settings, namely the Optimistic case, the Moderate case and the Pessimistic case, to validate the assumptions that the homolog GO information is useful to tackle the problems of data scarcity and data unavailability. To formally define the three cases, we first define the following sets:
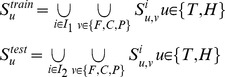 |
(7) |
where 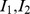 denote the training set and the test set,
denote the training set and the test set, 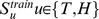 denotes the target GO term set and the homolog GO term set of the training data,
denotes the target GO term set and the homolog GO term set of the training data, 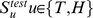 denotes the target GO term set and the homolog GO term set of the test data. Based on the notations, we can formally define the three cases as follows:
denotes the target GO term set and the homolog GO term set of the test data. Based on the notations, we can formally define the three cases as follows:
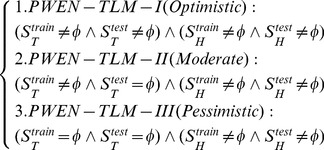 |
(8) |
From the formula, we can see that both the training set and the test set abound in target GO information in the Optimistic case, the test set contains no target GO information in the Moderate case, and neither the training set nor the test set contains target GO information in the Pessimistic case. In the Moderate case, we substitute the homolog GO information 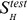 for the missing target GO information
for the missing target GO information 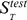 . In the Pessimistic case, we use the homolog GO information (
. In the Pessimistic case, we use the homolog GO information (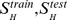 ) alone.
) alone.
We conduct model estimation and mode selection by two-level cross validation. The outer 3-fold cross validation is conducted for model estimation and the inner 2-fold cross validation is conducted to derive the weights of individual SVM classifiers. For the outer 3-fold cross validation, the dataset is randomly divided into three nearly-even disjoint subsets that have the same distributions as the original dataset (stratified cross validation). For each outer fold, one subset is used as test set and the other two subsets are merged as training set, which repeat three times until all data are estimated. Within each outer fold, 2-fold inner cross validation is further conducted for weight derivation on the training set.
HIV-1 protein can be catalogued as Env, Gag, Nef, Pol, Rev, Tat, Vif, Vpr and Vpu [21]. For the sake of critical assessment of model performance, we also conduct several independent tests by treating one catalogue of HIV-1 proteins (e.g. Env) as independent test set and the other catalogues of HIV-1 proteins (e.g. Gag, Nef, Pol, Rev, Tat, Vif, Vpr, Vpu) are merged as training set. In such a way, the independent test is more challenging because the test data (e.g. Env) have no corresponding training data in the training set (e.g. Gag, Nef, Pol, Rev, Tat, Vif, Vpr, Vpu). Wide variance between the test set and the training set helps conduct more critical performance estimation on the proposed model.
The model performance is measured by Receiver Operating Characteristic (ROC) AUC (Area Under Curve) (ROC-AUC), Precision recall curve AUC (PR-AUC), Specificity (SP), Sensitivity (SE) and MCC (Matthews correlation coefficient). The performance metrics SP, SE and MCC can be calculated through confusion matrix M. By means of the intermediate variables defined as formula (9), we can calculate SP, SE and MCC for each label (SPl, SEl and MCCl) by formula (10), and further calculate the overall accuracy (Acc) and the overall MCC (MCC) by formula (11).
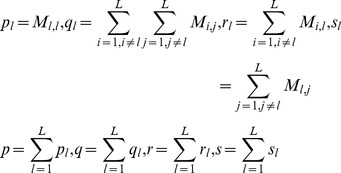 |
(9) |
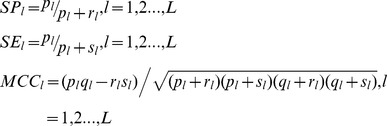 |
(10) |
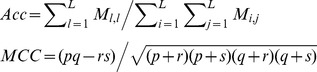 |
(11) |
where the confusion matrix 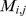 records the counts that class
records the counts that class  are classified to class
are classified to class 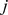 and L denotes the number of labels. AUC is calculated based on the weighted SVM decision values.
and L denotes the number of labels. AUC is calculated based on the weighted SVM decision values.
Results
Data and Materials
The interactions between HIV-1 and human proteins are taken from the database available at http://www.ncbi.nlm.nih.gov/projects/RefSeq/HIVInteractions/ [21]. In order to acquire corresponding gene ontology annotations, we map the protein accessions to Uniprot accessions via the id mapping file available at ftp://ftp.uniprot.org/pub/databases/uniprot/current_release/knowledgebase/idmapping/idmapping.dat.gz. After removing duplicate PPIs and putative PPIs, we totally get 3,638 PPIs including 539 Env PPIs, 487 Gag PPIs, 349 Nef PPIs, 272 Pol PPIs, 278 Rev PPIs, 1,101 Tat PPIs, 126 Vif PPIs, 338 Vpr PPIs and 148 Vpu PPIs. All the PPIs are treated as positive data. As far, there is no gold-standard negative data available for model training and model assessment. How to construct negative data is still a challenging problem for PPI prediction. At present, the common practice to generate negative data is random sampling from the huge protein-protein pair space exclusive of those experimentally derived PPIs. Unbiased as it is, random sampling would probably introduce a certain level of noise. For the reason, the work [33] proposed to exclude those subcellular co-localized proteins out of the negative data (hereinafter called exclusiveness of subcellular co-localized proteins), based on the common sense that subcellular co-localization is the premise of protein-protein physical interaction. But even so, the method still received criticism that the information about protein subcellular localization is likely to dominate the prediction and thus yields bias. In this work, we will compare the two methods of negative data sampling and investigate whether or not exclusiveness of subcellular co-localized proteins yields model bias. For simplicity of reference, we call S1 the dataset with negative data exclusive of subcellular co-localized proteins, and S2 the dataset with randomly sampled negative data. Dataset S1 and S2 both contain 3,638 positive data and 3,638 negative data.
How to determine the ratio of positive data to negative data is a second concern to be addressed. The work [7], [8] solved the problem by introducing different ratio of positive data to negative data (e.g. 1∶1, 1∶100) to train the model. Actually, the true ratio is hard to determine and pooling so large a negative data makes little sense to computational modelling. Contrarily, the adverse effect is that extremely unbalanced training data would yield a highly biased model. For the reason, we construct a negative data with the same size as the positive data. To randomly select a quality and representative negative data is a hard and important problem to computational biologists, though maybe not so appealing to experimental biologists. For reliable computational modelling, experimental evidences of negatome should be collected and made available to academic use.
Model comparison is a third concern for the reasons: (1) there is no standard benchmark data available for model evaluation and comparison; (2) some positive data are outdated and some novel positive data are included; (3) random sampling of negative data yields different training data; (4) there are no identical data partition of cross validation, etc. Hence, what we can do is to conduct critical assessment on the proposed model and conduct a rough comparison with other models for biologists’ reference.
Model Performance Evaluation
Cross validation performance evaluation
Dataset S1 totally contains 7,672 data including 3,638 positive data and 3,638 negative data. The ROC curve for 3-fold cross validation on dataset S1 is shown in Figure 1, where the ROC curves are drawn for the three cases. In the Optimistic case, PWEN-TLM achieves ROC-AUC score 0.9326, a little better than SMLR (ROC-AUC score 0.919) [7] that combined 16 catalogs of feature information including gene ontology. From Figure 1, we can see that PWEN-TLM performs the best in the Optimistic case (AUC = 0.9326), the second in the Pessimistic case (AUC = 0.8735) and the worst in the Moderate case (AUC = 0.8156). The relatively small performance difference between the Optimistic case and the Pessimistic case (ROC-AUC score difference = 0.0591) demonstrates that PWEN-TLM still works soundly when the target GO information is not available, and thus the homolog GO information can be treated as an effective substitute for the potentially unavailable target GO information. If the protein pair to be predicted contains novel protein, we can choose the model that is trained for the Pessimistic case. The results that PWEN-TLM performs the worst in the Moderate case can be explained that the heterogeneous distribution between 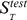 and
and 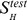 deteriorates the model performance. The performance deterioration reveals that data unavailability is an important concern to be addressed for computational modelling. The data integration model SMLR
[7] did not deliberately dwell on the problem of data unavailability.
deteriorates the model performance. The performance deterioration reveals that data unavailability is an important concern to be addressed for computational modelling. The data integration model SMLR
[7] did not deliberately dwell on the problem of data unavailability.
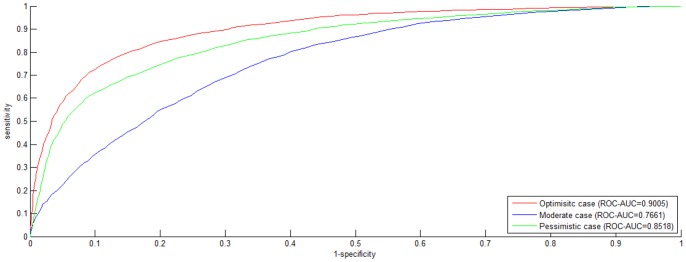
Figure 1. ROC curve on S1 dataset.
The negative data is constructed by the negative data sampling method of exclusiveness of subcellular co-localized proteins. The ROC curves in red, blue and green indicate the performance for the Optimistic case, the Moderate case and the Pessimistic case, respectively.
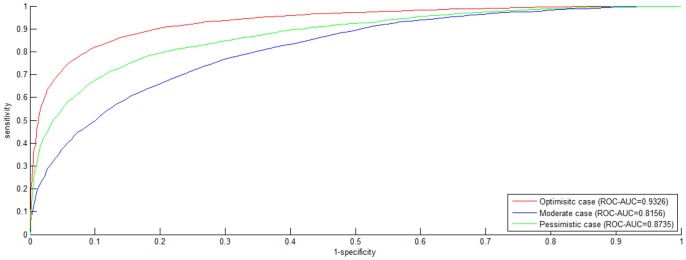
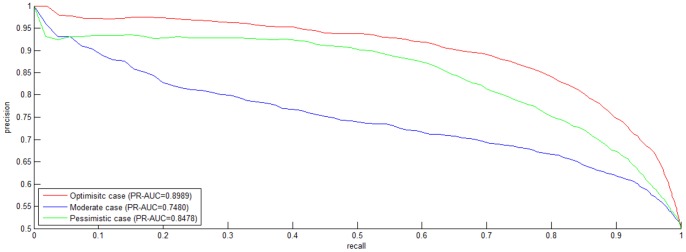
Dyer et al. [8] adopted PR-AUC (AUC of Precision-Recall Curve) as the performance metric of HIV-human PPI prediction. In their work, the best PR-AUC score among different ratios of positive data to negative data is 0.707. As compared to ROC Curve, Precision-Recall Curve is more suited to highly skewed (extremely unbalanced) data [34]. For comparison, we also plot Precision-Recall Curve and indicate the corresponding PR-AUC score in Figure 2. As shown in Figure 2, PWEN-TLM achieves PR-AUC score 0.9361, 0.8172 and 0.8799 in the Optimistic case, the Moderate case and the Pessimistic case, respectively. The PR-AUC scores demonstrate that PWEN-TLM significantly outperforms the baseline model (PR-AUC score 0.707) [7]. By comparing Figure 1 and Figure 2, we can see that there is little difference between ROC-AUC score and PR-AUC score. The reason is that dataset S1 is not skewed but perfectly balanced with 1∶1 ratio of positive data to negative data. Skewed training data is prone to yield a biased model.
Figure 2. Precision-Recall (PR) curve on S1 dataset.
The negative data is constructed by the negative data sampling method of exclusiveness of subcellular co-localized proteins. The PR curves in red, blue and green indicate the performance for the Optimistic case, the Moderate case and the Pessimistic case, respectively.
ROC curve plots the true positive rate against the false positive rate and Precision-Recall curve plots the precision against recall. Both the curves focus on the reliability of positive predictions, but the negative class is largely ignored. For 2-class classification, predictive balance is an important aspect of model performance. Highly biased predictions are not reliable. From the point of view of biomedical research, true protein-protein non-interaction (i.e. true negative) also provides much insight into functional proteomics and drug research. Hence we also report SP, SE, MCC and Accuracy for comprehensive survey of model performance. As shown in Table 1, PWEN-TLM achieves good predictive balance in the Optimistic case (Acc = 85.62%, MCC = 0.7446) and in the Pessimistic case (Acc = 80.22%, MCC = 0.6605). But PWEN-TLM shows bias towards the positive class in the Moderate case (Acc = 66.22%, MCC = 0.4606, positive SP = 0.6015, negative SE = 0.3631). Comparatively, AUC scores do not detect the bias (ROC-AUC score = 0.8156, PR-AUC score = 0.8172), implying that the performance metrics of SP, SE, MCC and Accuracy are important to model estimation. Summarizing all the performance metrics, we can see that PWEN-TLM performs well in the Optimistic case and in the Pessimistic case. If the target GO information of the test data is available, we choose the model trained in the Optimistic case; otherwise, we choose the model trained in the Pessimistic case.
Table 1. Cross validation performance estimation on dataset S1.
| PWEN-TLM-I (Optimistic) | PWEN-TLM-II (Moderate) | PWEN-TLM-III (Pessimistic) | |||||||
| SP | SE | MCC | SP | SE | MCC | SP | SE | MCC | |
| Positive (interacting) | 0.8774 | 0.8282 | 0.7439 | 0.6015 | 0.9612 | 0.5733 | 0.8160 | 0.7804 | 0.6589 |
| Negative (non-interacting) | 0.8373 | 0.8843 | 0.7471 | 0.9036 | 0.3631 | 0.4604 | 0.7896 | 0.8241 | 0.6631 |
| [AUC;Acc;MCC] | [0.9326; 85.62%; 0.7446] | [0.8155; 66.22%; 0.4606] | [0.8735;80.22%;0.6605] | ||||||
To attenuate the noise from the homolog GO information, we explicitly investigate the importance of the three aspects of gene ontology (molecular function, cellular component, biological process) to HIV-human PPI prediction. As illustrated in Figure 3, the target GO information and the homolog GO information contribute equivalently to the model performance in the Optimistic case. In the Moderate case, the target GO information unexpectedly makes less contribution than the homolog GO information. The result is not surprising, because we substitute the homolog GO information  for the missing target GO information
for the missing target GO information 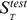 to derive the weights of the target GO information. The heterogeneous distribution between
to derive the weights of the target GO information. The heterogeneous distribution between 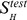 and
and 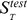 unjustly decreases the importance of the target GO information. The three aspects of gene ontology unexceptionally make equivalent contributions to the model performance in all the three cases. The GO information about cellular component does not predominate the contributions to model performance, indicating that the negative data constructed by exclusiveness of subcellular co-localized proteins does not yield predictive bias as worried about.
unjustly decreases the importance of the target GO information. The three aspects of gene ontology unexceptionally make equivalent contributions to the model performance in all the three cases. The GO information about cellular component does not predominate the contributions to model performance, indicating that the negative data constructed by exclusiveness of subcellular co-localized proteins does not yield predictive bias as worried about.
Figure 3. Individual SVM weight distribution on S1 dataset.
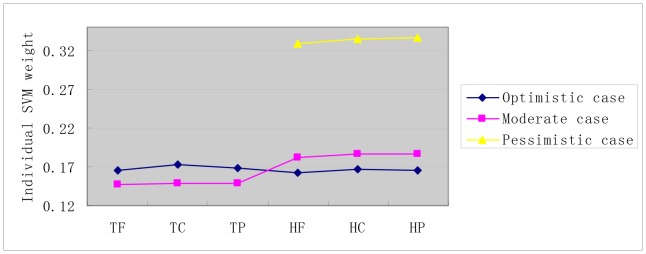
The negative data is constructed by the negative data sampling method of exclusiveness of subcellular co-localized proteins. The horizontal axis is the combination of two sets {T, H} and {F,C, P}. T denotes the target protein, H denotes the homolog protein; F denotes molecular function, C denotes cellular component and P denotes biological process.
Dataset S2 similarly contains 7,672 data including 3,638 positive data and 3,638 negative data, with the exception to dataset S1 that the negative data are randomly sampled. The ROC curve and the PR curve are plotted in Figure 4 and Figure 5. Comparing Figure 1 with Figure 4 and Figure 2 with Figure 5, we can see that dataset S1 achieves higher ROC-AUC score and PR-AUC score than dataset S2 for all the three cases. The highest difference of ROC-AUC score is 0.0495 and the highest difference of PR-AUC score is 0.0692. Table 2 demonstrates the performance metrics of SP, SE, MCC and Accuracy on dataset S2. Comparing Table 1 and Table 2, we can see that dataset S1 demonstrates much better predictive balance than dataset S2, with highest MCC difference 0.1053. The results demonstrate that exclusiveness of subcellular co-localized proteins is more reliable to construct a reliable and unbiased classifier than random sampling.
Figure 4. ROC curve on S2 dataset.
The negative data is constructed by by the negative data sampling method of random sampling. The ROC curves in red, blue and green indicate the performance for the Optimistic case, the Moderate case and the Pessimistic case, respectively.
Figure 5. Precision-Recall (PR) curve on S2 dataset.
The negative data is constructed by by the negative data sampling method of random sampling. The PR curves in red, blue and green indicate the performance for the Optimistic case, the Moderate case and the Pessimistic case, respectively.
Table 2. Cross validation performance estimation on dataset S2.
| PWEN-TLM-I (Optimistic) | PWEN-TLM-II (Moderate) | PWEN-TLM-III (Pessimistic) | |||||||
| SP | SE | MCC | SP | SE | MCC | SP | SE | MCC | |
| Positive (interacting) | 0.8413 | 0.7988 | 0.6926 | 0.5832 | 0.9560 | 0.5515 | 0.7811 | 0.7622 | 0.6175 |
| Negative (non-interacting) | 0.8085 | 0.8494 | 0.6966 | 0.8780 | 0.3167 | 0.4143 | 0.7678 | 0.7864 | 0.6204 |
| [AUC;Acc;MCC] | [0.9005;82.41%;0.6393] | [0.7661;63.63%;0.4258] | [0.8518;77.43%;0.6188] | ||||||
The weight distribution for the three aspects of gene ontology is illustrated in Figure 6. Comparing Figure 3 with Figure 6, we can see that there is little difference of weight distribution between dataset S1 and dataset S2.
Figure 6. Individual SVM weight distribution on S2 dataset.
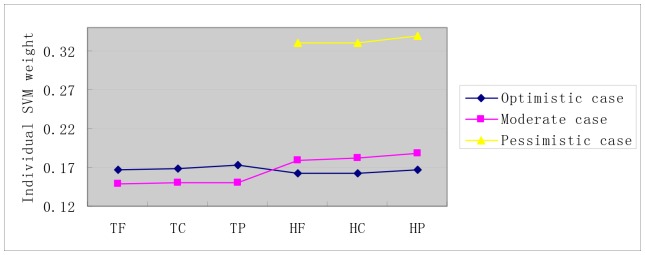
The negative data is constructed by by the negative data sampling method of random sampling. The horizontal axis is the combination of two sets {T, H} and {F,C, P}. T denotes the target protein, H denotes the homolog protein; F denotes molecular function, C denotes cellular component and P denotes biological process.
Independent test performance evaluation
The HIV-human PPI database [18] is catalogued into 9 categories (539 Env PPIs, 487 Gag PPIs, 349 Nef PPIs, 272 Pol PPIs, 278 Rev PPIs, 1,101 Tat PPIs, 126 Vif PPIs, 338 Vpr PPIs and 148 Vpu PPIs). To evaluate the generalization ability, we simply use one catalog of PPI (e.g. Env PPI) as independent test set and other catalogs of PPIs (e.g. Gag, Nef, Pol, Rev, Tat, Vif, Vpr, Vpu) are merged together as positive training set. The corresponding negative training set is derived for each catalog of HIV protein with the constraints: (1) the negative PPIs and the positive PPIs are of the same size; (2) the human proteins that are subcellular co-localized with the HIV proteins are excluded; (3) the human proteins are randomly sampled. Constraint (2) is based on the above experimental conclusion that exclusiveness of subcellular co-localized proteins yields unbiased and better performance. We don’t conduct independent test for the Moderate case because of its poor performance in the cross validation performance evaluation.
The experimental results of the independent test are shown in Table 3. We can see that PWEN-TLM can recognize most catalogs of HIV-human PPIs with high recall rate except one small Pol PPIs (272 PPIs, Optimistic 51.84%, Pessimistic 54.04%) and one large Tat PPIs (1,101 PPIs, Optimistic 52.04%, Pessimistic 55.77%). As compared to the generally small overlap between experimental host-pathogen PPIs and predicted host-pathogen PPIs, e.g. 10% overlap between siRNA screen and predictions [7] and 5.29% recall rate (57 PPIs were computationally recognized out of the 1,078 experimental PPIs) [10], the results are considerably promising. From the results, we also see that the Optimistic case is unsurprisingly better than the Pessimistic case, because the target GO information is available. Nevertheless, PWEN-TLM still works well in the Pessimistic case. The independent test again validates the assumption that the homolog GO information alone is sufficient to train a satisfactory HIV-human PPI classifier.
Table 3. Independent test performance estimation.
| env | gag | nef | pol | rev | tat | vif | vpr | vpu | |
| 539 | 487 | 349 | 272 | 278 | 1,101 | 126 | 338 | 148 | |
| PWEN-TLM-I (Optimistic) | 68.46% | 81.34% | 88.54% | 51.84% | 64.75% | 52.04% | 88.89% | 80.77% | 66.89% |
| PWEN-TLM-III (Pessimistic) | 67.53% | 65.91% | 77.36% | 54.04% | 52.88% | 55.77% | 87.30% | 81.66% | 66.89% |
Novel PPI Prediction
Overlap analysis of predicted interactions between PWEN-TLM and the existing models
Overlap analysis of predicted interactions between different computational models is of significance to reveal the confidence and complementariness of predictions. In this work, we investigate the overlap of predictions between PWEN-TLM and the latest bi-clustering method [20], for the reason that bi-clustering has found several supporting evidences from the recent literature. In bi-clustering method, there are 180 predicted interactions, among which there are 80 interactions overlapped with the work [6]. As pointed out in the work [20], some predicted interactions have been validated by the recent literatures, e.g. env_gp120:CASP8[83.33%] [35], env_gp120:CD86[83.33%] [36], env_gp120: NOS3[74.67%] [37], env_gp120:SOD2[88.89%] [38,], env_gp120:SRC[78%] [39], env_gp41:MAPK1[77.78%] [40], Gag_Pr55:MAPK1[71.43%] [41], Tat:TNFSF[86.30%]) [42]. The square bracketed percentage following the protein pair denotes the confidence level of predictions.
We apply PWEN-TLM to validate the 180 predicted interactions for overlap analysis. Among the HIV-1 proteins, the protein env_gp120 (Envelope surface glycoprotein gp120, NP_579894.2) has no reviewed entry in the UniprotKB database (http://www.uniprot.org/uniprot/). The target GO information of protein env_gp120 can not be retrieved from the database and thus is treated as novel protein in our model. For reliable training, env_gp120 is not included in the training data. Thus the training data is more stringent than that of bi-clustering method, because it contains no interaction patterns between env_gp120 and human proteins. The 180 interactions predicted by bi-clustering method are treated as test data without overlap with the training data.
The experimental results show that PWEN-TLM predicts 132 interactions in the Optimistic case (File S1) and 165 interactions in the Pessimistic case (File S2). Comparing the results of the two cases, we find that PWEN-TLM can not recognize most env_gp120 interactions in the Optimistic case, but PWEN-TLM behaves contrarily very well in the Pessimistic case. The results are not surprising because the unreviewed env_gp120 is treated as novel protein (the target GO information is treated as null and only the homolog GO information takes effect). In the Pessimistic case, PWEN-TLM correctly recognizes all the literature-validated interactions (env_gp120:CASP8[81.63%];env_gp120:CD86[83.51%];env_gp120:NOS3[84.34%];env_gp120:SOD2[70.78%];env_gp120:SRC[79.54%];env_gp41:MAPK1[89.88%];Tat:TNFSF[90.90%]) except Gag_Pr55:MAPK1 [20]. The results once again validate our model assumption that the homolog GO information can be effectively exploited to compensate for data scarcity and data unavailability. Especially, we can safely draw the conclusion that the homolog GO information alone is sufficient to train a satisfactory model for HIV-human PPI prediction. We can see that PWEN-TLM has less demanding data constraint and hardly fails to work even in the worst case (the Pessimistic case). As long as GO annotated homologs can be retrieved, PWEN-TLM can convincingly predict the protein pairs that contain novel proteins. It is noted that although the model is trained without env_gp120 interaction patterns, the env_gp120-related interactions are still soundly recognized, which implies that PWEN-TLM has good generalization ability.
Besides the validation of the 180 predicted interactions, we also validate against PWEN-TLM the 80 overlapped interactions between the two work [6], [20]. The results show that PWEN-TLM predicts 46 interactions in the Optimistic case (File S3) and 61 interactions in the Pessimistic case (File S4). From the results, we can see that PWEN-TLM narrows down the predictions and thus is relatively more conservative than the bi-clustering method [20]. Conservative prediction has the merit of low false positive rate but meanwhile has the demerit of missing some true interactions (e.g. Gag_Pr55:MAPK1). From the 8 literature-validated interactions, only one unrecognized interaction is acceptable.
Predicted interactions with peripheral human proteins
In addition to validating the interactions predicted by the existing models, we also independently apply PWEN-TLM to detect novel HIV-human PPIs for further biological research. To narrow down the scope of potential HIV-targeted human proteins, we first statistically investigate the way that HIV proteins attack the human PPI network. Some diseases, like lung squamous cell carcinoma [43], are prone to attack the densely-connected human proteins (hub proteins). Here we attempt to acquire the knowledge about the behaviour that HIV-1 attacks the human PPI network. We can calculate the degree distribution of the HIV-targeted human proteins from HPRD database (http://hprd.org/) [44]. The degree distribution of the HIV-targeted human proteins in human PPI network is plotted in Figure 7, where the horizontal axis denotes the protein degree and the vertical axis denotes the number of proteins possessing that degree. From Figure 7, we can intuitively see that the number of HIV-targeted human proteins exponentially decreases with protein degree. It can be inferred from the figure that the HIV proteins are prone to target the peripheral human proteins. For the sake, we choose the peripheral human proteins as test candidates. For each type of HIV proteins, we randomly choose 400 distinct human proteins with lowest degree (e.g. degree = 1, 2, 3) that do not occur in dataset S1. The predicted results are shown in File S5 (Optimistic case) and File S6 (Pessimistic case). Since literature could offer very sparse direct information about the interactions we are concerned about, we analyse the predicted interactions based on the study of gene ontology.
Figure 7. Degree distribution of the HIV-targeted human proteins in human PPI network.
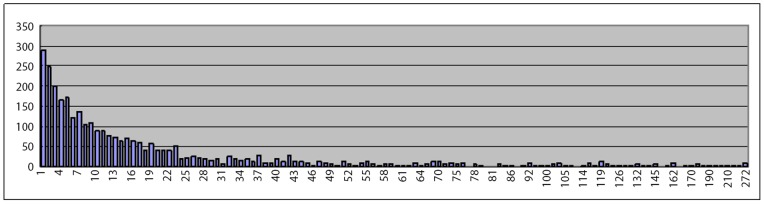
The horizontal axis denotes protein degree and the vertical axis denotes the number of proteins that possess that degree.
Interactions with env_gp160
Among the 400 human proteins, PWEN-TLM predicts 64 interactions with env_gp160 (P04578) in the Optimistic case (File S5) and 66 interactions in the Pessimistic case (File S6). After filtering the weak interactions (probability within [0.5, 0.6]), there are 44 interactions in the Pessimistic case and 45 interactions in the Optimistic case. Take the Optimistic case for example, Table 4 clusters the interacting human partners according to GO terms (see Table 4 Main cluster of interacting human partners). From Table 4, we can see that env_gp160 mainly interacts with the host membrane proteins (GO:0016020, GO:0016021, GO:0005886), and the interacting human partners are mainly involved in the biological processes of metabolic process (GO:0044267), post-translational modification (GO:0043687, GO:0006486), transport (GO:0006810), host immune response (GO:0006954, GO:0045087), etc. From the aspect of molecular functions, env_gp160 mainly affects host protein binding activity (GO:0005515), transferase activity (GO:0016740), etc. From the analysis of gene ontology, we can see that the interactions with env_gp120 may affect the metabolic process, molecule transfer, binding activity of the host proteins and may also activate the host immune response.
Table 4. Predicted interactions between env_gp160 and human proteins.
| GOcategory | Predicted interacting human partners | |||
| GO term | GO description | Rate | Main cluster of interacting human partners | |
| Biological process | GO:0044267 | cellular proteinmetabolic process | 23% | Q9NR34[0.91];Q5I7T1[0.67];Q9NYU2[0.92];Q8N3T1[0.71];Q9NY97[0.86];O43173[0.70];Q13454[0.72];Q9BV94[0.81];O60476[0.91];Q8IUC8[0.70] |
| GO:0043687 | post-translationalproteinmodification | 23% | Q9NR34[0.91];Q5I7T1[0.67];Q9NYU2[0.92];Q8N3T1[0.71];Q9NY97[0.86];O43173[0.70];Q13454[0.72];Q9BV94[0.81];O60476[0.91];Q8IUC8[0.70] | |
| GO:0006486 | proteinglycosylation | 18% | Q9NR34[0.91];Q5I7T1[0.67];Q9NYU2[0.92];Q8N3T1[0.71];Q9NY97[0.86];O43173[0.70];O60476[0.91];Q8IUC8[0.70] | |
| GO:0006457 | protein folding | 16% | Q9UDY4[0.79];Q9NYU2[0.92];O14967[0.87];O60884[0.79];P30414[0.78];Q14696[0.81];Q9BV94[0.81] | |
| GO:0006810 | transport | 9% | P13866[0.75];Q01650[0.86];Q13454[0.72];>O75947[0.73] | |
| GO:0006954 | inflammatoryresponse | 9% | O43916[0.87];Q96E93[0.78];Q9NYK1[0.62];Q9H293[0.62] | |
| GO:0045087 | innate immuneresponse | 7% | Q96E93[0.78];Q9NYK1[0.62];Q9NY25[0.61] | |
| Cellular component | GO:0016020 | membrane | 77% | O43916[0.87];Q86Z14[0.72];Q9NR34[0.91];Q9UDY4[0.79];O75509[0.63];P13866[0.75];Q96E93[0.78];Q8IXI1[0.82];Q5I7T1[0.67];O14967[0.87];Q8N3T1[0.71];Q01650[0.86];P41732[0.63];O75096[0.61];O60884[0.79];Q9NY97[0.86];P30414[0.78];O43173[0.70];P06126[0.82];Q9NYK1[0.62];P04062[0.79];Q13454[0.72];P43626[0.66];Q9P035[0.89];Q9NY25[0.61];O75947[0.73];Q9H293[0.62];Q6UW60[0.69];P23435[0.65];Q9BV94[0.81];O14548[0.71];O60476[0.91];[0.70];P09669[0.77] |
| GO:0016021 | integral tomembrane | 57% | O43916[0.87];Q86Z14[0.72];Q9NR34[0.91];O75509[0.63];P13866[0.75];Q96E93[0.78];Q8IXI1[0.82];Q5I7T1[0.67];O14967[0.87];Q8N3T1[0.71];Q01650[0.86];P41732[0.63];O75096[0.61];Q9NY97[0.86];O43173[0.70];P06126[0.82];Q9NYK1[0.62];Q13454[0.72];P43626[0.66];Q9P035[0.89];Q9NY25[0.61];Q6UW60[0.69];O60476[0.91];Q8IUC8[0.70];P09669[0.77] | |
| GO:0005886 | plasma membrane | 32% | Q86Z14[0.72];Q9UDY4[0.79];O75509[0.63];P13866[0.75];Q96E93[0.78];Q8IXI1[0.82];Q5I7T1[0.67];Q01650[0.86];O75096[0.61];P06126[0.82];Q9NYK1[0.62];P43626[0.66];Q9NY25[0.61];Q14696[0.81] | |
| GO:0000139 | Golgi membrane | 18% | O43916[0.87];Q9NR34[0.91];Q8N3T1[0.71];Q9NY97[0.86];O43173[0.70];Q9NYK1[0.62];O60476[0.91];Q8IUC8[0.70] | |
| Molecular function | GO:0016787 | hydrolase activity | 20% | Q9NR34[0.91];Q8IXI1[0.82];O00754[0.85];P04062[0.79];P04746[0.63];Q9NTJ4[0.74];Q9NRW3[0.67];Q6UW60[0.69];O60476[0.91] |
| GO:0016740 | transferase activity | 18% | O43916[0.87];Q08188[0.81];Q5I7T1[0.67];Q9NYU2[0.92];Q8N3T1[0.71];Q9NY97[0.86];O43173[0.70];Q8IUC8[0.70] | |
| GO:0005515 | protein binding | 18% | Q9UDY4[0.79];O75509[0.63];P13866[0.75];Q8IXI1[0.82];O75096[0.61];P13667[0.87];P06126[0.82];P43626[0.66] | |
| GO:0051082 | unfolded proteinbinding | 11% | Q9UDY4[0.79];Q9NYU2[0.92];O14967[0.87];O14657[0.61];O60884[0.79] | |
Illustrations:
[1] Rate denotes that the cluster of interacting human proteins possessing the same corresponding GO term accounts for the total predicted env_gp160-interacting human proteins;
Interactions with Rev
Among the 400 human proteins, PWEN-TLM predicts 37 interactions with Rev (P04618) in the Optimistic case and 54 interactions in the Pessimistic case (probability>0.6). From Table 5, we can see that Rev mainly interacts with the host nucleus proteins (GO:0005634) and cytoplasm proteins (GO:0005737), and participates in the biological processes of viral reproduction (GO:0010467, GO:0019083, GO:0016032, GO:0006355), viral mRNA translation (GO:0016071, GO:0006413, GO:0006414), etc. These predicted interactions indicate that Rev plays important roles in viral mRNA transcription and mRNA translation into viral proteins.
Table 5. Predicted interactions between Rev and human proteins.
| GOcategory | Predicted interacting human partners | |||
| GO term | GO description | Rate | Main cluster of interacting human partners | |
| Biological process | GO:0010467 | gene expression | 14% | Q9P2I0[0.87];P46781[0.90];P62899[0.90];Q9Y3U8[0.86];P62280[0.82] |
| GO:0019083 | viral transcription | 11% | P46781[0.90];P62899[0.90];Q9Y3U8[0.86];P62280[0.82] | |
| GO:0016071 | mRNA metabolicprocess | 11% | P46781[0.90];P62899[0.90];Q9Y3U8[0.86];P62280[0.82] | |
| GO:0016032 | viral reproduction | 11% | P46781[0.90];P62899[0.90];Q9Y3U8[0.86];P62280[0.82] | |
| GO:0006355 | regulation oftranscription,DNA-dependent | 11% | Q9NRC8[0.61];O00472[0.60];Q92925[0.64];Q13342[0.65] | |
| GO:0006413 | translationalinitiation | 11% | P46781[0.90];P62899[0.90];Q9Y3U8[0.86];P62280[0.82] | |
| GO:0006414 | translationalelongation | 11% | P46781[0.90];P62899[0.90];Q9Y3U8[0.86];P62280[0.82] | |
| Cellular component | O:0005634 | nucleus | 41% | Q9H1A4[0.69];Q9P2I0[0.87];Q99877[0.82];Q8IX01[0.62];Q8WWL7[0.70];Q9NRC8[0.61];O00472[0.60];Q9NYP9[0.66];Q92925[0.64];Q9H668[0.66];Q13342[0.65];Q15003[0.67];Q8NDV3[0.74];Q13601[0.76];O15523[0.63] |
| GO:0005737 | cytoplasm | 38% | Q96C10[0.74];P09972[0.63];Q9UBB4[0.76];Q08188[0.68];P46781[0.90];Q06210[0.70];Q01650[0.75];Q9NRC8[0.61];Q9NYP9[0.66];Q9Y3U8[0.86];Q13342[0.65];Q15003[0.67];Q13601[0.76];O15523[0.63] | |
| GO:0005829 | cytosol | 30% | Q9H1A4[0.69];P09972[0.63];Q9UBB4[0.76];Q8IXI1[0.72];P46781[0.90];Q06210[0.70];Q01650[0.75];P62899[0.90];Q9Y3U8[0.86];Q9BU89[0.66];P62280[0.82] | |
| GO:0005730 | nucleolus | 22% | P46781[0.90];Q8WWL7[0.70];Q9NRC8[0.61];O00472[0.60];Q9H668[0.66];Q9Y3U8[0.86];Q13342[0.65];Q13601[0.76] | |
| Molecular function | GO:0005515 | protein binding | 35% | Q96C10[0.74];P09972[0.63];Q9UBB4[0.76];P13866[0.64];Q9P2I0[0.87];Q8IXI1[0.72];P46781[0.90];Q8WWL7[0.70];Q9NRC8[0.61];P06126[0.70];Q9H668[0.66];Q15003[0.67];Q9BU89[0.66] |
| GO:0003723 | RNA binding | 24% | Q96C10[0.74];Q9P2I0[0.87];Q9Y6V7[0.63];P46781[0.90];Q8IX01[0.62];P62899[0.90];Q13601[0.76];P62280[0.82];O15523[0.63] | |
| GO:0003677 | DNA binding | 16% | 96C10[0.74];Q99877[0.82];Q9H668[0.66];Q13342[0.65];Q8NDV3[0.74];O15523[0.63] | |
Interactions with Vpr
Similarly, the predicted interactions with Vpr (Q77YF9) are shown in Table 6 (18 predicted interactions with probability greater than 0.6). From the results we can see that Vpr mainly affects the host cell cycle (GO:0007049, GO:0051301, GO:0007067, GO:0030261, GO:0007126) and the regulation of DNA transcription (GO:0006355). The predicted interactions are consistent with our prior knowledge about HIV-1 Vpr proteins.
Table 6. Predicted interactions between Vpr and human proteins.
| GOcategory | Predicted interacting human partners | |||
| GO term | GO description | Rate | Main cluster of interacting human partners | |
| Biological process | GO:0007049 | cell cycle | 22% | Q9H1A4[0.63];Q8WWL7[0.72];Q15003[0.61];Q8NDV3[0.82] |
| GO:0051301 | cell division | 17% | Q9H1A4[0.63];Q8WWL7[0.72];Q15003[0.61] | |
| GO:0007067 | mitosis | 11% | Q9H1A4[0.63];Q15003[0.61] | |
| GO:0022904 | respiratory electrontransport chain | 17% | O75947[0.64];O14548[0.74];P09669[0.77] | |
| GO:0030261 | chromosomecondensation | 11% | Q15003[0.61];Q8NDV3[0.82] | |
| GO:0006355 | regulation oftranscription,DNA-dependent | 11% | P58012[0.61];Q13342[0.67] | |
| GO:0007126 | meiosis | 11% | Q8WWL7[0.72];Q8NDV3[0.82] | |
| Cellular component | GO:0005634 | nucleus | 50% | Q9H1A4[0.63];Q9UDY4[0.62];Q99877[0.64];Q8WWL7[0.72];P58012[0.61];Q9H668[0.61];Q13342[0.67];Q15003[0.61];Q8NDV3[0.82] |
| GO:0016020 | membrane | 44% | Q9UDY4[0.62];Q96E93[0.61];Q8IXI1[0.64];P30414[0.69];P04062[0.64];O75947[0.64];O14548[0.74];P09669[0.77] | |
| GO:0005730 | nucleolus | 22% | Q9UDY4[0.62];Q8WWL7[0.72];Q9H668[0.61];Q13342[0.67] | |
| GO:0005737 | cytoplasm | 22% | Q9UDY4[0.62];Q13342[0.67];Q15003[0.61];Q5SW79[0.68] | |
| Molecular function | GO:0005515 | protein binding | 39% | Q9UDY4[0.62];Q8IXI1[0.64];Q8WWL7[0.72];P58012[0.61];Q9H668[0.61];Q15003[0.61];Q5SW79[0.68] |
| GO:0003677 | DNA binding | 28% | Q99877[0.64];P58012[0.61];Q9H668[0.61];Q13342[0.67];Q8NDV3[0.82] | |
| GO:0003700 | sequence-specific DNAbinding transcriptionfactor activity | 11% | P58012[0.61];Q13342[0.67] | |
Interactions with other HIV-1 proteins
The predicted interactions with other HIV-1 proteins (Gag, Pol, Tat, Vpu, Nef, Vif) are shown in File S5 and File S6. The experimental results show that Gag mainly interacts with the human proteins that participate in the biological processes of signal transduction (GO:0007165, the interacting partners include O75509[0.67]; Q9NYK1[0.69]; Q9NY25[0.63]; O60609[0.61]), innate immune response (GO:0045087, Q96E93[0.64]; Q9NYK1[0.69]; Q9NY25[0.63]), apoptotic process (GO:0006915, P09972[0.67];O75509[0.67];Q8IXI1[0.81]), etc. Tat mainly affects the regulation of transcription (GO:0006355, Q9NRC8[0.67]; O00472[0.74]; Q92925[0.72]; Q13342[0.80]), host cell defense response to virus (GO:0051607, Q96C10[0.67]; Q9NYK1[0.70]; Q9NRW3[0.69]), etc. Vpu mainly interacts with the human proteins of transport activity (GO:0006810, P13866[0.62]; O75947[0.86]), receptor activity (GO:0004872, Q96E93[0.70]; P22897[0.61]), cell death (GO:0008219, Q9UBB4[0.82]; P04062[0.78]), etc. Full interactions are shown in File S5 and File S6.
Discussion
Data scarcity, data unavailability and negative data sampling are the three major concerns to be addressed for the computational reconstruction of HIV-human PPI networks. At present feature-level data integration is still the major effective method to compensate for data scarcity, but potential unavailability of some feature information is likely to make the existing data integration methods fail to work. In this work, we are motivated to develop a less data-demanding computational model for HIV-human PPI prediction that hardly fails to work in most cases. We investigate the assumption that the homolog GO information is useful to well tackle the problems of data scarcity and data unavailability. To fulfil the motivation and assumption, we propose a probability weighted ensemble transfer learning model for HIV-human PPI prediction (PWEN-TLM). In this model, gene ontology is the only feature information used for model training and model evaluation. The target GO information and the homolog GO information are separately extracted to cope with data unavailability, and the three aspects of gene ontology are further separated to evaluate their contributions to the model performance. The contributions are measured in terms of weights by ROC-AUC performance metric of the individual classifiers. The weights of the homolog GO information play the role of enhancing positive knowledge transfer and depressing negative knowledge transfer.
To validate the assumption that the homolog GO information is effective to enrich or substitute for the target GO information, we conduct three experimental settings, namely the Optimistic case, the Moderate case and the Pessimistic case. The latter two cases take into account the unavailability of the target GO information. 3-fold cross validation and independent test are used to evaluate the model performance. The performance measured by multiple metrics (ROC-AUC, PR-AUC, MCC, SP, SE and Accuracy) show that PWEN-TLM performs well in the Optimistic case and in the Pessimistic case. The sound performance in the Optimistic case demonstrates that the homolog GO information is useful to solve the problem of data scarcity by enriching the target GO information. The good performance in the Pessimistic case shows that the homolog GO information is an effective substitute for the target GO information to solve the problem of data unavailability.
Negative data sampling is another important concern to be addressed for HIV-human PPI prediction. In this work, we have compared exclusiveness of subcellular co-localization to random sampling. We find that the GO information about cellular components makes equivalent contributions to the model performance as the GO information about biological processes and molecular functions does. This result shows that exclusiveness of subcellular co-localized proteins outperforms random sampling without introducing model bias.
Lastly, we apply PWEN-TLM to novel HIV-human PPIs detection. The overlap analysis of the predictions between PWEN-TLM and the existing models show that PWEN-TLM can recognize most of the literature-validated interactions and is relatively more conservative than the bi-clustering method. We also report some novel interactions for further biological research. The analysis based on gene ontology shows that the information revealed by the predicted interactions is consistent with our prior knowledge about the HIV-1 proteins.
Supporting Information
Text file contains the overlapped predictions between PWEN-TLM and Bi-clustering [20] ( Optimistic case ).
(TXT)
Text file contains the overlapped predictions between PWEN-TLM and Bi-clustering [20] ( Pessimistic case ).
(TXT)
Text file contains the overlapped predictions among PWEN-TLM , Bi-clustering [20] and the method [6] ( Optimistic case ).
(TXT)
Text file contains the overlapped predictions among PWEN-TLM , Bi-clustering [20] and the method [6] ( Pessimistic case ).
(TXT)
Text file contains the predictions between HIV-1 and peripheral human proteins ( Optimistic case ).
(TXT)
Text file contains the predictions between HIV-1 and peripheral human proteins ( Pessimistic case ).
(TXT)
Acknowledgments
Thanks for the helpful comments from the anonymous reviewers.
Funding Statement
This author has no support or funding to report.
References
- 1. von Mering C, Krause R, Snel B, Cornell M, Oliver SG, et al. (2002) Comparative assessment of large-scale datasets of protein-protein interactions. Nature 417: 399–403. [DOI] [PubMed] [Google Scholar]
- 2. Edwards A, Kus B, Jansen R, Greenbaum D, Greenblatt J, et al. (2002) Bridging structural biology and genomics: assessing protein interaction data with known complexes. Trends Genet 18: 529–536. [DOI] [PubMed] [Google Scholar]
- 3. Wu X, Zhu L, Guo J, Zhang D, Lin K (2006) Prediction of yeast protein-protein interaction network: insights from the Gene Ontology and annotations. Nucleic Acids Res 34: 2137–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. DeBodt S, Proost S, Vandepoele K, Rouzé P, Peer Y, et al. (2009) Predicting protein-protein interactions in Arabidopsis thaliana through integration of orthology, gene ontology and co-expression. BMC Genomics 10: 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shen J, Zhang J, Luo X, Zhu W, Yu K, et al. (2007) Predicting protein–protein interactions based only on sequences information. Proc Natl Acad Sci U S A 104: 4337–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tastan O, Qi Y, Carbonell J, Klein-Seetharaman J (2009) Prediction of interactions between HIV- 1 and human proteins by information integration. In: Proceedings of the Pacific Symposium on Biocomputing (PSB-2009): 516–527. [PMC free article] [PubMed] [Google Scholar]
- 7. Qi Y, Tastan O, Carbone J, Klein-Seetharaman, Weston J, et al. (2010) Semi-supervised multi-task learning for predicting interactions between HIV-1 and human proteins. Bioinformatics 26: i645–i652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dyer M, Muralib T, Sobrala B (2011) Supervised learning and prediction of physical interactions between human and HIV proteins. Infect Genet Evol 11: 917–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dyer M, Murali T, Sobral B (2007) Computational prediction of host-pathogen protein-protein interactions. Bioinformatics 23: i159–i166. [DOI] [PubMed] [Google Scholar]
- 10. Wuchty S (2011) Computational Prediction of Host-Parasite Protein Interactions between P. falciparum and H. sapiens. PLoS ONE 6: e26960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Miller J, Lo RS, Ben-Hur A, Desmarais C, Stagljar I, et al. (2005) Large-scale identification of yeast integral membrane protein interactions. Proc Natl Acad Sci U S A 102: 12123–12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lin N, Wu B, Jansen R, Gerstein M, Zhao H, et al. (2004) Information assessment on predicting protein-protein interactions. BMC Bioinformatics 5: 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Patil A, Nakamura H (2005) Filtering high-throughput protein-protein interaction data using a combination of genomic features. BMC Bioinformatics 6: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Maetschke S, Simonsen M, Davis M, Ragan MA (2012) Gene Ontology-driven inference of protein–protein interactions using inducers. Bioinformatics 28: 69–75. [DOI] [PubMed] [Google Scholar]
- 15. Qi Y, Bar-Joseph Z, Klein-Seetharaman J (2006) Evaluation of different biological data and computational methods for use in protein interaction prediction. Proteins 63: 490–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tirosh I, Barkai N (2005) Computational verification of protein-protein interactions by orthologous co-expression. BMC Bioinformatics 6: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yu H, Luscombe N, Lu H, Zhu X, Xia Y, et al. (2004) Annotation transfer between genomes: Protein-protein interologs and protein-dna regulogs. Genome Res 1: 1107–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Doolittle J, Gomez S (2010) Structural similarity-based predictions of protein interactions between HIV-1 and Homo sapiens. Virology J 7: 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yu J, Guo M, Needham C, Huang Y, Cai L, et al. (2010) Simple sequence-based kernels do not predict protein-protein interactions. Bioinformatics 26: 2610–2614. [DOI] [PubMed] [Google Scholar]
- 20. Mukhopadhyay A, Maulik U, Bandyopadhyay S (2012) A Novel Biclustering Approach to Association Rule Mining for Predicting HIV-1–Human Protein Interactions. PLoS One 7: e32289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fu W, Sanders-Beer BE, Katz KS, Maglott DR, Pruitt KD, et al. (2009) Human immunodeficiency virus type 1, human protein interaction database at NCBI. Nucleic Acids Res (Database Issue) 37: D417–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pan S, Yang Q (2010) A Survey on Transfer Learning. IEEE Transactions on Knowledge and Data Engineering 22: 1345–1359. [Google Scholar]
- 23. Tu Y, Tolovitzky G, Klein U (2002) Quantitative noise analysis for gene expression microarray experiments. Proc Natl Acad Sci U S A 99: 14031–14036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mei S, Wang F, Zhou S (2011) Gene ontology based transfer learning for protein subcellular localization. BMC Bioinformatics 12: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mei S (2012) Multi-kernel transfer learning based on Chou’s PseAAC formulation for protein submitochondria localization. J Theor Biol 293: 121–130. [DOI] [PubMed] [Google Scholar]
- 26. Mei S (2012) Multi-label Multi-kernel Transfer Learning for Human Protein Subcellular Localization. PLoS One 7: e37716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, et al. (2003) The SWISS-PROT Protein Knowledgebase and Its Supplement TrEMBL. Nucleic Acids Res 31: 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Altschul S, Madden T, Schaffer A, Zhang J, Zhang Z, et al. (1997) Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Barrell D, Dimmer E, Huntley R, Binns D, O’Donovan C, et al. (2009) The GOA database in 2009–an integrated Gene Ontology Annotation resource. Nucleic Acids Res (Database Issue) 37: D396–D403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dong J, Adam K, Ching Y (2005) Fast SVM Training Algorithm with Decomposition on Very Large Data Sets. IEEE Trans Pattern Anal Mach Intell 27: 603–18. [DOI] [PubMed] [Google Scholar]
- 31.Platt J (1999) Probabilistic outputs for support vector machines and comparison to regularized likelihood methods. In Advances in Large Margin Classifiers. MIT Press.
- 32. Faraggi D, Reiser B (2002) Estimation of the area under the ROC curve. Stat Med 21: 3093–106. [DOI] [PubMed] [Google Scholar]
- 33. Ben-Hur A, Noble W (2006) Choosing negative examples for the prediction of protein-protein interactions. BMC Bioinformatics 7: S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davis J, Goadrich M (2006) The relationship between precision-recall and ROC curves. In: Proceedings of the 23rd International Conference on Machine Learning.
- 35. Vashistha H, Husain M, Kumar D, Singhal PC (2009) Tubular cell HIV-1 gp120 expression induces caspase 8 activation and apoptosis. Ren Fail 31: 303–312. [DOI] [PubMed] [Google Scholar]
- 36. Wang X, Chen DG (2009) Recombinant murine cytomegalovirus vector activates human monocytederived dendritic cells in a NF-kappaB dependent pathway. Mol Immunol 46: 3462–3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jiang J, Fu W, Wang X, Lin PH, Yao Q, et al. (2010) HIV gp120 induces endothelial dysfunction in tumour necrosis factor-alpha-activated porcine and human endothelial cells. Cardiovasc Res 87: 366–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Saha RN, Pahan K (2007) Differential regulation of Mn-superoxide dismutase in neurons and astroglia by HIV-1 gp120: Implications for HIV-associated dementia. Free Radic Biol Med 42: 1866–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cheung R, Ravyn V, Wang L, Ptasznik A, Collman RG (2008) Signaling mechanism of HIV-1 gp120 and virion-induced IL-1beta release in primary human macrophages. J Immunol 180: 6675–6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhou C, Lu L, Tan S, Jiang S, Chen YH (2011) HIV-1 Glycoprotein 41 Ectodomain Induces Activation of the CD74 Protein-mediated Extracellular Signal-regulated Kinase/Mitogen-activated Protein Kinase Pathway to Enhance Viral Infection. J Biol Chem 286: 44869–44877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gupta P, Singhal PK, Rajendrakumar P, Padwad Y, Tendulkar AV, et al. (2011) Mechanism of host cell MAPK/ERK-2 incorporation into lentivirus particles:characterization of the interaction between MAPK/ERK-2 and proline-richdomain containing capsid region of structural protein Gag. J Mol Biol 410: 681–697. [DOI] [PubMed] [Google Scholar]
- 42. Gibellini D, De Crignis E, Ponti C, Borderi M, Clo A, et al. (2010) HIV-1 Tat protein enhances RANKL/M-CSF-mediated osteoclast differentiation. Biochem Biophys Res Commun 401: 429–434. [DOI] [PubMed] [Google Scholar]
- 43. Vidal M, Cusick M, Barabási A (2011) Interactome Networks and Human Disease. Cell 144: 986–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Keshava-Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, et al. (2009) Human Protein Reference Database–2009 Update. Nucleic Acids Res (Database Issue) 37: D767–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Text file contains the overlapped predictions between PWEN-TLM and Bi-clustering [20] ( Optimistic case ).
(TXT)
Text file contains the overlapped predictions between PWEN-TLM and Bi-clustering [20] ( Pessimistic case ).
(TXT)
Text file contains the overlapped predictions among PWEN-TLM , Bi-clustering [20] and the method [6] ( Optimistic case ).
(TXT)
Text file contains the overlapped predictions among PWEN-TLM , Bi-clustering [20] and the method [6] ( Pessimistic case ).
(TXT)
Text file contains the predictions between HIV-1 and peripheral human proteins ( Optimistic case ).
(TXT)
Text file contains the predictions between HIV-1 and peripheral human proteins ( Pessimistic case ).
(TXT)


