Abstract
Plakoglobin (γ-catenin) is a homolog of β-catenin with dual adhesive and signaling functions. Plakoglobin participates in cell-cell adhesion as a component of the adherens junction and desmosomes whereas its signaling function is mediated by its interactions with various intracellular protein partners. To determine the role of plakoglobin during tumorigenesis and metastasis, we expressed plakoglobin in the human tongue squamous cell carcinoma (SCC9) cells and compared the mRNA profiles of parental SCC9 cells and their plakoglobin-expressing transfectants (SCC9-PG). We observed that the mRNA levels of SATB1, the oncogenic chromatin remodeling factor, were decreased approximately 3-fold in SCC9-PG cells compared to parental SCC9 cells. Here, we showed that plakoglobin decreased levels of SATB1 mRNA and protein in SCC9-PG cells and that plakoglobin and p53 associated with the SATB1 promoter. Plakoglobin expression also resulted in decreased SATB1 promoter activity. These results were confirmed following plakoglobin expression in the very low plakoglobin expressing and invasive mammary carcinoma cell line MDA-MB-231 cells (MDA-231-PG). In addition, knockdown of endogenous plakoglobin in the non-invasive mammary carcinoma MCF-7 cells (MCF-7-shPG) resulted in increased SATB1 mRNA and protein. Plakoglobin expression also resulted in increased mRNA and protein levels of the metastasis suppressor Nm23-H1, a SATB1 target gene. Furthermore, the levels of various SATB1 target genes involved in tumorigenesis and metastasis were altered in MCF-7-shPG cells relative to parental MCF-7 cells. Finally, plakoglobin expression resulted in decreased in vitro proliferation, migration and invasion in different carcinoma cell lines. Together with the results of our previous studies, the data suggests that plakoglobin suppresses tumorigenesis and metastasis through the regulation of genes involved in these processes.
Introduction
Metastasis is a multi-step process that begins when tumor cells acquire the ability to degrade the basement membrane and move from the primary site of tumor formation to distant sites throughout the body, culminating in the formation of secondary tumors at these new sites. It is the formation of these secondary tumors that is the major cause of cancer-related deaths. In epithelial tissues, the abnormal proliferation, migration and invasion of constituent cells are limited by intercellular adhesive complexes, which tether neighboring cells to one another and maintain normal tissue architecture and function [1]–[5].
The main adhesive complexes in epithelia are the cadherin-based adherens junction and desmosomes [6]–[7]. Cadherins are single-pass transmembrane glycoproteins that make homotypic extracellular interactions with cadherin proteins on neighboring cells and intracellularly interact with catenin proteins [5]. At the adherens junction, E-cadherin interacts with either β-catenin or γ-catenin (plakoglobin), which then interact with α-catenin, an actin binding protein, which tethers the cadherin-catenin complex to the actin cytoskeleton [5]. Similarly, at the desmosome, the desmosomal cadherins (desmocollins and desmogleins) are tethered to the intermediate filament cytoskeleton through interactions with plakoglobin and desmoplakin [6]–[7].
β-catenin and plakoglobin are structural and functional homologs and members of the armadillo family of proteins with dual functions in cell-cell adhesion and cell signaling [8]–[10]. Both proteins interact with E-cadherin, Axin and APC and both are involved in the Wnt signaling pathway through their interactions with the TCF/LEF transcription factors. Despite their structural similarities and common interacting partners, β-catenin and plakoglobin appear to have different signaling activities and regulate tumorigenesis in opposite manners. While β-catenin-TCF/LEF complexes are transcriptionally active, several studies have demonstrated that plakoglobin-TCF complexes are inefficient in binding to DNA [11]–[13]. Conversely, plakoglobin, but not β-catenin, interacts with p53 and regulates gene expression independent of TCF [14]. Furthermore, β-catenin has well-documented oncogenic signaling activities as the terminal component of the Wnt signaling pathway, whereas plakoglobin has typically been associated with tumor/metastasis suppressor activities [14]–[22].
To determine the role of plakoglobin in tumorigenesis and metastasis, we previously expressed physiological levels of plakoglobin in the plakoglobin-null SCC9 cell line, a human squamous cell carcinoma cell line derived from the tongue. Plakoglobin expression in SCC9 cells (SCC9-PG) resulted in a mesenchymal (transformed)-to-epidermoid (normal) phenotypic transition that was concurrent with the increased levels of N-cadherin, decreased levels of β-catenin and the formation of desmosomes [15]. We subsequently performed proteomic and transcription microarray experiments to identify potential genes and proteins whose levels were differentially expressed following plakoglobin expression. These studies identified several tumor and metastasis suppressors and oncogenes whose levels were increased and decreased, respectively, in SCC9-PG cells. Among these differentially expressed genes was the global regulator of gene expression, Special AT-Rich Sequence Binding Protein 1 (SATB1).
SATB1 was initially identified as a DNA-binding protein that was highly expressed in the thymus [23]–[24]. This protein was shown to have a high affinity for binding to base-unpairing regions (BURs), which are genomic DNA sequences with high unfolding potential, containing clusters of sequences (approximately 20–40 base pairs long) with a bias in G and C distribution, with one DNA strand contains only A, T and C residues [23], [25]–[27]. Importantly, since BUR sequences are thought to be found all throughout the genome and since SATB1 demonstrated a specificity for these BUR sequences, it became evident that SATB1 could, through its interactions with different BUR sequences in different gene promoters, cause the looping of chromatin [26], [28]–[30]. These chromatin loops could, in turn, potentially result in the close physical proximity and coordinated regulation of genes that would otherwise remain silent. In addition to forming these chromatin loops, SATB1 was shown to recruit different chromatin remodeling enzymes to the gene loci close to the BURs and as a result altered gene expression [31]–[34].
SATB1, through the regulation of gene expression, has been shown to promote in vitro tumorigenesis and metastasis in various cell lines, including breast, lung, ovarian, colorectal, liver, laryngeal, glioma and melanoma [34]–[42]. Specifically, SATB1 has been shown to induce the expression of tumor and metastasis-promoting genes while suppressing the expression of metastasis suppressor genes [34], [39], [43]–[44]. Interestingly, our microarray experiments showed that SATB1 expression was decreased 3-fold in SCC9-PG cells compared to SCC9 cells [14], suggesting that plakoglobin may play a role in regulating the SATB1 gene and thus, may have an effect on a the expression of a wide range of tumorigenesis and metastasis associated genes.
In the present study, we examined the role of plakoglobin in regulating the expression of genes involved in tumorigenesis and metastasis. We showed that plakoglobin, in coordination with p53, associated with the SATB1 promoter and downregulated its expression. Decreased levels of SATB1 mRNA were accompanied by its decreased protein levels in squamous and mammary epithelial cell lines expressing plakoglobin. Furthermore, plakoglobin expression led to an increase and a decrease in the protein levels of a subset of SATB1 repressed and activated target genes, respectively. Concurrent with these transcriptional changes, plakoglobin expression resulted in decreased cell growth and in vitro migration and invasion. Taken together, our data suggests that plakoglobin suppresses tumorigenesis and metastasis (at least in vitro) through the regulation of genes involved in these processes.
Materials and Methods
Cell culture and conditions
All tissue culture reagents were purchased from Invitrogen (Burlington, Canada) unless stated otherwise. SCC9, SCC9-PG, MDA-231, MDA-231-PG, MCF-7 and MCF-7-shPG cells have all been described previously [14]–[15], [45].
Construction of SATB1-luciferase reporter constructs
The SATB1 promoter was cloned from SCC9 genomic DNA by PCR and ligated into the pBV-Luc vector at KpnI and SacI sites, respectively. The primer sequences used for the cloning reaction were CAGTggtaccGCCA-GGGCGACTCTAGAG (forward, starting at base pair 14 in the SATB1 gene) and AGCTgagctcCACTTCAAAACTTGACAGCACATA (reverse, ending at base pair 1222 in the SATB1 gene). The constructed luciferase plasmid was then used for reporter assays.
RNA isolation and RT-PCR
RNA was isolated from 150 mm confluent cultures using the RNeasy Plus Mini Kit (QIAGEN, Valencia, CA) according to the manufacturer's protocol. Following isolation, RNA was pre-treated with RNase-free DNaseI and reverse transcribed using the RevertAid H Minus First Strand cDNA Synthesis Kit (Fermentas, Burlington, ON, Canada). Polymerase chain reaction (PCR) was performed (Fermentas, Burlington, ON, Canada) on the amplified cDNA. The sequences of the primers used are outlined in Table 1. RT-PCR products were resolved on 2% agarose gels and visualized by ethidium bromide staining. qRT-PCR was performed using PerfeCta SYBR Green FastMix reagent (Quanta Biosciences) as per the manufacturer's instructions.
Table 1. Primer sequences and PCR conditions for reverse transcribed genes.
| Gene | Primers | Size (bp) | Annealing temperature | Ref. |
| RT-PCR | ||||
| ABL1 | Sense: 5′- GTATTTCACAGAGCACGCCT-3′; Antisense: 5′- GAGGTGATGTGCTGTAAGA-3′ | - | 60°C | [34] |
| BRMS1 | Sense: 5′-GGAAGAAGGCACCTCTGGTTT-3′; Antisense: 5′- GCTGCCCTAGCCTTTTTGATG-3′ | - | 60°C | [34] |
| CLDN1 | Sense: 5′- CCCCAGTGGAGGATTTACTCCTA-3′; Antisense: 5′- GCAATGTGCTGCTCAGATTCA-3′ | - | 60°C | [34] |
| ERBB2 | Sense: 5′- AGGGCAGTTACCAGTGCCAATATC-3′; Antisense: 5′- TCCAGAGTCTCAAACACTTGGAGC-3′ | - | 60°C | [34] |
| KISS1 | Sense: 5′- CCATTAGAAAAGGTGGCCTCTGT-3′; Antisense: 5′- AGGAGGCCCAGGGATTCTAG-3′ | - | 60°C | [34] |
| MMP3 | Sense: 5′- CGATGCAGCCATTTCTGATAAG-3′; Antisense: 5′- GCCTGGCTCCATGGAATTT-3′ | - | 60°C | [34] |
| SNAI1 | Sense: 5′- CCCCAATCGGAAGCCTAACT-3′; Antisense: 5′- GGACAGAGTCCCAGATGAGCAT-3′ | - | 60°C | [34] |
| NME1 | Sense: 5′-CGCAGTTCAAACCTAAGCAGCAGCTGG-3′; Antisense: 5′- GATCCAGTTCTGAGCACAGCTCG-3′ | 483 | 60°C | [93] |
| NME2 | Sense: 5′-TGACCTGAAAGACCGACCAT-3′; Antisense: 5′-GAATGATGTTCCTGCCAACC-3′ | 193 | 55°C | [94] |
| SATB1 | Sense: 5′- TGCAAAGGTTGCAGCAACCAAAAGC-3′; Antisense: 5′- AACATGGATAATGTGGGGCGGCCT-3′ | 156 | 60°C | [34] |
| GAPDH | Sense: 5′-GAAGGTGAAGGTCGGAGTC-3′; Antisense: 5′-GAAGATGGTGATGGGATTTC-3′ | 220 | 60°C | [95] |
| ChIP | ||||
| NME1 | Sense: 5′-CAACTGTGAGCGTACCTTCAT-3′; Antisense: 5′-AACAAGGCGGAATCCTTTCTG-3′ | 102 | 53.6°C | - |
| SATB1 | Sense: 5′-GATCATTTGAACGAGGCAACTCA-3′; Antisense: 5′-CCTGCATTTTTGCACCTGTACT-3′ | 157 | 53.6°C | - |
For all primers, pre-denaturation was done at 95°C for 5 minutes. This was followed by 35 cycles of denaturation at 95°C for 30 seconds, annealing for 45 seconds, and extension at 72°C for 45 seconds.
Antibodies
A list of antibodies and their respective dilutions in specific assays is presented in Table 2.
Table 2. Antibodies and their respective dilutions in specific assays.
| Assays | |||||
| Antibodies | Species | WB | ChIP | IF | Source |
| β-Actin | Mouse | 1:2000 | - | - | Sigma, A-5441 |
| Anti-mouse IgG | Goat | - | 1:2000 | - | Sigma, M-5899 |
| BrdU | Mouse | - | - | 1:300 | Sigma, B-5002 |
| BRMS1 | Mouse | 1:200 | - | - | Santa Cruz, sc-101219 |
| c-Abl | Rabbit | 1:1000 | - | - | Santa Cruz, sc-131 |
| Claudin-1 | Mouse | 1:500 | - | - | Santa Cruz, sc-137121 |
| ErbB2 | Rabbit | 1:1000 | - | - | Upstate, 06–562 |
| Kiss1 | Rabbit | 1:500 | - | - | Santa Cruz, sc-15400 |
| MMP3 | Mouse | 1:100 | - | - | Calbiochem, Ab-1 |
| Nm23-H1/H2 | Rabbit | 1:200 | - | - | Chemicon, CBL-446 |
| p53 | Mouse | - | 1:100 | - | Santa Cruz, sc-126 |
| SATB1 | Rabbit | 1:1000 | - | - | Cell Signaling, L745 |
| Snail | Rabbit | 1:2000 | - | - | Abcam, ab17732 |
| Plakoglobin | Mouse | 1:500 | 1:100 | - | Transduction Laboratories, 610254 |
| 2° Antibodies | |||||
| Anti-mouse HRP, Light Chain specific | Goat | 1:5000 | - | - | Jackson, 115-005-174 |
| Anti-rabbit HRP, Light Chain specific | Goat | 1:5000 | - | - | Jackson, 211-002-177 |
| Alexa Fluor 488 | Goat | - | - | 1:1500 | Molecular Probes, A11035 |
Preparation of total cell extracts and Western blotting
Confluent 150 mm culture dishes were washed twice with cold phosphate-buffered saline (PBS), solubilized in hot SDS sample buffer (10 mM Tris-HCl pH 6.8, 2% (w/v) SDS, 50 mM dithiothreitol (DTT), 2 mM EDTA, 0.5 mM PMSF) and boiled for 10 minutes. Protein determination was done using Bradford (Pierce) assays according to the manufacturer's instructions. Seventy-five micrograms of total cellular protein were resolved by SDS-PAGE, transferred to nitrocellulose membranes and processed for immunoblotting and developed by standard ECL (Perkin Elmer, Woodbridge, Canada) procedures.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) experiments were performed as previously described [14], [46]. Confluent 150 mm cultures were trypsinized and 2×107 cells pelleted by centrifugation at 1500×g for 10 minutes. The cell pellets were then resuspended in growth media to which formaldehyde (Fisher) was added to a final concentration of 1% and incubated at room temperature for 10 minutes. To stop fixation, glycine was added to a final concentration of 125 mM. The cell suspension was then centrifuged at 1500×g at 4°C for 10 minutes. The resulting cell pellets were then washed twice with PBS containing 1 μg/ml aprotinin and leupeptin and 1 mM PMSF, after which they were resuspended in cell lysis buffer (10 mM HEPES pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM DTT and 0.49 mM PMSF) and incubated on ice for 15 minutes. NP-40 was then added (final concentration of 0.6%) after which the samples were vortexed for 10 seconds at high speed and subsequently centrifuged at 18,000×g for 30 seconds. The resulting pellets were then resuspended in sonication buffer (1% SDS, 10 mM EDTA, 50 mM Tris pH 8, 0.49 mM DTT and 0.02 μg/ml aprotinin and leupeptin) and left on ice for 10 minutes. The samples were then sonicated (Branson Sonifier 450) for 1 minute at 20% output for a total of four times.
The sonicated chromatin samples were then diluted ten-fold in chromatin dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris pH 8, 167 mM NaCl) after which 50 μl was removed (Input). Forty μl Protein A/G agarose beads (Calbiochem) were added and the samples were pre-cleaned on a rocker-rotator at 4°C for 2 hours. Following incubation, the samples were centrifuged briefly and the resulting supernatant (pre-cleaned chromatin) was split into equal aliquots and processed for immunoprecipitation. Each aliquot was incubated with 5 μg antibodies and 40 μl pre-cleaned (by overnight incubation with 4 μg Salmon Sperm DNA and BSA) Protein A/G agarose beads overnight at 4°C on a rocker-rotator.
Following immunoprecipitation, the samples were centrifuged for 10 minutes at 420×g at 4°C, after which the resulting supernatants were removed. The beads were then subjected to six 5 minute washes in each of the four following wash buffers: W1 (1% SDS, 1% Triton X-100, 2 mM Tris pH 8, 167 mM NaCl), W2 (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris pH 8, 500 mM NaCl), W3 (250 mM LiCl, 1% NP-40, 1% sodium deoxycholate, 10 mM Tris pH 8, 1 mM EDTA) and W4 (10 mM Tris pH 8 and 1 mM EDTA). Following the washes, the protein-DNA complexes were eluted off the beads by incubation in elution buffer (1% SDS and 50 mM NaHCO3) for 15 minutes at room temperature on a rocker-rotator. Following elution, 1 μg RNase and NaCl (final concentration 300 mM) were added to the samples, which were then incubated at 65°C for 4 hours. Next, Tris (pH 6.8) and EDTA (final concentrations of 40 mM and 10 mM, respectively) and 4 μg proteinase K were added to the samples and the samples were incubated at 45°C for 2 hours. The samples were then purified using a PCR Purification Kit (QIAGEN, Valencia, CA) and processed for PCR.
Luciferase reporter assay
Confluent 35 mm cultures were transfected with 4 μg of various luciferase reporter plasmids. SATB1 promoter activity was analyzed by using a reporter construct downstream of the full SATB1 promoter [47] together with 1 μg of a plasmid encoding β-galactosidase. Forty-eight hours post-transfection, luciferase and β-galactosidase activities were measured. To assess activity from the NME1 promoter, cells were transfected with a reporter plasmid downstream of the NME1 promoter, a kind gift of Dr. Shimian Qu, Vanderbilt University, Nashville, TN, USA [48]. Each experiment was repeated at least 3 times and the mean with standard deviation was calculated. Statistical analysis was performed using a Student's t-test.
Cell growth and proliferation assays
To measure growth, 5×104 cells for each cell line were plated in triplicate in a 24-well plate. At 3, 5 and 7 days after plating, cultures were trypsinized and the cells were counted. Cell proliferation was assessed by performing BrdU incorporation experiments. For each cell line, 5×104 cells were plated on glass coverslips and allowed to proliferate for 6 days at which times they were incubated with BrdU (100 μM; Sigma B-5002) for 24 hours. To detect BrdU-labeled cells, coverslips were first prefixed by the addition of 3.7% formaldehyde directly to the culture media at a 1∶1 ratio (volume). Coverslips were then rinsed, fixed with 3.7% formaldehyde for 15 minutes and permeabilized with 0.5% Triton X-100 for 5 minutes. Coverslips were then washed with PBS and incubated in 2N HCl for 1 hour at room temperature followed by two 5 minute washes with 100 mM sodium borate (pH 8.5). Subsequently, coverslips were blocked for 1 hour with 4.0% goat serum and 50 mM NH4Cl4 in PBS containing 0.2% BSA (PBS-BSA) and incubated with a mouse monoclonal anti-BrdU antibody (1∶300; BD 347583) for 1 hour at room temperature followed by a 20 minute incubation with Alexa Fluor 488 (Molecular Probes, A11035) secondary antibodies. All antibodies were diluted in PBS-BSA. Nuclei were counterstained for 5 min with DRAQ5 (1∶40,000 in PBS; Biostatus). Coverslips were mounted in elvanol containing 0.2% (w/v) paraphenylene diamine (PPD) and viewed using a 63X objective of an LSM510 META (Zeiss) laser scanning confocal microscope.
Transwell cell migration and invasion assays
For cell migration assays, 2×105 cells were resuspended in 0.5 ml serum-free media containing 0.1% BSA and plated in the upper chamber of transwells (3 μm pore, 6.5 μm diameter; BD Biosciences, MD, USA). Normal media containing 10% FBS (0.75 ml) was added to the lower chamber. Cultures were incubated at 37°C for 12 or 48 hours to allow cell migration at which time the inserts were removed from the chambers, gently submerged in PBS to remove the unattached cells, fixed and stained using Diff Quick (IHC World, MD, USA). Following staining, membranes were cut, mounted on slides using permount (Fisher, Canada), viewed under an inverted microscope using a 20X objective and photographed. The migrated cells on the underside of the membrane were counted in five random fields for each transwell filter from the photographs.
Matrigel invasion assays were performed according to the manufacturer's protocol (BD Bioscience). For each cell line, 5×105 cells in 0.6 ml serum-free media containing 0.1% BSA were plated in the top compartment of Matrigel-coated invasion chambers (8 μm pore membrane). Fibroblast conditioned media (0.75 ml) was added to the bottom chambers and plates were incubated at 37°C in 5% CO2. Forty-eight hours later, the membranes were recovered, fixed, stained with Diff Quick, viewed under an inverted microscope using a 20X objective and photographed. The invaded cells were counted in five random fields for each membrane.
Each assay was repeated 3 independent times. The numbers of migrated/invaded cells were calculated using the ImageJ Cell Counter program and averaged.
Results
Plakoglobin regulates SATB1 expression
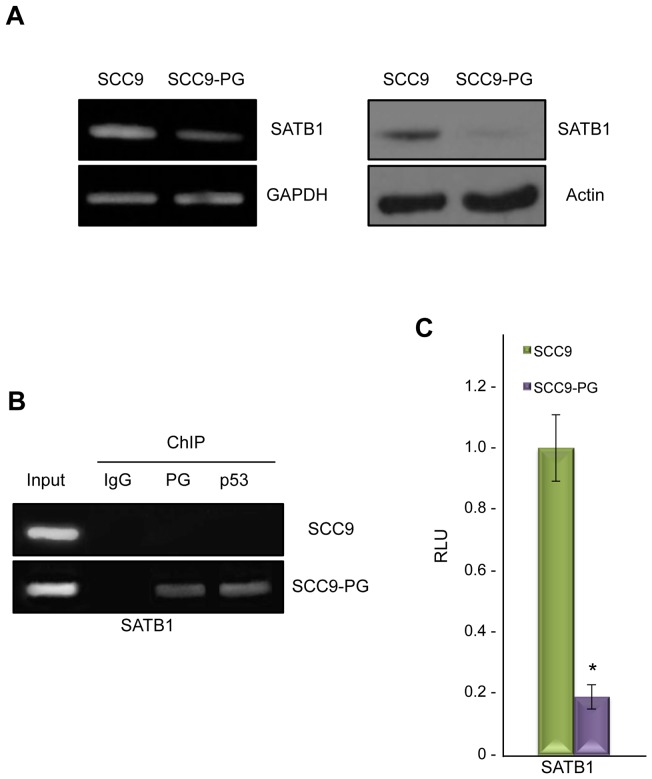
We previously observed that plakoglobin expression in the plakoglobin-null SCC9 squamous carcinoma cell line (SCC9-PG) resulted in a mesenchymal-to-epidermoid phenotypic transition that coincided with the formation of stable cell-cell junctions [15]. Further characterization of SCC9-PG cells using transcription microarray experiments revealed that the expression of the SATB1 gene was decreased over 3-fold in SCC9 cells following plakoglobin expression [14]. To confirm this observation, we first performed RT-PCR experiments and observed a notable decrease in SATB1 mRNA in SCC9-PG cells compared to SCC9 cells (Fig. 1A, left). In agreement with this result, western blot analysis revealed that while SATB1 protein was expressed in SCC9 cells, its levels were significantly decreased and barely detectable in SCC9-PG cells (Fig. 1A, right).
Figure 1. Plakoglobin associates with and suppresses the SATB1 promoter in SCC9-PG cells.
A. (Left) Total cellular RNA was isolated from SCC9 and SCC9-PG cells, reverse transcribed and processed for PCR using primers specific to SATB1 and GAPDH. (Right) Equal amounts of total cellular proteins from SCC9 and SCC9-PG cells were resolved by SDS-PAGE and processed for immunoblotting with antibodies to SATB1 and Actin. B. SCC9 and SCC9-PG cells were formaldehyde fixed and processed for chromatin immunoprecipitation. Following sonication, extracts were immunoprecipitated using control IgG, plakoglobin and p53 antibodies. Following extensive washes, immunoprecipitated DNA was separated from the immune complexes and purified using standard DNA purification protocols. The purified DNA was then processed for PCR using SATB1 primers. As a positive control, total cellular DNA (Input) was amplified using the same primers. C. SCC9 and SCC9-PG cells were transfected with luciferase reporter constructs under the control of a 1.2 kb sequence of the SATB1 promoter. Luciferase activities were measured 48 hours post-transfection. The levels of luciferase activities from the vector and SATB1 reporter constructs were determined from a minimum of three independent transfections and normalized for transfection efficiency by co-transfection with a β-galactosidase expression vector. The SATB1 promoter activity was normalized to the corresponding vector activity for each cell line and then normalized to SCC9 (*p<0.01). PG, plakoglobin; RLU, Relative Light Units.
To determine whether plakoglobin regulates the SATB1 gene, we performed chromatin immunoprecipitation (ChIP) experiments using plakoglobin antibodies and nuclear extracts from SCC9 and SCC9-PG cells. The isolated DNA was then processed for PCR using primers specific to the SATB1 promoter (Table 1). These experiments showed that plakoglobin associated with the SATB1 promoter in SCC9-PG cells, but not in SCC9 cells (Fig. 1B). ChIP experiments using control IgG antibodies produced negative results. Since plakoglobin interacts with and regulates gene expression in conjunction with p53 [14], we also performed the ChIP experiments using p53 antibodies, which demonstrated that while p53 associated with the SATB1 promoter in SCC9-PG cells, this association was absent in SCC9 cells (Fig. 1B).
The association of plakoglobin and p53 with the SATB1 promoter and the decreased levels of SATB1 mRNA and protein in SCC9-PG cells suggested that plakoglobin and p53 may function as negative regulators of the SATB1 promoter. To test this hypothesis, luciferase reporter assays were conducted using luciferase reporter constructs downstream of a 1.2 kb SATB1 promoter fragment [47]. Consistent with the role of plakoglobin in the negative regulation of the SATB1 promoter, the luciferase activity of the reporter constructs was significantly decreased (over 5-fold) in SCC9-PG cells compared to SCC9 cells (Fig. 1C).
Plakoglobin regulates SATB1 in mammary epithelial cell lines
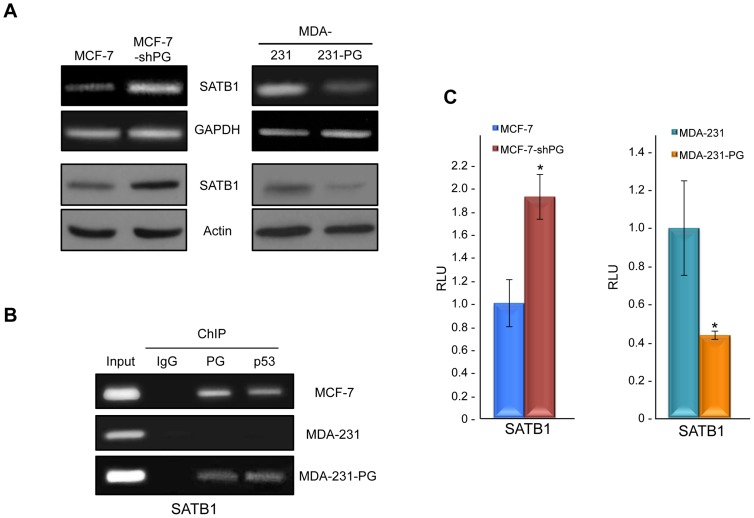
In addition to SCC9 cells, we also examined the role of plakoglobin in regulating SATB1 in MCF-7 and MDA-MB-231 (MDA-231) mammary epithelial cell lines, since SATB1 has previously been shown to play a major role in the regulation of breast cancer progression and metastasis [34]. As such, we set out to determine whether the results from SCC9-PG could be extended to breast cancer cell lines. To do so, we took two approaches: first, we knocked down plakoglobin in MCF-7 cells (MCF-7-shPG), which are non-invasive and express considerable levels of wild-type plakoglobin [14], [19], [49] and second, we expressed plakoglobin in MDA-231 cells (MDA-231-PG), which are highly invasive and express very low levels of endogenous plakoglobin [49]. RT-PCR and western blot experiments showed that knockdown of plakoglobin in MCF-7 cells resulted in increased levels of both SATB1 mRNA and protein. In contrast, plakoglobin expression in MDA-231 cells resulted in a decrease in both SATB1 mRNA and protein, although SATB1 protein was still detectable in MDA-231-PG cells (Fig. 2A).
Figure 2. Plakoglobin suppresses SATB1 in mammary epithelial cell lines.
A. (Top) Total cellular RNA was isolated from MCF-7, MCF-7-shPG, MDA-231 and MDA-231-PG cells, reverse transcribed and processed for PCR using primers specific to SATB1 and GAPDH. (Bottom) Equal amounts of total cellular proteins from these cells were resolved by SDS-PAGE and processed for immunoblotting with antibodies to SATB1 and Actin.B. MCF-7, MDA-231 and MDA-231-PG cells were formaldehyde fixed and processed for chromatin immunoprecipitation as described in Fig. 1B. The purified DNA was then processed for PCR using SATB1 primers. As a positive control, total cellular DNA (Input) was amplified using the same primers. C. MCF-7, MCF-7-shPG, MDA-231 and MDA-231-PG cells were transfected with luciferase reporter constructs and processed as described in Fig. 1C. The SATB1 promoter activity was normalized to the corresponding vector activity for each cell line and then normalized to MDA-231 or MCF-7, respectively (*p<0.01). PG, plakoglobin; RLU, Relative Light Units.
ChIP experiments showed that similar to SCC9-PG cells, both plakoglobin and p53 associated with the SATB1 promoter in MCF-7 cells. Furthermore, both proteins associated with the SATB1 promoter in MDA-231-PG cells, but not MDA-231 cells (Fig. 2B). To further demonstrate that plakoglobin and p53 negatively regulate the SATB1 promoter, we performed luciferase assay experiments using the SATB1-luciferase reporter constructs in MCF-7, MCF-7-shPG, MDA-231 and MDA-231-PG cells. The results of these experiments were consistent with those from SCC9-PG cells: luciferase activity in MDA-231-PG cells was decreased (over 2-fold) compared to MDA-231 cells, whereas activity in MCF-7-shPG cells was induced (approximately 2-fold) compared to MCF-7 cells (Fig. 2C). Taken together, the results from these experiments suggest that plakoglobin and p53 negatively regulate SATB1 expression.
Plakoglobin associates with and activates the NME1 promoter
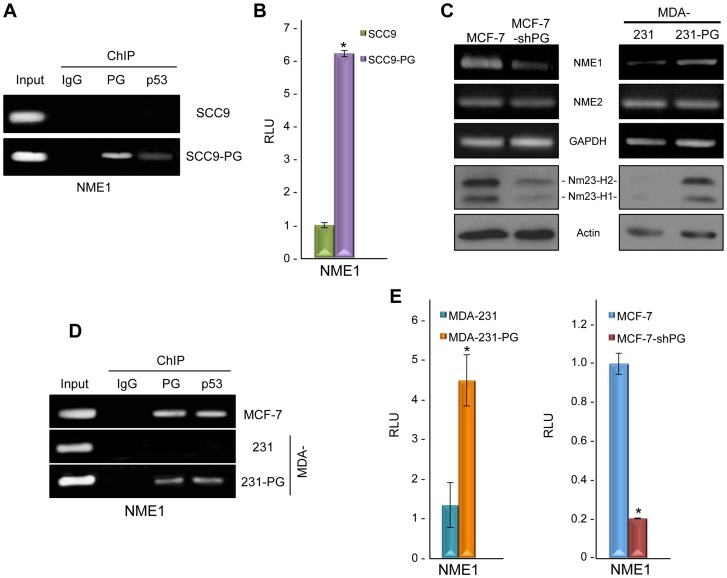
It has been suggested that the metastasis suppressor Nm23-H1 is a potential target of SATB1 [34]. Furthermore, we previously showed that plakoglobin expression in SCC9 cells resulted in increased Nm23-H1 and -H2 protein levels as well as increased Nm23-H1 (NME1), but not Nm23-H2 (NME2) gene expression [19]. Therefore, we set out to determine if the increased expression of NME1 in SCC9-PG cells were simply due to decreased SATB1 levels or whether plakoglobin actively promoted the expression of NME1. In order to do so, we performed ChIP experiments using plakoglobin antibodies and primers specific to the NME1 promoter. Plakoglobin associated with the NME1 promoter in SCC9-PG cells, but not SCC9 cells (Fig. 3A). Similar ChIP experiments were performed using p53 antibodies, which demonstrated that while p53 associated with the NME1 promoter in SCC9-PG cells, this association was absent in SCC9 cells (Fig. 3A). ChIP experiments using control IgG antibodies produced negative results.
Figure 3. Plakoglobin associates with and activates NME1.
A. SCC9 and SCC9-PG cells were processed for chromatin immunoprecipitation using control IgG, plakoglobin and p53 antibodies as described in Fig. 1B. The purified DNA was then processed for PCR using NME1 primers. As a positive control, total cellular DNA (Input) was amplified using the same primers. B. SCC9 and SCC9-PG cells were transfected with luciferase reporter constructs under the control of a 2 kb sequence of the NME1 promoter. Luciferase activities were measured 48 hours post-transfection. The levels of luciferase activities from the vector and NME1 reporter constructs were determined from a minimum of three independent transfections and normalized for transfection efficiency by co-transfection with a β-galactosidase expression vector. The NME1 promoter activity was normalized to the corresponding vector activity for each cell line and then normalized to SCC9 (*p<0.01). PG, plakoglobin; RLU, Relative Light Units. C. (Top) Total cellular RNA was isolated from MCF-7, MCF-7-shPG, MDA-231 and MDA-231-PG cells, reverse transcribed and processed for PCR using primers specific to NME1, NME2 and GAPDH. (Bottom) Equal amounts of total cellular proteins from these cells were resolved by SDS-PAGE and processed for immunoblotting with antibodies to Nm23-H1, -H2 and Actin. D. MCF-7, MDA-231 and MDA-231-PG cells were processed for chromatin immunoprecipitation using control IgG, plakoglobin and p53 antibodies and the purified DNA processed for PCR using NME1 primers. As a positive control, total cellular DNA (Input) was amplified using the same primers. E. MCF-7, MCF-7-shPG, MDA-231 and MDA-231-PG cells were transfected with luciferase reporter constructs as described in Fig. 3B. The NME1 promoter activity was normalized to the corresponding vector activity for each cell line and then normalized to MDA-231 or MCF-7, respectively (*p<0.01). PG, plakoglobin; RLU, Relative Light Units.
To confirm the role of plakoglobin in the regulation of NME1 expression, luciferase assays were done using luciferase reporter constructs downstream of a 2 kb NME1 promoter fragment [48]. In these experiments, luciferase activity was induced approximately 6-fold in SCC9-PG cells compared to SCC9 cells (Fig. 3B), demonstrating that plakoglobin expression resulted in increased NME1 promoter activity. Taken together, these data suggest that plakoglobin actively regulates the NME1 gene through its associations with the NME1 promoter, while it also downregulates SATB1 levels, which may in turn result in increased NME1 expression.
Plakoglobin regulates NME1 in mammary epithelial cell lines
We subsequently performed RT-PCR and western blot experiments to examine the levels of Nm23-H1 mRNA and protein in the mammary epithelial cell lines to confirm that plakoglobin-mediated regulation of NME1 was not specific to squamous cell lines. Knockdown of plakoglobin in MCF-7 cells resulted in a notable decrease in Nm23-H1 mRNA, which was accompanied by a corresponding decrease in the levels of Nm23-H1 and -H2 protein (Fig. 3C). In contrast, the levels of both Nm23-H1 mRNA and protein were increased considerably in MDA-231-PG cells compared to parental MDA-231 cells (Fig. 3C). We also performed the RT-PCR experiments using primers specific to the Nm23-H2 (NME2) gene and observed that plakoglobin expression had no effect on NME2 expression, since the levels of Nm23-H2 mRNA were not different between MCF-7 and MCF-7-shPG and MDA-231 and MDA-231-PG cells, respectively (Fig. 3C). These results were consistent with the lack of NME2 induction following plakoglobin expression in SCC9-PG cells [19].
Next, ChIP experiments were conducted with chromatin from MCF-7, MDA-231 and MDA-231-PG cells using plakoglobin and p53 antibodies. The results from these experiments showed that plakoglobin and p53 associated with the NME1 promoter in both MCF-7 and MDA-231-PG cells, but not MDA-231 cells (Fig. 3D). In addition, luciferase reporter assays using these cell lines were performed to determine the role of plakoglobin in the regulation of the NME1 promoter. While minimal luciferase activity was observed in MDA-231 cells, promoter activity was induced over 3-fold in MDA-231-PG cells (compared to parental MDA-231 cells; Fig. 3E). In contrast, luciferase activity was decreased by ∼5-fold in MCF-7-shPG cells compared to MCF-7 cells (Fig. 3E). Taken together, these results suggest that plakoglobin and p53 positively regulate the expression of the NME1 gene and that plakoglobin expression has no effect on the NME2 gene.
Changes in SATB1 target gene expression in response to plakoglobin levels
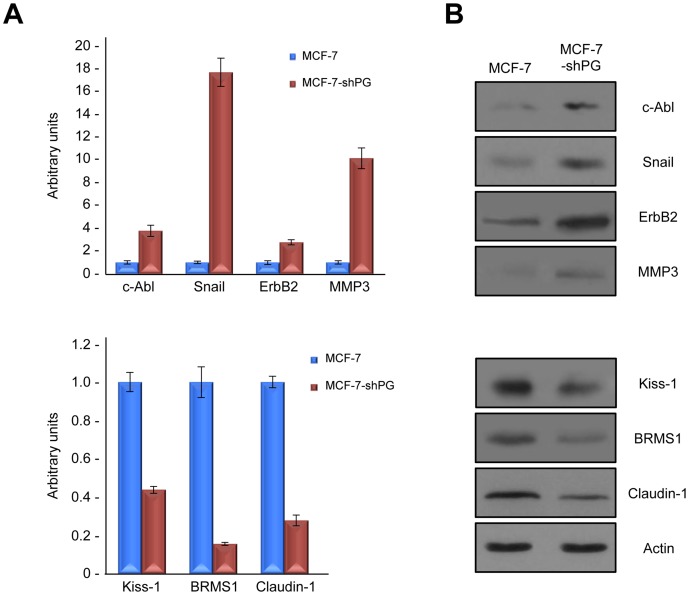
Since SATB1 is a major global regulator of gene expression, we argued that the alteration in SATB1 levels based on plakoglobin expression would result in alterations in the expression of various SATB1 target genes in addition to Nm23. More specifically, we focused on a select number of SATB1 target genes that are known to participate in tumorigenesis and metastasis (e.g. tumor/metastasis suppressors BRMS1, Kiss1, Claudin-1; tumor/metastasis promoters c-Abl, MMP3, ErbB2 and Snail). We performed qRT-PCR experiments and observed that the levels of c-Abl, Snail, ErbB2 and MMP3 mRNA were all increased in MCF-7-shPG cells, compared to MCF-7 cells. Consistent with the increased mRNA levels, western blot experiments showed that protein levels of these tumor/metastasis promoters were also increased in MCF-7-shPG cells (Fig. 4A-B, top). Furthermore, the mRNA and protein levels of tumor/metastasis suppressors BRMS1, Kiss1 and Claudin-1 were decreased in MCF-7-shPG cells relative to MCF-7 cells (Fig. 4A-B, bottom).
Figure 4. Plakoglobin knockdown changes the levels of SATB1 target genes.
A. Total cellular RNA was isolated from MCF-7 and MCF-7-shPG cells, reverse transcribed and processed for PCR using primers specific to SATB1 target genes c-Abl, MMP3, ErbB2, Snail, BRMS1, Kiss1 and Claudin-1. B. Equal amounts of total cellular proteins from these cells were resolved by SDS-PAGE and processed for immunoblotting with antibodies to c-Abl, MMP3, ErbB2, Snail, BRMS1, Kiss1 and Claudin-1. PG, plakoglobin.
Plakoglobin suppresses cancer cell growth, migration and invasion
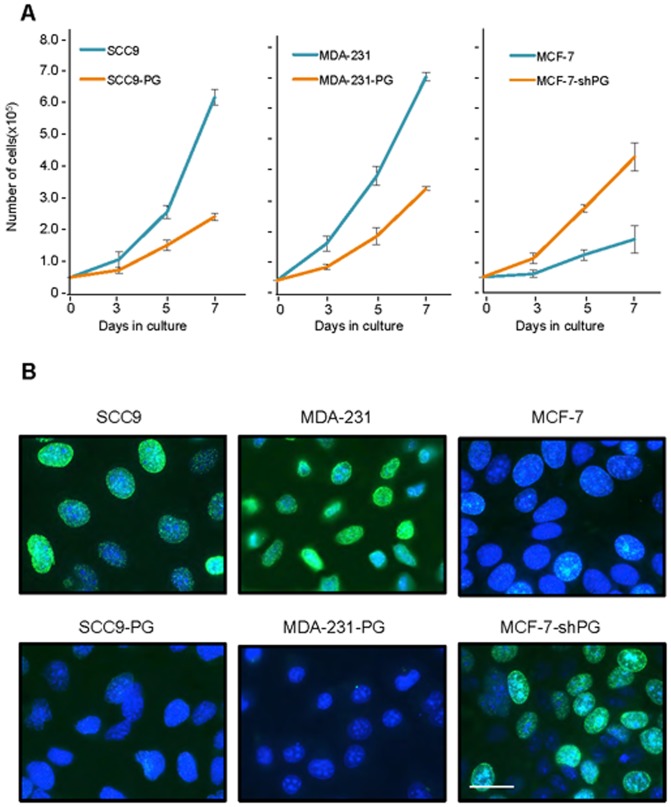
The results so far suggested that plakoglobin plays a role in promoting the expression of various genes involved in suppressing tumor growth, migration and invasion and suppressing the expression of genes that promote these processes. In order to determine whether plakoglobin's regulation of gene expression results in a biologically discernable effect on the in vitro growth and the migratory and invasive properties of cells, MCF-7 and MCF-7-shPG cells we processed for growth, migration and invasion assays as described in Materials and Methods. The results of the growth assay showed a significant increase (∼2.5-fold) in the growth of MCF-7-shPG relative to MCF-7 cells (Fig. 5A). As additional controls, we also assessed the growth rate of SCC9, MDA-231 and their plakoglobin expressing transfectants SCC9-PG and MDA-231-PG, respectively. In contrast to MCF-7-shPG, the growth rate of SCC9-PG cells was reduced ∼2.5-fold relative to parental SCC9 cells, whereas MDA-231-PG cells showed a 2-fold reduction in growth relative to parental MDA-231 cells (Fig. 5A) These results are consistent with our previous observations in SCC9 and MDA-231 cells [15], [49].
Figure 5. Plakoglobin decreases in vitro cell growth and proliferation.
A. Replicate cultures of SCC9, SCC9-PG, MDA-231, -231-PG, MCF-7 and MCF-7-shPG cells were established at single cell density and cells were counted at 3, 5 and 7 days. Each time point represents the average of three independent experiments. The absence of error bars at some time points is due to the small differences among the experiments. B. SCC9, SCC9-PG, MDA-231, -231-PG, MCF-7 and MCF-7-shPG cells were plated on glass coverslips and allowed to grow for 6 days at which time BrdU was added to the cell cultures for 24 hours. BrdU incorporation was then assessed by immunofluorescence staining using BrdU antibodies. Nuclei were countersatined with DRAQ5 and cells viewed using a 63X objective of an LSM510 META (Zeiss) laser scanning confocal microscope. Bar, 20 μm.
We then used BrdU labeling to verify if the differences observed at the end of the 7-day growth assay among different cell lines with various levels of plakoglobin expression were due to differences in cell proliferation. Cells from various cell lines were plated and allowed to grow for 6 days at which time they were labeled with BrdU for 24 hours and processed for confocal microscopy as described in Materials and Methods. The results showed that SCC9 and MDA-231 cells were highly proliferative as almost all cells displayed BrdU incorporation. In contrast, we detected very little or no BrdU incorporation in the plakoglobin expressing MCF-7, SCC9-PG and MDA-231-PG cells (Fig. 5B), whereas there was significant BrdU incorporation in the plakoglobin knockdown MCF-7-shPG cells (Fig. 5B).
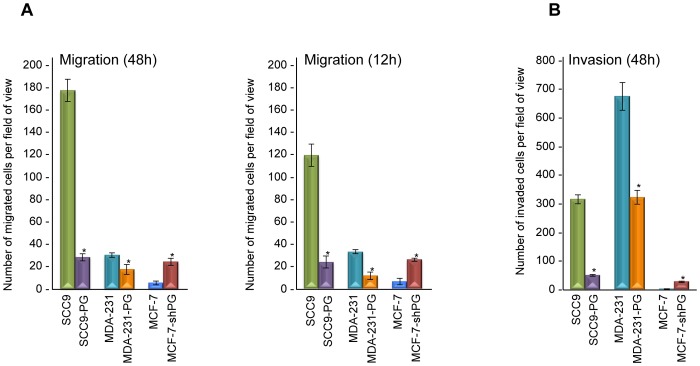
The migratory properties of the various cell lines were assessed using transwell chambers. Cells were allowed to migrate through transwell filters for 48 hours, after which the migrated cells were fixed and counted. Consistent with our previous observations, MDA-231-PG cells displayed ∼40% less migration than MDA-231 cells (Fig. 6A; [49]). Similarly, SCC9 cells were approximately 10-fold more migratory than SCC9-PG cells, whereas MCF-7-shPG cells showed a 4-fold increase in migration compared to MCF-7 cells (Fig. 6A). To rule out the possibility that the increased migration in SCC9, MDA-231 and MCF-7-shPG could be due to their higher cell proliferation rate, we repeated the migration assays for 12 hours, since our growth data showed that none of the cell lines had a doubling time less than 24 hours (Fig. 5A). The results of these experiments were consistent with those of the 48 hours assays and showed that SCC9, MDA-231 and MCF-7-shPG cells were considerably more migratory than their plakoglobin-expressing counterparts (SCC9-PG, MDA-231-PG, MCF-7; Fig. 6A).
Figure 6. Plakoglobin decreases in vitro migration and invasion.
A. Forty-eight- and twelve-hour Transwell migration assays were performed in triplicates for SCC9, SCC9-PG, MDA-231, MDA-231-PG, MCF-7 and MCF-7-shPG cell lines. The membranes were fixed, stained, cut and mounted on slides and viewed under an inverted microscope. B. Forty-eight-hour matrigel invasion assays were performed as described in A using matrigel coated transwell membranes. The number of migrated/invaded cells in five random fields for each membrane were calculated using the ImageJ Cell Counter program and averaged. Histograms represent the average ± SD of the number of migrated/invaded cells for each cell line. *p<0.01). PG, plakoglobin.
The invasive properties of the various cell lines were assessed using matrigel-coated transwell chambers. Similar to the migration experiments, cells were allowed to migrate through the matrigel matrix, after which the invaded cells were fixed and counted. These experiments showed that in addition to being more migratory, SCC9, MCF-7-shPG and MDA-231 cells were more invasive than SCC9-PG, MCF-7 and MDA-231-PG cells (approximately 6-, 7- and 2-fold, respectively; Fig. 6B; [49]). Taken together, these results suggest that plakoglobin, regulates the expression of genes involved in cell growth, migration and invasion concurrent with a suppression of in vitro migration and invasion.
Discussion
In the present study, we have further investigated the underlying mechanisms for plakoglobin's role in tumorigenesis and metastasis [also see 14, 49]. Our data showed that plakoglobin associated with the promoter of the oncogenic DNA binding protein SATB1 and downregulated its expression. The decreased expression of SATB1 following plakoglobin expression was associated with its decreased protein levels and in turn, altered expression of SATB1 target genes with an overall effect of decreased cell growth and in vitro migration and invasion. Conversely, knockdown of plakoglobin in MCF-7 cells resulted in the upregulation of SATB1 and increased cell proliferation, migration and invasion.
SATB1's ability to regulate gene expression was initially identified in the thymus, where several studies showed that it was essential for T-cell development and differentiation [24], [30], [50]. These studies demonstrated that SATB1 regulates gene expression by organizing target gene loci into distinct domains/chromatin loop structures and by recruiting different chromatin remodeling enzymes to promote gene expression and T-cell differentiation. Since then, SATB1 has been established as a contributing factor to the development and progression of many different types of cancer, including breast, lung, prostate, colon and ovarian [34]–[42]. SATB1 has also been shown to participate in the epidermis differentiation as SATB1−/− mice showed defects in epidermal differentiation [51]. These defects were associated with the improper activation of genes found within the epidermal differentiation complex locus, to which SATB1 was shown to bind. Other studies have demonstrated that SATB1 regulates the expression of at least 10% of genes in both T-cells and non T-cells, including genes involved in apoptosis, cell-extracellular matrix attachment, cellular metabolism, calcium signaling and the Wnt, Notch, and TGF-β pathways, suggesting that it plays a role in the global regulation of gene expression [32], [52].
SATB1 has been suggested to regulate gene expression in conjunction with β-catenin as part of the Wnt signaling pathway [44], [53]–[54], since during T-cell differentiation, SATB1 associates with and recruits p300/CBP histone acetyltransferase and β-catenin to the promoters of Wnt target genes, resulting in the increased expression of genes such as IL-2 and MYC [44]. SATB1 also associated with the major breakpoint region (mbr) in the 3′-UTR of the BCL2 gene and promoted the expression of this anti-apoptotic protein, whose expression is also regulated by β-catenin, through the induction of c-Myc and E2F1 [43], [55]–[57]. We previously showed that plakoglobin is also able to regulate the levels of Bcl-2 through the modulation of the signaling activity of β-catenin [58]. The data presented here clearly demonstrates that plakoglobin associates with the SATB1 promoter and downregulates its expression. Taken together, these observations suggest that plakoglobin may regulate Wnt/β-catenin and SATB1 signaling in multiple ways. First, plakoglobin downregulates the expression of SATB1, which would result in the decreased expression of SATB1 target genes. The decreased levels of SATB1 may also alter/reduce β-catenin recruitment to its target promoters and therefore reduce the expression of those genes. Second, nuclear plakoglobin decreases the interaction between β-catenin and TCF and results in inhibition of TCF/β-catenin signaling [13], [58]. Third, expression of physiological levels of plakoglobin results in decreased levels of β-catenin [15], [59]. Finally, plakoglobin associates with and inhibits the expression of the MYC promoter [60], a β-catenin and SATB1 target gene [44], [61]–[62].
More recent studies have suggested that SATB1 plays a role in breast tumorigenesis and metastasis. Indeed, SATB1 expression in SATB1 deficient SKBR3 breast cancer cells resulted in increased tumor growth and a more migratory and invasive phenotype that was concurrent with increased expression of tumor/metastasis promoter genes such as c-Abl, Snail, MMP3, TGFβ-1, ErbB2 and decreased expression of tumor/metastasis suppressors including Nm23, Claudin-1, Kiss1, BRMS1, KAI1. Conversely, knockdown of SATB1 in the highly invasive MDA-231 cells had the opposite effect: tumor/metastasis promoting genes were downregulated whereas inhibitors of these processes were upregulated [34].
Plakoglobin also appears to have a role in regulating tumorigenesis and metastasis through the modulation of gene expression. We recently showed that plakoglobin interacts with the transcription factor p53 and regulates the expression of SFN, the gene encoding the tumor suppressor 14-3-3σ [14]. Furthermore, we showed that p53-transcriptional activity is enhanced in the presence of plakoglobin and that mutant p53 proteins may, in association with plakoglobin, be functional in regulating their wild-type target genes. In the current study, we have identified SATB1 as another target gene of plakoglobin and p53, as ChIP experiments clearly demonstrated an association of both proteins with the SATB1 promoter (Figs. 1–2). However, as opposed to SFN, SATB1 is negatively regulated by p53 and plakoglobin. While we have shown that plakoglobin and p53 interact with one another [14], whether these interactions are direct or involve other cofactors is not clear and warrants further investigation. Furthermore, although plakoglobin is known to associate with TCF/LEF and regulate gene expression [9], [13], [57], [60], neither the human SATB1 nor the NME1 genes contain potential TCF/LEF binding sites, therefore it is likely that plakoglobin-mediated regulation of these genes is independent of TCF/LEF. It was previously shown that p63 is a transcriptional activator of SATB1 during epidermal differentiation [51], however, to the best of our knowledge, the present work is the first to show that p53 also regulates SATB1 expression, albeit opposite to p63. What other co-factors are involved in the regulation of p53 and plakoglobin target genes and to what extent these co-factors differ based on whether the complex is activating or repressing gene expression remains unknown and warrants further investigation.
Along with repressing SATB1 expression, plakoglobin appears to regulate the expression of (at least a subset of) potential SATB1 target genes, including the metastasis suppressor Nm23-H1 [63]. Since its initial discovery, a total of ten Nm23 isoforms (Nm23-H1 to -H10) have been identified in humans, with Nm23-H1 and -H2 being the best studied and characterized [64]–[65]. Nm23-H1 has diverse biological functions including nucleoside diphosphate kinase (NDPK), protein histidine kinase and 3′–5′ exonuclease activities, all of which may potentially contribute to its metastasis suppressor function [66]–[70]. In addition, both Nm23-H1 and -H2 are capable of binding to DNA and regulating gene expression [71]–[77]. Previous studies have shown that exogenous expression of Nm23 in cells lacking its expression resulted not only in decreased migration and invasion, but also in decreased cell proliferation and inhibition of anchorage independent growth [78]–[82]. Furthermore, Nm23 proteins reduced telomerase activity [83] and promoted cell-cell adhesion [84], cell-cycle arrest, and apoptosis [75], as well as DNA-repair following U.V. and ionizing radiation [85]–[86]. These results suggest that Nm23 proteins may also suppress tumor formation in addition to metastasis.
We previously showed that Nm23-H1 mRNA and protein as well as Nm23-H2 protein were upregulated in SCC9-PG cells and that plakoglobin and Nm23 interacted in both the soluble and cytoskeleton-associated pools of cellular proteins [19]. In the present study, we further characterized the role of plakoglobin in the regulation of the NME1 gene and showed that plakoglobin and p53 associated with the NME1 promoter and activated its expression (Fig. 3). The association of plakoglobin with the NME1 promoter is novel and consistent with a previous report that showed decreased Nm23-H1 mRNA levels following plakoglobin knockdown in breast cancer cells [87]. Furthermore, it has been suggested that NME1 is a transcriptional target of p53, since its mRNA and protein levels were higher in breast and colon carcinoma cell lines expressing active p53 [88]–[89]. These observations are strongly supported by our data, which clearly showed that p53 associated with the NME1 promoter. Taken together with our previous results, these data suggest that plakoglobin can increase the levels of its potential target genes through different mechanisms, including direct regulation of gene expression (Nm23-H1) and protein-protein interactions that result in increased protein levels (Nm23-H2; [19]).
In addition to NME1, we also observed alterations in the mRNA and protein levels of other SATB1 target genes. More specifically, knockdown of plakoglobin in MCF-7 cells resulted in the increased mRNA and protein levels of the tumor/metastasis promoters c-Abl, Snail, ErbB2 and MMP3 and the decreased levels of tumor/metastasis suppressors BRMS1, Kiss1 and Claudin-1. Whether plakoglobin may alter the expression of these SATB1 target genes by altering the expression of SATB1 itself and/or by associating with the promoters of these target genes and promoting/repressing their expression requires further investigation.
We showed that the biological consequence of plakoglobin expression in null/low plakoglobin expressing cells was decreased cell growth and in vitro migration and invasion (Figs. 5–6). These results were in agreement with other studies that have previously shown that plakoglobin suppresses these processes and promotes a more “epithelial” phenotype, consistent with its role as a tumor (and metastasis) suppressor [15], [20], [49], [87].
Increasing evidence suggests that plakoglobin regulates tumorigenesis independent of its cell-cell adhesion function. Plakoglobin regulates the expression of genes such as MYC, DSC2 and SFN [14], [60], [90] and also suppresses Ras-mediated oncogenesis through increased HDAC4 mRNA levels [91]. In addition to regulation of gene expression, plakoglobin has been shown to act as a tumor/metastasis suppressor by modulating Rho, Fibronectin and Vitronectin-dependent Src signaling [21], [92].
Our findings are significant in that they clearly point to a role of plakoglobin in regulating a variety of genes that are involved in tumor progression and metastasis. Our data also suggests that plakoglobin may regulate a number of genes under normal cellular conditions (i.e. in the absence of cell stress or activation of different growth pathways), implying that plakoglobin may be a “basal” and more global type of regulator of gene expression. As such, our results have larger implications in that plakoglobin may have a potential as a new therapeutic target for the treatment of various cancers.
Funding Statement
This work is supported by the Canadian Breast Cancer Foundation- Prairies/NWT Chapter (MP) and by the Killam Trusts Izaak Walton Killam Memorial Graduate Scholarship and the University of Alberta Dissertation Fellowship (ZA). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jeanes A, Gottardi CJ, Yap AS (2008) Cadherins and cancer: how does cadherin dysfunction promote tumor progression? Oncogene 27: 6920–6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Makrilia N, Kollias A, Manolopoulos L, Syrigos K (2009) Cell adhesion molecules: role and clinical significance in cancer. Cancer Invest 27: 1023–1037. [DOI] [PubMed] [Google Scholar]
- 3. Yilmaz M, Christofori G (2010) Mechanisms of motility in metastasizing cells. Mol Cancer Res 8: 629–642. [DOI] [PubMed] [Google Scholar]
- 4. Saito M, Tucker DK, Kohlhorst D, Niessen CM, Kowalczyk AP (2012) Classical and desmosomal cadherins at a glance. J Cell Sci 125: 2547–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nelson WJ, Dickinson DJ, Weis WI (2013) Roles of cadherins and catenins in cell-cell adhesion and epithelial cell polarity. Prog Mol Biol Transl Sci 116: 3–23. [DOI] [PubMed] [Google Scholar]
- 6. Yonemura S (2011) Cadherin-actin interactions at adherens junctions. Curr Opin Cell Biol 23: 515–522. [DOI] [PubMed] [Google Scholar]
- 7. Kowalczyk AP, Green KJ (2013) Structure, function, and regulation of desmosomes. Prog Mol Biol Transl Sci 116: 95–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Peifer M, McCrea PD, Green KJ, Wieschaus E, Gumbiner BM (1992) The vertebrate adhesive junction proteins beta-catenin and plakoglobin and the Drosophila segment polarity gene armadillo form a multigene family with similar properties. J Cell Biol 118: 681–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhurinsky J, Shtutman M, Ben-Ze'ev A (2000a) Plakoglobin and beta-catenin: protein interactions, regulation and biological roles. J Cell Sci 113 (Pt 18): 3127–3139. [DOI] [PubMed] [Google Scholar]
- 10. Aktary Z, Pasdar M (2012) Plakoglobin: role in tumorigenesis and metastasis. Int J Cell Biol 2012: 189521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Simcha I, Shtutman M, Salomon D, Zhurinsky J, Sadot E, et al. (1998) Differential nuclear translocation and transactivation potential of beta-catenin and plakoglobin. J Cell Biol 141: 1433–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhurinsky J, Shtutman M, Ben-Ze'ev A (2000b) Differential mechanisms of LEF/TCF family-dependent transcriptional activation by beta-catenin and plakoglobin. Mol Cell Biol 20: 4238–4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Miravet S, Piedra J, Miro F, Itarte E, Garcia de Herreros A, et al. (2002) The transcriptional factor TCF-4 contains different binding sites for beta-catenin and plakoglobin. J Biol Chem 277: 1884–1891. [DOI] [PubMed] [Google Scholar]
- 14. Aktary Z, Kulak S, Mackey J, Jahroudi N, Pasdar M (2013) Plakoglobin interacts with the transcription factor p53 and regulates the expression of 14-3-3sigma. J Cell Sci 126: 3031–3042. [DOI] [PubMed] [Google Scholar]
- 15. Parker HR, Li Z, Sheinin H, Lauzon G, Pasdar M (1998) Plakoglobin induces desmosome formation and epidermoid phenotype in N-cadherin-expressing squamous carcinoma cells deficient in plakoglobin and E-cadherin. Cell Motil Cytoskeleton 40: 87–100. [DOI] [PubMed] [Google Scholar]
- 16. Winn RA, Bremnes RM, Bemis L, Franklin WA, Miller YE, et al. (2002) gamma-Catenin expression is reduced or absent in a subset of human lung cancers and re-expression inhibits transformed cell growth. Oncogene 21: 7497–7506. [DOI] [PubMed] [Google Scholar]
- 17. Rieger-Christ KM, Ng L, Hanley RS, Durrani O, Ma H, et al. (2005) Restoration of plakoglobin expression in bladder carcinoma cell lines suppresses cell migration and tumorigenic potential. Br J Cancer 92: 2153–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yin T, Getsios S, Caldelari R, Kowalczyk AP, Muller EJ, et al. (2005) Plakoglobin suppresses keratinocyte motility through both cell-cell adhesion-dependent and -independent mechanisms. Proc Natl Acad Sci U S A 102: 5420–5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Aktary Z, Chapman K, Lam L, Lo A, Ji C, et al. (2010) Plakoglobin interacts with and increases the protein levels of metastasis suppressor Nm23-H2 and regulates the expression of Nm23-H1. Oncogene 29: 2118–2129. [DOI] [PubMed] [Google Scholar]
- 20. Bailey CK, Mittal MK, Misra S, Chaudhuri G (2012) High motility of triple-negative breast cancer cells is due to repression of plakoglobin gene by metastasis modulator protein SLUG. J Biol Chem 287: 19472–19486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Franzen CA, Todorovic V, Desai BV, Mirzoeva S, Yang XJ, et al. (2012) The desmosomal armadillo protein plakoglobin regulates prostate cancer cell adhesion and motility through vitronectin-dependent Src signaling. PLoS One 7: e42132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim W, Kim M, Jho EH (2013) Wnt/beta-catenin signalling: from plasma membrane to nucleus. Biochem J 450: 9–21. [DOI] [PubMed] [Google Scholar]
- 23. Dickinson LA, Joh T, Kohwi Y, Kohwi-Shigematsu T (1992) A tissue-specific MAR/SAR DNA-binding protein with unusual binding site recognition. Cell 70: 631–645. [DOI] [PubMed] [Google Scholar]
- 24. de Belle I, Cai S, Kohwi-Shigematsu T (1998) The genomic sequences bound to special AT-rich sequence-binding protein 1 (SATB1) in vivo in Jurkat T cells are tightly associated with the nuclear matrix at the bases of the chromatin loops. J Cell Biol 141: 335–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kohwi-Shigematsu T, Kohwi Y (1990) Torsional stress stabilizes extended base unpairing in suppressor sites flanking immunoglobulin heavy chain enhancer. Biochemistry 29: 9551–9560. [DOI] [PubMed] [Google Scholar]
- 26. Kohwi-Shigematsu T, Poterlowicz K, Ordinario E, Han HJ, Botchkarev VA, et al. (2013) Genome organizing function of SATB1 in tumor progression. Semin Cancer Biol 23: 72–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bode J, Kohwi Y, Dickinson L, Joh T, Klehr D, et al. (1992) Biological significance of unwinding capability of nuclear matrix-associating DNAs. Science 255: 195–197. [DOI] [PubMed] [Google Scholar]
- 28. Cai S, Han HJ, Kohwi-Shigematsu T (2003) Tissue-specific nuclear architecture and gene expression regulated by SATB1. Nat Genet 34: 42–51. [DOI] [PubMed] [Google Scholar]
- 29. Cai S, Lee CC, Kohwi-Shigematsu T (2006) SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat Genet 38: 1278–1288. [DOI] [PubMed] [Google Scholar]
- 30. Kumar PP, Bischof O, Purbey PK, Notani D, Urlaub H, et al. (2007) Functional interaction between PML and SATB1 regulates chromatin-loop architecture and transcription of the MHC class I locus. Nat Cell Biol 9: 45–56. [DOI] [PubMed] [Google Scholar]
- 31. Yasui D, Miyano M, Cai S, Varga-Weisz P, Kohwi-Shigematsu T (2002) SATB1 targets chromatin remodelling to regulate genes over long distances. Nature 419: 641–645. [DOI] [PubMed] [Google Scholar]
- 32. Kumar PP, Purbey PK, Ravi DS, Mitra D, Galande S (2005) Displacement of SATB1-bound histone deacetylase 1 corepressor by the human immunodeficiency virus type 1 transactivator induces expression of interleukin-2 and its receptor in T cells. Mol Cell Biol 25: 1620–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wen J, Huang S, Rogers H, Dickinson LA, Kohwi-Shigematsu T, et al. (2005) SATB1 family protein expressed during early erythroid differentiation modifies globin gene expression. Blood 105: 3330–3339. [DOI] [PubMed] [Google Scholar]
- 34. Han HJ, Russo J, Kohwi Y, Kohwi-Shigematsu T (2008) SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature 452: 187–193. [DOI] [PubMed] [Google Scholar]
- 35. Li QQ, Chen ZQ, Xu JD, Cao XX, Chen Q, et al. (2010) Overexpression and involvement of special AT-rich sequence binding protein 1 in multidrug resistance in human breast carcinoma cells. Cancer Sci 101: 80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhao XD, Ji WY, Zhang W, He LX, Yang J, et al. (2010) Overexpression of SATB1 in laryngeal squamous cell carcinoma. ORL J Otorhinolaryngol Relat Spec 72: 1–5. [DOI] [PubMed] [Google Scholar]
- 37. Chen H, Takahara M, Oba J, Xie L, Chiba T, et al. (2011) Clinicopathologic and prognostic significance of SATB1 in cutaneous malignant melanoma. J Dermatol Sci 64: 39–44. [DOI] [PubMed] [Google Scholar]
- 38. Xiang J, Zhou L, Li S, Xi X, Zhang J, et al. (2012) AT-rich sequence binding protein 1: Contribution to tumor progression and metastasis of human ovarian carcinoma. Oncol Lett 3: 865–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tu W, Luo M, Wang Z, Yan W, Xia Y, et al. (2012) Upregulation of SATB1 promotes tumor growth and metastasis in liver cancer. Liver Int 32: 1064–1078. [DOI] [PubMed] [Google Scholar]
- 40. Nodin B, Hedner C, Uhlen M, Jirstrom K (2012) Expression of the global regulator SATB1 is an independent factor of poor prognosis in high grade epithelial ovarian cancer. J Ovarian Res 5: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chu SH, Ma YB, Feng DF, Li ZQ, Jiang PC (2012) Correlation between SATB1 and Bcl-2 expression in human glioblastoma multiforme. Mol Med Rep. [DOI] [PubMed]
- 42. Huang B, Zhou H, Wang X, Liu Z (2013) Silencing SATB1 with siRNA inhibits the proliferation and invasion of small cell lung cancer cells. Cancer Cell Int 13: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ramakrishnan M, Liu WM, DiCroce PA, Posner A, Zheng J, et al. (2000) Modulated binding of SATB1, a matrix attachment region protein, to the AT-rich sequence flanking the major breakpoint region of BCL2. Mol Cell Biol 20: 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Notani D, Gottimukkala KP, Jayani RS, Limaye AS, Damle MV, et al. (2010) Global regulator SATB1 recruits beta-catenin and regulates T (H)2 differentiation in Wnt-dependent manner. PLoS Biol 8: e1000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lam L, Hu X, Aktary Z, Andrews DW, Pasdar M (2009) Tamoxifen and ICI 182,780 increase Bcl-2 levels and inhibit growth of breast carcinoma cells by modulating PI3K/AKT, ERK and IGF-1R pathways independent of ERalpha. Breast Cancer Res Treat 118: 605–621. [DOI] [PubMed] [Google Scholar]
- 46. Peng Y, Jahroudi N (2003) The NFY transcription factor inhibits von Willebrand factor promoter activation in non-endothelial cells through recruitment of histone deacetylases. J Biol Chem 278: 8385–8394. [DOI] [PubMed] [Google Scholar]
- 47. Lei L, Lu L, Xiang L, Xue-song W, De-pei L, et al. (2010) Epigenetic repression of SATB1 by polycomb group protein EZH2 in epithelial cells. Chin Med Sci J 25: 199–205. [DOI] [PubMed] [Google Scholar]
- 48. Qu S, Long J, Cai Q, Shu XO, Cai H, et al. (2008) Genetic polymorphisms of metastasis suppressor gene NME1 and breast cancer survival. Clin Cancer Res 14: 4787–4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lam L, Aktary Z, Bishay M, Werkman C, Kuo CY, et al. (2012) Regulation of subcellular distribution and oncogenic potential of nucleophosmin by plakoglobin. Oncogenesis 1: e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Alvarez JD, Yasui DH, Niida H, Joh T, Loh DY, et al. (2000) The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes Dev 14: 521–535. [PMC free article] [PubMed] [Google Scholar]
- 51. Fessing MY, Mardaryev AN, Gdula MR, Sharov AA, Sharova TY, et al. (2011) p63 regulates Satb1 to control tissue-specific chromatin remodeling during development of the epidermis. J Cell Biol 194: 825–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Notani D, Ramanujam PL, Kumar PP, Gottimukkala KP, Kumar-Sinha C, et al. (2011) N-terminal PDZ-like domain of chromatin organizer SATB1 contributes towards its function as transcription regulator. J Biosci 36: 461–469. [DOI] [PubMed] [Google Scholar]
- 53. Purbey PK, Singh S, Notani D, Kumar PP, Limaye AS, et al. (2009) Acetylation-dependent interaction of SATB1 and CtBP1 mediates transcriptional repression by SATB1. Mol Cell Biol 29: 1321–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Burute M, Gottimukkala K, Galande S (2012) Chromatin organizer SATB1 is an important determinant of T-cell differentiation. Immunol Cell Biol 90: 852–859. [DOI] [PubMed] [Google Scholar]
- 55. Zhang J, Ma C, Han X, Durrin LK, Sun Y (2006) The bcl-2 major breakpoint region (mbr) possesses transcriptional regulatory function. Gene 379: 127–131. [DOI] [PubMed] [Google Scholar]
- 56. Ma C, Zhang J, Durrin LK, Lv J, Zhu D, et al. (2007) The BCL2 major breakpoint region (mbr) regulates gene expression. Oncogene 26: 2649–2657. [DOI] [PubMed] [Google Scholar]
- 57. Li Q, Dashwood WM, Zhong X, Nakagama H, Dashwood RH (2007) Bcl-2 overexpression in PhIP-induced colon tumors: cloning of the rat Bcl-2 promoter and characterization of a pathway involving beta-catenin, c-Myc and E2F1. Oncogene 26: 6194–6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li L, Chapman K, Hu X, Wong A, Pasdar M (2007) Modulation of the oncogenic potential of beta-catenin by the subcellular distribution of plakoglobin. Mol Carcinog 46: 824–838. [DOI] [PubMed] [Google Scholar]
- 59. Salomon D, Sacco PA, Roy SG, Simcha I, Johnson KR, et al. (1997) Regulation of beta-catenin levels and localization by overexpression of plakoglobin and inhibition of the ubiquitin-proteasome system. J Cell Biol 139: 1325–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Williamson L, Raess NA, Caldelari R, Zakher A, de Bruin A, et al. (2006) Pemphigus vulgaris identifies plakoglobin as key suppressor of c-Myc in the skin. EMBO J 25: 3298–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. He TC, Sparks AB, Rago C, Hermeking H, Zawel L, et al. (1998) Identification of c-MYC as a target of the APC pathway. Science 281: 1509–1512. [DOI] [PubMed] [Google Scholar]
- 62. MacDonald BT, Tamai K, He X (2009) Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell 17: 9–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Steeg PS, Bevilacqua G, Kopper L, Thorgeirsson UP, Talmadge JE, et al. (1988) Evidence for a novel gene associated with low tumor metastatic potential. J Natl Cancer Inst 80: 200–204. [DOI] [PubMed] [Google Scholar]
- 64. Thakur RK, Yadav VK, Kumar P, Chowdhury S (2011) Mechanisms of non-metastatic 2 (NME2)-mediated control of metastasis across tumor types. Naunyn Schmiedebergs Arch Pharmacol 384: 397–406. [DOI] [PubMed] [Google Scholar]
- 65. Marino N, Nakayama J, Collins JW, Steeg PS (2012) Insights into the biology and prevention of tumor metastasis provided by the Nm23 metastasis suppressor gene. Cancer Metastasis Rev 31: 593–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wagner PD, Steeg PS, Vu ND (1997) Two-component kinase-like activity of nm23 correlates with its motility-suppressing activity. Proc Natl Acad Sci U S A 94: 9000–9005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lacombe ML, Milon L, Munier A, Mehus JG, Lambeth DO (2000) The human Nm23/nucleoside diphosphate kinases. J Bioenerg Biomembr 32: 247–258. [DOI] [PubMed] [Google Scholar]
- 68. Fan Z, Beresford PJ, Oh DY, Zhang D, Lieberman J (2003) Tumor suppressor NM23-H1 is a granzyme A-activated DNase during CTL-mediated apoptosis, and the nucleosome assembly protein SET is its inhibitor. Cell 112: 659–672. [DOI] [PubMed] [Google Scholar]
- 69. Steeg PS, Horak CE, Miller KD (2008) Clinical-translational approaches to the Nm23-H1 metastasis suppressor. Clin Cancer Res 14: 5006–5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Novak M, Jarrett SG, McCorkle JR, Mellon I, Kaetzel DM (2011) Multiple mechanisms underlie metastasis suppressor function of NM23-H1 in melanoma. Naunyn Schmiedebergs Arch Pharmacol 384: 433–438. [DOI] [PubMed] [Google Scholar]
- 71. Postel EH, Berberich SJ, Flint SJ, Ferrone CA (1993) Human c-myc transcription factor PuF identified as nm23-H2 nucleoside diphosphate kinase, a candidate suppressor of tumor metastasis. Science 261: 478–480. [DOI] [PubMed] [Google Scholar]
- 72. Postel EH, Berberich SJ, Rooney JW, Kaetzel DM (2000) Human NM23/nucleoside diphosphate kinase regulates gene expression through DNA binding to nuclease-hypersensitive transcriptional elements. J Bioenerg Biomembr 32: 277–284. [DOI] [PubMed] [Google Scholar]
- 73. Ma D, Xing Z, Liu B, Pedigo NG, Zimmer SG, et al. (2002) NM23-H1 and NM23-H2 repress transcriptional activities of nuclease-hypersensitive elements in the platelet-derived growth factor-A promoter. J Biol Chem 277: 1560–1567. [DOI] [PubMed] [Google Scholar]
- 74. Postel EH (2003) Multiple biochemical activities of NM23/NDP kinase in gene regulation. J Bioenerg Biomembr 35: 31–40. [DOI] [PubMed] [Google Scholar]
- 75. Cervoni L, Egistelli L, Eufemi M, d'Abusco AS, Altieri F, et al. (2006) DNA sequences acting as binding sites for NM23/NDPK proteins in melanoma M14 cells. J Cell Biochem 98: 421–428. [DOI] [PubMed] [Google Scholar]
- 76. Thakur RK, Kumar P, Halder K, Verma A, Kar A, et al. (2009) Metastases suppressor NM23-H2 interaction with G-quadruplex DNA within c-MYC promoter nuclease hypersensitive element induces c-MYC expression. Nucleic Acids Res 37: 172–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Choudhuri T, Murakami M, Kaul R, Sahu SK, Mohanty S, et al. (2010) Nm23-H1 can induce cell cycle arrest and apoptosis in B cells. Cancer Biol Ther 9: 1065–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lee HY, Lee H (1999) Inhibitory activity of nm23-H1 on invasion and colonization of human prostate carcinoma cells is not mediated by its NDP kinase activity. Cancer Lett 145: 93–99. [DOI] [PubMed] [Google Scholar]
- 79. Khan MH, Yasuda M, Higashino F, Haque S, Kohgo T, et al. (2001) nm23-H1 suppresses invasion of oral squamous cell carcinoma-derived cell lines without modifying matrix metalloproteinase-2 and matrix metalloproteinase-9 expression. Am J Pathol 158: 1785–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Suzuki E, Ota T, Tsukuda K, Okita A, Matsuoka K, et al. (2004) nm23-H1 reduces in vitro cell migration and the liver metastatic potential of colon cancer cells by regulating myosin light chain phosphorylation. Int J Cancer 108: 207–211. [DOI] [PubMed] [Google Scholar]
- 81. Jung S, Paek YW, Moon KS, Wee SC, Ryu HH, et al. (2006) Expression of Nm23 in gliomas and its effect on migration and invasion in vitro. Anticancer Res 26: 249–258. [PubMed] [Google Scholar]
- 82. McDermott WG, Boissan M, Lacombe ML, Steeg PS, Horak CE (2008) Nm23-H1 homologs suppress tumor cell motility and anchorage independent growth. Clin Exp Metastasis 25: 131–138. [DOI] [PubMed] [Google Scholar]
- 83. Kar A, Saha D, Purohit G, Singh A, Kumar P, et al. (2012) Metastases suppressor NME2 associates with telomere ends and telomerase and reduces telomerase activity within cells. Nucleic Acids Res 40: 2554–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Bago R, Pavelic J, Maravic Vlahovicek G, Bosnar MH (2009) Nm23-H1 promotes adhesion of CAL 27 cells in vitro. Mol Carcinog 48: 779–789. [DOI] [PubMed] [Google Scholar]
- 85. Zhang Q, McCorkle JR, Novak M, Yang M, Kaetzel DM (2011) Metastasis suppressor function of NM23-H1 requires its 3′–5′ exonuclease activity. Int J Cancer 128: 40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Jarrett SG, Novak M, Dabernat S, Daniel JY, Mellon I, et al. (2012) Metastasis suppressor NM23-H1 promotes repair of UV-induced DNA damage and suppresses UV-induced melanomagenesis. Cancer Res 72: 133–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Holen I, Whitworth J, Nutter F, Evans A, Brown HK, et al. (2012) Loss of plakoglobin promotes decreased cell-cell contact, increased invasion, and breast cancer cell dissemination in vivo. Breast Cancer Res 14: R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Chen SL, Wu YS, Shieh HY, Yen CC, Shen JJ, et al. (2003) P53 is a regulator of the metastasis suppressor gene Nm23-H1. Mol Carcinog 36: 204–214. [DOI] [PubMed] [Google Scholar]
- 89. Rahman-Roblick R, Roblick UJ, Hellman U, Conrotto P, Liu T, et al. (2007) p53 targets identified by protein expression profiling. Proc Natl Acad Sci U S A 104: 5401–5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tokonzaba E, Chen J, Cheng X, Den Z, Ganeshan R, et al. (2013) Plakoglobin as a Regulator of Desmocollin Gene Expression. J Invest Dermatol. [DOI] [PMC free article] [PubMed]
- 91.Yim JH, Baek JH, Lee CW, Kim MJ, Yun HS, et al. (2013) Identification of HDAC4 as a target of gamma-catenin that regulates the oncogenic K-Ras-mediated malignant phenotype of Rat2 cells. Biochem Biophys Res Commun. [DOI] [PubMed]
- 92. Todorovic V, Desai BV, Patterson MJ, Amargo EV, Dubash AD, et al. (2010) Plakoglobin regulates cell motility through Rho- and fibronectin-dependent Src signaling. J Cell Sci 123: 3576–3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ayabe T, Tomita M, Matsuzaki Y, Ninomiya H, Hara M, et al. (2004) Micrometastasis and expression of nm23 messenger RNA of lymph nodes from lung cancer and the postoperative clinical outcome. Ann Thorac Cardiovasc Surg 10: 152–159. [PubMed] [Google Scholar]
- 94. Syed V, Mukherjee K, Lyons-Weiler J, Lau KM, Mashima T, et al. (2005) Identification of ATF-3, caveolin-1, DLC-1, and NM23-H2 as putative antitumorigenic, progesterone-regulated genes for ovarian cancer cells by gene profiling. Oncogene 24: 1774–1787. [DOI] [PubMed] [Google Scholar]
- 95. Nakanishi K, Kumaki F, Hiroi S, Mukai M, Ikeda E, et al. (2006) Mre11 expression in atypical adenomatous hyperplasia and adenocarcinoma of the lung. Arch Pathol Lab Med 130: 1330–1334. [DOI] [PubMed] [Google Scholar]