Abstract
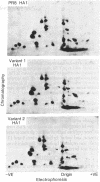
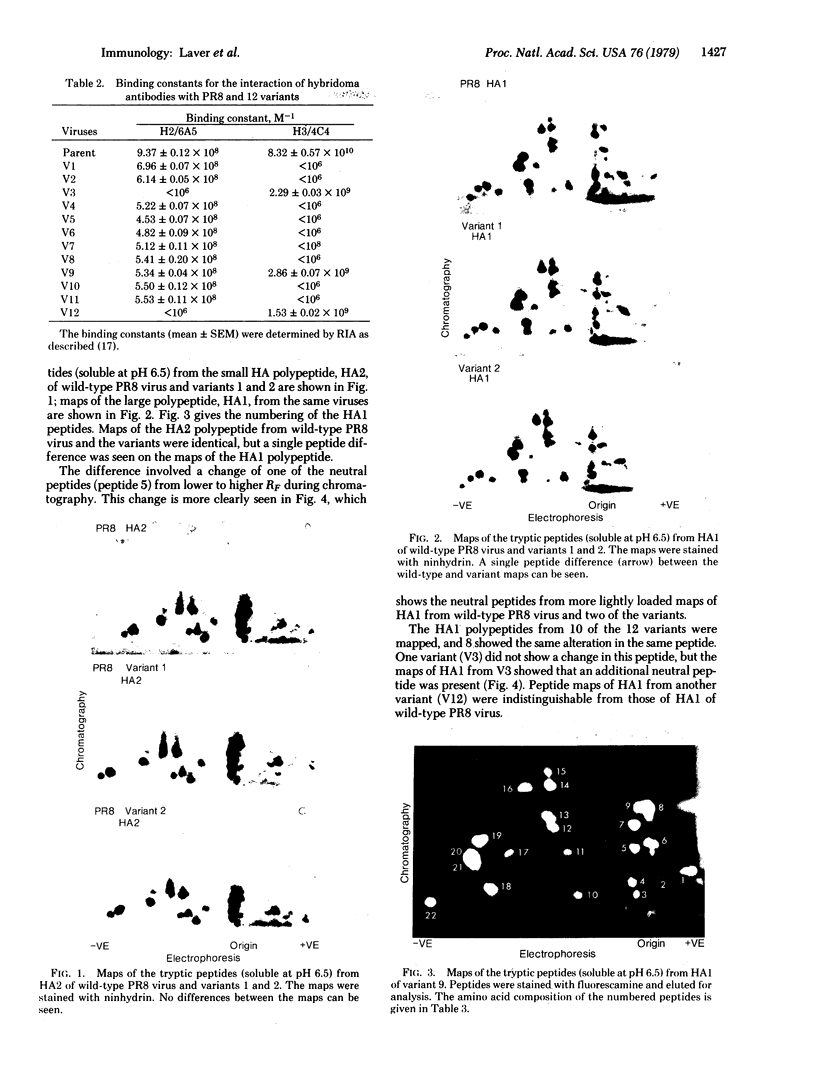
Variants of A/PR/8/34 (HON1) influenza virus, having hemagglutinin molecules with probably a single altered antigenic determinant, were isolated by growing the virus in the presence of the monoclonal hybridoma antibody PEG-1. The variants were analyzed by peptide mapping and characterized antigenically by using PEG-1 and four other monoclonal hybridoma antibodies to PR8 hemagglutinin. Peptide maps of the large hemagglutinin polypeptide, HA1, from 8 out of 10 variants showed a single changed peptide. This peptide from two of the variants was analyzed, and in each case a serine residue in the wild-type hemagglutinin was replaced by leucine in the variant. Although these eight variants showed identical peptide maps, one could be discriminated antigenically from the others with one of the hybridomas. (The peptide maps represented about one-third of the HA1 molecule.) Of the other two variants, one gave the same HA1 map as the wild type, but could be distinguished antigenically from wild-type virus by two of the hybridomas. The other was unique, and could be distinguished, both antigenically and by peptide mapping, from the other variants.
Since a large number of the variants selected with PEG-1 showed the same peptide change, it is likely that this alteration in amino acid sequence (serine to leucine) was responsible for the inability of the variants to bind PEG-1 monoclonal antibody. We do not know, however, whether the changed amino acids were located within the antigenic sites or whether the change occurred somewhere else in the hemagglutinin molecule and altered the determinants through conformational changes.
Keywords: antigenic variants, hemagglutinin, peptide maps, amino acid changes
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ARCHETTI I., HORSFALL F. L., Jr Persistent antigenic variation of influenza A viruses after incomplete neutralization in ovo with heterologous immune serum. J Exp Med. 1950 Nov 1;92(5):441–462. doi: 10.1084/jem.92.5.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazekas de St Groth, Webster R. G. Disquisitions of Original Antigenic Sin. I. Evidence in man. J Exp Med. 1966 Sep 1;124(3):331–345. doi: 10.1084/jem.124.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhard W., Webster R. G. Antigenic drift in influenza A viruses. I. Selection and characterization of antigenic variants of A/PR/8/34 (HON1) influenza virus with monoclonal antibodies. J Exp Med. 1978 Aug 1;148(2):383–392. doi: 10.1084/jem.148.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAMRE D., LOOSLI C. G., GERBER P. Antigenic variants of influenza A virus (PR8 strain). IV. Serological characteristics of a second line of variants developed in mice given polyvalent vaccine. J Exp Med. 1958 Jun 1;107(6):845–855. doi: 10.1084/jem.107.6.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprowski H., Gerhard W., Croce C. M. Production of antibodies against influenza virus by somatic cell hybrids between mouse myeloma and primed spleen cells. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2985–2988. doi: 10.1073/pnas.74.7.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laver W. G., Downie J. C., Webster R. G. Studies on antigenic variation in influenza virus. Evidence for multiple antigenic determinants on the hemagglutinin subunits of A-Hong Kong-68 (H3 N2) virus and the A-England-72 strains. Virology. 1974 May;59(1):230–244. doi: 10.1016/0042-6822(74)90218-9. [DOI] [PubMed] [Google Scholar]
- Laver W. G., Webster R. G. Selection of antigenic mutants of influenza viruses. Isolation and peptide mapping of their hemagglutination proteins. Virology. 1968 Feb;34(2):193–202. doi: 10.1016/0042-6822(68)90230-4. [DOI] [PubMed] [Google Scholar]
- Laver W. G., Webster R. G. Studies on the origin of pandemic influenza. 3. Evidence implicating duck and equine influenza viruses as possible progenitors of the Hong Kong strain of human influenza. Virology. 1973 Feb;51(2):383–391. doi: 10.1016/0042-6822(73)90437-6. [DOI] [PubMed] [Google Scholar]
- Udenfriend S., Stein S., Böhlen P., Dairman W., Leimgruber W., Weigele M. Fluorescamine: a reagent for assay of amino acids, peptides, proteins, and primary amines in the picomole range. Science. 1972 Nov 24;178(4063):871–872. doi: 10.1126/science.178.4063.871. [DOI] [PubMed] [Google Scholar]
- Virelizier J. L., Allison A. C., Schild G. C. Antibody responses to antigenic determinants of influenza virus hemagglutinin. II. Original antigenic sin: a bone marrow-derived lymphocyte memory phenomenon modulated by thymus-derived lymphocytes. J Exp Med. 1974 Dec 1;140(6):1571–1578. doi: 10.1084/jem.140.6.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley D. C., Skehel J. J. Crystallization and x-ray diffraction studies on the haemagglutinin glycoprotein from the membrane of influenza virus. J Mol Biol. 1977 May 15;112(2):343–347. doi: 10.1016/s0022-2836(77)80149-6. [DOI] [PubMed] [Google Scholar]
- Wiley D. C., Skehel J. J., Waterfield M. Evidence from studies with a cross-linking reagent that the haemagglutinin of influenza virus is a trimer. Virology. 1977 Jun 15;79(2):446–448. doi: 10.1016/0042-6822(77)90371-3. [DOI] [PubMed] [Google Scholar]
- Wrigley N. G., Laver W. G., Downie J. C. Binding of antibodies to isolated haemagglutinin and neuraminidase molecules of influenza virus observed in the electron microscope. J Mol Biol. 1977 Jan 25;109(3):405–421. doi: 10.1016/s0022-2836(77)80020-x. [DOI] [PubMed] [Google Scholar]