Abstract
Prostate cancer is the most frequently diagnosed cancer in males in developed countries. To identify common prostate cancer susceptibility alleles, we genotyped 211,155 SNPs on a custom Illumina array (iCOGS) in blood DNA from 25,074 prostate cancer cases and 24,272 controls from the international PRACTICAL Consortium. Twenty-three new prostate cancer susceptibility loci were identified at genome-wide significance (P < 5 × 10−8). More than 70 prostate cancer susceptibility loci, explaining ~30% of the familial risk for this disease, have now been identified. On the basis of combined risks conferred by the new and previously known risk loci, the top 1% of the risk distribution has a 4.7-fold higher risk than the average of the population being profiled. These results will facilitate population risk stratification for clinical studies.
Epidemiological studies provide strong evidence for genetic predisposition to prostate cancer. Most susceptibility loci identified thus far are common, low-penetrance variants found through genome-wide association studies (GWAS; reviewed in ref. 1). Fifty-four loci have been identified so far1–6.
Because the risks associated with common susceptibility alleles are modest (per-allele odds ratios, ORs, ranging from 1.10–1.25), it is likely that other predisposition loci for prostate cancer have been missed by previous studies and that such loci should be detectable by studies with larger sample sizes7. Here, we report the findings from an extensive follow-up of GWAS conducted as part of a collaborative study with the Breast Cancer Association Consortium (BCAC), Ovarian Cancer Association Consortium (OCAC) and The Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA) as part of the COGS initiative.
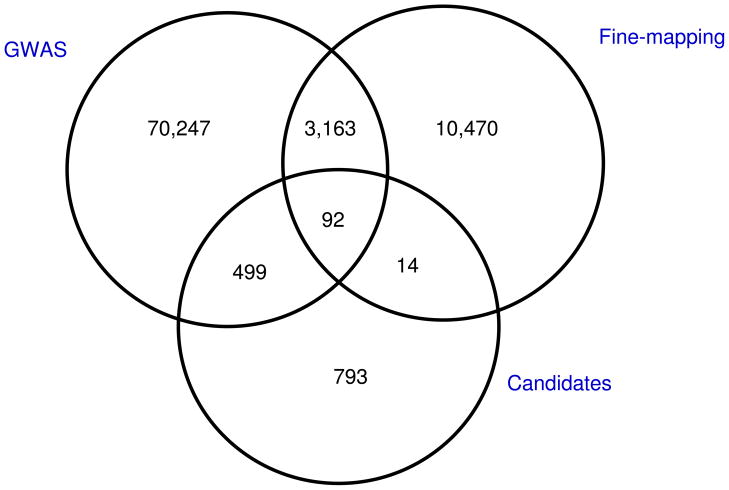
We first conducted a meta-analysis of 4 GWAS conducted in populations of European ancestry that included 11,085 cases and 11,463 controls: UK/Australia, Cancer Genetic Markers of Susceptibility (CGEMS); Cancer of the Prostate in Sweden (CAPS) and the Breast and Prostate Cancer Cohort Consortium (BPC3). Genotype data from these GWAS were imputed using the HapMap 2 CEU panel (Utah residents of Northern and Western European ancestry) as a reference, and combined tests of association were then performed for ~2.6 million SNPs (Online Methods). From this meta-analysis, we selected 74,001 SNPs showing evidence of association with overall prostate cancer, aggressive prostate cancer or prostate cancer diagnosed at <55 years of age (Online Methods). Specifically, we included all SNPs with significant association at P < 0.01 for overall prostate cancer. These SNPs were genotyped as part of a custom array that included 211,155 SNPs (the iCOGS chip), 85,278 of which were specifically chosen for their potential relevance to prostate cancer (74,001 were from GWAS top hits as described, 13,739 were from fine mapping of known susceptibility regions at the time of the chip design and 1,398 were from candidate gene studies in key pathways (for example, hormone metabolism, HOX genes, the cell cycle and DNA repair; Fig. 1 and Online Methods); some SNPs were in more than one category). The results of the GWAS component are presented here. The details of the iCOGS array can be found on the COGS website (see URLs).
Figure 1.
The iCOGS array was used for the genotyping of 25,074 prostate cancer cases and 24,272 controls from 32 studies participating in the PRACTICAL Consortium (Online Methods). Of these, 39,337 samples of European ancestry and 1,192 of African-American or mixed African origin passed quality control and did not overlap with the GWAS sample sets. Only the results from samples of European ancestry are reported here (19,662 prostate cancer cases and 19,715 controls; Supplementary Table 1 and Supplementary Note). Of the 201,598 SNPs that passed quality control, 72,157 were selected for replication of the combined GWAS (Online Methods).
Associations between SNP genotypes and prostate cancer were evaluated by logistic regression, adjusted for study and six principal components. Evidence for association was assessed using a 1-degree-of-freedom test for trend in risk by allele dose. When considering those SNPs not selected for association with prostate cancer, there was little evidence of inflation in the test statistics (λ= 1.136, equivalent to λ1,000 = 1.008). There was, however, clear evidence of an excess of significant association for SNPs selected for replication of the prostate cancer GWAS (Supplementary Fig. 1).
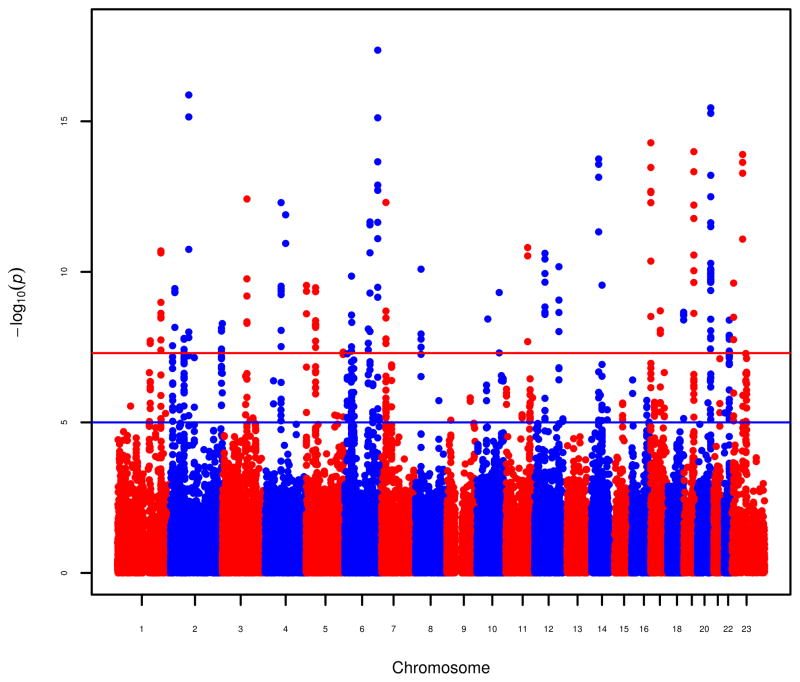
Results from the iCOGS replication stage were then combined with those from the GWAS to provide overall tests for association. After exclusion of SNPs in regions containing previously known loci associated with prostate cancer, 23 SNPs in 23 regions showed evidence of association in the combined GWAS and iCOGS replication stage analysis at P < 5 × 10−8 (Fig. 2 and Table 1). There was no strong evidence for heterogeneity in the per-allele ORs between studies (Supplementary Fig. 2). All alleles are common (minor allele frequencies of 8–50%; Table 1) and conferred estimated per-allele ORs from 1.06–1.15. All but two of the autosomal SNPs associated with prostate cancer risk showed a pattern of association consistent with a log-additive model, as observed for most common cancer susceptibility alleles. For rs11902236 on chromosome 2, the estimated OR in the iCOGS replication stage for the heterozygote genotype was 1.04 (95% confidence interval (CI) = 0.99–1.08), which is smaller than expected under a log-additive model (P = 0.05), and, for rs7141529 on chromosome 14, the estimated OR in the iCOGS replication stage for the heterozygote genotype was 1.16 (95% CI = 1.10–1.21), which is greater than expected under a log-additive model (P = 0.004).
Figure 2.
Table 1.
Summary results for 23 SNPs identified as associated in European ancestry samples
| Marker | Chr | Position Allele | MAF1 | Per allele OR2 (95%CI) | Stage 3 | P-value | Candidate genes | |
|---|---|---|---|---|---|---|---|---|
| Stage | Combined | |||||||
| rs1218582 | 1 | 153100807 AG | .45 | 1.06 (1.03–1.09) | Combined GWAS | 6.0×10−5 | 1.95×10−8 | KCNN3 |
| iCOGS Replication Stage | 5.2×10−5 | |||||||
| rs4245739 | 1 | 202785465 AC | .25 | .91 (.88–.95) | Combined GWAS | 2.6×10−5 | 2.01×10−11 | MDM4 |
| iCOGS Replication Stage | 1.7×10−7 | |||||||
| rs11902236 | 2 | 10035319 GA | .27 | 1.07 (1.03–1.10) | Combined GWAS | 1.8×10−5 | 2.84×10−8 | |
| iCOGS Replication Stage | 1.1×10−4 | TAF1B:GRHL1 | ||||||
| rs3771570 | 2 | 242031537 GA | .15 | 1.12 (1.08–1.17) | Combined GWAS | 2.0×10−2 | 5.22×10−9 | |
| iCOGS Replication Stage | 2.9×10−8 | FARP2 | ||||||
| rs7611694 | 3 | 114758314 AC | .41 | .91 (.88–.93) | Combined GWAS | 7.1×10−4 | 3.80×10−13 | |
| iCOGS Replication Stage | 6.3×10−11 | SIDT1 | ||||||
| rs1894292 | 4 | 74568022 GA | .48 | .91(.89–.94) | Combined GWAS | 9.8×10−5 | 5.02×10−13 | |
| iCOGS Replication Stage | 9.9×10−10 | AFM,RASSF6 | ||||||
| rs6869841 | 5 | 172872032 GA | .21 | 1.07 (1.04–1.11) | Combined GWAS | 1.2×10−4 | 4.63×10−8 | |
| iCOGS Replication Stage | 7.8×10−5 | FAM44B (BOD1) | ||||||
| rs3096702 | 6 | 32300309 GA | .40 | 1.07 (1.04–1.10) | Combined GWAS | 1.0×10−4 | 4.78×10−9 | |
| iCOGS Replication Stage | 1.1×10−5 | NOTCH4 | ||||||
| rs2273669 | 6 | 109391882 AG | .15 | 1.07 (1.03–1.11) | Combined GWAS | 1.6×10−7 | 7.91×10−9 | |
| iCOGS Replication Stage | 1.0×10−3 | ARMC2, SESN1 | ||||||
| rs1933488 | 6 | 153482772 AG | .41 | .89 (.87–.92) | Combined GWAS | 1.9×10−5 | 4.34×10−18 | |
| iCOGS Replication Stage | 2.6×10−14 | RSG17 | ||||||
| rs12155172 | 7 | 20961016 GA | .23 | 1.11 (1.07–1.15) | Combined GWAS | 3.1×10−5 | 4.95×10−13 | |
| iCOGS Replication Stage | 3.5×10−9 | SP8 | ||||||
| rs11135910 | 8 | 25948059 GA | .16 | 1.11 (1.07–1.16) | Combined GWAS | 7.1×10−5 | 8.16×10−11 | |
| iCOGS Replication Stage | 2.6×10−7 | EBF2 | ||||||
| rs3850699 | 10 | 104404211 AG | .29 | .91 (.89–.94) | Combined GWAS | 3.5×10−3 | 4.87×10−10 | |
| iCOGS Replication Stage | 2.6×10−8 | TRIM8 | ||||||
| rs11568818 | 11 | 101906871 AG | .44 | .91 (.88–.94) | Combined GWAS | 1.0×10−2 | 1.56×10−11 | |
| iCOGS Replication Stage | 2.4×10−10 | MMP7 | ||||||
| rs1270884 | 12 | 113169954 GA | .49 | 1.07 (1.04–1.10) | Combined GWAS | 4.1×10−7 | 6.75×10−11 | |
| iCOGS Replication Stage | 1.2×10−5 | TBX5 | ||||||
| rs8008270 | 14 | 52442080 GA | .18 | .89 (.86–.93) | Combined GWAS | 3.1×10−7 | 1.78×10−14 | |
| iCOGS Replication Stage | 9.4×10−9 | FERMT2 | ||||||
| rs7141529 | 14 | 68196497 AG | .50 | 1.09 (1.06–1.12) | Combined GWAS | 3.3×10−3 | 2.77×10−10 | |
| iCOGS Replication Stage | 1.3×10−8 | RAD51L1 | ||||||
| rs684232 | 17 | 565715 AG | .36 | 1.10 (1.07–1.14) | Combined GWAS | 4.7×10−6 | 5.17×10−15 | |
| iCOGS Replication Stage | 2.2×10−10 | VPS53, FAM57A | ||||||
| rs11650494 | 17 | 44700185 GA | .08 | 1.15 (1.09–1.22) | Combined GWAS | 1.5×10−3 | 1.97×10−9 | |
| iCOGS Replication Stage | 3.4×10−7 | HOXB13 | ||||||
| rs7241993 | 18 | 74874961 GA | .30 | .92 (.89–.95) | Combined GWAS | 1.6×10−3 | 2.19×10−9 | |
| iCOGS Replication Stage | 3.6×10−7 | SALL3 | ||||||
| rs2427345 | 20 | 60449006 GA | .37 | .94 (.91–.97) | Combined GWAS | 4.4×10−4 | 3.64×10−8 | |
| iCOGS Replication Stage | 2.1×10−5 | GATAS, CABLES2 | ||||||
| rs6062509 | 20 | 61833007 AC | .30 | .89 (.86–.92) | Combined GWAS | 1.6×10−4 | 3.57×10−16 | |
| iCOGS Replication Stage | 4.1×10−13 | ZGPAT | ||||||
| rs2405942 | X | 9774135 AG | .21 | .88 (.83–.92) | Combined GWAS | 1.4×10−4 | 2.37×10−10 | |
| iCOGS Replication Stage | 2.8×10−7 | SHROOM2 | ||||||
Allele frequency of the second allele in iCOGS replication stage.
Per-allele OR in iCOGS replication stage for the second allele
Combined GWAS: Stage 1&2 UK, CGEMS, CAPS, BPC3
Aggressive disease was defined as that with Gleason score ≥ 8, prostate-specific antigen (PSA) >100 ng/ml, disease stage of ‘distant’ (outside the pelvis) or death from prostate cancer. When aggressive disease was thus defined, three of the SNPs (rs3771570, rs2273669 and rs1270884) showed a significant difference in per-allele OR between aggressive and non-aggressive disease, in each case with a higher OR for non-aggressive disease and little or no association with aggressive disease (Supplementary Table 2). A similar pattern of association with respect to aggressive disease has been observed for SNPs in the KLK3 region8. The majority of SNPs, however, showed clear association when analysis was restricted to aggressive disease (for example, 13 SNPs showed significant associations at P < 0.01 and 16 at P < 0.05), and, for 22 of the 23 SNPs, the estimated ORs were in the same direction for aggressive and non-aggressive disease. Two SNPs, rs6869841 and rs1270884, were associated with PSA levels (Supplementary Table 3). Two of the SNPs showed a significantly higher OR in cases with a first- or second-degree relative with prostate cancer (rs3771570 and rs11135910; Supplementary Table 4). Six SNPs showed a trend in OR with respect to age at diagnosis, with a higher OR at younger ages (rs3771570, rs7611694, rs6869841, rs3096702, rs684232 and rs7241993; Supplementary Table 5). This age effect has been seen previously for four prostate cancer susceptibility SNPs 9.
We have also conducted an analysis of possible pathway enrichment for the previously reported susceptibility regions and those newly reported by extracting all genes overlapping a 500-kb or a 1-Mb window flanking each lead SNP (72 regions, 589 or 960 genes, respectively). GeneGo pathway enrichment analysis was used to identify any canonical pathways that were over-represented within this gene set. The most strongly associated pathways identified (false discovery rate < 0.05) were cell adhesion and extracellular matrix (ECM) remodeling (P = 1.31 × 10−6 to 3.6 × 10−9) and transcriptional regulation by the androgen receptor (P = 3.5 × 10−6 to 3.5 × 10−8). WNT, FGF and IGF signaling also showed significant levels of enrichment (P = 1.69 × 10−4 to 9.41 × 10−5).
The overall inflation in the test statistics for those SNPs selected for GWAS replication suggests that the number of susceptibility loci may be much larger. To address this possibility more formally, we identified 22,662 SNPs selected for replication of the prostate cancer GWAS that were uncorrelated (r2 < 0.1 for any pair) and examined the directions of the estimated ORs in the iCOGS replication data set. The estimated effects were in the same direction as in the GWAS for 12,278 SNPs and in the opposite direction for 10,384 SNPs. On the basis of this analysis, 1,894 (95% CI = 1,600–2,188) selected SNPs reflect true associations with disease.
We have found 23 new loci associated with prostate cancer, 16 of which are associated with aggressive as well as non-aggressive disease, although none of the new loci are associated exclusively with the latter. This finding is, however, notable, as aggressive disease requires radical treatment, and, previously, the loci associated with prostate cancer were associated exclusively with non-aggressive disease, which is less likely to require clinical intervention.
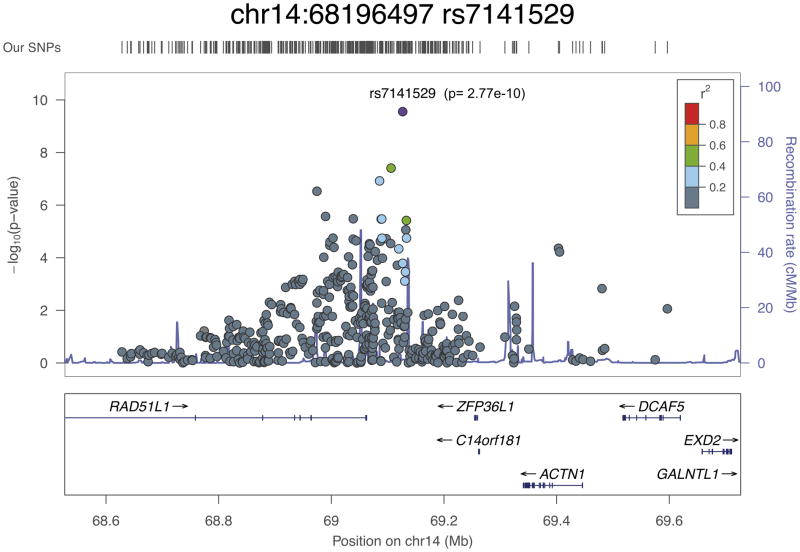
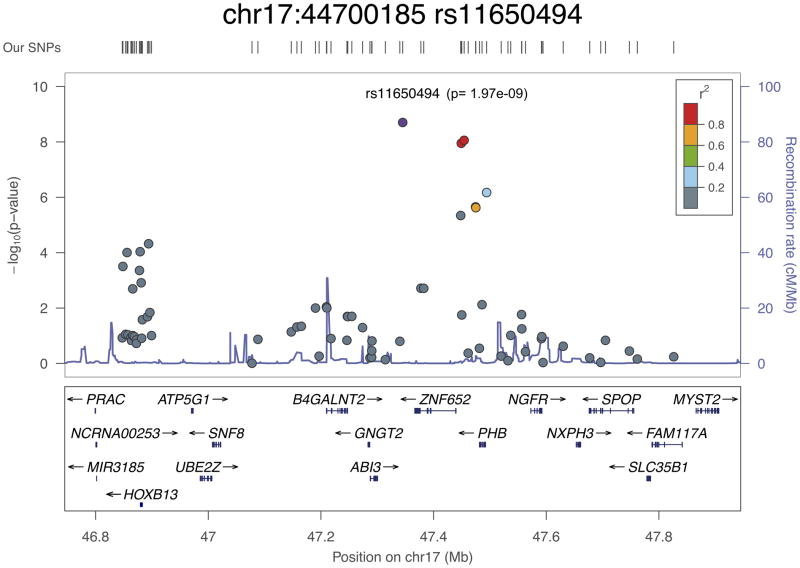
All of the newly associated loci lie in linkage disequilibrium (LD) blocks that include plausible causative genes (Fig. 3a–d and Supplementary Fig. 3). LD regions vary greatly in the genome; here, we defined LD blocks as regions with SNPs with r2 > 0.2 or took a 500-kb window around the lead SNPs. The list of genes in these 23 new susceptibility regions is given in Supplementary Table 6. Fifteen of the 23 SNPs are either intronic (12 SNPs) or in the promoter region of a gene (3 SNPs). As described below, there are data in the literature that suggest that two of the newly associated SNPs impart direct functional effects that result in allele-specific alterations to the expression of the associated genes. This raises the possibility that these SNPs could themselves represent causative variants, although further fine-mapping studies and analysis of expression in primary prostate tissue would be needed to confirm this.
Figure 3.
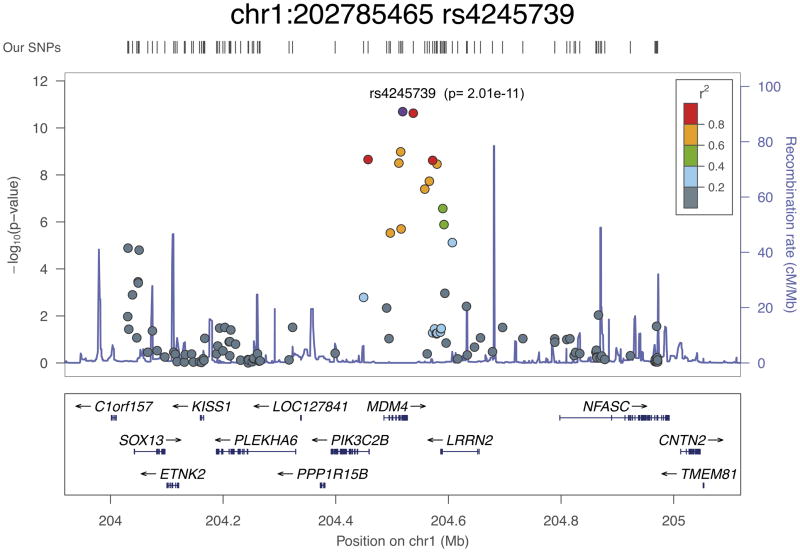
Of the new loci identified in this study, SNP rs4245739 at 1q32 is situated in the 3′ UTR of the MDM4 gene, 32 bp downstream of the stop codon. MDM4 is a negative regulator of TP53, thereby acting to inhibit cell cycle arrest and apoptosis, and is frequently overexpressed in a number of tumor types. rs4245739 is correlated with rs7556371 (r2 = 0.89), which showed some evidence of association with prostate cancer in a candidate gene study10, and rs1380576 (r2 = 0.86), has previously been reported to be associated with prostate cancer aggressiveness in a case-only analysis of candidate SNPs in the TP53 pathway11. rs4245739 has been shown to create an illegitimate binding site for miR-191 that results in the downregulation of MDM4 expression12; this is in agreement with our analysis using mirsnpscore13, which predicted that the risk allele creates a binding site for miR-191, miR-887 and miR-3669. However, rs4245739 is also highly correlated with a number of other MDM4 variants that overlap functional elements identified by the Encyclopedia of DNA Elements (ENCODE) Project13,14. Other analyses using the iCOGS array have found that rs4245739 and correlated SNPs are associated with estrogen receptor (ER)-negative breast cancer15 and breast cancer in BRCA1 mutation carriers16. In addition, the risk allele (C) of rs4245739 has been associated with increased aggressiveness in individuals with ovarian cancer17.
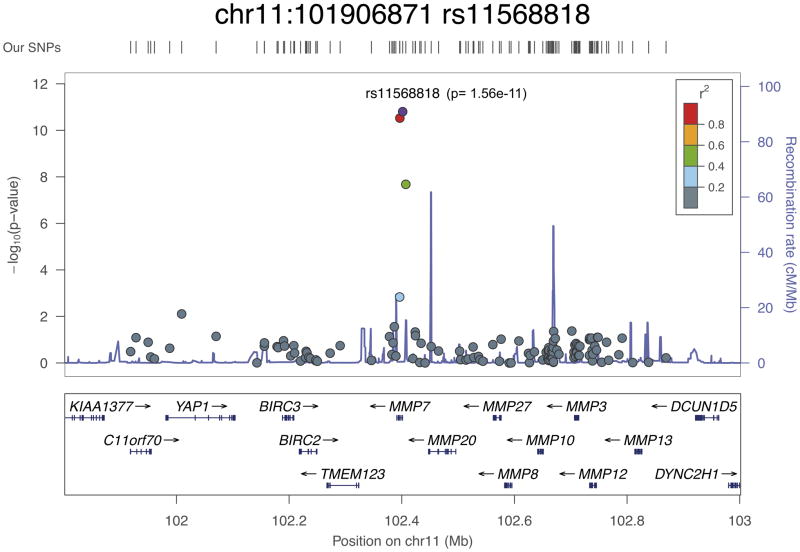
rs11568818 at 11q22 lies within a small LD region containing a single gene, MMP7, encoding a matrix metalloproteinase. Matrix metalloproteinases are implicated in metastasis, and elevated MMP7 expression itself has been reported as a potential biomarker for metastatic prostate cancer and poor disease prognosis18. This SNP is situated 181 bp upstream of the transcriptional start site in the promoter region, within an area of high sequence conservation that overlaps strong DNase hypersensitivity and transcription factor binding sites13,14. rs11568818 itself has been established as a functional promoter variant, with the risk allele (A) having been shown to create a binding site for the FOXA2 transcription factor and result in higher MMP7 expression19. Increased expression of MMP7 may represent a plausible mechanism responsible for the greater prostate cancer risk associated with this SNP; rs11568818 is correlated at r2 > 0.5 with only four other variants and seems to be the most likely candidate for a causal variant.
rs7141529 at 14q24 lies within the last intron of the longest isoform of RAD51B (also known as RAD51L1). Members of the RAD51 family are involved in the repair of double-stranded DNA breaks by homologous recombination, and their loss is potentially oncogenic. A variant in RAD51B (rs999737) has previously been associated with breast cancer20, and a second breast cancer susceptibility locus in intron 7 has also been identified in the iCOGS replication stage study21. However, there is no correlation between rs7141529 and any of the breast cancer–associated SNPs.
rs11650494 is located at 17q21, a gene-dense locus that contains several genes that have been proposed as potential prostate cancer susceptibility or somatically altered genes, including HOXB13, PRAC, SPOP and ZNF652. rs11650494 is highly correlated with a number of other variants that overlap functional motifs identified in the ENCODE Project13,14. This signal appears to center around the ZNF652 gene, with rs11650494 itself situated downstream of the gene within a long noncoding RNA (lincRNA) sequence. rs7210100 in intron 1 of ZNF652 has previously been identified as a prostate cancer susceptibility gene in African-American men22; however, this variant is rare among individuals of European ancestry, and the correlation between rs11650494 and rs7210100 is modest in the YRI (Yoruba from Ibadan, Nigeria) population (r2 = 0.22), suggesting that rs11650494 represents an independent or European-specific prostate cancer risk association. In addition, ZNF652 has been reported to be highly expressed in the majority of prostate tumors and is associated with higher risk of relapse23.
The HOXB13, PRAC and SPOP genes are all situated approximately 500 kb upstream or downstream of rs11650494; however, all are considered candidate prostate cancer genes, and, therefore, the possibility of a trans-regulatory element or locus control region associated with the rs11650494 association signal cannot be excluded. HOXB13 is one of a cluster of homeobox domain–containing genes at this locus.
These genes are essential for vertebrate embryonic development, and HOXB13 is important for normal prostate development and is a key regulator of the response to androgens24. A rare variant in HOXB13 (rs138213197; encoding a p.Gly84Glu alteration) has recently been shown to significantly increase prostate cancer risk, occurring in families with multiple cases of prostate cancer25, and HOXB13 expression levels have been proposed as a marker of prostate cancer26. Analysis of 1,927 cases and 987 control samples from the CAPS study in which both rs11650494 and rs138213197 were genotyped showed that these SNPs are not correlated (r2 = 0.001) and that the OR for rs11650494 was not altered by adjustment for rs138213197 (Supplementary Table 7). SPOP encodes a protein that may modulate the transcriptional repression activities of death-associated protein 6 (encoded by DAXX), which interacts with histone deacetylase, core histones and other histone-associated proteins. SPOP is reported to be frequently mutated in prostate tumors, and it has been suggested that SPOP mutations may anchor a distinct genetic subtype of ETS fusion–negative prostate cancers27.
In addition to the presence of plausible candidate genes, most of the 23 newly associated loci harbor several transcription factor binding sites within their LD regions.
With the identification of these new loci, 77 susceptibility loci for prostate cancer have now been identified. On the basis of an overall twofold familial relative risk for the first-degree relatives of prostate cancer cases and on the assumption that SNPs combine multiplicatively, the new loci reported here, together with those already known, explain approximately 30% of the familial risk of prostate cancer. Taking into consideration these SNPs and this risk model, the top 1% of men in the highest risk stratum have a 4.7-fold greater risk relative to the population average, and the top 10% of men have a 2.7-fold greater risk. For comparison, the former risk estimate is similar to that conferred by deleterious mutations in BRCA2 (ref. 28), and such mutation carriers are undergoing targeted screening in trials, for example, in the IMPACT (Identification of Men with a genetic predisposition to ProstAte Cancer: Targeted screening in men at higher genetic risk and controls) Study (see URLs). The SNP-based prostate cancer risk profile now available should therefore be able to distinguish men at a clinically meaningful level of risk. To evaluate the combined effect of the loci associated with prostate cancer risk, we included 68 of the known loci in a logistic regression (59 which were on iCOGS and 9 for which a surrogate with r2 > 0.76 was available). The parameters from this model were used to generate polygenic risk scores (Online Methods). On the basis of these scores, the estimated risk for men in the top 1% of the risk distribution was 4.4-fold greater than the population average risk (Supplementary Table 8), very close to the theoretical estimate predicted under a simple polygenic model (4.7-fold). Furthermore, under a polygenic genetic risk model29, an unaffected man aged 50 who has a father with prostate cancer diagnosed at 60 years of age would have a predicted lifetime risk of prostate cancer from his family history alone of just over 20%. However, if family history is taken into consideration along with the explicit effects of all known common prostate cancer susceptibility alleles, this predicted risk would rise to just over 60% if he were in the top 1% of the known polygenic risk score distribution (A. Antoniou, personal communication). Such differences in predicted risks will be important for facilitating risk stratification in targeted screening and prevention programs.
URLs
COGs website, http://ec.europa.eu/research/health/medical-research/cancer/fp7-projects/cogs_en.html;
IMPACT Study, http://www.impact-study.co.uk/;
SNAP plots from the University of Michigan, http://csg.sph.umich.edu/locuszoom/;
SNPTEST, https://mathgen.stats.ox.ac.uk/genetics_software/snptest/snptest.html;
MACH 1.0, http://www.sph.umich.edu/csg/abecasis/MACH/;
PRACTICAL, http://ccge.medschl.cam.ac.uk/consortia/practical/index.html;
GeneGo (now Thomson Reuters), http://thomsonreuters.com/products_services/science/systems-biology/;
CGEMS Project, http://dceg.cancer.gov/research/how-we-study/genomic-studies/cgems-summary;
BPC3, http://epi.grants.cancer.gov/BPC3/cohorts.html;
CAPS, http://ki.se/ki/jsp/polopoly.jsp?d=13809&a=29862&l=en;
ONLINE METHODS
GWAS analysis
Primary genotype data were obtained for three prostate cancer GWAS (CGEMS, UK/Australia stages 1 and 2, and CAPS). Standard quality control was performed on all scans; all individuals with low call rate (<95%), extremely high or low heterozygosity (P < 1 × 10−5) and non-European ancestry (>15% non-European component by multidimensional scaling using the three HapMap 2 populations (European (CEU), Asian (CHB and JPT) and African (YRI)) as a reference) were excluded. SNPs with call rate < 95%; call rate < 99% and MAF < 5%, or MAF < 1% and SNPs whose genotype frequencies departed from Hardy-Weinberg equilibrium at P < 1 × 10−6 in controls or P < 1 × 10−12 in cases were excluded. For BPC3, quality control was performed as previously described30. Genotypes in all four GWAS were imputed for ~2.6 million SNPs using the HapMap phase 2 CEU population as a reference. UK/Australia stages 1 and 2 and CGEMS were imputed using MACH 1.0 (see URLs) for auto-somal markers and IMPUTE v1 (ref. 31) for chromosome X markers. Imputation for the BPC3 study used MACH 1.0. The CAPS study used IMPUTE v1. We included imputed data from a SNP in the combined analysis if the estimated correlation between the genotype scores and the true genotypes (r2) was >0.3 (MACH) or if the quality information was >0.3 (IMPUTE).
For UK stages 1 and 2 and CGEMS, the imputed genotype probabilities were used to derive a 1-degree-of-freedom association score statistic and its corresponding variance for each SNP. The test statistic for UK/Australia stage 2 was stratified by population as previously described32. In the BPC3 study, estimated β values and standard errors were calculated for each component study, including one principal component as a covariate to adjust for population structure using ProbABEL33, and the results were combined to generate overall β values and standard errors using a fixed-effects meta-analysis. CAPS used SNPTEST (see URLs) to estimate β values and standard errors. We converted the results from all studies into test scores and variances and hence derived a combined χ2 trend statistic for each SNP (equivalent to the Mantel extension test or as in a fixed-effects meta-analysis) in R. All studies were approved by the appropriate national ethics committees, and informed consent was obtained.
SNP selection
SNPs were selected for the iCOGS array separately by each consortium. Each consortium was given a share of the array: nominally, 25% of the SNPs each for BCAC, PRACTICAL and OCAC and 17.5% for CIMBA; 7.5% were of general interest (COMMON area). In practice, the allocations were larger as a result of overlaps. In each consortium, the allocation was divided into three categories for GWAS replication, fine-mapping and candidate SNPs. The GWAS replication category consisted of a series of lists for each analysis (see the PRACTICAL website for a full description of the lists).
In general, we considered only SNPs with an Illumina design score of 0.8 or greater (some OCAC and CIMBA SNPs with lower design scores were included). Where possible, preference was given to SNPs previously genotyped by Illumina (design score = 1.1). For each category, we defined a series of ranked lists of SNPs. For the GWAS SNPs, these were merged in the following way to generate a single list. We selected SNPs in priority order from each list according to predefined weightings. When a SNP (or a surrogate) was selected on the basis of more than one list, the SNP counted toward the tally for each list. For each SNP, we preferentially accepted the SNP if it had a design score of 1.1 (meaning it had previously been genotyped on an Illumina platform). If this was not the case, we sought SNPs with r2 = 1 with the chosen SNP and selected the SNP with the best design score. If no such SNP was available, we selected SNPs with r2 > 0.8 with the chosen SNP and selected the SNP with the best design score. We excluded SNPs that were in strong LD with a previously selected SNP (r2 > 0.9). However, for SNPs that were highly significant in each list (P < 0.00001), we required two surrogate SNPs. The candidate lists were merged in the same way, giving equal weight to lists from each study. The only differences were that (i) there was no provision for additional surrogates and (ii) SNPs were excluded if there was an existing surrogate at r2 = 1.
To merge the three categories, we first included all the selected fine-mapping SNPs and then included SNPs from the merged GWAS and candidate lists in priority order. COMMON SNPs were selected in a similar way.
Finally, lists from each of the constituent consortia were merged, in priority order and in proportion to the allocated share of each consortium. SNPs selected by one consortium and subsequently selected by another counted toward both lists. The process continued until the maximum 240,000 attempted beadtypes had been reached. The final list comprised 220,123 SNPs. Of these, 211,155 were successfully manufactured on the array.
iCOGS genotyping
Samples for the iCOGS replication stage were drawn from 32 studies participating in the PRACTICAL Consortium. The majority of studies were population-based or hospital-based case-control studies or were nested case-control studies, but some studies selected samples by age or oversampled for cases with a family history of disease; in the latter instance only, one case per family was genotyped (Supplementary Table 1 and Supplementary Note). Studies were required to provide ~2% of samples in duplicate.
Genotyping was conducted using a custom Illumina Infinium array (iCOGS) in seven centers, of which five were used for PRACTICAL samples. Genotypes were called using Illumina’s proprietary GenCall algorithm. Initial calling used a cluster file generated with 270 samples from HapMap 2. To generate the final calls, we first selected a subset of 3,018 individuals, including samples from each of the genotyping centers, each of the participating consortia and each major ancestry group. Only plates with consistently high call rates in the initial calling were used. We also included 380 samples of European, Asian or African ancestry genotyped as part of the HapMap Project and 1000 Genomes Project and 160 samples that were known positive controls for rare variants on the array. This subset was used to generate a cluster file that was then applied to call the genotypes for the remaining samples. We also investigated two other calling algorithms: Illumnus34 and GenoSNP35. All three algorithms were >99% concordant in their calling for 91% of the SNPs on the array. However, manual inspection of a sample of the discrepant SNPs indicated that the calls from GenCall were almost invariably superior (generally because Illumnus or GenoSNP attempted to call SNPs that clustered poorly). Therefore, only the genotypes called by GenCall have been used in the analyses reported here.
Quality control
We excluded individuals for any of the following reasons: genotypically not male XY (XX or XXY); overall call rate < 95%; low or high heterozygosity (P < 1 × 10−6, separately for individuals of European and African-American ancestry); not concordant with previous genotyping within PRACTICAL; genotypes for the duplicate sample that appeared to be from a different individual; and cryptic duplicates where the phenotypic data indicated that the individuals were different. We searched for cryptic duplicates both within each study and between studies from the same country. For known and cryptic duplicates, the sample with the lower call rate was excluded. We attempted to identify first-degree relative pairs using identity-by-state estimates based on data from ~37,000 uncorrelated SNPs. For apparent first-degree relative pairs, we removed the control from a case-control pair, otherwise, the individual with the lower call rate. For all analyses presented here, we also excluded 6,766 individuals who were included in any of the GWAS to allow the GWAS and iCOGS replication stages to be combined.
Ancestry outliers were identified by multidimensional scaling, combining the iCOGS replication stage data with those from the three HapMap 2 populations, based on a subset of 37,000 uncorrelated markers that passed quality control (including ~1,000 selected as ancestry-informative markers). Most studies included individuals predominantly of single, European ancestry, and individuals with >15% minority ancestry were excluded. One study (SCCS) primarily contained individuals of African-American ancestry, and two studies, FHCRC and MOFFITT, contained substantial fractions of individuals of both African-American and European ancestry. After exclusion of ancestry outliers, we used principal-components analysis to correct for inflation. Principal-components analyses were carried out separately for the European and African-American subgroups on the basis of a subset of 37,000 uncorrelated SNPs. We included the first six principal components as covariates in both the European and African-American subgroups. Addition of further principal components did not reduce inflation further. Only the European data are reported here.
We excluded SNPs with call rates of <95%. We also excluded SNPs that deviated from Hardy-Weinberg equilibrium in controls at P < 1 × 10−7, on the basis of a stratified 1-degree-of-freedom test in which the deviations were summed across strata36. We also excluded SNPs for which the genotypes were discrepant in more than 2% of duplicate samples, across all COGS consortia. The final analyses were based on data from 201,598 SNPs.
Genotype intensity cluster plots were examined manually (Supplementary Fig. 4) for SNPs in each new region in which an association at genome-wide significance was obtained, and SNPs eliminated in the clustering were judged to be poor.
Statistical analysis
For each SNP, we estimated a per-allele log(OR) and standard error by logistic regression, including study and principal components as covariates. Overall significance levels were obtained by combining the estimates from the combined GWAS and the iCOGS replication stage using a fixed-effects meta-analysis. Tests of homogeneity of the ORs across strata and populations were assessed using likelihood ratio tests. Modification of the ORs by disease aggressiveness and family history was assessed using a case-only analysis. Modification of the ORs by age was examined using a case-only analysis assessing the association between age and SNP genotype in the cases using polytomous regression. The associations between SNP genotypes and PSA levels were assessed using linear regression, after log transformation of PSA levels to correct for the positively skewed distribution of PSA levels (ng/ml). Analyses were performed in R, principally using GenABEL37, SNPTEST, ProbABEL33 and Stata.
The contribution of the known SNPs to the familial risk of prostate cancer, under a multiplicative model, was computed using the formula
where λ0 is the observed familial risk to first-degree relatives of prostate cancer cases, assumed to be 2, and λk is the familial relative risk due to locus k, given by
where pk is the frequency of the risk allele for locus k, qk = 1 − pk, and rk is the estimated per-allele OR.
Inflation
We estimated the inflation for each analysis on the basis of the 45th percentile of the test statistic for SNPs not selected by PRACTICAL and not in the COMMON fine-mapping regions. The inflation was 1.136 for the subgroup of European ancestry and 1.001 for the subgroup of African-American ancestry. Inflation was converted to an equivalent inflation for a study with 1,000 cases and 1,000 controls (λ1,000) by adjusting for effective study size, namely
where nk and mk were the number of cases and controls, respectively, for study k.
Estimation of the number of associated loci
To estimate the total number of newly associated loci selected for the iCOGS replication stage, we identified a set of 22,662 SNPs selected for replication of the GWAS and not selected for fine mapping to exclude previously known loci that were uncorrelated (r2 < 0.1 for any pair). We then determined the number of loci for which the estimated effect sizes in the iCOGS replication were in the same direction as in the combined GWAS or in the opposite direction. Similar results were obtained using cutoffs of r2 < 0.05 and r2 < 0.2.
Pathway analysis
GeneGo pathway enrichment was used to determine whether any canonical pathway was significantly enriched with false discovery rate < 0.05.
Supplementary Material
Acknowledgments
Acknowledgments are detailed in the Supplementary Note.
Footnotes
Note: Supplementary information is available in the online version of the paper.
AUTHOR CONTRIBUTIONS
R.A.E. and D.F.E. designed the study. R.A.E. is principal investigator of PRACTICAL. D.F.E. is Scientific Director of the COGS initiative. Z.K.-J. is co-investigator of PRACTICAL. R.A.E., D.F.E., Z.K.-J. and A.A.A.O. wrote the manuscript; the following named coauthors commented on the manuscript. A.A.A.O. and D.F.E. performed the statistical analyses; S.B. collated the data set. J.D. managed the database. Z.K.-J., E.J. Saunders, D.A.L. and M.T. coordinated sample collation and quality control for iCOGS PRACTICAL genotyping. S.J.-L. carried out pathway analysis and constructed regional plots, and T. Dadaev, K.G., M. Guy, R.A.W., E.J. Sawyer and A.M. managed the UKGPCS database and manifests for genotyping. C.L., A.M.D., C.B., D. Conroy, M.J.M., S.A., E.D., A. Lee, D.C.T., F.B. and D.V. carried out iCOGS PRACTICAL genotyping and set quality control standards. M. Ghoussaini selected the iCOGS PRACTICAL SNPs for fine-scale mapping. K.M. and A. Lophatananon collected some of the UKGPCS samples and controls. F.C.H., D.E.N. and J.L.D. are joint principal investigators of ProtecT. B.E.H. and L.L.M. are principal investigators of MEC; C.A.H. and F.S. are co-investigators. S.I.B. and D.A. are principal investigators of the PLCO study; G.A. is the principal investigator for the St. Louis screening center for PLCO; and S.J.C. and M.Y. led the genotyping for PLCO. S.G., R.B.H. and W.R.D. provided samples for PLCO. D.J.H. directs and P. Kraft coordinates data collection and management/analysis for HPFS. M.W. is the principal investigator of CPCS1 and CPCS2. B.G.N., S.F.N. S.E.B., P. Klarskov and M.A.R. have collected samples and data, and contributed to genotyping in this study. J.L.S. is principal investigator of the Fred Hutchinson–based study; E.A.O. collaborated on the study; L.M.F. and S.K. coordinated data collation; and E.M.K. and D.M.K. coordinated the preparation of samples. L.C.-A. is principal investigator of the Utah study; C.T. is the analyst; and R.A.S. is the surgeon. S.L. is a co-investigator of the BPC3 Consortium. H.G. is principal investigator of the CAPS and STHM1 study; J.A., M.A., F.W., S.L.Z. and J.X. have contributed to sample collection, clinical data retrieval, analyses and molecular work. S.A.I. is principal investigator of the USC study, and E.M.J. is principal investigator of SFPCS; M.C. Stem and R.C. led the genotyping of both studies. A.D.J. and A. Shahabi were both involved in genotype data production for the USC and SFPCS studies. A.S.K. is principal investigator of WUGS. B.D. and G.C. collected and collated clinical data and performed sample selection. M.R.T. is the principal investigator of the IPO-Porto study; S.M. and P.P. collected familial and molecular data on cases. L.B.S. and W.J.B. are the principal investigators of SCCS; L.B.S., W.J.B., W.Z. and Q.C. were responsible for the original collection of the samples. W.Z. and Q.C. coordinated sample retrieval, DNA extraction and genotyping. L.B.S. oversaw the assembly of the phenotype data. J.B. and J.A.C. are principal investigators of the Queensland study with input from A.B.S., F.L. and S.S. coordinated the data collation. K.A.C. and E.L. provided imputed data for genotyping in carriers of the mutation encoding the p.Gly84Glu alteration in the HOXB13 region. G.G.G., J.L.H., D.R.E. and G.S. are principal investigators of the Australian studies; M.C. Southey manages the molecular work. J.S. is principal investigator of the Tampere study; T.W. collected and collated clinical data and performed sample selection. T.L.J.T. coordinated sample collection. H.B. is principal investigator of the ESTHER study; D.R. and C.S. contributed to design and data collection; and H.M. is study coordinator. J.Y.P. is principal investigator of the Moffitt study; T.A.S. and H.-Y.L. are contributors to this study. R. Kaneva is principal investigator of the PCMUS study; C.S. provided the samples in the PCMUS study; V.M. oversaw the data collation. C.C. and J.L. are principal investigators of the Poland study; C.C. and D.W. collated the samples. C.M. and W.V. are principal investigators of the Ulm study; A.E.R. identified and collected clinical material, processed samples, undertook genotyping and/or collated data. E.R. is principal investigator of EPIC; F.C., R. Kaaks and D. Campa are investigators in Germany. T.J.K. is principal investigator of the EPIC-Oxford cohort and collected clinical material. R.C.T. collated data. K.-T.K. is principal investigator of the EPIC-Norfolk study. S.N.T. and D.S. are principal investigators of the Mayo Clinic study; S.K.M. coordinated data collation. M.M.G. provided samples for the ACS study. P.D.P.P. and N.P. provided samples for the East Anglia SEARCH study. C.S.C. gave advice about results and contributed to the manuscript. A.C.A. undertook risk prediction analysis for clinical application. D.P.D., A.H., R.A.H., V.S.K., C.C.P., N.J.V.A., C.J.W., A.T., T. Dudderidge, C.O., A.A., A.C., J.V. and A. Siddiq identified and collected clinical material. Other members of the UK Genetic Prostate Cancer Study Collaborators/British Association of Urological Surgeons’ Section of Oncology, the UK ProtecT Study Collaborators and the PRACTICAL Consortium (membership lists provided in the Supplementary Note) collected clinical samples, assisted in genotyping and provided data management. Members of the COGS–Cancer Research UK GWAS–ELLIPSE (part of GAME-ON) Initiatives, the Australian Prostate Cancer Bioresource, the UK Genetic Prostate Cancer Study Collaborators/British Association of Urological Surgeons’ Section of Oncology, the UK ProtecT Study Collaborators, the PRACTICAL Consortium and CSC collected clinical samples and/or assisted in genotyping and/or provided data management and/or discussion of the data.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Goh CL, et al. Genetic variants associated with predisposition to prostate cancer and potential clinical implications. J Intern Med. 2012;271:353–365. doi: 10.1111/j.1365-2796.2012.02511.x. [DOI] [PubMed] [Google Scholar]
- 2.Akamatsu S, et al. Common variants at 11q12, 10q26 and 3p11.2 are associated with prostate cancer susceptibility in Japanese. Nat Genet. 2012;44:426–429. doi: 10.1038/ng.1104. [DOI] [PubMed] [Google Scholar]
- 3.Gudmundsson J, et al. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat Genet. 2009;41:1122–1126. doi: 10.1038/ng.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu J, et al. Genome-wide association study in Chinese men identifies two new prostate cancer risk loci at 9q31.2 and 19q13.4. Nat Genet. 2012;44:1231–1235. doi: 10.1038/ng.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amin Al Olama A, et al. A meta-analysis of genome-wide association studies to identify prostate cancer susceptibility loci associated with aggressive and non-aggressive disease. Hum Mol Genet. 2013;22:408–415. doi: 10.1093/hmg/dds425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gudmundsson J, et al. A study based on whole-genome sequencing yields a rare variant at 8q24 associated with prostate cancer. Nat Genet. 2012;44:1326–1329. doi: 10.1038/ng.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park JH, et al. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat Genet. 2010;42:570–575. doi: 10.1038/ng.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kote-Jarai Z, et al. Identification of a novel prostate cancer susceptibility variant in the KLK3 gene transcript. Hum Genet. 2011;129:687–694. doi: 10.1007/s00439-011-0981-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kote-Jarai Z, et al. Multiple novel prostate cancer predisposition loci confirmed by an international study: the PRACTICAL Consortium. Cancer Epidemiol Biomarkers Prev. 2008;17:2052–2061. doi: 10.1158/1055-9965.EPI-08-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koutros S, et al. Pooled analysis of phosphatidylinositol 3-kinase pathway variants and risk of prostate cancer. Cancer Res. 2010;70:2389–2396. doi: 10.1158/0008-5472.CAN-09-3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun T, et al. Single-nucleotide polymorphisms in p53 pathway and aggressiveness of prostate cancer in a Caucasian population. Clin Cancer Res. 2010;16:5244–5251. doi: 10.1158/1078-0432.CCR-10-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wynendaele J, et al. An illegitimate microRNA target site within the 3′ UTR of MDM4 affects ovarian cancer progression and chemosensitivity. Cancer Res. 2010;70:9641–9649. doi: 10.1158/0008-5472.CAN-10-0527. [DOI] [PubMed] [Google Scholar]
- 13.ENCODE Project Consortium. A user’s guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Closas M, et al. Genome-wide association studies identify four ER negative–specific breast cancer risk loci. Nat Genet. 2013 Mar 27; doi: 10.1038/ng.2561. published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Couch FJ, et al. Genome-wide association study in BRCA1 mutation carriers identifies novel loci associated with breast and ovarian cancer risk. PLoS Genet. 2013;9:e1003212. doi: 10.1371/journal.pgen.1003212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Volinia S, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szarvas T, et al. Elevated serum matrix metalloproteinase 7 levels predict poor prognosis after radical prostatectomy. Int J Cancer. 2011;128:1486–1492. doi: 10.1002/ijc.25454. [DOI] [PubMed] [Google Scholar]
- 19.Richards TJ, et al. Allele-specific transactivation of matrix metalloproteinase 7 by FOXA2 and correlation with plasma levels in idiopathic pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. 2012;302:L746–L754. doi: 10.1152/ajplung.00319.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas G, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat Genet. 2009;41:579–584. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michailidou K, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013 Mar 27; doi: 10.1038/ng.2563. published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haiman CA, et al. Genome-wide association study of prostate cancer in men of African ancestry identifies a susceptibility locus at 17q21. Nat Genet. 2011;43:570–573. doi: 10.1038/ng.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Callen DF, et al. Co-expression of the androgen receptor and the transcription factor ZNF652 is related to prostate cancer outcome. Oncol Rep. 2010;23:1045–1052. doi: 10.3892/or_00000731. [DOI] [PubMed] [Google Scholar]
- 24.Norris JD, et al. The homeodomain protein HOXB13 regulates the cellular response to androgens. Mol Cell. 2009;36:405–416. doi: 10.1016/j.molcel.2009.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ewing CM, et al. Germline mutations in HOXB13 and prostate-cancer risk. N Engl J Med. 2012;366:141–149. doi: 10.1056/NEJMoa1110000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edwards S, et al. Expression analysis onto microarrays of randomly selected cDNA clones highlights HOXB13 as a marker of human prostate cancer. Br J Cancer. 2005;92:376–381. doi: 10.1038/sj.bjc.6602261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barbieri CE, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Breast Cancer Linkage Consortium. Cancer risks in BRCA2 mutation carriers. The Breast Cancer Linkage Consortium. J Natl Cancer Inst. 1999;91:1310–1316. doi: 10.1093/jnci/91.15.1310. [DOI] [PubMed] [Google Scholar]
- 29.Macinnis RJ, et al. A risk prediction algorithm based on family history and common genetic variants: application to prostate cancer with potential clinical impact. Genet Epidemiol. 2011;35:549–556. doi: 10.1002/gepi.20605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schumacher FR, et al. Genome-wide association study identifies new prostate cancer susceptibility loci. Hum Mol Genet. 2011;20:3867–3875. doi: 10.1093/hmg/ddr295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marchini J, et al. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 32.Eeles RA, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 33.Aulchenko YS, Struchalin MV, van Duijn CM. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics. 2010;11:134. doi: 10.1186/1471-2105-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teo YY, et al. A genotype calling algorithm for the Illumina BeadArray platform. Bioinformatics. 2007;23:2741–2746. doi: 10.1093/bioinformatics/btm443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giannoulatou E, et al. GenoSNP: a variational Bayes within-sample SNP genotyping algorithm that does not require a reference population. Bioinformatics. 2008;24:2209–2214. doi: 10.1093/bioinformatics/btn386. [DOI] [PubMed] [Google Scholar]
- 36.Haldane JB, Slater E. Assortative mating. Eugen Rev. 1946;38:103. [PMC free article] [PubMed] [Google Scholar]
- 37.Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.