Abstract
Due to the heterogeneous nature of breast cancer and the widespread use of single-gene studies, there is limited knowledge of multi-gene, locus-specific DNA methylation patterns in relation to molecular subtype and clinical features. We, therefore, quantified DNA methylation of 70 candidate gene loci in 140 breast tumors and matched normal tissues and determined associations with gene expression and tumor subtype. Using Sequenom’s EpiTYPER platform, approximately 1,200 CpGs were interrogated and revealed six DNA methylation patterns in breast tumors relative to matched normal tissue. Differential methylation of several gene loci was observed within all molecular subtypes, while other patterns were subtype-dependent. Methylation of numerous gene loci was inversely correlated with gene expression, and in some cases, this correlation was only observed within specific breast tumor subtypes. Our findings were validated on a larger set of tumors and matched adjacent normal tissue from The Cancer Genome Atlas dataset, which utilized methylation data derived from both Illumina Infinium 27 and 450 k arrays. These findings highlight the need to control for subtype when interpreting DNA methylation results, and the importance of interrogating multiple CpGs across varied gene regions.
Electronic supplementary material
The online version of this article (doi:10.1007/s10549-013-2738-0) contains supplementary material, which is available to authorized users.
Keywords: Epigenetic, Methylation, Breast cancer, Illumina, BRCA1, Basal-like
Introduction
Breast cancer is one of the most prevalent and well-studied forms of cancer. Despite abundant research, knowledge of the molecular basis of breast cancer subtypes is still incomplete, due in large part to the heterogeneous nature of the disease. Aberrant patterns of DNA methylation are consistently observed in human cancers [1, 5, 7], and increasing attention is being placed on the varied roles DNA methylation can play in gene expression regulation and DNA–protein interactions [9, 10, 25].
Much of the progress that has been made in the characterization of altered DNA methylation patterns in breast cancer has used a candidate-gene approach, and has consistently shown numerous methylated genes in breast cancer cell lines and tumors such as RASSF1, RARB, ESR1, BRCA1, CCND2, and CDKN2A [2, 8, 20]. Recently, “genome-wide” methylation studies have found DNA methylation patterns associated with molecular subtypes of breast cancer; namely lower overall levels of methylation in basal-like tumors, and higher levels of methylation in a subset of luminal B tumors [3, 12]. A sizeable number of these observed methylated loci have also been shown to be associated with decreased gene expression [2, 19, 22, 23].
The Cancer Genome Atlas (TCGA) breast consortium (2012) reported five methylation groups defined by breast tumor sample clustering; groups 1–4 were enriched for ER+, PR+ tumors, while group 5 had the lowest levels of methylation and was enriched for triple-negative, basal-like tumors. Group 3 tumors had the highest levels of methylation and were enriched for the luminal B subtype [3]. Nevertheless, each of the five methylation groups described by the TCGA were represented by an admixture of multiple tumor subtypes. Overall, previous studies have had limited descriptions of methylation patterns and their relation to subtype, and few have explored the similarities and differences between methylation patterns at different loci in relation to methylation in matched normal breast tissues.
Therefore, the purpose of this study was to quantify DNA methylation in a set of 70 candidate genes from n = 140 breast tumors and matched normal tissues, and to test associations with gene expression, and breast tumor subtype (e.g., Basal-like, HER2-enriched, Luminal A and B tumors). In parallel, we also sought to determine if two different detection assays, Sequenom’s EpiTYPER MassARRAY and the Illumina Infinium platforms, provided comparable methylation values for identical CG loci. In contrast with the approach used to define methylation groups by the TCGA consortium, we a priori stratified our methylation analyses based on PAM50 subtype calls from Agilent microarrays previously run in the UNC tumors. Subsequently, we statistically validated our findings in the TCGA dataset by molecular subtype. We took care to insure that our validation in the TCGA dataset was as equivalent as possible to the UNC dataset by only analyzing those TCGA samples for which Agilent microarray data were used to determine relative gene expression and to make the PAM50 calls.
We observed six distinct patterns of DNA methylation within our candidate gene loci in breast tumors relative to molecular subtype and matched normal tissue. These methylation patterns (MPs) have unique distributions, either by virtue of tumor subtype, and/or their level of methylation in matched normal breast tissue. Methylation patterns interrogated by MassARRAY in the UNC dataset were validated in matched CGs in tumor and normal breast tissues obtained from TCGA using the Illumina Infinium platform. Many of the gene loci analyzed were inversely associated with gene expression in breast tumors, and often novel or stronger correlations were observed when the data were stratified by molecular subtype. Importantly, correlations of methylation with gene expression were independent of methylation pattern group membership. These results may help to further our understanding of the genetic and epigenetic contributions to breast cancer heterogeneity.
Methods
UNC sample and previous gene expression data accrual
University of North Carolina (UNC) breast tissue samples consisted of n = 140 specimens, n = 83 tumors, and n = 57 paired normal breast tissues, collected in accordance with Biomedical Institutional Review Board approval through the UNC Office of Human Research Ethics. All breast tissues for this methylation study were collected from fresh frozen samples. All tumors had greater than 50 % tumor cells, and on average 70 % tumor epithelium, as determined by pathological/histological analysis. Adjacent matched normal tissues from the ipsilateral breast were processed in the same manner as the tumors.
Additionally, oligonucleotide gene expression microarrays (Agilent Technologies, Santa Clara, CA, USA) [13] had previously been performed on these samples prior to this study and deposited in the Gene Expression Omnibus (GEO) under the accession number GSE35629. The PAM50 algorithm [15] was used to assign molecular subtypes of n = 83 breast tumors, consisting of 29 % Luminal A, 28 % Luminal B, 27 % Basal-like, 12 % HER2-enriched, and 2 % Normal-like, as previously described [15]. The two Normal-like tumors were excluded from all subsequent analyses. Clinical and demographic data, PAM50 molecular subtypes, and GEO accession numbers for the UNC sample set are listed in Online Resource 1.
Finally, Lowess normalized log2 ratios (Cy5 sample/Cy3 control) of the 70 genes interrogated for methylation in this study were median-centered prior to generating relative gene expression values. Multiple probes for the same gene were collapsed by averaging before median-centering. Subsequently, gene expression values were correlated with percent methylation values for the CpG units interrogated on the MassARRAY platform (Table 1, 2).
Table 1.
Correlations between CpG methylation and gene expression
| Methylation Pattern | MassARRAY gene name | #UNC Tumorsa | Average correlation | Low-range correlation | High-range correlation | p value range | TCGA match or # bp from amplicon | MassARRAY CpGID | Illumina match probe name | #TCGA Tumors | Illumina probe correlation | Correlation p value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MP1 | KRT5 | 51 | −0.27 | 0.00 | −0.43 | (0.0016–0.9986) | Match | KRT5_cpg20 | cg04254916 | 455 | −0.10 | 0.042 |
| MP1 | KRT17 | 56 | −0.35 | −0.24 | −0.41 | (0.0014–0.0656) | * | * | * | * | * | * |
| MP1 | MIA | 39 | −0.59 | −0.50 | −0.66 | (0.0000–0.0003) | 267 | MIA | cg25152942 | 455 | −0.54 | 0.000 |
| MP2 | SFN | 78 | −0.44 | −0.40 | −0.46 | (0.0000–0.0002) | Match | SFN_cpgl | cg03421300 | 455 | −0.39 | 0.000 |
| MP2 | TNF | 79 | −0.42 | −0.31 | −0.48 | (0.0000–0.0044) | Match | TNF_cpg9 | cg11484872 | 454 | −0.28 | 0.000 |
| MP3 | ACTB | 76 | −0.32 | −0.25 | −0.35 | (0.0014–0.0275) | * | * | * | * | * | * |
| MP3 | GRB7 | 75 | −0.46 | −0.33 | −0.53 | (0.0000–0.0036) | −16 | GRB7 | cg17740645 | 455 | −0.36 | 0.000 |
| MP4 | CST6 | 63 | −0.35 | −0.31 | −0.40 | (0.0011–0.0116) | Match | CST6_cpgl2 | cg15887846 | 455 | −0.37 | 0.000 |
| MP4 | CXCL6 | 42 | −0.37 | −0.30 | −0.42 | (0.0044–0.0467) | −37 | CXCL6 | cg25432696 | 447 | −0.33 | 0.000 |
| MP4 | CYP1B1 | 40 | −0.31 | −0.10 | −0.45 | (0.0011–0.5292) | −168 | CYP1B1 | cg25856383 | 455 | −0.23 | 0.000 |
| MP4 | DAPK1 | 52 | −0.36 | −0.30 | −0.45 | (0.0007–0.0274) | −30 | DAPK1 | cg19734228 | 455 | −0.34 | 0.000 |
| MP4 | EGFR | 74 | −0.43 | −0.29 | −0.49 | (0.0000–0.0115) | Match | EGFR_cpg8 | cg03860890 | 455 | −0.38 | 0.000 |
| MP4 | EREG | 47 | −0.31 | −0.26 | −0.38 | (0.0069–0.0677) | Match | EREG_cpgl8 | cg04941721 | 455 | −0.10 | 0.038 |
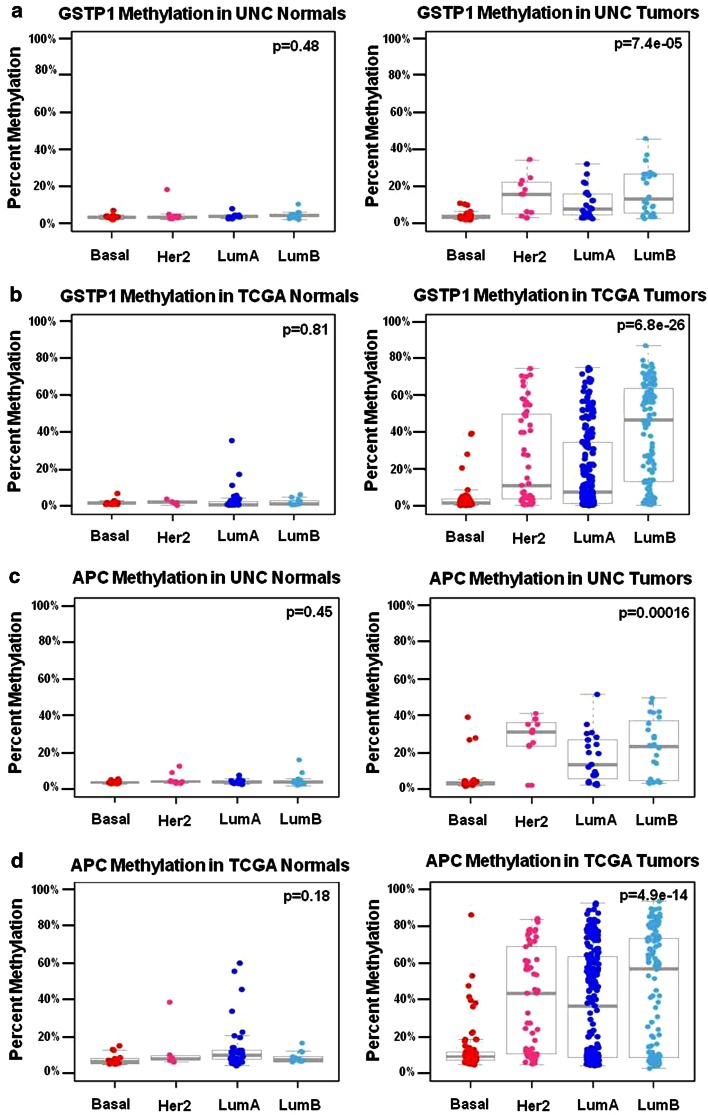
| MP4 | GSTP1 | 79 | −0.56 | −0.44 | −0.65 | (0.0000–0.0000) | Match | GSTPl_cpgl3 | cg04920951 | 455 | −0.58 | 0.000 |
| MP4 | HDAC9 | 42 | −0.33 | −0.25 | −0.36 | (0.0154–0.0980) | Match | HDAC9_cpg3 | cg12081743 | 455 | −0.09 | 0.046 |
| MP4 | KLK10 | 51 | −0.49 | −0.42 | −0.57 | (0.0000–0.0018) | Match | KLK10_cpgl7 | cg15910208 | 455 | −0.24 | 0.000 |
| MP4 | RUNX3 | 79 | −0.33 | −0.18 | −0.43 | (0.0001–0.1014) | Match | RUNX3_cpg14 | cg22737001 | 455 | −0.31 | 0.000 |
| MP4 | SFRP1 | 51 | −0.37 | −0.26 | −0.42 | (0.0017–0.0602) | Match | SFRP1 | cg22418909 | 454 | −0.40 | 0.000 |
| MP4 | SNAI2 | 51 | −0.22 | −0.12 | −0.43 | (0.0014–0.3977) | 183 | SNAI2 | cg24593475 | 455 | −0.11 | 0.019 |
| MP4 | VIM | 78 | −0.27 | −0.21 | −0.36 | (0.0012–0.0629) | 229 | VIM | cg12874092 | 455 | −0.32 | 0.000 |
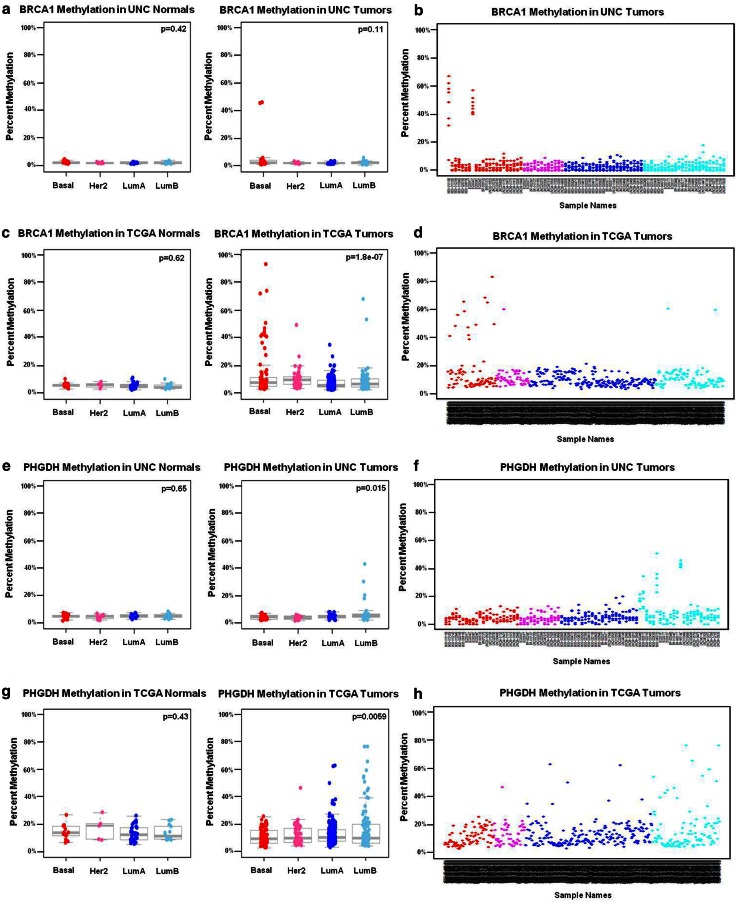
| MP5 | BRCA1 | 79 | −0.35 | −0.24 | −0.41 | (0.0002–0.0331) | Match | BRCA1_cpg5 | cg04658354 | 455 | −0.41 | 0.000 |
| MP5 | CDKN2A | 79 | −0.20 | −0.13 | −0.35 | (0.0012–0.2437) | Match | CDKN2A_cpg23 | cg03079681 | 455 | −0.12 | 0.008 |
| MP5 | PHGDH | 79 | −0.30 | −0.25 | −0.34 | (0.0018–0.0235) | Match | PHGDH_cpg15 | cg26791905 | 455 | −0.21 | 0.000 |
Pearson correlation coefficients were calculated in the UNC dataset based on averaged log2 expression values from oligo DNA microarray (Agilent) with percent methylation values for CpG units averaged across the entire amplicon. Gene loci with significant correlations between methylation and gene expression above ±0.20 are listed, with average correlation, low-range, high-range correlations, and p value range. Pearson correlation coefficients in the TCGA dataset were based on averaged log2 expression values from oligo DNA microarray (Agilent) versus percent methylation value for the single closest CpG illumina probe
a In some cases, the number of UNC tumors used for correlation analysis was less than the total number interrogated for methylation analysis because samples were limited to tumors that had both methylation and gene expression data
* No Illumina CG probe corresponding to CpGs in our massARRAY amplicon was available in the published TCGA methylation dataset used for this study
Table 2.
Parsing by molecular subtype reveals major contributors to gene expression correlations in the TCGA dataset
| Methylation pattern | MassARRAY gene name | MassARRAY CpG ID | CpGs from amplicon | Overall probe correlation | Basal (n = 81) | HER2 (n = 53) | Lum A (n = 209) | Lum B (n = 112) | Matched normal (n = 56) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Correlation | p value | Correlation | p value | Correlation | p value | Correlation | p value | Correlation | p value | |||||
| MP1 | KRT5 | KRT5_cpg20 | Match | −0.10 | −0.44 | 0.000 | −0.23 | 0.091 | 0.06 | 0.408 | 0.15 | 0.126 | −0.64 | 0.000 |
| MP1 | MIA | MIA | 4 CpGs 5′ | −0.54 | −0.26 | 0.019 | −0.23 | 0.102 | −0.20 | 0.004 | −0.02 | 0.818 | −0.43 | 0.001 |
| MP2 | SFN | SFN_cpg1 | Match | −0.39 | −0.51 | 0.000 | −0.58 | 0.000 | −0.33 | 0.000 | −0.53 | 0.000 | −0.66 | 0.000 |
| MP2 | TNFa | TNF_cpg9 | Match | −0.28 | −0.24 | 0.028 | −0.19 | 0.164 | −0.21 | 0.002 | −0.30 | 0.002 | −0.17 | 0.219 |
| MP3 | GRB7 | GRB7 | 2 CpGs 3′ | −0.36 | −0.41 | 0.000 | −0.67 | 0.000 | −0.34 | 0.000 | −0.40 | 0.000 | 0.17 | 0.200 |
| MP4 | CST6 | CST6_cpg12 | Match | −0.37 | −0.08 | 0.464 | −0.35 | 0.009 | −0.25 | 0.000 | −0.50 | 0.000 | −0.45 | 0.001 |
| MP4 | CXCL6 | CXCL6 | 2 CpGs 3′ | −0.33 | −0.24 | 0.033 | −0.51 | 0.000 | −0.24 | 0.001 | −0.18 | 0.056 | 0.09 | 0.523 |
| MP4 | CYP1B1 | CYP1B1 | 13 CpGs 5′ | −0.23 | −0.23 | 0.040 | −0.20 | 0.157 | −0.11 | 0.102 | −0.28 | 0.003 | 0.07 | 0.620 |
| MP4 | DAPK1 | DAPK1 | 1 CpG 3′ | −0.34 | −0.31 | 0.004 | −0.25 | 0.077 | −0.08 | 0.251 | −0.17 | 0.077 | −0.25 | 0.065 |
| MP4 | EGFR | EGFR_cpg8 | Match | −0.38 | −0.01 | 0.950 | −0.28 | 0.045 | −0.29 | 0.000 | −0.11 | 0.235 | −0.13 | 0.356 |
| MP4 | EREG | EREG_cpg18 | Match | −0.10 | −0.18 | 0.112 | −0.20 | 0.142 | −0.10 | 0.158 | 0.02 | 0.846 | −0.09 | 0.507 |
| MP4 | GSTP1 | GSTPl_cpg13 | Match | −0.58 | −0.14 | 0.226 | −0.69 | 0.000 | −0.42 | 0.000 | −0.53 | 0.000 | −0.14 | 0.287 |
| MP4 | HDAC9 | HDAC9_cpg3 | Match | −0.09 | 0.00 | 0.980 | −0.17 | 0.211 | 0.10 | 0.158 | −0.ll | 0.230 | −0.13 | 0.354 |
| MP4 | KLK10 | KLK10_cpg17 | Match | −0.24 | −0.09 | 0.416 | −0.10 | 0.463 | −0.21 | 0.002 | 0.00 | 0.962 | 0.03 | 0.847 |
| MP4 | RUNX3 | RUNX3_cpg14 | Match | −0.31 | 0.15 | 0.168 | −0.33 | 0.015 | −0.17 | 0.012 | −0.34 | 0.000 | 0.06 | 0.637 |
| MP4 | SFRP1 | SFRP1 | l CpG 3′ | −0.40 | −0.09 | 0.417 | −0.24 | 0.078 | −0.08 | 0.251 | −0.17 | 0.081 | 0.15 | 0.282 |
| MP4 | SNAI2 | SNAI2 | 4 CpGs 3′ | −0.11 | 0.25 | 0.025 | −0.38 | 0.005 | −0.06 | 0.379 | −0.11 | 0.229 | 0.13 | 0.345 |
| MP4 | VIM | VIM | 31 CpGs 3′ | −0.32 | −0.08 | 0.477 | −0.01 | 0.917 | −0.25 | 0.000 | −0.29 | 0.002 | −0.18 | 0.190 |
| MP5 | BRCA1 | BRCA1_cpg5 | Match | −0.41 | −0.63 | 0.000 | −0.40 | 0.003 | −0.08 | 0.280 | −0.58 | 0.000 | −0.03 | 0.848 |
| MP5 | CDKN2A | CDKN2A_cpg23 | Match | −0.12 | −0.32 | 0.003 | −0.15 | 0.273 | −0.18 | 0.009 | 0.06 | 0.565 | 0.26 | 0.051 |
| MP5 | PHGDH | PHGDH_cpg15 | Match | −0.21 | 0.12 | 0.269 | 0.06 | 0.651 | −0.17 | 0.012 | −0.37 | 0.000 | 0.20 | 0.132 |
Pearson correlation coefficients were calculated in the TCGA dataset based on averaged log2 expression values from oligo DNA microarray (Agilent) with percent methylation values for a single IIlumina Infinium probe. If the probe was a direct match for an Epityper CpG, the CpG number is listed. If the probe was not a direct match for an Epityper CpG, the distance and direction from the amplicon is listed. Correlation values are listed for the overall dataset (including all subtypes), and by individual tumor subtype (Basal, HER2 enriched, LumA, LumB), and matched normal breast tissue. coefficients > + or − 0.2 and p values < 0.05
Candidate gene selection
The candidate genes selected for this study were carefully chosen due to their pivotal roles in cancer biology in general, and/or because they represent PAM50 genes such as MIA, PHGDH, KRT5, GRB7, EGFR, and CDH3. For example, we chose to interrogate methylation in “BRCA1 related” genes (such as BRCA1 and BRCA2), genes involved in epithelial–mesenchymal transition (such as VIM, TWIST, and CDH1), genes which direct methylation metabolism or histone modifications (such as DNMT3b and HDAC9), or genes that previous studies have repeatedly identified as being significantly methylated in breast cancer (such as RASSF1, APC, CCND2, PTEN, and RARB).
DNA extraction and sodium bisulfite conversion
DNA extraction was performed on the UNC n = 140 sample set using either the Qiagen Puregene® Core Kit A or the Qiagen DNAeasy® Blood & Tissue Kit (Qiagen, Germantown, MD, USA). Sodium bisulfite (NaBi) conversion of genomic DNA extracted from breast tissue was carried out using the EZ DNA Methylation-Direct Kit (Zymo Research, Irvine,CA, USA) as previously described [21].
Quantification of DNA methylation using mass spectrometry
Mass spectrometry was used to quantify percent methylation for 70 candidate gene loci on the SEQUENOM MassARRAY platform using the EpiTYPER® T complete reagent kit as previously described [21]. Custom primers were designed for amplicons representing 70 genes with a total coverage of approximately 1,200 CGs. PCR was carried out on 5–10 ng of NaBi-converted DNA using NaBi conversion specific primers (Online Resource 2), in 5 μl volumes with PCR conditions as previously described [21]. As per the EpiTYPER protocol, shrimp alkaline phosphatase was used to dephosphorylate unincorporated dNTPs. Finally, RNase-A was added in the T-cleavage reaction, rendering methylated and unmethylated CG containing fragments subsequently quantified by mass spectrometry.
The EpiTYPER® software identifies methylated versus unmethylated CGs based on detection of a 16-Dalton mass shift between the two peaks. The software then calculates the percent methylation based on the relative ratio of methylated to unmethylated CGs within a margin of 5 % methylation confidence interval [4, 6, 16]. In some cases, fragments resulting from the T-cleavage reaction may contain more than one CpG dinucleotide, and are thus referred to as “CpG units.” Percent methylation of such CpG units are calculated as previously described [4]. Some values for CpG containing fragments fall near or outside the 1,000–8,000 Dalton window in which the MassArray platform performs accurate percent methylation, and thus calculations are assigned an “N/A” as these values cannot be quantified reliably.
Hierarchical clustering in the UNC tumor set
Nucleic acids derived from the UNC tumors and matched normal breast tissues had previously been used for separate molecular studies, including the gene expression analysis described below. Therefore, there was limited DNA remaining from the n = 81 UNC tumor dataset to perform the methylation assays. The result was that we were able to quantify methylation for 33 gene loci in 81 tumors (UNC set A), and an additional 37 genes in a subset of 53 of the 81 tumors (UNC set B). As complete data are needed for clustering analysis, we performed unsupervised hierarchical clustering (HCA) separately on these two distinct gene/tumor sets (Online Resource 3). HCA of MassARRAY methylation data in the UNC tumor/matched normal dataset, followed by validation of methylation patterns in TCGA tumors and matched normal tissues revealed the six methylation patterns described herein.
Independent validation in TCGA breast tumor and normal samples
Methylation and gene expression data accession from TCGA
The MassARRAY methylation findings from the UNC study of breast cancer patients were compared with a publically available, open-access dataset of invasive breast adenocarcinoma from The Cancer Genome Atlas (TCGA). Each tumor and adjacent normal tissue specimen (if available) was embedded and a histologic section was obtained for review. A board-certified pathologist reviewed each H&E-stained case to confirm that the tumor specimen was histologically consistent with breast adenocarcinoma and the adjacent normal specimen contained no tumor cells, in accordance with TCGA protocol requirements [3]. DNA methylation data were generated using the Illumina Infinium Meth27K or Meth450K platform and presented as β values, with 0 indicating 0 % DNA methylation and β values of 1 indicating 100 % DNA methylation. Methylation data from 21,986 CpG sites from 813 breast tumors and 123 adjacent non-tumor breast tissue samples was obtained from the TCGA Data Portal (https://tcga-data.nci.nih.gov/docs/publications/brca_2012/) in the file BRCA.methylation.27k.450k.zip (Data freeze: November 11, 2011). In order to insure equivalent comparisons between UNC and TCGA samples, only those tumors with PAM50 subtype calls from Agilent arrays were utilized for this study, leaving 455 tumor tissue samples and 70 matched normal samples (Online Resources 4 and 5).
Inclusion of data for specific CpG sites were chosen based on proximity to the CpG units that were interrogated by MassArray. Data for CpG sites with direct matches with MassArray amplicons were included in the dataset, and are labeled in Tables 1 and 2 by the CpG unit they correspond to in the MassArray amplicon. If there was no direct match for the CpG unit in the TCGA dataset, then CpG sites closest to the MassArray amplicon were included, with the base pair distance from the MassArray amplicon listed in Table 2.
Statistical analyses
Unsupervised hierarchical clustering based on complete linkage and Euclidean distance of percent methylation values in the UNC dataset was performed and displayed using MeV (version 4.8.1) of the TM4 software suite [17] (Fig. 1a–d). Relative gene expression in both UNC and TCGA sample sets was measured by normalized log2 ratios (Cy5 sample/Cy3 control) for each of the 70 genes interrogated in this study. In the cases where there were multiple probes per gene, log2 values were averaged. The Pearson r statistic was used to correlate relative gene expression and percent methylation in the UNC dataset, and by Illumina β methylation values in the TCGA dataset (Tables 1, 2). Pearson correlation values greater than (+ or −0.2) with a p value equal or less than 0.05 were considered significantly correlated. In order to validate each of the six unique methylation pattern features observed in the UNC tumor/matched normal pairs, ANOVA was used to assess differences in mean percent methylation or β values in the UNC and TCGA datasets, respectively (Figs. 2, 3, 4, 5, 6). Finally, R (http://www.R-project.org) was used to plot the contributors to significant inverse correlations of methylation with gene expression (Figs. 7, 8).
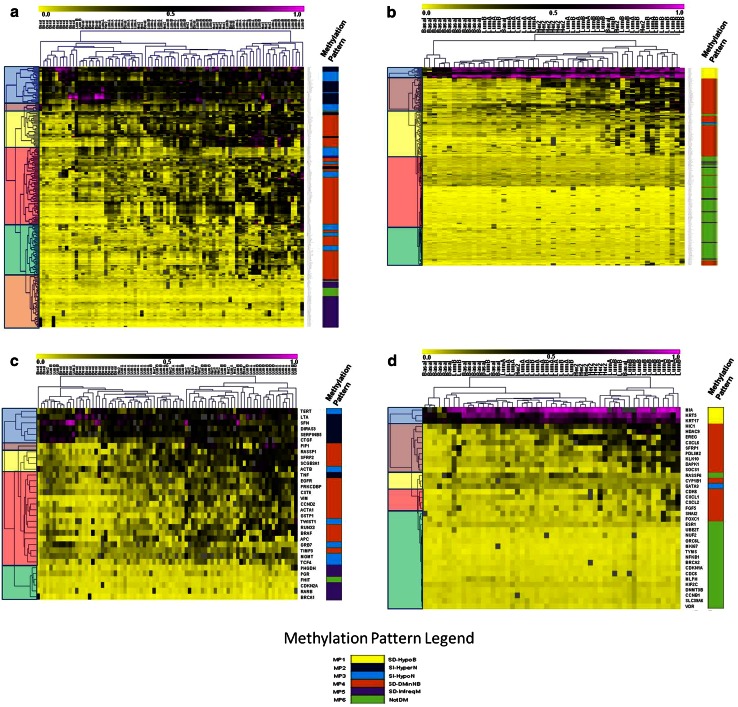
Fig. 1.
a–d Unsupervised hierarchical clustering analysis of candidate loci methylation in UNC datasets identifies six methylation patterns. The clustergram is highlighted on the left to display the major clada for each dataset. The colored bar on the right of the clustergram displays the methylation pattern group for either each CpG unit or average methylation per gene (MP1 = yellow, MP2 = dark blue, MP3 = light blue, MP4 = orange, MP5 = purple, and MP6 = green). Hierarchical clustering analysis (HCA) by CpG unit of a 81 tumors and b 53 tumors reveal enrichment of methylation patterns for each cluster. HCA of averaged methylation per locus for c 81 tumors and 33 genes and d 53 tumors and 37 genes show similar clustering groups and methylation patterns compared to clustergrams based on individual CpG units. See online resource 3 for a detailed listing of rows (genes, CpG IDs) and columns (tumor subytpe)
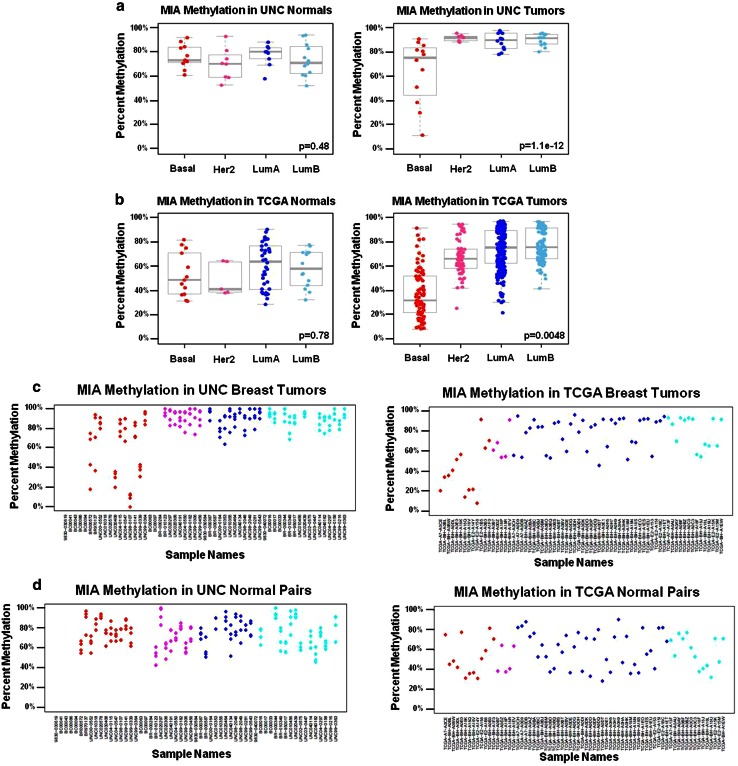
Fig. 2.
a–d MP1 gene loci display hypermethylation in normal tissue and all tumor subtypes with a subset of basal tumors displaying a hypomethylated phenotype. Box plots display percent methylation distributions in normal breast tissue and matched tumors (n = 57 matched pairs for the UNC dataset and n = 70 matched pairs for the TCGA dataset) where the upper and lower whiskers represent 1.5 times the interquartile range (IQR). Molecular subtype is listed on the horizontal axis and percent methylation on the vertical axis. Each dot represents the average percent methylation by MassARRAY across the amplicon for the UNC dataset, or for the β values of the closest MIA Illumina probe (cg25152942) in the TCGA dataset, respectively. Tumors are grouped by PAM50 molecular subtypes assigned from previous oligoarray analysis (Basal = red, HER2-enriched = pink, Luminal A = dark blue, and Luminal B = light blue), while normal tissues are grouped by the molecular subtype of the matched tumor. The MP1 “SD-HypoB” locus pattern was recapitulated in TCGA breast samples by t test of methylation differences between basal and non-basal tumors significant for tumors in both a the UNC dataset and b the TCGA dataset, while no significant difference was observed in matched normal tissue in either dataset. MIA methylation in c UNC breast tumors and matched normal tissue and d TCGA breast tumors and matched normal tissue are displayed in scatterplots. (Note: similar or overlapping percent methylation values for each CpG within an amplicon by MassARRAY will appear as one “dot” in the UNC scatterplots). T test p values for the basal vs. non-basal test are provided in the bottom right of each figure
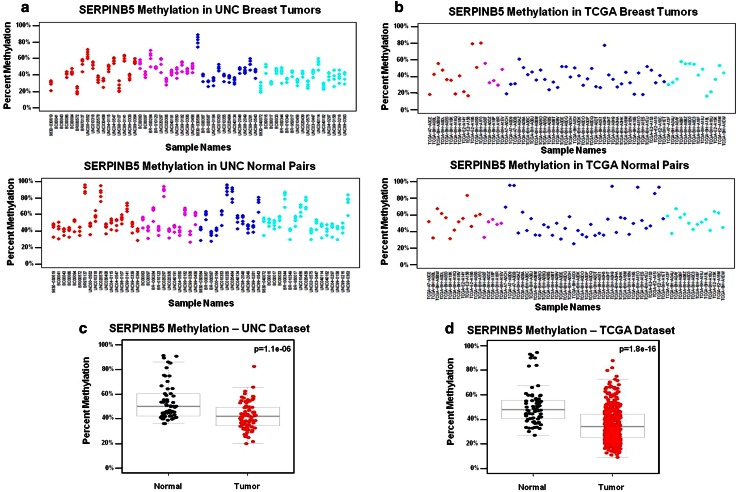
Fig. 3.
a–d MP2 gene loci display subtype-independent differential methylation pattern with tumors exhibiting lower methylation compared to normal tissue. SERPINB5 methylation in a UNC breast tumors and matched normal tissue and b TCGA breast tumors and matched normal tissue are displayed in scatterplots of individual CpG units in the UNC dataset, and by β values for matched SERPINB5 probe cg20837735. MP2 “SI-HyperN” gene loci display significantly lower average percent methylation in tumor samples vs. matched normal tissue in both the c UNC dataset and were recapitulated in d the TCGA dataset. T test p values for methylation differences between tumor vs. normal samples are provided in the top right of each box plot
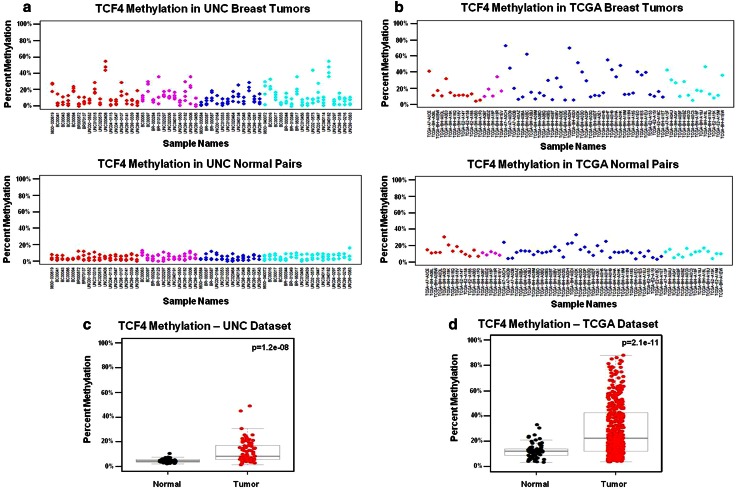
Fig. 4.
a–d MP3 gene loci display subtype-independent differential methylation with tumors exhibiting higher methylation compared to normal tissue. MP3 gene loci are distinguished from MP2 loci by relative hypomethylation in matched normal tissues, which are subtype-independent; e.g., “SI-HypoN.” TCF4 methylation patterns for a UNC breast tumors and matched normal tissue and b TCGA breast tumors and matched normal tissue are displayed in scatterplots of individual CpG units in the UNC dataset, and by β values for matched TCF4 Illumina probe cg08491964. MP3 “SI-HypoN” gene loci display significantly higher average percent methylation in tumors compared to matched normal tissue in both the c UNC dataset and d the TCGA datasets. T test p values for methylation differences between tumor vs. normal samples are provided in the top right of each box plot
Fig. 5.
a–d MP4 gene loci display a hypomethylated phenotype in basal tumors and differential methylation in non-basal HER2, LumA, and LumB tumors. Box plots show percent methylation distributions in normal breast tissue and matched breast tumors. MP4, subtype-dependent, differentially methylated in non-basal tumors “SD-DMinNB” patterns were validated in the TCGA tumor and matched normal sample set. A significant difference by ANOVA was observed in average percent methylation of GSTP1 between molecular subtypes in both a the UNC tumor dataset and b the TCGA tumor dataset for the matched GSTP1 cg04920951 probe, while no significant difference was observed in matched normal tissue in either dataset. APC also demonstrated an MP4 methylation locus pattern, but unlike GSTP1, APC methylation was not associated with gene expression in either the c UNC or d TCGA dataset. (Boxplots shown are of averaged percent methylation across the MassARRAY amplicon in the UNC samples, and averaged β values for three matched APC probes; cg21634602, cg20311501, and cg16970232, respectively). ANOVA p values for testing methylation differences between molecular subtype are provided in the top right of each box plot
Fig. 6.
a–h MP5 gene loci display subtype-dependent methylation patterns with infrequent methylation. MP5 loci were subtype-dependent and infrequently methylated “SD-InfreqM.” Only two tumors were methylated at the BRCA1 locus in the UNC samples and no significant differences were observed by ANOVA between molecular subtypes in a UNC tumors and matched normal breast tissues, with percent CpG methylation values averaged for the entire amplicon. Frequency of methylation is displayed in a scatterplot of b the entire UNC dataset (n = 81 tumors), where each CpG unit in the amplicon is plotted. (Note: similar or overlapping percent methylation values for each CpG within an amplicon by MassARRAY will appear as one dot in the UNC scatterplots). A significant difference was observed in β values for the BRCA1-matched cg08993267 Illumina probe between molecular subtypes in the c TCGA-matched tumor normal dataset (sample size n = 70). Frequency of methylation is displayed in a scatterplot of d the entire TCGA dataset (n = 455 tumors). A significant difference in percent methylation was observed in PHGDH between molecular subtypes in e the UNC dataset and recapitulated in g the TCGA dataset (PHGDH probe cg26791905). Methylation frequency is displayed in scatterplot; f the entire UNC tumor dataset and h the entire TCGA tumor dataset. ANOVA p values for testing methylation differences between molecular subtypes is provided in the top right of each boxplot
Fig. 7.
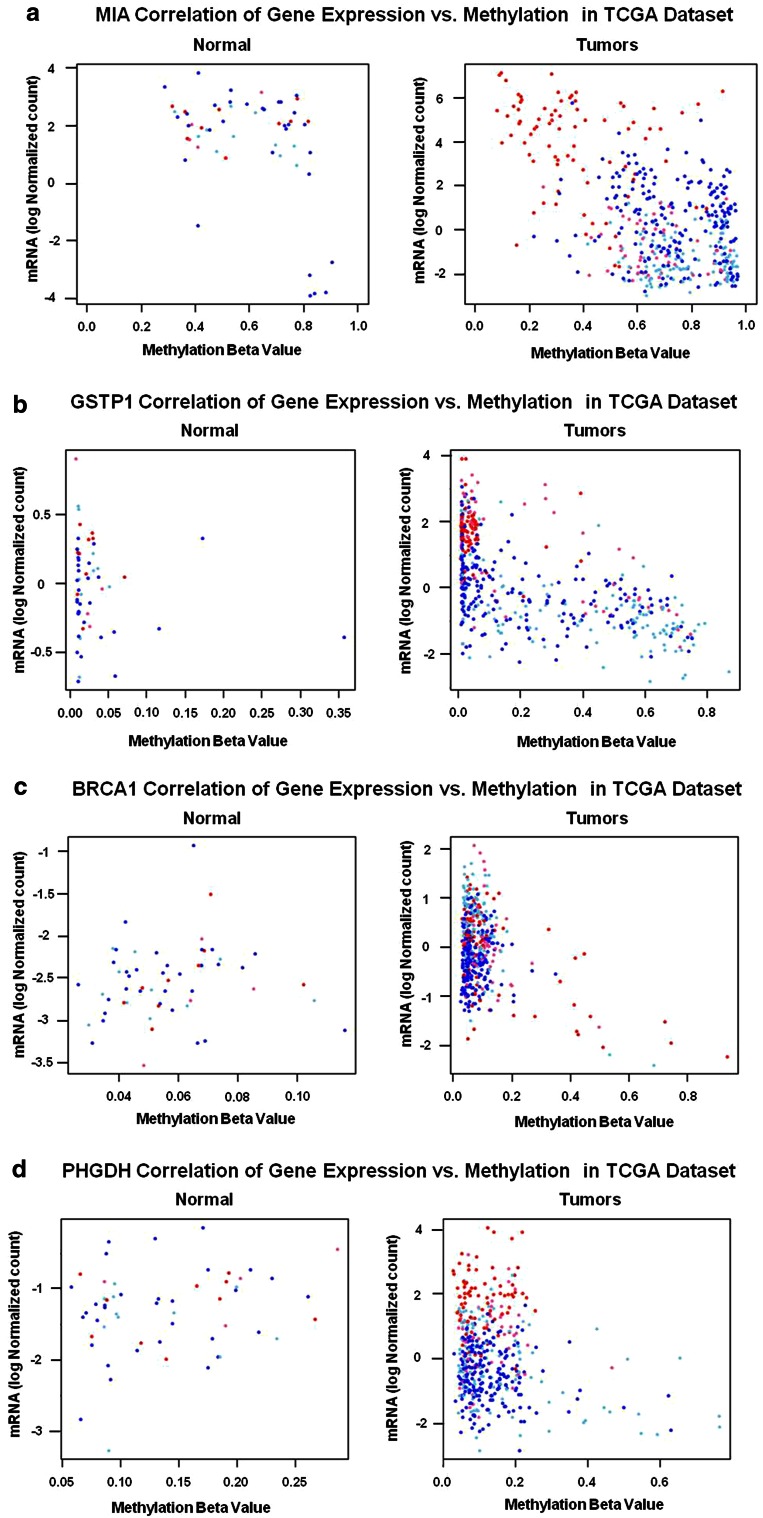
a–d Plotting contributors of significant inverse correlations with gene expression. Methylation beta values were plotted against mRNA (logbase2 normalized values) in the TCGA dataset, with each data point representing a tumor (n = 455 tumors). Tumor subtype is displayed by the color of each data point (Basal = red, HER2-enriched = pink, Luminal A = dark blue, and Luminal B = light blue). a MIA methylation correlation with gene expression in tumors is driven by the subset of basal tumors with methylation β values < 0.5, and by the six outlier Lumina A matched normal samples. When both the relatively hypomethylated subset of basal tumors and outlier normal samples were removed, correlations were no longer significant. b GSTP1 also displayed significant overall correlation between methylation and gene expression as well as significant correlations in all subtypes except Basal tumors. c BRCA1 overall correlation between methylation and gene expression was driven mainly by Basal and Luminal B tumors. d PHGDH overall correlation was driven by the significant correlation in Luminal B tumors
Fig. 8.
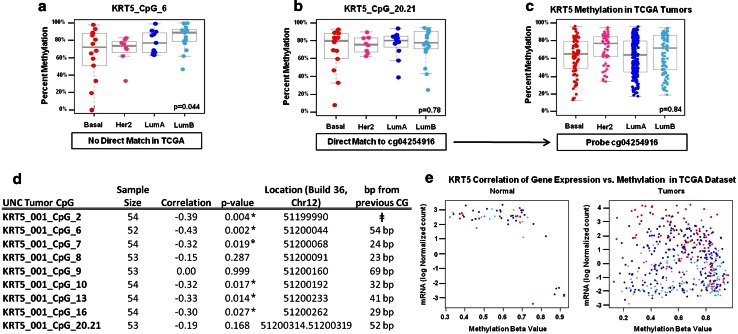
a–e The KRT5 interrogated locus shows high methylation variability. MassARRAY methylation data for the KRT5 gene locus in UNC tumors reveals heterogeneity throughout the 439-bp amplicon. a CpG number 6 in the KRT5 amplicon was significantly (p = 0.04) differentially methylated by ANOVA between tumor subtypes in the UNC samples, but did not have a direct probe match in the TCGA dataset. b CpG unit 20.21 was not differently methylated by ANOVA in the UNC samples, yet was the only CpG unit for which the corresponding c TCGA KRT5 Illumina probe cg04254916 was available. The non-significant ANOVA finding at KRT CpG_20.21 was confirmed in the TCGA (e.g., ANOVA was not significant in either the UNC or TCGA samples at this specific CpG unit). To further illustrate the heterogeneity observed in the KRT5 amplicon, d correlation analysis between individual CpGs and gene expression reveal CpGs as close as 23 bp apart have strikingly different correlation values. While many CpGs in the amplicon were significantly inversely correlated to gene expression, several CpGs were not, including CpG 20.21, which is consistent with e the matching TCGA probe not significantly associated with gene expression. ǂ Values for CpG fragments falling near or outside the mass Dalton detection window cannot be reliably quantified and are, therefore, excluded by the MassARRAY Epityper analytical software. These include KRT5 CpGs 1, 3, 4, 5, 11, 12, 14, 15, 17, 18, 19, and 22. * Significant correlation between methylation and gene expression by individual CG
Results
Unsupervised clustering of methylation data reveals distinct methylation patterns in breast cancer subtypes
Unsupervised hierarchical clustering of DNA methylation data within candidate gene loci was performed on the two UNC datasets and revealed six distinct methylation patterns relative to breast cancer subtype and matched normal breast tissues. Consensus clustering was not possible when attempting to validate methylation patterns in the TCGA dataset due to a lack of equivalence between methylated loci in the UNC and TCGA samples. The methylation data used for this validation study were derived from both 27 and 450 k Illumina Infinium platforms that, once normalized and filtered by the TCGA investigators, resulted in methylation data for only ~22,000 probes covering the entire human genome (BRCA.methylation.27k.450k.zip) [3]. Therefore, this publically available methylation data file had far fewer methylation probes than the ~480,000 CG sequences originally interrogated. We were, therefore, fortunate to have been able to match 61 (see Online Resource 6), corresponding Illumina CG probes in the published TCGA dataset relative to the 1,200 CGs interrogated in the UNC dataset. Specifically, MassARRAY is more of a fine mapping platform which allows interrogation of many consecutive CpG sequences within a single amplicon, while the Illumina platform has a “genome wide” application, and consequently interrogates fewer CpGs per gene. Using the available TCGA methylation data described, observed methylation patterns were statistically validated in the TCGA by hypothesis testing of each of six unique pattern features.
Methylation pattern 1 (MP1) gene loci were subtype-dependent (SD) and characterized by a subset of relatively hypomethylated basal-like tumors “SD-HypoB.” This group included MIA, KRT17, and KRT5, (Fig. 1b,d) which were hypermethylated in all normal tissues and tumor subtypes, except for a subset of basal-like tumors that were relatively hypomethylated as exemplified by MIA (Fig. 2). MP2 gene loci such as SFN, SERPINB5, and DIRAS3 (Fig. 1a,c) were differentially methylated across all subtypes, and thus methylation patterns were subtype-independent (SI). In addition, MP2 loci were differentially methylated in tumors, had high methylation levels in normal tissue that typically ranged from 30 to 60 % (Fig. 3), and, therefore, were referred to as “SI-HyperN.” Differential methylation for MP3 gene loci such as GRB7, TCF4, MGMT, TWIST, and TERT was also independent of subtype; however, this pattern was distinguished by hypomethylation in matched normal tissues, in contrast to the hypermethylation in normal tissues observed at MP2 loci (Fig. 4). Therefore, we describe MP3 loci as “SI-HypoN” (Fig. 4).
MP4 gene loci were hypomethylated in the majority of basal-like tumors, and differentially methylated across non-basal-like subtypes (e.g., HER2-enriched and Luminal A and B tumors), with relative hypomethylation in matched normal breast tissue (Figs. 1a–c, 5). Therefore, these subtype-dependent, differentially methylated in non-basal-like tumor loci were designated as “SD-DMinNB.” MP5 genes such as PHGDH, PGR, CDKN2A, RARB, and BRCA1 were infrequently methylated at the loci interrogated, reaching a level of 20 % methylation or higher in fewer than 15 % of all tumor samples (Figs. 1a,b, 6). These subtype-dependent, infrequently methylated loci (designated SD-InfreqM) were hypomethylated in matched normal breast tissues. Finally, MP6 gene loci were not differentially methylated (NotDM), and, therefore, uninformative (Fig. 1a–d). Thus MP6 loci were excluded from further analyses (see Online Resource 2).
Correlations between gene expression and DNA methylation are concordant between MassARRAY and Illumina platforms and vary by breast cancer subtype
Many of the amplicons analyzed in the UNC dataset showed significant inverse correlations between DNA methylation and gene expression (Table 1). Each CpG unit was correlated with the log2 gene expression value; therefore, correlations are displayed from a low–high range, as well as an overall correlation based on average methylation over the entire amplicon. While the UNC dataset was not large enough to examine correlations of DNA methylation and gene expression by subtype, the TCGA dataset was large enough to enable stratified analysis. Many of the CpG units analyzed revealed varying correlations between DNA methylation and gene expression that were subtype-dependent, including MIA, DAPK1, KLK10, BRCA1, and PHGDH (Table 2).
With few exceptions, methylation correlations with gene expression in the UNC dataset were comparable to corresponding IIlumina probes from TCGA, particularly for those gene loci having the least variable methylation throughout the amplicon (Table 1). Low methylation variability for all CGs interrogated within an amplicon is evidenced by concordance of average, low, and high range significant Pearson r and p values listed in Table 1, and by clustering of CGs within the same gene locus (Fig. 1a,b). We also observed several loci in normal tissues with significant correlations (Table 2).
To investigate the major contributors to significant correlations, we plotted methylation by log2 expression values for several genes in TCGA tumors and matched normal pairs (Fig. 7). Figure 7a demonstrates that the subset of hypomethylated basal-like tumors at the MP1 MIA locus drives the significant correlation in tumors. Notably, when the subset of basal-like tumors with methylation β < 0.5 were removed, the correlation was no longer significant. Likewise, when the six high methylation outlier matched normal samples from Luminal A tumors were removed, MIA methylation was no longer correlated with the gene expression in normal tissues (data not shown). Additionally, these plots show that the non-basal-like tumors for GSTP1, the basal-like tumors for BRCA1, and the Luminal B tumors for PHGDH drive the respective significant correlations at these loci.
Discordant correlations between the UNC and TCGA datasets include KRT5, EREG, and HDAC9, all loci with variable CpG methylation across the amplicon. For example, KRT5 did not achieve significance after hypothesis testing of the MP1 pattern because the matched probe available corresponded only to the highly variable CpG 20.21 in the MassARRAY amplicon (Fig. 8). Examination of each KRT5 CpG interrogated by EpiTYPER show KRT5_CpG6 is significant for the MP1 pattern (Fig. 8a–c), while CpG 20.21 is not. In this case, the Illumina platform was not truly discordant, but rather faithfully reflects the variable methylation at this specific CpG.
Discussion
We studied the DNA methylation of 70 amplicons in 81 breast tumors and describe six locus-specific methylation patterns in relation to tumor subtype and matched normal breast tissues. These patterns were successfully validated in a larger TCGA dataset of n = 455 tumors and n = 70 matched normal breast tissues. We found that differential methylation was either subtype-dependent or subtype-independent (e.g., differential methylation occurs in all subtypes). For example, methylation patterns (MP) 1, MP4, and MP5 are differentially methylated in a subtype-dependent manner, whereas MP2 and MP3 loci were differentially methylated across all subtypes.
Importantly, methylation is CpG locus-dependent and may vary greatly over short bp distances as exemplified by the KRT5 amplicon (Fig. 8). For this reason, not all CpG units within the same amplicon cluster together, and can segregate as “outlier” CpGs such as MGMT_001_7.8.9, CST6_001_10, and HDAC9_001_1 (Online Resource 3). Conversely, other loci such as MIA and VIM are more homogeneously methylated over longer distances. For example, the closest corresponding MIA and VIM CG Illumina probes were ~250 bp away from the EpiTyper amplicon, yet these validation probes nevertheless had highly similar methylation values with interrogated CpGs in the UNC dataset, despite their distance from the target CpG of measure (Tables 1, 2). Thus, the specific CpG locus is critically important in any comparison between methylation platforms, and in correlative analyses with gene expression. Overall, Illumina CG probes having direct matches with interrogated MassArray CGs were highly comparable. While the Illumina platform provides good genome-wide coverage for most genes, the EpiTYPER MassARRAY platform has the distinct advantage of quantifying an average 15–40 consecutive CpGs per amplicon, thereby enabling the identification of highly heterogeneous and informative loci that might otherwise go undetected.
Historically, DNA methylation has been considered noteworthy when associated with changes in gene expression. Indeed, the TCGA consortium identified 490 methylated genes inversely correlated with gene expression in their Group 3 breast tumors, samples populated with hypermethylated genes and enriched for luminal B tumors [3]. Of particular interest is our finding that multiple methylation patterns were represented within the TCGA Group 3 tumors such as MIA, DIRAS3, and GSTP1, loci with MP1 (SD-HypoB), MP2 (SI-HyperN), and MP4 (SD-DMinNB) patterns, respectively. We also found the MIA and GSTP1 loci, (but not the DIRAS3 locus), reported by the TCGA consortium were associated with gene expression. Moreover, our analyses relative to subtype and matched normal tissue (Table 2) allowed us to identify specific contributors to significant correlations. For example, the subset of hypomethylated basal-like tumors for MIA and the non-basal-like tumors for the GSTP1 loci, respectively, drive the significant inverse correlations of methylation with gene expression (Fig. 7). When identified contributors were removed from the analyses, including the six outlier luminal A matched normal samples for KRT5, MIA, SFN, and CST6, all correlations became insignificant. High methylation/low expression findings in outlier matched normal breast may have been due to field effects in these six samples.
Distinct methylation patterns may or may not be associated with gene expression as exemplified by MP4 loci GSTP1 and APC (Fig. 5). Overall, methylation of many CpGs was associated with lower log2 expression levels (e.g., BRCA1 and GSTP1); however, we also observed the reverse at the MIA locus; e.g., lower methylation was associated with higher gene expression (Fig. 7). As proof of principle, we were encouraged that correlation plots of gene expression and methylation (Table 2; Fig. 7) confirmed past studies showing that BRCA1 methylation is associated with decreased gene expression [14, 24], and preferentially methylated in ER-negative and basal-like breast cancer [11, 18]. Whereas previous studies have used DNA methylation data to cluster breast tumor samples with similar DNA methylation patterns, here we utilized methylation data to identify and describe gene loci that have distinct patterns of methylation between the four subtypes of breast tumors and normal tissues.
In summary, percent methylation values obtained from MassARRAY in the UNC dataset were recapitulated in the TCGA using the Illumina Infinium platform, as were methylation patterns MP1–MP6. Importantly, MP1–MP6 were revealed when comparing CG specific methylation in both tumors and matched normal breast tissues, and when stratifying methylation by PAM50 tumor subtype. Depending on the locus, methylated loci may or may not be correlated with gene expression, regardless of membership within a particular methylation pattern. Moreover, methylation can be exquisitely locus specific and may vary greatly within short base pair distances. We describe six methylation patterns (MPs) found within our candidate loci; however, future studies of other loci are likely to yield additional, distinctive patterns by breast cancer subtype. Further investigations of the variable frequency of the methylation patterns described herein, together with their contributions to altered gene expression, may ultimately shed light on their role as passengers or drivers of carcinogenesis. Given the contributions of MIA, KRT5, KRT17, and PHGDH in defining the PAM50 basal-like subtype, future studies will explore the mechanisms by which these differentially methylated loci are associated with altered gene expression, and the impact such changes may have on breast cancer progression and prognosis.
Acknowledgments
We wish to acknowledge the TCGA Research Network and publically available TCGA breast cancer datasets. Without this rich resource we would not have been able to validate our findings. We are grateful for the support of the KL2RR025746 to T. Swift-Scanlan from the National Center for Research Resources, the NIH/NINR T32NR007091-17 to S.A. Bardowell, the NIH/NCI Breast SPORE CA058823 to C.M. Perou, and the Susan G. Komen Foundation KG090180 to T. Swift-Scanlan.
Disclosures
The experiments described in this study comply with the current laws of the country in which they were performed. C.M.P is an equity stock holder, and Board of Director Member, of BioClassifier LLC and University Genomics. C.M.P is also listed as an inventor on a patent application on the PAM50 molecular assay. The other authors declare no competing interest.
Abbreviations
- TCGA
The Cancer Genome Atlas
- MP
Methylation pattern
- UNC
University of North Carolina
- GEO
Gene expression omnibus
- NaBi
Sodium bisulfite
- HCA
Hierarchical clustering analysis
- SD
Subtype-dependent
- SI
Subtype-independent
- HypoB
Hypomethylated in basal tumors
- HyperN
Hypermethylated in normals
- HypoN
Hypomethylated in normals
- DMinNB
Differentially methylated in non-basal tumors
- InfreqM
Infrequently methylated
- NotDM
Not differentially methylated
Electronic Supplementry Materials
References
- 1.Baylin SB, Esteller M, Rountree MR, Bachman KE, Schuebel K, Herman JG. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum Mol Genet. 2001;10:687–692. doi: 10.1093/hmg/10.7.687. [DOI] [PubMed] [Google Scholar]
- 2.Belinsky SA. Gene-promoter hypermethylation as a biomarker in lung cancer. Nature reviews. Cancer. 2004;4:707–717. doi: 10.1038/nrc1432. [DOI] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas N Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coolen MW, Statham AL, Gardiner-Garden M, Clark SJ. Genomic profiling of CpG methylation and allelic specificity using quantitative high-throughput mass spectrometry: critical evaluation and improvements. Nucleic Acids Res. 2007;35:e119. doi: 10.1093/nar/gkm662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Das PM, Singal R. DNA methylation and cancer. J Clin Oncol. 2004;22:4632–4642. doi: 10.1200/JCO.2004.07.151. [DOI] [PubMed] [Google Scholar]
- 6.Ehrich M, Bocker S, van den Boom D. Multiplexed discovery of sequence polymorphisms using base-specific cleavage and MALDI-TOF MS. Nucleic Acids Res. 2005;33:e38. doi: 10.1093/nar/gni038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esteller M, Herman JG. Cancer as an epigenetic disease: DNA methylation and chromatin alterations in human tumours. J Pathol. 2002;196:1–7. doi: 10.1002/path.1024. [DOI] [PubMed] [Google Scholar]
- 8.Fackler MJ, Malone K, Zhang Z, Schilling E, Garrett-Mayer E, Swift-Scanlan T, Lange J, Nayar R, Davidson NE, Khan SA, Sukumar S. Quantitative multiplex methylation-specific PCR analysis doubles detection of tumor cells in breast ductal fluid. Clin Cancer Res. 2006;12:3306–3310. doi: 10.1158/1078-0432.CCR-05-2733. [DOI] [PubMed] [Google Scholar]
- 9.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 10.Gardner KE, Allis CD, Strahl BD. Operating on chromatin, a colorful language where context matters. J Mol Biol. 2011;409:36–46. doi: 10.1016/j.jmb.2011.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hasan TN, Leena Grace B, Shafi G, Syed R. Association of BRCA1 promoter methylation with rs11655505 (c.2265C>T) variants and decreased gene expression in sporadic breast cancer. Clin Transl Oncol. 2013;15:555–562. doi: 10.1007/s12094-012-0968-y. [DOI] [PubMed] [Google Scholar]
- 12.Holm K, Hegardt C, Staaf J, Vallon-Christersson J, Jonsson G, Olsson H, Borg A, Ringner M. Molecular subtypes of breast cancer are associated with characteristic DNA methylation patterns. Breast Cancer Res. 2010;12:R36. doi: 10.1186/bcr2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, Livasy C, Carey LA, Reynolds E, Dressler L, Nobel A, Parker J, Ewend MG, Sawyer LR, Wu J, Liu Y, Nanda R, Tretiakova M, Ruiz Orrico A, Dreher D, Palazzo JP, Perreard L, Nelson E, Mone M, Hansen H, Mullins M, Quackenbush JF, Ellis MJ, Olopade OI, Bernard PS, Perou CM. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu Y, Chu A, Turker MS, Glazer PM. Hypoxia-induced epigenetic regulation and silencing of the BRCA1 promoter. Mol Cell Biol. 2011;31:3339–3350. doi: 10.1128/MCB.01121-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, Quackenbush JF, Stijleman IJ, Palazzo J, Marron JS, Nobel AB, Mardis E, Nielsen TO, Ellis MJ, Perou CM, Bernard PS. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Radpour R, Kohler C, Haghighi MM, Fan AX, Holzgreve W, Zhong XY. Methylation profiles of 22 candidate genes in breast cancer using high-throughput MALDI-TOF mass array. Oncogene. 2009;28:2969–2978. doi: 10.1038/onc.2009.149. [DOI] [PubMed] [Google Scholar]
- 17.Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J. TM4 microarray software suite. Methods Enzymol. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 18.Stefansson OA, Jonasson JG, Olafsdottir K, Hilmarsdottir H, Olafsdottir G, Esteller M, Johannsson OT, Eyfjord JE. CpG island hypermethylation of BRCA1 and loss of pRb as co-occurring events in basal/triple-negative breast cancer. Epigenetics. 2011;6:638–649. doi: 10.4161/epi.6.5.15667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suijkerbuijk KP, Fackler MJ, Sukumar S, van Gils CH, van Laar T, van der Wall E, Vooijs M, van Diest PJ. Methylation is less abundant in BRCA1-associated compared with sporadic breast cancer. Ann Oncol. 2008;19:1870–1874. doi: 10.1093/annonc/mdn409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swift-Scanlan T, Vang R, Blackford A, Fackler MJ, Sukumar S. Methylated genes in breast cancer: associations with clinical and histopathological features in a familial breast cancer cohort. Cancer Biol Ther. 2011;11:853–865. doi: 10.4161/cbt.11.10.15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ulirsch J, Fan C, Knafl G, Wu MJ, Coleman B, Perou CM, Swift-Scanlan T. Vimentin DNA methylation predicts survival in breast cancer. Breast Cancer Res Treat. 2013;137:383–396. doi: 10.1007/s10549-012-2353-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilcox CB, Baysal BE, Gallion HH, Strange MA, DeLoia JA. High-resolution methylation analysis of the BRCA1 promoter in ovarian tumors. Cancer Genet Cytogenet. 2005;159:114–122. doi: 10.1016/j.cancergencyto.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 23.Wu JM, Fackler MJ, Halushka MK, Molavi DW, Taylor ME, Teo WW, Griffin C, Fetting J, Davidson NE, De Marzo AM, Hicks JL, Chitale D, Ladanyi M, Sukumar S, Argani P. Heterogeneity of breast cancer metastases: comparison of therapeutic target expression and promoter methylation between primary tumors and their multifocal metastases. Clin Cancer Res. 2008;14:1938–1946. doi: 10.1158/1078-0432.CCR-07-4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu J, Huo D, Chen Y, Nwachukwu C, Collins C, Rowell J, Slamon DJ, Olopade OI. CpG island methylation affects accessibility of the proximal BRCA1 promoter to transcription factors. Breast Cancer Res Treat. 2010;120:593–601. doi: 10.1007/s10549-009-0422-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell. 2012;22:9–20. doi: 10.1016/j.ccr.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]