Abstract
Camelpox is considered as emerging public health problem during this decade due to increased reported cases and outbreaks in camels. Camelpox is a contagious, often sporadic, and notifiable skin disease of camelids and is socio-economically significant as it incurs considerable loss in terms of morbidity, mortality, loss of weight and reduction in milk yield and confined to camel-rearing countries. The causative agent, camelpox virus (CMLV) is genetically closely related to variola virus and has gained much attention from researchers due to its recent emergence in human. The virus carrying genes responsible for host immune evasion mechanisms owing to the threat posed by potential bio-warfare agents. Although the disease can be diagnosed based on clinical features, the similar confounding skin lesions necessitate identification, detection and differentiation of the CMLV by molecular techniques. Vaccines are available in some countries and the available live attenuated vaccine provides long-lasting immunity. Further, novel highly sensitive and specific techniques would be useful in the identification of emerging and re-emerging virus, thereby therapeutic, prophylactic, preventive measures would be applied in time to curtail further spread of camelpox like other zoonotic diseases. This review provide overview of the camelpox particularly on its epidemiology, pathogenesis and biology of the disease, diagnostic approaches and control measures.
Keywords: Camelpox virus, Biology, Epidemiology, Diagnosis, Prophylaxis, Control measures
Introduction
Camelpox is an economically important, contagious, often sporadic, and notifiable to Office Internationale des Epizootics (World organization for animal health—WOAH) skin disease of camelids [35]. The causative agent, Camelpox virus (CMLV) is closely related to Variola virus (VARV), the causative agent of smallpox. Although camelpox has presumably existed for millennia, its causative agent was not isolated until the early 1970s, during the opening phase of the global smallpox eradication campaign [74, 75]. The disease is restricted to camels and is enzootic in almost every region, where camels are reared except Australia. According to the UN Food and Agriculture Organization (FAO), the total world camel population is ≈25 million (faostat.fao.org). Camelpox is confined to camel-rearing belts particularly in developing countries and causes economic impact due to considerable loss in terms of morbidity, mortality, loss of weight and reduction in milk yield. Effective control of any disease warrants a prophylactic as well as a rapid, specific, and sensitive diagnostic assay (s) and molecular epidemiological studies. The virus has attracted researchers due to its close genetic relatedness to VARV and carrying genes responsible for host immune evasion. Recent emergence of zoonotic camelpox outbreaks in India is a serious public health concern [13]. This review article comprehend a note on camelpox, and CMLV particularly on its epidemiology, pathogenesis and biology of the disease, diagnostic approaches and control measures.
Etiology
Camelpox virus (CMLV), the causative agent of camelpox, belongs to the genus Orthopoxvirus (OPV), of the subfamily Chordopoxvirinae of the family Poxviridae [59]. The other members of the genus include several pathogens of veterinary and zoonotic importance. These are VARV, Monkeypox virus (MPXV), Vaccinia virus (VACV), Buffalopox virus (BPXV, a variant of VACV), Cowpox virus (CPXV), Ectromelia virus (ECTV), Rabbitpox virus (RPXV), Taterapox virus, the North American OPVs (Volepox virus, Raccoonpox virus and Skunkpox virus), and an unclassified OPV species, Uasin Gishu disease virus [33, 35]. Parapox- and papilloma-viruses also cause a similar kind of infections in camelids. There are numerous CMLV strains that have been isolated from different outbreaks in different parts of the camel rearing countries [33].
The identification of CMLV agent was an alarm when it was described as smallpox-like disease during smallpox eradication campaign [11], which led to the discovery of the CMLV. CMLV is one of the least studied members of OPVs till recently. It is quite difficult to distinguish the CMLV from the prototypic VACV with respect to size, shape, structure, physico-chemical properties and replication mechanisms [42, 76]. OPVs are large and the average size of CMLV virion is 224 × 389 nm [24, 59]. Like OPVs, camelpox virion consists of an envelope, outer membrane, two lateral bodies and a core. Cryo-electron tomography of VAVC revealed brick-shaped particles with slightly rounded edges and dimensions of ~360 × 270 × 250 nm [22]. The outer layer was consistent with a lipid membrane (5–6 nm thick), below which two lateral bodies were found. The internal core contains electron dense coils of presumptive DNA–protein complexes and surrounded by two layers with thickness of ~18–19 nm [22]. The inner layer was consistent with a lipid membrane and the outer layer was discontinuous, formed by a periodic palisade built by the side interaction of T-shaped protein spikes that were anchored in the lower membrane and were arranged into small hexagonal crystallites [22]. The growth kinetics in human embryonic lung (HEL) fibroblast cells indicates that CMLV is different from VACV and CPXV [31].
CMLV, like other OPVs, shows variable responses to physical/chemical agents. In general, CMLV is sensitive to both acidic (pH 3–5) and alkaline (pH 8.5–10) conditions [25]. Like other poxviruses, CMLV is susceptible to various disinfectants. The virus can be destroyed either by autoclaving or boiling for 10 min and ultraviolet rays in a few minutes [21]. The difference in the physico-chemical properties of different CMLV strains has been reviewed earlier [33]. The CMLV haemagglutonates cockerel erythrocytes. In general, pox virions show high environmental stability (tolerance to temperature, pH and chemicals) and can remain contagious over several months [72].
Genome and biology of virus
CMLV genome consists of a single linear double-stranded DNA molecule terminated by a hairpin loop that replicates in the cytoplasm [59]. The genome is AT-rich (66.9 %) having cross-links that join the two DNA strands at both ends. Each end of DNA strand has long inverted tandem repeats that form single-stranded loops. The central region of the genome contains genes that are highly conserved amongst all sequenced OPVs [37]. Like other poxviruses, the genes are tightly packed with little non-coding sequences. The sequencing of full-genome of CMLV strains revealed that CMLV is closest to VARV, sharing genes involved in basic replication and host related functions and probably, they may share a common ancestor [1, 42]. The molecular details about the genome structure and phylogenetic analysis of some selected genes indicate that CMLV is clearly distinct from VARV and VACV. Genomic differences between CMLV and other OPVs are located in terminal regions. In this region, open reading frame (ORF) co-linearity and average amino acid identity decreases (82 % to VACV) due to small and large nucleotide insertions, deletions, and translocations. CMLV is similar to other OPVs in overall genome structure and composition, but CMLV genome lacks homologues of VARV (C1L, E7L, A26L, A27L, A39L, A42R, B2L, B3L and B4L), and VACV (K6L, A25L, A40R, A52R and A53R) genes as reviewed earlier [15].
CMLV appears to share biological features with other OPVs mainly VARV. Both CMLV and VARV are restricted to a single host and induce similar disease course [24, 88]. Earlier, CMLV was shown to share strong similarities with VARV as they both had a narrow host range. They were indistinguishable in terms of pock formation on chorioallantoic membrane (CAM) of embryonated eggs, growth in cells and low or absence of pathogenicity in various animal models [10–12]. Serological studies demonstrated the cross-antigenicity amongst VACV, VARV, CPXV and CMLV, but not with parapoxviruses (PPVs) and Avipoxvirus infections [11, 25]. Camels have been used as animal models of camelpox infection, mainly for evaluating the efficacy of camelpox vaccines [43, 62, 91]. The two models of camelpox (moderate and fatal), have been successfully used to evaluate vaccine pathogenicity and efficacy [89]. There have been several attempts to infect animals other than camels with CMLV in order to define its host range and develop animal models of camelpox. CMLV could exhibit different growth properties on CAM of embryonated eggs, cell cultures and also in laboratory animals. Many authors have compared the growth behaviour of numerous CMLV strains in various cell cultures, embryonated eggs and animals [33, 71]. In general, cells derived from camel, lamb, calf, pig, monkey, chicken, hamster and mouse enable the propagation of CMLV strains. Both, transformed and primary human cells are permissive to CMLV replication. However, cell monolayers derived from horse, rabbit and dog lead to a poor replication of CMLV for most of the strains [25, 71, 76, 81]. Other than camels, the species that have been infected successfully are monkeys and infant mice [11].
Epidemiology
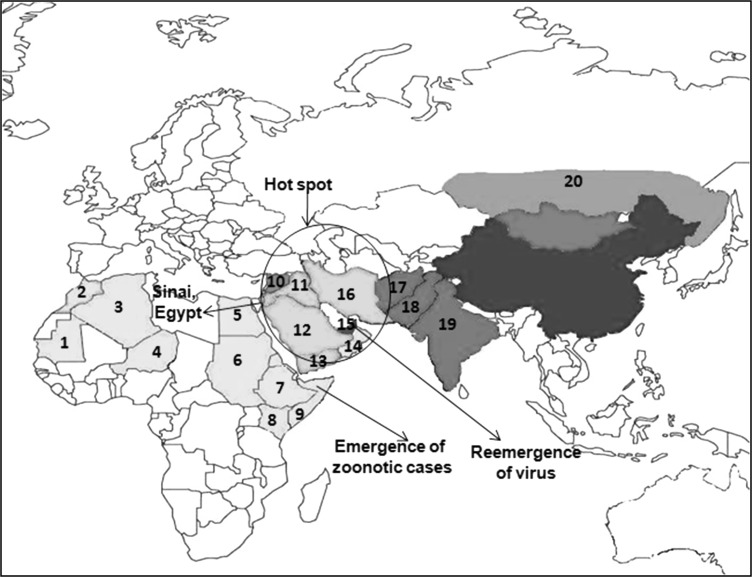
Camelpox is one of the most common contagious OPV diseases of the Old-World (both Camelus dromedarius and C. bactrianus) and the new-world camelids [35]. CMLV is considered to naturally infect solely the old world camelids [88]. The disease occurs throughout the camel-breeding areas of Africa, north of the equator, the Middle East and Asia, as the camels are used for nomadic pastoralism, transportation, racing, and production of milk, wool and meat purposes [14, 88]. Infections are commonly encountered in the herds of the nomadic pastoralists in the semi-desert zones. It occurs in almost every country in which camel husbandry is practiced apart from the introduced dromedary camel in Australia and tylopoda (llama and related species) in South America [35]. The disease has been reported initially in Punjab and Rajaputana (India) [53, 88] and later from many other countries. The disease is endemic in the Middle East (Iran, Iraq, Saudi Arabia, United Arab Emirates (UAE) and Yemen), in Asia (India, Afghanistan and Pakistan), in Africa (Algeria, Egypt, Kenya, Mauretania, Niger, Somalia and Morocco, Ethiopia, Oman, Sudan) and in the southern parts of former USSR [16, 20, 33, 43, 55, 71]. Recently, the first outbreak of camelpox has also been reported in two provinces named Hama and Duma in Syria [5]. The geographical distribution of camelpox in different parts of the world is depicted in the map (Fig. 1).
Fig. 1.
Geographical distribution of camelpox in the world. 1 Mauritania 2 Morocco 3 Algeria 4 Niger 5 Egypt (Sinai) 6 Sudan 7 Ethiopia 8 Kenya 9 Somalia 10 Syria 11 Iraq 12 Saudi Arabia 13 Yemen 14 Oman 15 United Arab Emirates 16 Iran 17 Afghanistan 18 Pakistan 19 India 20 Sothern parts of former Soviet Union (Russia)
The disease is socio-economically significant as it incurs considerable loss in terms of morbidity, mortality, loss of weight and reduction in milk yield [16]. The disease mostly affects young calves aged 2–3 years in a herd with fatal severe form (generalized form) causing high mortality occasionally due to waning of acquired immunity after 5–8 months [64], Various studies have demonstrated that the incidence of camelpox outbreaks increased during rainy seasons [88] with the appearance of more severe form of the disease, while milder form occurs during the dry season [47, 90]. The incidence and case fatality rate (CFR) are mostly higher in male camels than females. The mortality in adult animals ranged from 10 to 28 % and in young animals, it is 25–100 %. Further, the mortality is influenced by the presence of inter-current diseases (like trypanosomosis), stress, age, the nutritional status of the animal and virus virulence. Outbreaks are often temporal due to the movement of camels for grazing and watering and it results in mixing of the herds and the introduction of new camels into a herd [7]. In a recent investigation of a CMLV outbreak in Eastern Saudi Arabia [95], a atypical minute pock-like skin lesion (AMPL) persisted on 42.9 % of convalescent camels (8.8 % of herd) for more than a year after the onset of clinical signs and live CMLV was recovered from AMPL homogenates. They concluded that, the small, often missed AMPL on infected animals or CMLV survival in the persistent skin lesions may play a key persistence mechanism in previously infected camel herds during inter-epizootic periods. A high prevalence of CMLV antibodies in camels has also been reported. Recovered animals become life-long immune to re-infection and there is no chronic carrier state. In general, the course and the outcome of camelpox may vary depending on age, sex and the circulating CMLV strains differing in virulence [5, 39, 45, 52]. Thus, the risk factors associated with higher incidence of camelpox include the average age of the animals (<4 years), the rainy season of the year, the introduction of new camels in a herd and the common watering sources [47].
Pathogenesis
The CMLV enters commonly through skin. However, the oro-nasal infection is also reported. After local replication and development of a primary skin lesion, the virus spreads to local lymph nodes leads to a leukocyte-associated viremia, which may be associated with pyrexia. During this period, virus can be isolated from various tissues, including the skin, turbinates, lungs and also lymphoid organs. Widespread secondary skin lesions appear a few days after the onset of viremia, and new lesions continue to appear for 2–3 days, at that time the viremia subsides. CMLV and VARV cause illness in a single host species and both viruses are distinguishable. CMLV has rarely cause disease in man. Similarly, VARV is unable to cause disease in camels, although camels immunized with VARV are resistant to subsequent infection with CMLV [10]. The virus have also been found to be non-pathogenic to sheep, goats, rabbits, guinea pigs, rats, hamsters, and mice when inoculated by intra-dermal route [15]. The CMLV is host specific and does not infect other animal species, including cattle, sheep and goats [5].
Camelpox can produce severe disease, suggesting CMLV may interfere with the host response to infection. Like other OPVs, CMLV encode multiple genes that antagonize or affect the antiviral host immune response by interfering with the interferon (IFN) response, key pro-inflammatory cytokines [(Interleukin-IL-1b, IL-18 and tumor necrosis factors (TNFs)], chemokines and the complements [33]. A number of immune interfere strategies used by the virus have been extensively reviewed [61, 66]. CMLV contains genes encoding for specific protein, which can modulate or evade host immune responses, host cell apoptosis and cell or tissue tropism. They are chemokine-binding protein, TNF Receptor II crmB, complement binding protein, protein kinase inhibitors, signal transducer and activator of transcription (STAT) 1-inhibitor [60], serine proteinase inhibitors, CD47-like protein, IL-1/Toll-like receptor inhibitor [19], IFN inhibitor [66], IFN-γ receptor, and IFN-α/β binding protein [58]. Similarly, CMLV encodes homologues of pox viral proteins of vaccinia, myxoma virus and rabbit fibroma virus, which are known to affect virulence or host range. Proteins encoded by open reading frames (ORF) 31L, 188R and 200R have similarity to serpins that have anti-fusion or anti-apoptotic activity and involved in the inflammation [82]. Proteins encoded by ORFs 32L and 55L are similar to VAVC proteins K3L and E3L that mediate resistance to IFN [79]. Protein 6L is closely related to an uncharacterized human protein of family UPF0005 [86] and possibly it regulates apoptosis in CMLV-infected cells [15, 28]. Protein 201R contains a signal peptide, a RGD motif, which mediates the binding of proteins to cell surface integrins [4]. VARV, CPXV, and CMLV encode soluble interferon gamma receptors (IFN-gRs) that counteract the activity of the cytokine and possess broad species specificity. This novel property of the IFN-gR probably helped all these OPVs to replicate in several species [3]. Recently, it has been demonstrated that CMLV expressed a novel protein inhibiting apoptosis (v-GAAP) and a novel virulence factor, the schlafen-like protein 176R-(v-slfn-57 kDa) [40, 41], which is expressed both early and late phase of infection and play a role in the modulation of the innate and adaptive immune responses against pathogens [36, 38]. CMLV may utilize several ways to alter or shut down the host immune response. These mechanisms have been described in vitro simulating in vivo environment [33].
Transmission
The CMLV is transmitted by either direct or indirect contact via a contaminated environment. The direct transmission occurs in between infected and susceptible animals either by inhalation or through skin abrasion; although a mechanical transmission may play a role [88]. The affected camels may shed the virus through scab materials and secretions like milk, saliva and ocular and nasal discharges [70] in the environment such as in water. These becomes then the source of infection for susceptible animals [47]. The dried scabs shed from the pox lesions may contain live virus particles for 4 months and contaminate the environment [35]. The role of an arthropod vector in the transmission of the disease has also been suspected [5] and the tick population during the rainy season is probably involved in the spread of the disease [87, 90]. Among tick species, Hyalomma dromedarii have been found to be the predominant (90 %) species infesting camels. Further studies are needed to ensure the involvement of arthropods in the transmission of CMLV, but if confirmed, CMLV would be the first OPV transmitted via arthropods [33]. Like smallpox, camelpox is usually transmitted to airborne saliva droplets, but it can also spread through direct contact with skin lesions, and the virus can be transferred mechanically by ticks and other biting arthropods.
Zoonosis
Camelpox is a common cause of camel morbidity under traditional nomadic management system in developing countries but this disease is of less significant in developed countries, where it incurs less threat to man or animals. The threat CMLV poses to people whose well-being depends on the health of their camels makes the disease of considerable economic and public health importance [33]. CMLV is a zoonotic agent and mostly host specific [25] but recently an evidence has been documented from Somalia in smallpox-unvaccinated individuals [45, 52] and from India in smallpox-unvaccinated camel handlers or attendants [13]. Mild skin lesions in humans associated with camelpox have been reported [21], indicating camelpox may be of public health impact. Among the human cases, people drinking milk from camelpox-affected animals have been reported to develop ulcers on the lips and in the mouth, but these observations could not be visualised or laboratory confirmed [25]. However, under certain conditions, the virus could be pathogenic for human like that of cowpox and monkeypox [54] especially in immune-compromised individuals. However, no systematic epidemiological studies have been undertaken on human cases due to the lack of immunological surveys for specific camelpox antibodies among unvaccinated herds [7]. The self-limited nature of human infection with CMLV suggests that it could be used as a live smallpox vaccine earlier [33]. Recently, the first conclusive evidence of zoonotic CMLV infection in humans (unvaccinated smallpox individuals) associated with outbreaks in dromedarian camels in northwest region of India has been reported [13]. Further epidemiological studies of camelpox on these endemic regions are necessary to assess the circulation of CMLV, both in camels and humans in order to know its public health significance [33].
Clinical features
The disease is characterized by an incubation period of 9–13 days with an initial rise in temperature, followed by enlarged lymph nodes, skin lesions and prostration. The clinical manifestation of camelpox varies from mild local to severe systemic disease depending on the CMLV strains involved in the infection [88]. The typical skin lesion/rash will pass through all the stages of pox lesions progression, i.e. development of papules on labia, macules, papules, pustules, vesicles and scabs [33, 87, 90]. Skin lesions appear 1–3 days after the onset of fever with erythematous macules to papules and vesicles, and pustules and then crusts from ruptured pustules. In general, the lesion takes 4–6 weeks to heal. The lesion is usually localized in skin but occasionally, it leads to generalized form. The later form is frequently seen in young animals aged 2–3 years in a herd associated with weaning and poor nutrition. Eruptions are mainly localized on the head, nostrils, the margins of the ears and eyelids, as well as on the mucous membranes of the lips, the nose and also in the oral cavity. Later, lesions may extend to the neck, limbs, genitalia, mammary glands and perineum or scrotum [33]. In contrast, in the generalized form lesions may spread over the body, particularly on the head and the limbs with sometime swellings on the neck and abdomen and even multiple pox-like lesions can be found on the mucous membranes of the mouth, respiratory and digestive tracts and the consequences is more likely fatal [68]. The affected animals may show salivation, anorexia, lacrimation, mucopurulent nasal discharge and diarrhoea. Pregnant animals may abort and mortality in affected animals is due to septicaemia caused by secondary bacterial infections like Staphylococcus aureus [64, 88]. In contrast to smallpox, in which pustules occur only on the skin and the squamous epithelium of the oropharynx, severely affected camels also develop proliferative poxviral lesions in the bronchi and lungs [51].
The histopathology of skin lesions reveals characteristic cytoplasmic swelling, vacuolation and ballooning of the keratinocytes of the outer stratum spinosum of the epidermis [28, 30]. The rupture of these cells produces vesicles and localized oedema associated with perivascular cuffing of mononuclear cells, neutrophils and eosinophils. Marked epithelial hyperplasia may also occur at the borders of the skin lesions [92]. The lung lesions are usually characterized by hydropic degeneration, proliferation of bronchial epithelial cells associated with proliferative alveolitis and bronchiolitis infiltrated by macrophages, necrosis and fibrosis, which leads to obliteration of normal architecture [68, 69].
Diagnosis
The diagnosis of camelpox infection can be done based on clinical signs in affected animals. Following the appearance of clinical signs of the disease, tissue samples (skin or organ biopsies) are most useful to identify the infectious agent [33]. However, the confounding signs caused by contagious ecthyma (orf-parapox virus), papillomatosis and insect bites demand camelpox to be differentiated from these infections using laboratory-based diagnostic methods. It is necessary to apply more than one diagnostic techniques for confirmatory diagnosis as several diagnostic approaches are available [15]. Few complementary techniques might be advised for camelpox diagnosis namely transmission electron microscopy (TEM), virus isolation using cell culture, standard PCR assays, immune-histochemistry and demonstration of neutralizing antibodies [15]. However, the identity of the causative agent as CMLV must be confirmed by TEM, PCR and/or sequencing [35].
Transmission electron microscopy (TEM) and restriction enzyme analysis (REA) can be used to differentiate camelpox from other infections caused by OPV and PPVs [5, 37]. TEM is a reliable and rapid method to demonstrate the presence of OPVs in scabs or tissue samples, even though a relative high concentration of the virus in the sample is required [33]. This technique enables the differentiation between OPVs, which are brick-shaped, and PPVs, which are ovoid-shaped [24]. The camelpox antigen in the infected scabs and pox lesions can be identified by using immunohistochemistry technique [88]. However, the isolation of the virus using embryonated eggs and various cell lines such as HeLa, GMK-AH1, WISH, Vero [11], MA-104 and BHK21 cells as well as primary cell cultures like lamb testis and kidney, camel kidney, calf kidney and chicken embryo fibroblast [25] can be used for isolation study. However, Vero, MA-104 or Dubca cells, in which the virus replicates easily are generally preferred [69]. The infected cells should be monitored for cytopathic effects (CPE) for 10–12 days, which depend on the concentration of the virus. CPE includes the formation of multinucleated syncytia, rounding, ballooning and syncytia with degenerative changes. The isolation alone cannot be the golden standard diagnostic procedure, it must be followed by serology and/or PCR.
Similar to antigen detection methods, a battery of serological tests are available to identify CMLV antibodies. The conventional serological tests like haemagglutination, haemagglutination inhibition, neutralization [18], Indirect ELISA, complement fixation, and fluorescent antibody tests/assays have been described to detect CMLV antibodies [2, 25, 50, 54, 81]. Most of the conventional serological tests are time consuming, labour intensive, less sensitive and not rapid and therefore generally, not suitable for primary diagnosis but useful in secondary confirmatory testing and retrospective epidemiological studies.
To overcome the drawbacks associated with aforesaid tests, the recent molecular biology tools and techniques like PCR, real time PCR and loop mediated isothermal amplification (LAMP) assays have been used for the rapid and sensitive detection of CMLV DNA from clinical samples. DNA can be extracted either from cell culture or from clinical scab samples or tissue material using numerous commercial DNA extraction kits. Recently, a reliable low-cost two-step extraction method has been developed for isolating CMLV DNA from skin samples [94]. The PCR techniques have been developed targeting A-type inclusion body protein (ATIP) gene [56] and Haemagglutinin (HA) gene [23, 73] for specific detection of CMLV from scab materials/skin biopsy and infected Vero cell cultures [5]. The above genus-specific PCR yield product size specific for CMLV and thus, can be differentiated from other OPVs. A rapid PCR followed by restriction enzyme digestion allows differentiation of OPVs including CMLV [44]. An extra step consisting of a BglII or XbaI restriction digestion allows the unequivocal identification of the virus species [56, 57]. Similarly, species-specific primers within the HA ORF of OPVs have also been described for differentiation. Hence, the HA-PCR amplicon Taq I restriction fragment length polymorphism (RFLP) permits to differentiate between OPV species [73].
Further, PCR strategies targeting HA gene [73] and B2L gene [48] have also been developed for detection of CMLV and its differentiation from OPV and PPV infections in camels. In a similar direction, recently a PCR assay based on the C18L gene (encoding ankyrin repeat protein) has been developed, which yields a specific amplicon of 243 bp in CMLV suspected cases [8]. This assay was employed successfully for the direct detection and differentiation of CMLV from other OPVs, PPVs and capripoxviruses (CaPVs) in both cell culture samples and clinical specimens. Further, a duplex PCR based on the C18L and DNA polymerase (DNA pol) genes for specific and rapid detection and differentiation of CMLV from BPXV has also been developed [8, 77]. Similarly, a multiplex PCR for differentiating OPVs from CaPVs and PPVs targeting different genes has been reported [83]. These assays have the advantage of avoiding an extra step of restriction enzyme analysis (REA). This method will be an improved assay over the OPV-specific ATI- or HA- gene based assays for the simultaneous detection and differentiation of CMLV. As an improvement over conventional PCR approaches, the real-time PCR techniques targeting A36R gene using fluorescence resonance energy transfer (FRET) method [65] have been developed. Similarly, the real-time PCR targeting A13L, rpo18 and viral early transcription factor (VETF) genes [63] using melting curve analysis have been in use for rapid, highly sensitive and specific detection and quantitation of CMLV and other related OPVs. Recently, C18L gene based real-time PCR based on either SYBR green chemistry [8] or TaqMan hydrolysis probe [84] have also been optimized for specific detection of CMLV in clinical samples. Some of these PCR assays/methods are delineated in the OIE’s Manual of Diagnostic Tests and Vaccines for Terrestrial Animals [35]. As a field application diagnostic tool, a simple, rapid, specific and highly sensitive novel approach called as LAMP have also been developed targeting C18L gene [85] and evaluated using field clinical samples. This assay appears to be potential as rapid and sensitive diagnostic tool for its application in less equipped rural diagnostics laboratory settings in developing countries.
Antiviral therapy
Post-exposure therapeutic approaches for camelpox infection are not mentioned in the literature. However, application of antibiotics and administration of supplements may be useful to reduce the severity of the disease [88]. The use of antiviral drugs/agents may be of choice particularly in young camels as an alternative treatment. There are several classes of antiviral agents found useful for camelpox as applicable to other pox viral infections. There are potent antiviral molecules active in vitro and in vivo against poxviruses, including OPVs and could be envisaged for the treatment of camelpox [78, 80]. They include molecules belonging to the acyclic nucleoside phosphonate (ANP) family, i.e., cidofovir (Gilead, CA, USA) and its lipid derivative CMX001 (Chimerix Inc., NC, USA) [26, 46], and the compound ST-246 (SIGA Inc., OR, USA) [93]. Cidofovir and CMX001 are active against a broad range of DNA viruses including poxviruses. Both compounds target the viral DNA polymerase of OPVs and inhibit its functions [6]. Certain novel antiviral drugs are effective orally against pox viruses including CMLV targeting cellular enzymes [IMP dehydrogenase inhibitors, such as ribavirin, as well as the tyrosine kinase inhibitor (STI-571), also called imatinib mesylate, or Gleevec] and viral enzymes including inhibitors of viral morphogenesis (TTP-6171), viral release (ST-246), and viral DNA synthesis (ANPs analogues including HPMPC) [80]. ST-246 is a potent inhibitor for OPVs only. It targets the protein F13L of VACV, which is required for the wrapping of intracellular mature viruses and the production of extracellular enveloped viruses [30, 32, 93]. Numerous studies have also shown that ST-246 administered for 10–14 days at a dose of 100 mg/kg once per day protects OPV-infected animals from disease development. In the case of CMLV, the activity of the molecule (Cidofovir, CMX001 and ST-246) has only been evaluated in vitro and are potent inhibitors of CMLV replication [27, 33]. Nevertheless, CMX001 and ST-246 offer the advantage of being orally available which may render them more attractive for veterinary use [29].
Prophylaxis
The CMLV, which is reported to be closely related to Variola at molecular level, warrants bio-security and bio-safety measures especially at borders to contain this transboundary and emerging disease. Because of CMLV resembles Variola in its dependence on a single host, the disease could potentially be eliminated through a combination of surveillance, vaccination and quarantine [33]. Research has been oriented towards the development of prophylactic methods to contain the spread of camelpox in enzootic countries. However, the development of camelpox vaccines has been initiated after the worldwide eradication of smallpox. At that time, the use of VACV as a prophylactic agent for other orthopoxviral diseases of animals was not recommended, most probably due to the potential danger to non-vaccinated human contacts [43]. Because of concern that VACV could accidentally be spread from recently inoculated camels to unvaccinated humans or to animals, researchers began to focus on developing attenuated CMLV vaccines that could only infect camels [33].
There is a little information of literature on the production of vaccines against camelpox since its first inception about the concept of camelpox vaccine from former Soviet Union [17]. The knowledge of camelpox vaccine efficacy originates from field investigations using the commercialized CMLV-based vaccines. Of late, lacto-therapy (scarification of a mixture of milk and camelpox infected crusts) had been in use and practiced in Punjab (India), former USSR and Arabian Bedouin to control camelpox. A Saudi isolate of CMLV attenuated (Jouf-78 strain) in camel kidney cell cultures (CKCC) at passage level 78 was found to be safe and potent (at 103 TCID50) in camels by intradermally or subcutaneously [43]. This Jouf-78 strain is commercially manufactured as vaccine (OrthovacR) by Jordanian Vaccine Company (JOVAC) and is being currently used in many countries. A similar kind of vaccine for camelpox has been in use in Egypt, Morocco and Russia (apps.cfsph.iastate.edu). Further, in the UAE, Vero cell attenuated camelpox vaccine (Dubai camelpox attenuated vaccine) developed by Onderstepoort Biological Products (OBP) using isolate of UAE (strain CaPV298-2) [7, 67]- named as DucapoxR and produced by Highveld Biologicals, Republic of South Africa (RSA) [91]. This vaccine conferred immunity up to 6 years of vaccination in two animals. However, a booster vaccination is recommended in young animals vaccinated below 6 months. Further, the attenuated CMLV strain VD47/25, passaged 80 times in cell culture, has also been evaluated as camelpox vaccine in Mauritania [62]. Similarly, the formalin inactivated aluminium hydroxide adjuvanted camelpox vaccine (CMLV strain T8-1984), is available in Morocco, which reported to give protection only for 1 year [35]. This vaccine is manufactured and distributed by Biopharma and safe for young and adult camels and has been shown to induce CMLV neutralizing antibodies [34] but it requires booster as well annual vaccination for efficient protection. Both, “DucapoxR” and inactivated camelpox vaccines were found safe in pregnant camels [49]. However, there is imperative to have a thermostability of these poxviral vaccines, which would facilitate their use in hot, dry regions where the disease frequently occurs.
Immunity against camelpox is both humoral and cell mediated [35]. However, it is believed that circulating antibodies do not reflect the immune status of the animal [88]. Recovered animals become life-long immune to re-infection. Live attenuated vaccines provide protection for at least 6 years, probably longer [91], whereas, inactivated vaccine reported to provide 1 year of protection [35]. The prevention and control of sporadic cases of camelpox infection in camel husbandry is of prime importance in developing countries like India. Further, considering the increased incidence of camelpox not only in camel but also in human [13], studies on molecular epidemiology, specific diagnosis, and control measures are paramount importance in reducing the circulation of CMLV in camels, and also in humans as a public health aspect.
The diagnostic tests and vaccines are needed for control and an eradication of any infectious disease. Unfortunately, as with many public health problems, the challenge lies in bringing those tools to the affected animals. To eradicate camelpox, it would not be necessary to vaccinate all of the world’s world camels. Instead, the ‘‘ring vaccination’’ strategy could be employed, in which intensive surveillance to detect cases of disease, followed by vaccination of all surrounding contacts and continued monitoring to ensure that no more cases occurred [33]. Although, it has not been a recognized target for eradication efforts, and the toll of animal and human suffering from camelpox cannot be compared to the mass die-offs and famine caused by rinderpest, a deadly disease of cattle. Massive effort would not be required for camelpox, control and eradication, as it is confined to a specific region only. Researchers now focus toward the goal of ‘‘One Health’’ approaches to combat the diseases of zoonotic and public health important [9].
Concluding remarks
The disease was considered inconsequential till recently, but it is considered as emerging public health problem during this decade due to increased reported cases and outbreaks in camels. In this context, particular attention should be given to camelpox outbreaks in camels, as well as to the identification of any human infections. Considering these and the virus spreads through contaminated environments, studies on molecular epidemiology, improved diagnostic methods and prophylaxis and control measures in the developing countries are paramount importance in reducing the circulation of CMLV in camels, and also in humans. Effective prevention and control measures can be achieved through the use of proper diagnostic and prophylactic aids to curtail further spread of camelpox as described for most of the zoonotic diseases. In general, several factors that are related to human activities, environmental changes and virus could be the determinants of incidence and prevalence of the disease. To safeguard the public health from pathogens of zoonotic infections, application of skills, knowledge and resources of veterinary public health is essential. Further, the control measures for emerging and re-emerging pathogens are demanding, as there is population explosion. Novel highly sensitive and specific techniques comprising genomics and proteomics along with conventional methods would be useful in the identification of emerging and re-emerging pathogen or virus; thereby therapeutic, prophylactic, preventive measures would be applied in time.
Contributor Information
Vinayagamurthy Balamurugan, Email: balavirol@gmail.com.
Raj Kumar Singh, Email: rks_virology@rediffmail.com.
References
- 1.Afonso CL, Tulman ER, Lu Z, Zsak L, Sandybaev NT, Kerembekova UZ, Zaitsev VL, Kutish GF, Rock DL. The genome of camelpox virus. Virology. 2002;295(1):1–9. doi: 10.1006/viro.2001.1343. [DOI] [PubMed] [Google Scholar]
- 2.Al Hendi AB, Abuelzein EM, Gameel AA, Hassanein MM. A slow-spreading mild form of camelpox infection. J Vet Med B. 1994;41:71–73. doi: 10.1111/j.1439-0450.1994.tb00207.x. [DOI] [PubMed] [Google Scholar]
- 3.Alcami A, Smith GL. Vaccinia, cowpox, and camelpox viruses encode soluble gamma interferon receptors with novel broad species specificity. J Virol. 1995;69:4633–4639. doi: 10.1128/jvi.69.8.4633-4639.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alcami A, Symons JA, Khanna A, Smith GL. Poxviruses: capturing cytokines and chemokines. Semin Virol. 1998;8:419–427. doi: 10.1006/smvy.1997.0143. [DOI] [Google Scholar]
- 5.Al-Ziabi O, Nishikawa H, Meyer H. The first outbreak of camelpox in Syria. J Vet Med Sci. 2007;69(5):541–543. doi: 10.1292/jvms.69.541. [DOI] [PubMed] [Google Scholar]
- 6.Andrei G, Gammon DB, Fiten P, De Clercq E, Opdenakker G, Snoeck R, Evans DH. Cidofovir resistance in vaccinia virus is linked to diminished virulence in mice. J Virol. 2006;80:9391–9401. doi: 10.1128/JVI.00605-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Azwai SM, Carter SD, Woldehiwet Z, Wernery U. Serology of Orthopoxvirus cameli infection in dromedary camels: analysis by ELISA and western blotting. Comp Immunol Microbiol Infect Dis. 1996;19(1):65–78. doi: 10.1016/0147-9571(95)00023-2. [DOI] [PubMed] [Google Scholar]
- 8.Balamurugan V, Bhanuprakash V, Hosamani M, Kallesh DJ, Bina Chauhan, Venkatesan G, Singh RK. A polymerase chain reaction strategy for the diagnosis of camelpox. J Vet Diag Invest. 2009;21(2):231–237. doi: 10.1177/104063870902100209. [DOI] [PubMed] [Google Scholar]
- 9.Bary M, Babiuk S. Camelpox: target for eradication? Antiviral Res. 2011;92:164–166. doi: 10.1016/j.antiviral.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 10.Baxby D, Ramyar H, Hessami M, Ghaboosi B. A comparison of the response of camels to intradermal inoculation with camelpox and smallpox viruses. Infect Immun. 1975;11:617–621. doi: 10.1128/iai.11.4.617-621.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baxby D. Smallpox viruses from camels in Iran. Lancet. 1972;7786:1063–1065. doi: 10.1016/S0140-6736(72)92343-4. [DOI] [PubMed] [Google Scholar]
- 12.Baxby D. Differentiation of smallpox and camelpox viruses in cultures of human and monkey cells. J Hyg (London) 1974;72:251–254. doi: 10.1017/S0022172400023457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bera BC, Shanmugasundaram K, Barua S, Venkatesan G, Virmani N, Riyesh T, Gulati BR, Bhanuprakash V, Vaid RK, Kakker NK, Malik P, Bansal M, Gadvi S, Singh RV, Yadav V, Sardarilal, Nagarajan G, Balamurugan V, Hosamani M, Pathak KM, Singh RK. Zoonotic cases of camelpox infection in India. Vet Microbiol. 2011;152(1–2):29–38. doi: 10.1016/j.vetmic.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 14.Bett B, Jost C, Allport R, Mariner J. Using participatory epidemiological techniques to estimate the relative incidence and impact on livelihoods of livestock diseases amongst nomadic pastoralists in Turkana South District, Kenya. Prev Vet Med. 2009;90:194–203. doi: 10.1016/j.prevetmed.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 15.Bhanuprakash V, Prabhu M, Venkatesan G, Balamurugan V, Hosamani M, Pathak KML, Singh RK. Camelpox: epidemiology, diagnosis and control measures. Expert Rev Anti Infect Ther. 2010;8(10):1187–1201. doi: 10.1586/eri.10.105. [DOI] [PubMed] [Google Scholar]
- 16.Bhanuprakash V, Balamurugan V, Hosamani M, Venkatesan G, Chauhan B, Srinivasan VA, Chauhan RS, Pathak KM, Singh RK. Isolation and characterization of Indian isolates of Camelpox viruses. Trop Anim Health Prod. 2010;42(6):1271–1275. doi: 10.1007/s11250-010-9560-z. [DOI] [PubMed] [Google Scholar]
- 17.Borisovich Yu F. Camelpox. (Little-known contagious diseases of animals) etd by F. M. Orlov. Izdatel’stvo Kolos, USSR, 2nd edn., 32–42 (Ru) Vet Bull. 1973;44(9):139. [Google Scholar]
- 18.Boulter EA, Zwartouw HT, Titmuss DH, Maber HB. The nature of the immune state produced by inactivated vaccinia virus in rabbits. Am J Epidemol. 1971;94(6):612–620. doi: 10.1093/oxfordjournals.aje.a121360. [DOI] [PubMed] [Google Scholar]
- 19.Bowie A, Kiss-Toth E, Symons JA, Smith GL, Dower SK, O’Neill LA. A46R and A52R from vaccinia virus are antagonists of host IL-1 and toll-like receptor signaling. Proc Natl Acad Sci USA. 2000;97(18):10162–10167. doi: 10.1073/pnas.160027697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chauhan RS, Kaushik RK. Isolation of camelpox virus in India. Brit Vet J. 1987;143:581–582. doi: 10.1016/0007-1935(87)90050-9. [DOI] [PubMed] [Google Scholar]
- 21.Coetzer JAW. Poxviridae. In: Coetzer JAW, Tustin RC, editors. Infectious diseases of livestock. 2. Southern Africa: Oxford University Press; 2004. pp. 1265–1267. [Google Scholar]
- 22.Cyrklaff M, Risco C, Fernández JJ, Jiménez MV, Estéban M, Baumeister W, Carrascosa JL. Cryo-electron tomography of vaccinia virus. Proc Natl Acad Sci USA. 2005;102(8):2772–2777. doi: 10.1073/pnas.0409825102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Damaso CR, Esposito JJ, Condit RC, Moussatche N. An emergent poxvirus from humans and cattle in Rio de Janeiro state: cantagalo virus may derive from Brazilian small pox vaccine. Virology. 2000;277:439–449. doi: 10.1006/viro.2000.0603. [DOI] [PubMed] [Google Scholar]
- 24.Damon I. Poxviruses. In: Knipe DM, Howley PM, editors. Fields virology. 5. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 2947–2975. [Google Scholar]
- 25.Davies FG, Mungai JN, Shaw T. Characteristics of a Kenyan camelpox virus. J Hyg. 1975;7:381–385. doi: 10.1017/S002217240002444X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Clercq E, Sakuma T, Baba M, Pauwels R, Balzarini J, Rosenberg I, Holy A. Antiviral activity of phosphonylmethoxyalkyl derivatives of purine and pyrimidine. Antiviral Res. 1987;8:261–272. doi: 10.1016/S0166-3542(87)80004-9. [DOI] [PubMed] [Google Scholar]
- 27.Duraffour S, Andrei G, Snoeck R. Tecovirimat, a p37 envelope protein inhibitor for the treatment of smallpox infection. IDrugs. 2010;13:181–191. [PubMed] [Google Scholar]
- 28.Duraffour S, Matthys P, van Den Oord JJ, De Schutter T, Mitera T, Snoeck R, Andrei G. Study of camelpox virus pathogenesis in athymic nude mice. PLoS One. 2011;6:e21561. doi: 10.1371/journal.pone.0021561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duraffour S, Snoeck R, de Vos R, van Den Oord JJ, Crance JM, Garin D, Hruby DE, Jordan R, De Clercq E, Andrei G. Activity of the anti orthopoxvirus compound ST-246 against vaccinia, cowpox and camelpox viruses in cell monolayers and organotypic raft cultures. Antivir Ther. 2007;12:1205–1216. [PubMed] [Google Scholar]
- 30.Duraffour S, Snoeck R, Fiten P, Opdenakker G, Andrei G. Selection and characterization of (S)-1-[3-hydroxy-2-(phosphonomethoxypropyl)-2,6-diaminopurine [HPMPDAP] resistant Camelpox viruses. Antivir Res. 2009;82:A67–A68. doi: 10.1016/j.antiviral.2009.02.165. [DOI] [Google Scholar]
- 31.Duraffour S, Snoeck R, Krecmerova M, Van den OJ, de Vos R, Holy A, Crance JM, Garin D, De Clercq E, Andrei G. Activities of several classes of acyclic nucleoside phosphonates against camelpox virus replication in different cell culture models. Antimicrob Agents Chemother. 2007;51:4410–4419. doi: 10.1128/AAC.00838-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duraffour S, Vigne S, Garcel A, Jordan R, Hruby DE, Crance JM, Garin D, Andrei G, Snoeck R. Antiviral potency of ST-246 on the production of enveloped orthopoxviruses and characterization of ST-246 resistant vaccinia, cowpox and camelpox viruses. Antivir Res. 2008;78:A29–A30. doi: 10.1016/j.antiviral.2008.01.049. [DOI] [Google Scholar]
- 33.Duraffour S, Meyer H, Andrei G, Snoeck R. Camelpox virus. Antivir Res. 2011;92(2):167–186. doi: 10.1016/j.antiviral.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 34.El Harrak M, Loutfi C. La variole du dromadaire chez le jeune au Maroc. Isolement et identification du virus. Mise au point du vaccin et application à la prophylaxie. Revue d’élevage et de médecine vétérinaire des pays tropicaux. 2000;53:165–167. [Google Scholar]
- 35.Elliot H, Tuppurainen E. Camelpox. Manual of diagnostic tests and vaccines for terrestrial animals, Vol. 2, Chap. 2.9.2. 2008. p. 177–84.
- 36.Eskra L, Mathison A, Splitter G. Microarray analysis of mRNA levels from RAW264.7 macrophages infected with Brucella abortus. Infect Immun. 2003;71:1125–1133. doi: 10.1128/IAI.71.3.1125-1133.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fenner F, Wittek R, Dumbell KR. The orthopox viruses. New York: Academic Press Inc.; 1989. [Google Scholar]
- 38.Geserick P, Kaiser F, Klemm U, Kaufmann SH, Zerrahn J. Modulation of T cell development and activation by novel members of the Schlafen (slfn) gene family harbouring an RNA helicase-like motif. Intl Immunol. 2004;16:1535–1548. doi: 10.1093/intimm/dxh155. [DOI] [PubMed] [Google Scholar]
- 39.Gitao CG. An investigation of camelpox outbreaks in two principal camel (Camelus dromedarius) rearing areas of Kenya. Revue scientifique et technique/Office international des épizooties. 1997;16:841–847. doi: 10.20506/rst.16.3.1077. [DOI] [PubMed] [Google Scholar]
- 40.Gubser C, Bergamaschi D, Hollinshead M, Lu X, van Kuppeveld FJ, Smith GL. A new inhibitor of apoptosis from vaccinia virus and eukaryotes. PLoS Pathog. 2007;3:e17. doi: 10.1371/journal.ppat.0030017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gubser C, Goodbody R, Ecker A, Brady G, O’Neill LA, Jacobs N, Smith GL. Camelpox virus encodes a schlafen-like protein that affects orthopoxvirus virulence. J GenVirol. 2007;88:1667–1676. doi: 10.1099/vir.0.82748-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gubser C, Smith GL. The sequence of camelpox virus shows it is most closely related to variola virus, the cause of smallpox. J GenVirol. 2002;83(4):855–872. doi: 10.1099/0022-1317-83-4-855. [DOI] [PubMed] [Google Scholar]
- 43.Hafez SM, Al-Sukayran A, Dela Cruz D, Mazloum KS, Al-Bokmy AM, Al-Mukayyel A, Amjad AM. Development of a live cell culture camelpox vaccine. Vaccine. 1992;8(10):533–539. doi: 10.1016/0264-410X(92)90353-L. [DOI] [PubMed] [Google Scholar]
- 44.Huemer HP, Hönlinger B, Höpfl R. A simple restriction fragment PCR approach for discrimination of human pathogenic old world animal orthopoxvirus species. Can J Microbiol. 2008;54(2):159–162. doi: 10.1139/W07-129. [DOI] [PubMed] [Google Scholar]
- 45.Jezek Z, Kriz B, Rothbauer V. Camelpox and its risk to the human population. J Hyg Epidemiol Microbiol Immunol. 1983;27:29–42. [PubMed] [Google Scholar]
- 46.Kern ER, Hartline C, Harden E, Keith K, Rodriguez N, Beadle JR, Hostetler KY. Enhanced inhibition of orthopoxvirus replication in vitro by alkoxyalkyl esters of cidofovir and cyclic cidofovir. Antimicrob Agents Chemother. 2002;46:991–995. doi: 10.1128/AAC.46.4.991-995.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khalafalla AI, Ali YH. Observations on risk factors associated with some camel viral diseases of camels in Sudan. In: Proceedings of the 12th international conference of the Association of Institutions of Tropical Veterinary Medicine. Montpellier, France, 20–22 August 2007; pp. 101–105.
- 48.Khalafalla AI, Buttner M, Rziha HJ. Polymerase chain reaction (PCR) for rapid diagnosis and differentiation of Para and orthopox virus infections in camels. FAO/IAEA International Animal Production and Health in Developing Countries, Vienna, Austria. 6–10 Oct, 2003.
- 49.Khalafalla AI, El Dirdiri GA. Laboratory and field investigations of a live attenuated and an inactivated camelpox vaccine. J Cam Prac Res. 2003;10(2):191–200. [Google Scholar]
- 50.Khalafalla AI, Mohamed MEM, Agab H. Serological survey in camels of the Sudan for prevalence of antibodies to camelpox virus using ELISA technique. J Cam Prac Res. 1998;5:197–200. [Google Scholar]
- 51.Kinne J, Cooper JE, Wernery U. Pathological studies on camelpox lesions of the respiratory system in the United Arab Emirates (UAE) J Comp Pathol. 1998;118:257–266. doi: 10.1016/S0021-9975(07)80002-8. [DOI] [PubMed] [Google Scholar]
- 52.Kriz B. A study of camelpox in Somalia. J Comp Pathol. 1982;92:1–8. doi: 10.1016/0021-9975(82)90037-8. [DOI] [PubMed] [Google Scholar]
- 53.Leese S. Two diseases of young camels. J Trop Vet Sci. 1909;4:1. [Google Scholar]
- 54.Marennikova SS. The results of examinations of wildlife monkeys for the presence of antibodies and viruses of the pox group. Voprosy Virusolgii. 1975;3:321–326. [PubMed] [Google Scholar]
- 55.Marodam V, Nagendrakumar SB, Tanwar VK, Thiagarajan D, Reddy GS, Tanwar RK, Srinivasan VA. Isolation and identification of camelpox virus. Ind J Anim Sci. 2006;76:326–327. [Google Scholar]
- 56.Meyer H, Pfeffer M, Rziha HJ. Sequence alterations within and downstream of the A-type inclusion protein genes allow differentiation of Orthopoxvirus species by polymerase chain reaction. J Gen Virol. 1994;75:1975–1981. doi: 10.1099/0022-1317-75-8-1975. [DOI] [PubMed] [Google Scholar]
- 57.Meyer H, Ropp SL, Esposito JJ. Gene for A-type inclusion body protein is useful for a polymerase chain reaction assay to differentiate orthopoxviruses. J Virol Methods. 1997;64:217–221. doi: 10.1016/S0166-0934(96)02155-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moss B, Shisler JL. Immunology 101 at poxvirus U: immune evasion genes. Semin Immunol. 2001;13(1):59–66. doi: 10.1006/smim.2000.0296. [DOI] [PubMed] [Google Scholar]
- 59.Moss B. Poxviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields virology. 5. Philadelphia: Lippincot Williams & Wilkins; 2007. pp. 2905–2945. [Google Scholar]
- 60.Najarro P, Traktman P, Lewis JA. Vaccinia virus blocks gamma interferon signal transduction: viral VH1 phosphatase reverses Stat1 activation. J Virol. 2001;75(7):3185–3196. doi: 10.1128/JVI.75.7.3185-3196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nazarian SH, McFadden G. Immunomodulation by poxviruses. In: Schmidt AA, Weber A, Mercer O, editors. Poxviruses. Basel: Birkhäuser Verlag; 2007. pp. 273–296. [Google Scholar]
- 62.Nguyen BV, Guerre L, Saint-Martin G. Preliminary study of the safety and immunogenicity of the attenuated VD47/25 strain of camelpox virus. Rev Elev Med Vet Pays Trop. 1996;49:189–194. [PubMed] [Google Scholar]
- 63.Nitsche A, Ellerbrok H, Pauli G. Detection of orthopoxvirus DNA by real- time PCR and variola virus DNA by melting analysis. J Clin Microbiol. 2004;42(3):1207–1213. doi: 10.1128/JCM.42.3.1207-1213.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nothelfer HB, Wernery U, Czerny CP. Camelpox: antigen detection within skin lesions-immunohistochemistry as a simple method of etiological diagnosis. J Cam Prac Res. 1995;2(2):119–121. [Google Scholar]
- 65.Panning M, Asper M, Kramme S, Schmitz H, Drosten C. Rapid detection and differentiation of human pathogenic orthopox viruses by a fluorescence resonance energy transfer real- time PCR assay. Clin Chem. 2004;50:702–708. doi: 10.1373/clinchem.2003.026781. [DOI] [PubMed] [Google Scholar]
- 66.Perdiguero B, Esteban M. The interferon system and vaccinia virus evasion mechanisms. J Interferon Cytokine Res. 2009;29:581–598. doi: 10.1089/jir.2009.0073. [DOI] [PubMed] [Google Scholar]
- 67.Pfeffer M, Meyer H, Wernery U, Kaaden OR. Comparison of camelpox viruses isolated in Dubai. Vet Microbiolol. 1996;49:135–146. doi: 10.1016/0378-1135(95)00181-6. [DOI] [PubMed] [Google Scholar]
- 68.Pfeffer M, Neubauer H, Wernery U, Kaaden OR, Meyer H. Fatal form of camelpox virus infection. Vet J. 1998;155:107–109. doi: 10.1016/S1090-0233(98)80045-2. [DOI] [PubMed] [Google Scholar]
- 69.Pfeffer M, Wernery U, Kaaden OR, Meyer H. Diagnostic procedures for poxvirus infections in camelids. J Cam Prac Res. 1998;5:189–195. [Google Scholar]
- 70.Ramyar H, Hessami M. Isolation, cultivation and characterisation of camelpox virus. Zentralbl Veterinarmed B. 1972;19:182–189. doi: 10.1111/j.1439-0450.1972.tb00393.x. [DOI] [PubMed] [Google Scholar]
- 71.Renner-Muller IC, Meyer H, Munz E. Characterization of camelpoxvirus isolates from Africa and Asia. Vet Microbiol. 1995;45:371–381. doi: 10.1016/0378-1135(94)00143-K. [DOI] [PubMed] [Google Scholar]
- 72.Rheinbaden FV, Gebel J, Exner, et al. Environmental resistance, disinfection, and sterilization of poxviruses. In: Schmidt AA, Weber A, Mercer O, et al., editors. Poxviruses. Basel: Birkhäuser Verlag; 2007. pp. 397–405. [Google Scholar]
- 73.Ropp SL, Jin Q, Knight JC, Massung RF. Esposito. Polymerase chain reaction strategy for identification and differentiation of smallpox and other orthopoxviruses. J Clin Microbiol. 1995;33:2069–2076. doi: 10.1128/jcm.33.8.2069-2076.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Roslyakov AA. Comparative ultrastructure of viruses of camelpox, a pox-like disease of camels (‘‘auzdik’’) and contagious ecthyma of sheep. Vopr Virusol. 1972;17:26–30. [PubMed] [Google Scholar]
- 75.Sadykov RG. Cultivation of camelpox virus in chick embryos. In: Virusnye Bolezni Selskokhozyaystvenykh Zhivotnykh Part I (In Russian). Moscow; 1970. p. 55.
- 76.Sheikh Ali HM, Khalafalla AI, Nimir AH. Detection of Camelpox and vaccinia viruses by polymerase chain reaction. Trop Anim Health Prod. 2009;41(8):1637–1641. doi: 10.1007/s11250-009-9359-y. [DOI] [PubMed] [Google Scholar]
- 77.Singh RK, Balamurugan V, Hosamani M, Kallesh DJ, Bhanuprakash V. Sequence analysis of C18L gene of Buffalopox virus: PCR strategy for specific detection and its differentiation from orthopoxviruses. J Virol Methods. 2008;154(1–2):146–153. doi: 10.1016/j.jviromet.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 78.Smee DF. Progress in the discovery of compounds inhibiting orthopoxviruses in animal models. Antiviral Chem Chemother. 2008;19:115–124. doi: 10.1177/095632020801900302. [DOI] [PubMed] [Google Scholar]
- 79.Smith GL, Symons JA, Alcamı A. Poxviruses; interfering with interferon. Sem Virol. 1992;8:409–418. doi: 10.1006/smvy.1997.0145. [DOI] [Google Scholar]
- 80.Snoeck R, Andrei G, De Clercq E. Therapy of poxvirus infections. In: Mercer AA, Schmidt A, Weber O, editors. Poxviruses. Basel: Birkhauser; 2007. pp. 375–395. [Google Scholar]
- 81.Tantawi HH, El Dahaby H, Fahmy LS. Comparative studies on poxvirus strains isolated from camels. Acta Virol. 1978;22:451–457. [PubMed] [Google Scholar]
- 82.Turner PC, Musy PY, Moyer RW. Poxvirus serpins. In McFadden G, Austin, Landes RG, editors. Viroceptors, Virokines and related immune modulators encoded by DNA Viruses. TX, USA: RG Landes; 1995. p. 67–88.
- 83.Venkatesan G, Balamurugan V, Hasamani M, Singh RK, Bhanuprakash V. Multiplex PCR for detection and differentiation of pox viruses, New letter. Indian Veterinary Research Institute, Izatnagar, UP, India, 2009. 30 (2)–31 (1): 1–2.
- 84.Venkatesan G, Bhanuprakash V, Balamurugan V, Prabhu M, Pandey AB. TaqMan hydrolysis probe based real time PCR for detection and quantitation of camelpox virus in skin scabs. J Virol Methods. 2012;181(2):192–196. doi: 10.1016/j.jviromet.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 85.Venkatesan G, Bhanuprakash V, Balamurugan V, Singh RK, Pandey AB. Development of loop-mediated isothermal amplification assay for specific and rapid detection of camelpox virus in clinical samples. J Virol Methods. 2012;183(1):34–39. doi: 10.1016/j.jviromet.2012.03.019. [DOI] [PubMed] [Google Scholar]
- 86.Walter L, Marynen P, Szpirer J, Levan G, Gunther E. Identification of a novel conserved human gene, TEGT. Genomics. 1995;28:301–304. doi: 10.1006/geno.1995.1145. [DOI] [PubMed] [Google Scholar]
- 87.Wernery U, Kaaden OR, Ali M. Orthopox virus infections in dromedary camels in United Arab Emirates (UAE) during winter season. J Cam Pract Res. 1997;4(1):51–55. [Google Scholar]
- 88.Wernery U, Kaaden OR. Infectious diseases in camelids. Berlin: Blackwell; 2002. Camelpox; pp. 176–185. [Google Scholar]
- 89.Wernery U, Kinne J, Zachariah R. Experimental camelpox infection in vaccinated and unvaccinated guanacos. J Camel Pract Res. 2000;7(2):153–157. [Google Scholar]
- 90.Wernery U, Meyer H, Pfeffer M. Camelpox in the United Arab Emirates and its prevention. J Cam Pract Res. 1997;4(2):135–139. [Google Scholar]
- 91.Wernery U, Zachariah R. Experimental camelpox infection in vaccinated and unvaccinated dromedaries. Zentralbl Veterinarmed B. 1999;46(2):131–136. doi: 10.1111/j.0931-1793.1999.00250.x. [DOI] [PubMed] [Google Scholar]
- 92.Yager JA, Scott DW, Wilcock BP. Viral diseases of the skin. In: Jubb KVF, Kennedy PC, Palmer N, editors. Pathology of domestic animals. 4. San Diego: Academic Press; 1991. pp. 629–644. [Google Scholar]
- 93.Yang G, Pevear DC, Davies MH, Collett MS, Bailey T, Rippen S, Barone L, Burns C, Rhodes G, Tohan S, Huggins JW, Baker RO, Buller RL, Touchette E, Waller K, Schriewer J, Neyts J, DeClercq E, Jones K, Hruby D, Jordan R. An orally bioavailable anti poxvirus compound (ST-246) inhibits extracellular virus formation and protects mice from lethal orthopoxvirus challenge. J Virol. 2005;79:13139–13149. doi: 10.1128/JVI.79.20.13139-13149.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yousif AA, Al-Naeem AA, Al-Ali MA. A three-minute nonenzymatic extraction method for isolating PCR-quality camelpox virus DNA from skin. J Virol Methods. 2010;169(1):138–142. doi: 10.1016/j.jviromet.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 95.Yousif AA, Al-Naeem AA. Recovery and molecular characterization of live camelpox virus from skin 12 months after onset of clinical signs reveals possible mechanism of virus persistence in herds. Vet Microbiol. 2012;159:320–326. doi: 10.1016/j.vetmic.2012.04.022. [DOI] [PubMed] [Google Scholar]