The cohesin loading protein is required for nonhomologous end joining of double-strand DNA breaks
Abstract
DNA double strand breaks (DSBs) are mainly repaired via homologous recombination (HR) or nonhomologous end joining (NHEJ). These breaks pose severe threats to genome integrity but can also be necessary intermediates of normal cellular processes such as immunoglobulin class switch recombination (CSR). During CSR, DSBs are produced in the G1 phase of the cell cycle and are repaired by the classical NHEJ machinery. By studying B lymphocytes derived from patients with Cornelia de Lange Syndrome, we observed a strong correlation between heterozygous loss-of-function mutations in the gene encoding the cohesin loading protein NIPBL and a shift toward the use of an alternative, microhomology-based end joining during CSR. Furthermore, the early recruitment of 53BP1 to DSBs was reduced in the NIPBL-deficient patient cells. Association of NIPBL deficiency and impaired NHEJ was also observed in a plasmid-based end-joining assay and a yeast model system. Our results suggest that NIPBL plays an important and evolutionarily conserved role in NHEJ, in addition to its canonical function in sister chromatid cohesion and its recently suggested function in HR.
DNA double strand breaks (DSBs) pose a severe threat to genome integrity, but can also be a necessary part of normal cellular processes, such as meiosis and Ig class switch recombination (CSR). Depending on cell cycle phase and DSB structure, different strategies are used for repair. Homologous recombination (HR) depends on a homologous DNA template for repair, preferentially the identical sister chromatid, and is therefore mainly active during the S and G2 phases. Nonhomologous end joining (NHEJ), however, is active throughout the cell cycle and is the principal pathway during the G1 phase, when there is no immediate close template for homologous repair. The classical NHEJ pathway requires not only the key components of the NHEJ machinery, i.e., Ku70/Ku80, DNA-PKcs, Artemis, XLF (Cernunnos), XRCC4, and DNA ligase IV, but also several DNA damage sensors or adaptors, such as ATM, γH2AX, 53BP1, MDC1, RNF168, and the Mre11–Rad50–NBS1 complex.
Cohesin is an evolutionarily conserved multisubunit complex consisting of a heterodimer of two structural maintenance of chromosomes (SMC) proteins, SMC1A and SMC3, one kleisin protein RAD21 (MCD1 or SCC1) and SA (STAG1/2 or SCC3). The SMC proteins fold back on themselves in the hinge region to form long antiparallel coiled-coil arms, with the amino and carboxyl termini coming together to create head domains that contain ATPases. RAD21 bridges the two head domains to facilitate the formation of the proposed ring-like structure of the complex, and it also interacts with the SA subunit. The cohesin complex ensures correct chromosome segregation through cohesion between sister chromatids (Nasmyth and Haering, 2009). In addition to this canonical role, cohesin and its loading complex NIPBL/MAU2 have also been suggested to be important for regulation of gene expression and repair of DSBs through HR, presumably by facilitating proximity between the broken DNA ends and the repair template (Sjögren and Nasmyth, 2001; Vrouwe et al., 2007; Nasmyth and Haering, 2009). Smc1, the yeast SMC1A orthologue, has furthermore been suggested to coordinate the HR and NHEJ processes (Schär et al., 2004).
Cornelia de Lange syndrome (CdLS) is a developmental disorder characterized by growth retardation, severe intellectual disability, gastrointestinal abnormalities, malformations, of the upper limbs and characteristic facial dysmorphisms. Heterozygous loss-of-function mutations in NIPBL, encoding the cohesin loader NIPBL, are the major cause of CdLS (Liu and Baynam, 2010). In addition, mutations in the SMC1A, SMC3, PDS5B, RAD21, and HDAC8 encoding genes, all being part of the cohesion pathway, have been found in selected CdLS patients. The multisystem dysfunctions connected to the syndrome implicate defective gene regulation during fetal development and current evidence suggests that CdLS may be caused by alterations in cohesin chromatin-binding dynamics (Liu et al., 2009). In addition, cell lines established from CdLS patients have an increased sensitivity to DNA damage that has been suggested to be caused by defective HR-mediated repair (Vrouwe et al., 2007).
Here, we show an increased DNA damage sensitivity, especially after exposure to γ-rays, in B-lymphoblastoid (LCLs) and fibroblast cell lines (FBs) from NIPBL-deficient CdLS patients. However, we also observed that the majority of the patient and control cells studied were in the G1 phase of the cell cycle, where NHEJ is the principle DSB repair mechanism. We therefore investigated whether defective NHEJ could underlie the DNA damage sensitivity observed in the patient cells.
RESULTS AND DISCUSSION
Increased DNA damage sensitivity in CdLS cell lines
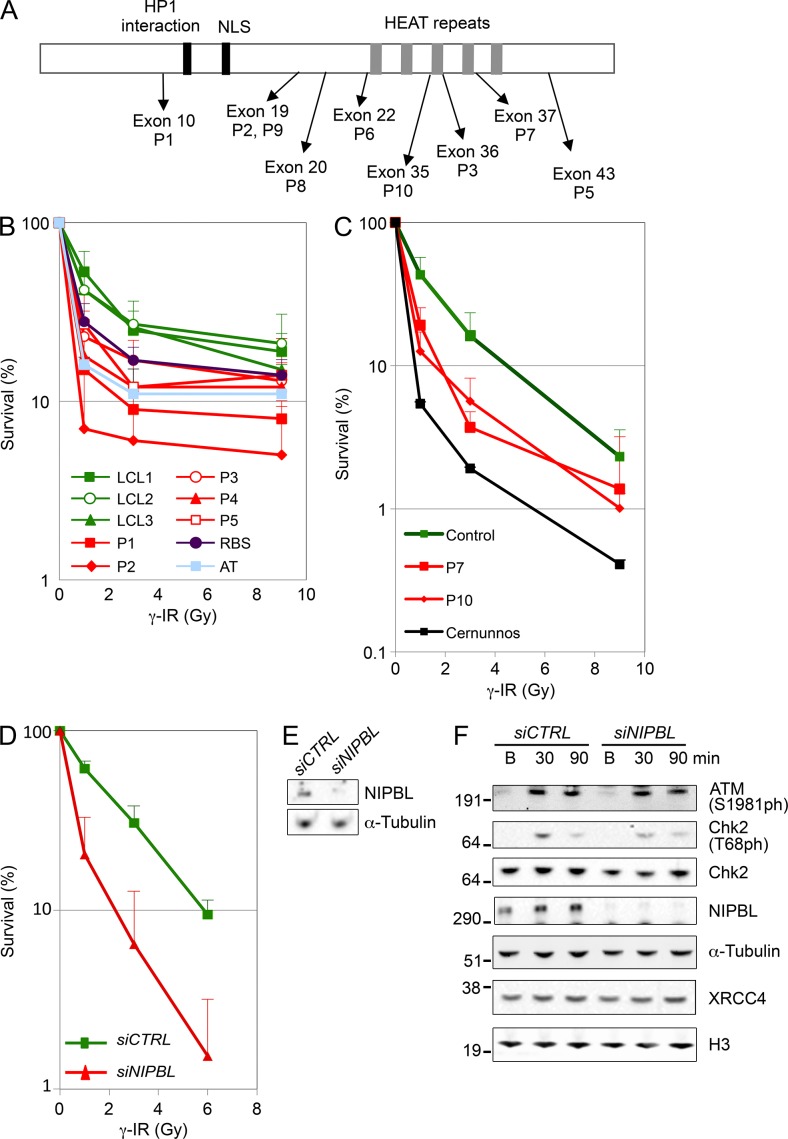
DNA damage sensitivity assays were performed using LCLs and FBs from 7 CdLS patients (P1-P3, P5, P7, P10 had defined NIPBL mutations, whereas P4 had no coding region mutation in NIPBL, SMC1A, SMC3 (Schoumans et al., 2007), MAU2, or PDS5B (Fig. 1 A and Table 1). For comparison, LCLs or FBs from healthy individuals, radiation-sensitive patients (ATM- or Cernunnos-deficient), and a Roberts syndrome (RBS) patient were also evaluated. RBS is caused by mutations in the gene encoding ESCO2, which is responsible for establishment of cohesion.
Figure 1.
NIPBL-deficient cells display increased DNA damage sensitivity. (A) Schematic representation of NIPBL (not to scale) with approximate localization of conserved motifs, and relative positioning of mutations identified in the CdLS patients included in this study. (B) LCLs from healthy controls and CdLS patients (P1-P3 and P5 had defined NIPBL mutations), as well as LCLs from patients deficient for ESCO2 (RBS) or ATM (AT) were exposed to γ-IR at indicated dosages, and survival was monitored after three population doublings using the MTS assay. Doubling times and significant differences in survival are indicated in Table 1. (C) FBs from patients deficient in NIPBL (P7 and P10), Cernunnos, or control FBs were exposed to γ-IR at indicated dosages and analyzed for survival by the colony formation assay. (D) Control FBs were transfected with control (siCTRL) or NIPBL siRNA (siNIPBL) and exposed to γ-IR at indicated dosages and analyzed for survival by the colony formation assay. (B–D) An average from ≥3 experiments for each cell type is shown, and error bars indicate SD. (E) Protein extracts were isolated from control FBs 48 h after transfection with siCTRL or siNIPBL, but before exposure to γ-IR for the colony formation assay shown in D, and were run on SDS gels. (F) Control FBs were transfected with siCTRL or siNIPBL, and 48 h after transfection and at indicated time points after exposure to γ-IR, protein extracts were isolated and run on SDS gels. (E and F) Indicated proteins were detected by Western blotting. Representative gels from three independent experiments are shown.
Table 1.
Patient and mutation information
| Age (years) | NIPBL mutationa | Doubling timeb | NIPBL RNA/Protein % of control | γ-IRc | Increased microhomology | |
| P1 | 3 | c.2494C>T; p.Arg832X | 46 h | 75/50 | 1 Gy**** 3 Gy**** 9 Gy*** | YES |
| P2 | 13 | c.4320+2T>A; p.Val1441_Lys1520del | 75 h | 60/50 | 1 Gy**** 3 Gy*** 9 Gy** | YES |
| P3 | 17 | c.6250_6255del; p.Val2084_Val2085del | 44 h | 90/60 | 1 Gy** 3 Gy* 9 Gy n.s. | YES |
| P4 | 14 | ND | 50 h | 60/50 | 1 Gy**** 3 Gy**** 9 Gy n.s. | NA |
| P5 | 13 | c.7306G>A; p.Ala2436Thr | 42 h | 150/70 | 1 Gy* 3 Gy*** 9 Gy n.s. | NA |
| P6 | 2 | c.4593T>A; p.Tyr1531X | 42 h | NA | NA | YES |
| P7 | 1 | c.6436A>C; p.Thr2146Pro | NA | NA/50 | 1 Gy* 3 Gy* 9 Gy n.s. | YES Plasmid |
| P8 | 4 | c.4321G>T; p.Val1441Leu | NA | NA | NA | YES |
| P9 | 1 | c.4320+5G>C; p.Val1414_Ala1440del | NA | NA | NA | YES |
| P10 | 10 | c.6242G>C; p.Gly2081Ala | NA | NA/40 | 1 Gy* 3 Gy n.s. 9 Gy n.s. | YES Plasmid |
All mutations described are de novo. ND, not detected.
Doubling times for control cell lines (LCL1-3) were between 23 to 32 h. NA, not analyzed.
Statistical comparison of γ-IR sensitivity was performed between P1-P5 and LCL2 and between P7 and P10 and control FB, *, P < 0.05, **, P < 0.01, ***, P < 0.005, ****, P < 0.001 (Student’s t test). n.s., not significant.
LCLs from patients (P1-P5) and controls (LCL1-LCL3) were exposed to increasing dosages of γ-irradiation (γ-IR). After the time required for three population doublings in the absence of DNA damage, the relative number of viable cells was determined using a MTS proliferation assay. Low doses of γ-IR caused a significantly reduced survival in all CdLS LCLs tested, compared with controls (Fig. 1 B and Table 1). The radiation sensitivity was confirmed by a colony formation assay in FBs from P7 and P10 after exposure to γ-IR (Fig. 1 C). We also treated the control FBs with NIPBL siRNA. This typically resulted in >70% reduction of the NIPBL protein levels (Fig. 1 E) and caused a significant increase in sensitivity to γ-IR as analyzed by the colony formation assay (Fig. 1 D). The general DNA damage response, however, can be activated properly in NIPBL knockdown cells, as measured by the level of phosphorylated ATM and Chk2 (Fig. 1 F). The P1-P5 cells were also found to be sensitive to the interstrand cross-linking agent mitomycin C and the topoisomerase II inhibitor etoposide to various degrees (unpublished data). Collectively, there was an increased DNA damage sensitivity in CdLS cells, especially to agents inducing DSBs.
Proper S-phase cohesion has been shown to be essential for HR-mediated repair of DSBs during G2 in yeast (Sjögren and Nasmyth, 2001). One explanation for the DNA damage sensitivity in cells from NIPBL-deficient CdLS patients could be defective S-phase established cohesion caused by insufficient cohesin loading. Although precocious sister chromatid separation (PSCS) has been shown in CdLS cells in one study (Kaur et al., 2005), metaphase spreads of the CdLS cell lines included in this study did not, in line with other studies (Vrouwe et al., 2007; Liu and Baynam, 2010), show any increased PSCS (unpublished data). Thus, the increased DNA damage sensitivity in CdLS cells is probably independent of the canonical role of the cohesin complex in cohesion.
DSB repair via NHEJ is impaired in NIPBL-deficient cells
When determining the cell cycle distribution, we found that the majority (62–88%) of control and NIPBL-deficient CdLS patient cells were in the G1 phase of the cell cycle at the time of γ-IR exposure (unpublished data). This raised the possibility that NIPBL may not only be important for HR, but also for NHEJ, as the latter is the predominant pathway used for repair of DSBs during G1. To further understand how DSB end joining is performed in CdLS cells and to investigate whether NIPBL is important for NHEJ, we used three experimental systems outlined below.
Altered pattern of switch recombination junctions in NIPBL-deficient B cells
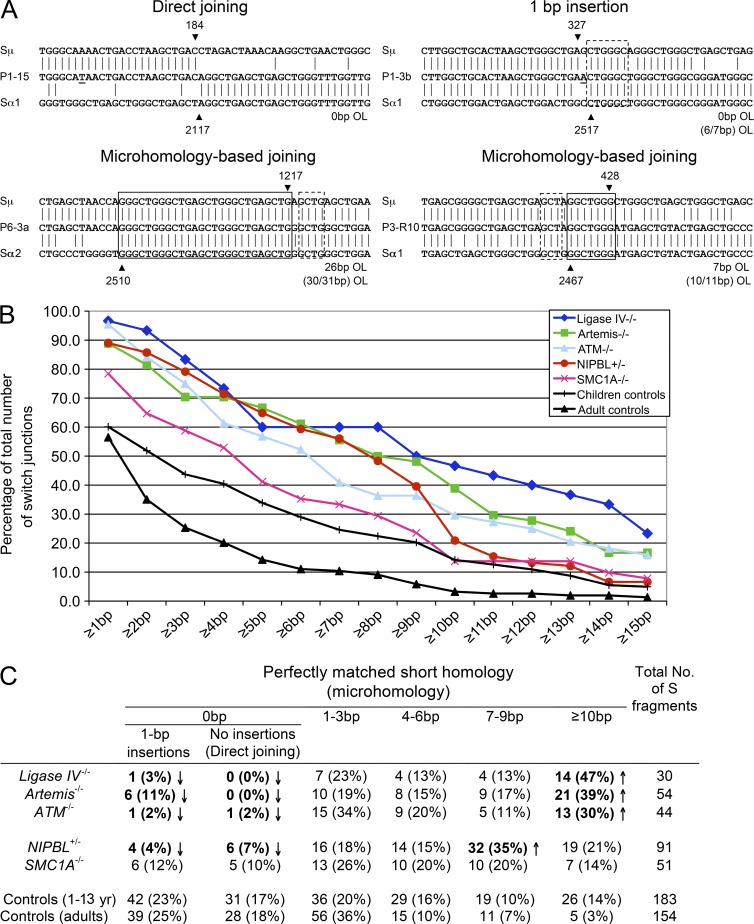
CSR is a physiological process that is initiated by a B cell–specific factor, activation-induced cytidine deaminase (AID), through DNA deamination, but relies on ubiquitously expressed DNA repair factors for subsequent DSB formation (mismatch and base excision repair factors) and joining of the broken DNA ends (NHEJ factors; Stavnezer et al., 2010). During CSR, DSBs are generated in the donor and acceptor switch (S) regions. These breaks are processed and repaired during the G1 phase of cell cycle, leading to recombination of the two S regions (Sharbeen et al., 2012). The nature of the recombination junctions reflect the DNA repair pathway used, and defects in the NHEJ machinery have previously been associated with characteristic abnormalities (Pan-Hammarström et al., 2005; Yan et al., 2007). We therefore sequenced CSR junctions from in vivo–switched B cells from 7 CdLS patients with NIPBL mutations (P1, P2, P3, P6, P8, P9, and P10, 1–17 yr old) and 6 age-matched controls (1–13 yr old) and compared them to those previously described in DNA ligase IV, Artemis, and ATM-deficient patients (Pan-Hammarström et al., 2005; Du et al., 2008). Altogether, 92 unique CSR junctions (91 Sμ–Sα and 1 Sμ–Sγ–Sα) from NIPBL-deficient patient B cells, representing independent CSR events, were PCR amplified and sequenced. 46 unique CSR junctions were also obtained from the controls, and this set of data was merged with our previously described data from control children (1–6 yr; 137 junctions; Du et al., 2008) as the two sets of controls were largely similar. As one CdLS patient with NIPBL mutation (P3) was 17 yr old at the time of sampling, and we have previously shown that there are some differences in the CSR pattern between adult and child controls, we also included our previously described adult controls (Du et al., 2008) for comparison (Fig. 2, B and C). There was a significantly reduced proportion of Sμ-Sα junctions in NIPBL-deficient B cells with direct end joining, i.e., no microhomology (MH; perfectly matched, short donor–acceptor sequence homology) and no insertions (nontemplate nucleotide additions) at the junctions (Fig. 2, B and C). The number of junctions with 1-bp insertions was also significantly reduced (Fig. 2, B and C). Conversely, a significantly increased proportion of the Sμ-Sα junctions from NIPBL-deficient patient B cells displayed MH of at least 1 bp (89 vs. 60 or 57% in children or adult control group, respectively; χ2 test; P < 0.001). The mean length of MH was dramatically increased in NIPBL-deficient patients (7.0 ± 5.1 bp) as compared with 3.9 ± 4.9 bp in children controls and 1.8 ± 3.2 bp in adult controls (Student’s t test, P = 4.7 × 10−6 or P = 8 × 10−15). The pattern of in vivo–generated CSR junctions from NIPBL-deficient patients thus largely resembles that observed in patients with NHEJ defects (DNA ligase IV or Artemis deficiency) or ATM deficiency, which are characterized by a lack of “direct end joining” and a shift toward MH usage (Fig. 2, B and C).
Figure 2.
Increased usage of microhomologies at Sμ–Sα junctions in NIPBL-deficient cells. (A) Sμ and Sα recombination junctions from patient and control B cells were amplified via nested PCR and sequenced. Four sequences of recombination junctions from NIPBL-deficient patients are shown, representing different types of joining, including direct joining, 1-bp insertion, and microhomology (MH)-based joining. The Sμ and Sα reference sequences are aligned above and below the recombination junction sequences. MHs and imperfect repeats are indicated by boxes (solid and dotted lines, respectively). OL, overlap. The Sμ and Sα breakpoints for each switch fragment are indicated by black arrowheads, and their positions in the reference sequences are indicated above or below the arrowheads. (B) The accumulated percentages of switch junctions with given lengths of MH are plotted. (C) The age-matched control data (1–13 yr; 44 junctions) derived from the present study were merged with the previously published children control data (1–6 yr; 137 junctions; Du et al., 2008). The CSR junctions from the adult controls were described previously (Du et al., 2008). Significant differences (as compared with both children and adult control groups) are highlighted with bold letters and indicated by arrows (χ2 test, P < 0.05). (A–C) DNA from each patient was tested in 10 separate PCR reactions for each experiment, and the experiment was repeated at least once.
We also observed an unusual sequence of an amplified Sμ-Sα fragment in NIPBL-deficient patient’s cells, with two large pieces of Sγ4 regions inserted between the Sμ and Sα regions, one of which is inverted (Sμ-Sγ4-Sγ4rev-Sα2). This type of junction, with inverted Sγ regions inserted at the breakpoints, has not been observed in any of the CSR junctions derived from control B cells.
We furthermore characterized 51 Sμ-Sα junctions from 4 CdLS patients with X-linked SMC1A mutations (S1-S4). These junctions showed a similar trend, albeit less dramatic, in shifting to MH-based end joining (Fig. 2, B and C).
Descriptions of the immunological phenotype of CdLS patients are scarce in the literature. No overt clinical manifestations of immunodeficiency, characterized by recurrent, severe, or unusual infections, were observed in any of the CdLS patients included in the study. The total Ig levels were largely normal in three patients where serum samples were available (P8, P9 and P10), although two patients (P8 and P9) had reduced or borderline low level of specific antibodies (IgG isotype) against Haemophilus influenzae, which may indicate a subtle defect in CSR. We subsequently reviewed three additional CdLS patients with defined NIPBL mutations from whom cryopreserved peripheral blood cells were available. Two patients (P12 and P13) showed a reduced or borderline percentage of class-switched CD19+CD27+IgM−IgD− memory B cells. Moreover, B cells from P11 and P12 showed a normal cell proliferation rate but a reduced ability to switch to IgE in response to cytokines in an in vitro CSR assay (unpublished data).
Collectively, deficiency of the cohesin loader NIBPL results in a moderately reduced efficiency of CSR but, more importantly, an abnormal pattern of recombination junctions. The skewing toward the MH-based alternative end joining (A-EJ) pathway implicates that the classical NHEJ is compromised in NIPBL-deficient cells.
Increased MH-based joining of blunt ends in NIPBL-deficient cells in an in vitro plasmid–based assay
We next used a previously described assay where linearized plasmids with defined blunt DNA ends were transiently transfected into FBs (Verkaik et al., 2002). The contribution of direct joining, where two blunt DNA ends are joined precisely, and 6-bp MH-based end joining can be estimated by restriction enzyme digestion or, more accurately, determined by direct sequencing. FBs from patients with deficiency of NIPBL (P7, P10) or Artemis and one unaffected individual were studied by this assay, and previously published data from Cernunnos-deficient cells were included for comparison (Du et al., 2012). As shown in Table 2, the proportion of direct joining was dramatically reduced in Cernunnos-deficient cells (4.4 vs. 73.0% in controls) and to a lesser, but significant degree, in NIPBL-deficient cells (45.8%, χ2 test; P = 6.10−6). Conversely, the proportion of 6-bp, MH-mediated end joining was significantly increased in Cernunnos- and NIPBL-deficient cells when compared with the controls (Table 2). A similar trend was observed in Artemis-deficient cells, although not to a significant level, which is consistent with a previous study using a similar construct (Noordzij et al., 2003). Deletions (nucleotide loss at the DNA ends), caused by resection of the DNA ends, could also be detected in ∼10–30% of recombination junctions in cells from controls and different groups of patients. Notably, there was a significant increase of junctions with deletions, especially deletions that are associated with short MH (typically 1–4 bp) in NIPBL-deficient cells (Table 2), suggesting that an alternative end joining pathway, involving DNA end resection and short sequence homologies is more active when the function of NIPBL is impaired. The plasmid-based assay was subsequently performed in G1-arrested FB cells from P7, and a similar end-joining defect was observed (Table 2).
Table 2.
Reduced direct joining and increased use of microhomology in a plasmid-based end-joining assay in NIPBL-deficient cellsa
| Direct joining | 6-bp MH | Deletion + MH | Deletion only | Insertion | Total no. of junctions | |
| % | % | % | % | % | ||
| Controlb | 73.0 | 6.3 | 7.9 | 11.9 | 0.8 | 126 |
| Artemis−/− p with control | 59.5 0.099 | 11.9 0.243 | 14.3 0.225 | 11.9 1.000 | 2.4 0.411 | 42 |
| Cernunnos−/− p with control | 4.4 7.5 × 10−20 | 80.9 3.8 × 10−26 | 14.7 0.139 | 0 3.1 × 10−3 | 0 0.461 | 68 |
| NIPBL+/−(P7+P10) p with control | 45.8 6.1 × 10−6 | 20.8 6.4 × 10−4 | 22.9 7.9 × 10−4 | 10.4 0.698 | 0 0.284 | 144 |
| NIPBL+/− (P7), G1 arrested p with control | 43.1 1.7 × 10−4 | 27.5 1.2 x10−4 | 21.6 0.011 | 7.8 0.429 | 0 0.523 | 51 |
| Control+ siCTRL p with control | 68.6 0.509 | 8.6 0.563 | 10.0 0.623 | 12.9 0.846 | 0 0.455 | 70 |
| Control + siNIPBL p with control p with siCTRL | 52.1 0.003 0.044 | 16.4 0.022 0.156 | 19.2 0.019 0.121 | 12.3 0.930 0.924 | 0 0.445 | 73 |
Linearized pDVG94 plasmid was transfected into FBs derived from NIPBL, Artemis and Cernunnos deficient patients and one control. At least two independent experiments are summarized.
Part of the control data (n = 106) as well as the junctions derived from Cernunnos deficient cells have been described previously (Du et al., 2012). The two sets of controls showed very similar pattern and were thus merged in the summary. Statistical analysis was performed using χ2 test.
To further investigate if the end-joining defect observed in the patient cell lines in this assay was specifically associated with NIPBL deficiency, we transfected the control FBs with siRNA against NIPBL 2 d before the transfection of linearized plasmids. As measured by quantitative real-time PCR assay, the expression of NIPBL was reduced by 20% and 65%, respectively after the treatment of scrambled control (siCTRL) or specific NIPBL siRNA (siNIPBL). As demonstrated in Table 2, the NIPBL knockdown cells showed a very similar end-joining pattern to that derived from the NIPBL-deficient patient cells, with significantly reduced direct joining and increased usage of 6-bp MH as well as deletions associated with short MH. Collectively, NIPBL deficiency appears to result in a reduced direct joining of blunt ends but an increased MH-based joining of resected DNA ends, in the G1 phase of the cell cycle in this plasmid-based assay.
Involvement in NHEJ is a conserved function of NIPBL
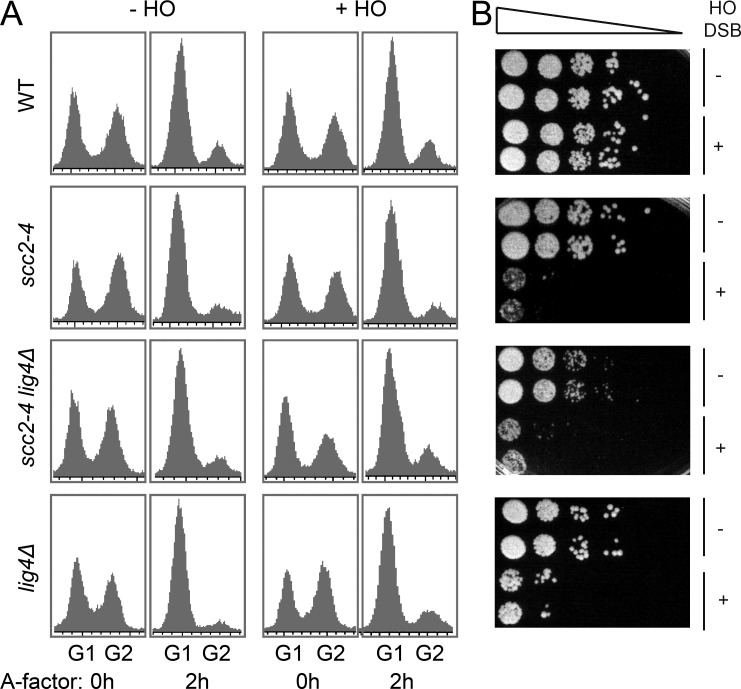
Lastly, we analyzed the importance of NIPBL for NHEJ using genetically modified Saccharomyces cerevisiae strains. The yeast NIPBL orthologue Scc2 is an essential protein, required for cell survival (Michaelis et al., 1997), thus we used a temperature-sensitive allele of SCC2 (scc2-4). Logarithmically growing wild-type, scc2-4, lig4Δ, or scc2-4 lig4Δ cells were arrested in G1 by addition of the A-factor pheromone (O’Reilly et al., 2012; Fig. 3 A). The temperature was then raised to the restrictive level for 30 min and the galactose-inducible HO endonuclease was activated by addition of galactose to create a sequence-specific DSB at the yeast MATα locus (Fig. 3 B). This DSB could be repaired solely via NHEJ, as the intrachromosomal regions normally used for its repair via HR were deleted and the DSB was induced on both sister chromatids (Moore and Haber, 1996). After 1 h of break induction, the cells were plated in serial dilutions for analysis of survival. This showed that Scc2 might be as important for NHEJ as DNA ligase IV (lig4Δ), a defined, conserved and essential NHEJ factor (Fig. 3 B). It is notable that scc2-4 alone and in combination with lig4Δ resulted in a slightly more severe defect than the lig4Δ alone, which could reflect that Scc2 has additional functions for DSB repair than DNA ligase IV. It is possible that although most of the cells are arrested in G1, there are some cells that still proceed into G2. Furthermore, occasionally (not measurable by pulse field gel electrophoresis), there might be a few cells with breaks only on one chromatid, and if these cells by chance enter into the G2 phase, they can theoretically still be repaired by HR. Thus, the slightly more severe defect observed in scc2-4 cells compared with the lig4Δ cells could be due to the fact that Scc2 is also required for HR, whereas DNA ligase IV only has a function in NHEJ.
Figure 3.
The involvement of NIPBL in NHEJ is evolutionarily conserved. (A and B) WT cells, cells expressing a temperature-sensitive allele of the yeast NIPBL orthologue SCC2 (scc2-4), and cells with a deletion of lig4Δ, either alone or in combination with the scc2-4, were arrested in G1 by addition of A-factor for 2 h and subsequently incubated at a restrictive temperature (32°C) for 30 min. A site-specific DSB was then induced by the galactose inducible HO enzyme for 60 min, as indicated, during continuous G1 arrest at a restrictive temperature. (A) The cell cycle distribution was determined by FACS analysis before addition of A-factor and on G1-arrested cells, 2 h after addition of A-factor. (B) After DSB induction, cells were plated in the absence of galactose at equal starting concentrations and in 6 10-fold subsequent dilution steps (indicated by triangle). Plates were left to recover for 72 h at 30°C (semipermissive temperature for scc2-4). One representative experiment from ≥3 is shown.
To further investigate the type of repair used in the surviving cells, a 600-bp region encompassing the HO cut site was amplified and subsequently sequenced in cells derived from single colonies. In the surviving WT cells, the HO-induced DSBs were, indeed, repaired by NHEJ with most of the recovered junctions having small deletions and insertions, as previously described (Moore and Haber, 1996; Fig. S1). However, most of the amplified sequences derived from the few colonies of surviving Scc2 or DNA ligase IV–inactivated cells are germline at the MATα locus (Fig. S1), suggesting that these surviving cells are probably those few cells that had no DSB induced at this locus. Altogether, our data clearly points to a role for NIPBL orthologues in the classical NHEJ.
Potential mechanisms for the involvement of NIPBL in NHEJ
By sequencing of Ig Sμ-Sα recombination joints, through analysis of DSB rejoining in a plasmid assay, and by a NHEJ assay in yeast, we demonstrated that NIPBL is important for repair of DSBs via the classical NHEJ pathway. NIPBL may, however, not be a component of the classical NHEJ machinery as such, which includes DNA-PKcs, Ku70/80, Cernunnos, Artemis, DNA ligase IV, and XRCC4, as deficiency of any of these factors will result in a more pronounced immunodeficiency syndrome due to a severe V(D)J recombination defect, another physiological process in lymphocytes that requires the classical NHEJ pathway (de Villartay, 2009). The CdLS patients with NIPBL mutations described here have no overt V(D)J recombination defect (unpublished data) but rather a CSR defect, which is more similar to patients with ATM- or NBS- deficiency as well as the 53BP1-, H2AX-, and MDC1- deficient mouse models (Stavnezer et al., 2010). Based on the current experimental evidence, partial loss of NIPBL is more likely to affect the regulation of the classical NHEJ pathway, or for the choice between the classical NHEJ and A-EJ.
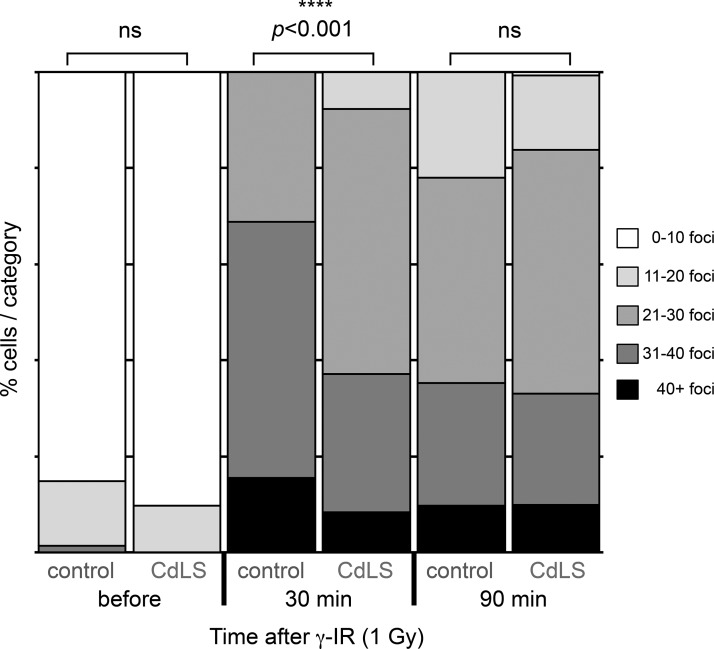
To date, no cohesin-independent function has been described for NIPBL. Thus, several mechanisms for the involvement of NIPBL in NHEJ, probably dependent on cohesin, can be proposed. RNAi-mediated inactivation of the cohesin component Rad21/SCC1 has been shown to result in defects in recruitment of the DNA damage response factor and mediator protein 53BP1 to DSBs (Watrin and Peters, 2009). Intriguingly, absence of 53BP1 has previously been shown to lead to increased resection around a DSB in the Ig S regions and a short-range repair via the MH-based A-EJ was consequently favored (Bothmer et al., 2010). As CdLS patients with deficient NIPBL have reduced levels of chromatin-bound cohesin (Liu et al., 2009), it is possible that 53BP1 recruitment is also reduced in patient cells and thus results in an increased rate of resection of DNA ends and higher degree of DSB repair via MH-mediated A-EJ. To test this hypothesis, we analyzed the 53BP1 foci formation in response to DSB in NIPBL-deficient LCLs. Control LCLs and cells from P1-P3 and P5 were exposed to γ-IR (1 Gy) and stained for 53BP1 at indicated time points (Fig. 4). In undamaged cells, the numbers of foci were low, with no apparent difference between control and patient cells. There was however a significant difference (Mann-Whitney U test, P = 6.85 × 10−8) in the number of foci formed 30 min after IR, between control LCLs and patient cells (Fig. 4). At a later time point (90 min), no significant difference was observed (Fig. 4). Thus, with an impaired function of NIPBL, the early recruitment of 53BP1 to DNA damage repair foci in response to DSBs was indeed reduced.
Figure 4.
53BP1 foci formation is reduced at an early time point in NIPBL-deficient LCLs. LCLs from healthy controls or LCLs deficient in NIPBL (P1-P3, P5) were exposed to γ-IR (1Gy) and harvested, cytospun, and fixed at the indicated time points. The cells were then stained with anti-53BP1 antibodies and 53BP1 foci were detected using a confocal microscope. Image stacks were exported to ImageJ and 53BP1 foci were quantified in ∼40 nuclei per LCL per time point in each experiment. The bars represent the relative distribution of 53BP1 foci at indicated time points before and after γ-IR. Two independent experiments were performed. Statistically significant differences are highlighted with stars (Mann-Whitney U test).
There is currently no patient with 53BP1 deficiency described; however, B cells from 53BP1−/− mice display greatly reduced CSR efficiency (Manis et al., 2004). Furthermore, similar to observations in NIPBL-deficient patient B cells, the pattern of CSR junctions in 53BP1−/− mouse B cells is also altered, with an increased usage of MH at the Sμ-Sα junctions (unpublished data), and unusually large insertions at the Sμ-Sγ junctions (Reina-San-Martin et al., 2007). It is also of note that recruitment of NIPBL to DSBs depends on MDC1 and RNF168, two DNA damage response factors that have previously been implicated in NHEJ during CSR (Oka et al., 2011). Interestingly, in RNF168-deficient patient B cells, where recruitment of 53BP1 to DSBs is reduced, a dramatically increased usage of MH was also observed at the Sμ-Sα junctions (Stewart et al., 2007). Thus, NIPBL may regulate NHEJ by recruiting 53BP1 to the DSB sites, but it is likely that interactions between NIPBL and the DNA damage response factors are complex and that factors in addition to 53PB1 may also influence the function of NIPBL in NHEJ.
Given that cohesin and NIPBL have been shown to regulate gene expression, an indirect effect caused by dysregulation of genes involved in the respective DSB repair pathway cannot be ruled out. We have thus performed real-time PCR assays on nonirradiated and γ-ray–irradiated NIPBL knockdown fibroblasts, but no significant changes in the expression of NHEJ genes (LIG4/DNA ligase IV, PRKDC/DNA-PKcs, and XRCC5/KU80) were observed (unpublished data). Furthermore, the expression at the protein level of another key NHEJ factor XRCC4 was not significantly affected in the NIPBL knockdown cells (Fig. 1 F). Cohesin has also been shown to co-localize with the CCCTC-binding factor (CTCF), both genome wide and in the Ig locus, and these interactions have been suggested to be important for CTCF insulator function and three-dimensional chromosomal interactions (Degner et al., 2011). Moreover, it was recently shown that cohesin influences the rearrangement of the T cell receptor α locus through histone methylation, recruitment of the Rag recombinase and long range enhancer/promoter interactions (Seitan et al., 2011). These types of interactions are known to be crucial for induction of germline transcripts encompassing the S regions, a requirement for accessibility of the Ig locus and thereby for CSR (Stavnezer et al., 2010). However, since defective germline transcription has no implication on the choice between classical NHEJ and MH-based A-EJ, it would be a less likely explanation for the defective NHEJ in NIPBL-deficient cells. Finally, although the expression level of NHEJ genes seems to be normal in NIPBL-deficient cells, it remains possible that NIPBL and/or cohesin are important for recruitment of AID, or AID-associated DSB repair factors, to the Ig locus. Failure in this recruitment might lead to the shift toward usage of the A-EJ pathway (Kracker et al., 2010). This possibility needs to be further investigated.
Our study suggests a role for NIPBL in NHEJ during CSR where the repetitive, intronic S regions are involved. Importantly, a similar function for NIPBL during DSB repair in other parts of the genome is plausible, which would have more severe effects, as these DSBs can also appear within gene-coding regions. Increased resection of DSBs stimulates the A-EJ pathway, which has been reported to augment the frequency of translocations between chromosomes (Yan et al., 2007). Thus, an impaired NHEJ may be the underlying reason for some of the developmental and neurological defects observed in these patients. It is also important to point out that CdLS causing NIPBL mutations are heterozygous loss-of-function mutations. A more severe NHEJ defect, which could lead to a more pronounced immunodeficiency, might be expected with a complete loss of the NIPBL protein. However, as knockout of NIPBL was reported to be incompatible with survival (Kawauchi et al., 2009), this question cannot be assessed. Further experiments are thus required to completely elucidate the functional role of NIPBL/cohesin in NHEJ.
MATERIALS AND METHODS
Patient and control materials.
Clinical phenotypes of CdLS patients included in this study have been described previously (Pié et al., 2010; Schoumans et al., 2007). LCLs were established via Epstein-Barr virus transformation. Cell lines (P1-P10) from patients with CdLS were compared with cell lines established from unaffected individuals, a patient diagnosed with Roberts syndrome, a Cernunnos−/− patient and an ataxia telangiectasia patient (AT153LA c.8977C>T). LCLs were grown in RPMI 1640 with 10% fetal calf serum, glutaMAX, sodium-pyruvate, penicillin (100 U/ml), and streptomycin (0.1 mg/ml). FBs (P7 and P10, one control, and Cernunnos−/−) were cultured in RPMI 1640/HAMS F-10 vol/vol 1:1 or DMEM supplemented as medium for LCLs (all from Invitrogen). Genomic DNA samples were prepared from peripheral blood from patients P1-P3, P6, P8, P9, and P10 with NIPBL mutations, and four patients with the following ages and mutations in SMC1A: S1, 2 yr, c.587G>A; S2, 5 yr, c. 3254A>G; S3, 8 mo, c.2455A>C; and S4, 9 yr, c.3568A>G. Peripheral lymphocytes were purified and stored using standard methods, from three additional patients with NIPBL mutations (P11, 1 yr, c.1982A>T; P12, 10 yr, 5′UTR, c.-321_-320delCCinsA; P13, 5 yr, c.771+1G>A). The institutional review board at the Karolinska Institutet approved the study.
Mutation analysis of PDS5B and KIAA0892 (MAU2).
Genomic DNA was isolated from peripheral blood lymphocytes from P4 using Puregene blood kit (Gentra Systems Inc.). PCR primer design and PCR amplifications were performed according to standard procedures, followed by direct sequencing of the coding regions and corresponding exon–intron boundaries of PDS5B and KIAA0892, using Big Dye Terminator cycle sequencing kit 3.1 (Applied Biosystems) and sequenced on an ABI genetic analyzer.
Cell sensitivity to γ-IR was determined with the MTS or colony formation assay.
For the MTS assay, cells were seeded at 1.5 × 105 cells/ml and exposed to γ-IR 1–9 Gy, using a 137Cs radiation source (IBL 677; CIS Bio International; 15 Gy/min dose rate), 800 ng/ml mitomycin C (Sigma-Aldrich) for 3 h, or etoposide 300 ng/ml (Sigma-Aldrich) for 36 h. Cells were then left to recover during the time required for three calculated population doublings. Cell viability was analyzed using Cell Titer 96 Aqueous One Solution MTS Proliferation Assay (Promega) as described by the manufacturer. Absorbance values are proportional to the number of viable cells and normalized to untreated controls. For colony formation assay, 125–4,000 cells were seeded in 6-well plates, incubated for 3–5 h and exposed to various doses of γ-IR as indicated. After 11–17 d recovery, cells were washed in PBS, fixed in methanol, dried, and stained using Giemsa stain (Sigma-Aldrich). Colonies exceeding 20 cells were counted.
Knockdown of NIPBL expression by siRNA.
2–3 × 105 FBs were transfected using DharmaFECT-1 transfection reagent and a pool of four siRNAs targeting NIPBL (Thermo Fisher Scientific), according to the manufacturer’s instructions. In brief, cells were incubated with 4 µl transfection reagent and 25 nM siRNA in 6-well plates for 48 h, when the cells were harvested or γ-irradiated. For the NHEJ plasmid assay, the siRNA treatment was repeated twice but otherwise performed as above. The effect of the siNIPBL transfection was detected at the RNA level by real-time PCR or at the protein level by Western blotting (see below). Total RNA was extracted using the RNeasy Mini kit (QIAGEN). cDNA synthesis was performed using 1 µg of total RNA, a NotI-d(T)18 primer and a cDNA synthesis kit (GE Healthcare) according to the manufacturer’s protocol. The amplification was performed using the Kapa SYBR Fast qPCR Master Mix kit (Kapa Biosystems) and the StepOne detection system (Applied Biosystems). For analyses of the expression of LIG4, PRKDC, and XRCC5, 500 ng of total RNA were used. The housekeeping genes, β-actin, GADPH, and HRPT1 were used for calculation of relative expression levels.
SDS-PAGE and Western blotting.
Protein extracts were prepared with standard procedures using lysis buffer (10 mM Tris- HCl, pH 8.2, 5 mM MgCl2, and 0.1% SDS supplemented with 1 mM PMSF, 10 mM DTT, 1x Protease Inhibitor Cocktail (Roche), 1 U DNase, and 10 mg/ml RNase). Protein extracts were analyzed by SDS PAGE and Western blotting. For detection of NIPBL, 3–8% Tris-Acetate gels in 1x Tricine buffer and, for ATM, Chk2 and XRCC4 and 4–12% Bis-Tris gels in 1x MOPS buffer were used (NuPAGE Invitrogen). Proteins were transferred to PVDF Immobilon-P membrane (Millipore) in 1x Tris-Glycine buffer supplemented with 20% methanol using standard procedures. For NIPBL detection, 0.05% SDS was added and blotting was performed for 15 h at 4°C to nitrocellulose membrane (Protean). Primary antibodies used were guinea pig anti-NIPBL peptide (Peptide Specialty Laboratories GmbH; peptide sequences available on request), mouse anti–α-tubulin (Sigma-Aldrich), mouse anti-phospho-ATM(Ser1981) (NOVUS Biological), rabbit anti-Chk2 (Abcam), rabbit anti–phospho-Chk2(Thr68) (Cell Signaling Technology), mouse anti-XRCC4 (Abnova), and rabbit anti-histone H3 (Abcam).
FACS analyses for determination of cell cycle phase distribution.
Asynchronously growing or G1-arrested cells were washed twice in ice-cold PBS and fixed in 70% ethanol at 4°C, subsequently incubated with RNase A 40 µg/ml (Sigma-Aldrich) and 20 µg/ml propidium iodide (Sigma-Aldrich) at 37°C for 30 min. Analysis was performed with a FACSCalibur system and the CellQuest software (BD).
Sequencing of switch recombination junctions.
Amplification, cloning, and sequencing of the Sμ-Sα fragments derived from in vivo–switched cells was performed using a previously described nested PCR assay (Pan-Hammarström et al., 2005). The switch recombination junctions were determined by aligning the switch fragment sequences with the reference Sμ, Sα1, or Sα2 sequences. Analysis of MH usage at the switch junctions was performed as described previously (Pan-Hammarström et al., 2005), with the recently suggested guidelines taken into account (Stavnezer et al., 2010).
The full sequences of the switch junctions from NIPBL and SMC1 patients, as well as controls, are shown in the Supplemental data.
Analysis of NHEJ via transfection of a linearized plasmid with defined DNA ends.
The assay was performed as previously described (Verkaik et al., 2002). 1 μg of the pDVG94 construct was digested by EcoRV and EcoR47III (Promega), resulting in blunt DNA ends with 6-bp direct repeats (ATCAGC). The linearized plasmids were transiently transfected into FBs (60–70% confluent) or G1 arrested cells (by overgrown the cells to confluent) using TurboFect (Fermentas) according to the manufacturer’s instruction. 48 h later, plasmid DNA was recovered using the DNEasy Blood and Tissue kit (QIAGEN). Recombination junctions were PCR amplified using the primers FM30 and DAR5 (Verkaik et al., 2002). The resulting PCR products were gel purified using QIAquick gel extraction kit (QIAGEN), cloned into pGEM-T vectors, and sequenced by an automated fluorescent sequencer (Macrogen).
NHEJ assay in yeast.
Wild-type cells or cells expressing a temperature-sensitive allele of the yeast NIPBL orthologue SCC2 (scc2-4; Michaelis et al., 1997), were arrested in G1 by A-factor (O’Reilly et al., 2012; INNOVAGEN, Sweden). The temperature was then raised to inactivate Scc2 function. A single site-specific DSB was induced by addition of 2% galactose activating the yeast pGAL-HO endonuclease. The gene for Ligase 4 (lig4)Δ was deleted using standard yeast protocols. Cells were counted and plated at equal concentration in 10-fold dilutions. Plates were left to recover for 72 h at a semipermissive temperature for scc2-4 (30°C). Surviving cells forming single colonies were picked and allowed to propagate to get sufficient amount of cells for preparation of genomic DNA. A 600-bp region encompassing the HO cut site was then PCR-amplified using the UP 5′-CCAAATTCACAGGATAGCG-3′ and DO 5′-GTCATCCGTCCCGTATAGCC-3′ primers, following the protocol for high fidelity amplification with the Phusion enzyme (Thermo Fisher Scientific). The PCR products were purified and sequenced using the UP 5′-CCAAATTCACAGGATAGCG-3′ primer. The sequences were subsequently aligned to the genomic MATα sequence.
Quantitation of 53BP1 foci.
3 × 105 cells/ml were seeded in 6-well plates and exposed to γ-IR (1 Gy) after 24 h. At the indicated time points, cells were washed in PBS, cytospun onto slides (700 rpm, 5 min), and fixed in 4% formaldehyde in PBS for 15 min at 22°C. The slides were incubated in 0.1% glycine/PBS for 30 min and blocked in 3% BSA, 10% goat serum, and 0.05% Triton X-100 in PBS. 53BP1 was detected using rabbit anti-53BP1 (ab36823; Abcam) and FITC-swine anti–rabbit antibodies (F0205; Dako). DNA was stained with DAPI and images were captured using a LSM510META confocal microscope (Carl Zeiss). Image stacks were exported to ImageJ (National Institutes of Health) and 53BP1 foci were quantified in ∼40 nuclei per LCL and time point. The Mann-Whitney U test was used for statistical analyses due to a non-Gaussian distribution of the number of foci in the material.
Online supplemental material.
A summary and representative sequences amplified from the MATα locus from surviving WT, Scc2 inactive, and DNA ligase IV–deleted yeast cells are shown in Fig. S1. Supplemental data shows the full sequences of the switch junctions from NIPBL and SMC1 patients, as well as controls. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20130168/DC1.
Supplementary Material
Acknowledgments
We are grateful to all the controls and patients that donated material and the French CdLS association (AFSCDL). We thank R. Gatti for kindly providing the A-T cell line AT153LA, M. Nordenskjöld and C. Höög for support, S. Carter for carefully reading the manuscript, and C. Liu and J. Lu for technical help.
This work was supported by a young investigator award from the Swedish Cancer Society to L. Ström and grants from the Swedish Research Council, the Swedish Cancer Society, the European Research Council (242551-ImmunoSwitch), the Jeansson’s foundation, the Norwegian Research Council, the Deutsche Forschungsgemeinschaft, the Spanish ISCIII-FIS (Ref. PS09-01422) and DGA/European Social Fund (Ref. B20).
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- A-EJ
- alternative end joining
- AID
- activation induced cytidine deaminase
- CdLS
- Cornelia de Lange Syndrome
- CSR
- class switch recombination
- DSB
- DNA double-strand break
- FB
- fibroblast
- γ-IR
- γ irradiation
- HR
- homologous recombination
- LCL
- B-lymphoblastoid cell line
- MH
- microhomology
- NHEJ
- nonhomologous end joining
References
- Bothmer A., Robbiani D.F., Feldhahn N., Gazumyan A., Nussenzweig A., Nussenzweig M.C. 2010. 53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination. J. Exp. Med. 207:855–865 10.1084/jem.20100244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Villartay J.P. 2009. V(D)J recombination deficiencies. Adv. Exp. Med. Biol. 650:46–58 10.1007/978-1-4419-0296-2_4 [DOI] [PubMed] [Google Scholar]
- Degner S.C., Verma-Gaur J., Wong T.P., Bossen C., Iverson G.M., Torkamani A., Vettermann C., Lin Y.C., Ju Z., Schulz D., et al. 2011. CCCTC-binding factor (CTCF) and cohesin influence the genomic architecture of the Igh locus and antisense transcription in pro-B cells. Proc. Natl. Acad. Sci. USA. 108:9566–9571 10.1073/pnas.1019391108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., van der Burg M., Popov S.W., Kotnis A., van Dongen J.J., Gennery A.R., Pan-Hammarström Q. 2008. Involvement of Artemis in nonhomologous end-joining during immunoglobulin class switch recombination. J. Exp. Med. 205:3031–3040 10.1084/jem.20081915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Peng R., Björkman A., Filipe de Miranda N., Rosner C., Kotnis A., Berglund M., Liu C., Rosenquist R., Enblad G., et al. 2012. Cernunnos influences human immunoglobulin class switch recombination and may be associated with B cell lymphomagenesis. J. Exp. Med. 209:291–305 10.1084/jem.20110325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur M., DeScipio C., McCallum J., Yaeger D., Devoto M., Jackson L.G., Spinner N.B., Krantz I.D. 2005. Precocious sister chromatid separation (PSCS) in Cornelia de Lange syndrome. Am. J. Med. Genet. A. 138:27–31 10.1002/ajmg.a.30919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi S., Calof A.L., Santos R., Lopez-Burks M.E., Young C.M., Hoang M.P., Chua A., Lao T., Lechner M.S., Daniel J.A., et al. 2009. Multiple organ system defects and transcriptional dysregulation in the Nipbl(+/-) mouse, a model of Cornelia de Lange Syndrome. PLoS Genet. 5:e1000650 10.1371/journal.pgen.1000650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kracker S., Imai K., Gardès P., Ochs H.D., Fischer A., Durandy A.H. 2010. Impaired induction of DNA lesions during immunoglobulin class-switch recombination in humans influences end-joining repair. Proc. Natl. Acad. Sci. USA. 107:22225–22230 10.1073/pnas.1012591108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Baynam G. 2010. Cornelia de Lange syndrome. Adv. Exp. Med. Biol. 685:111–123 [PubMed] [Google Scholar]
- Liu J., Zhang Z., Bando M., Itoh T., Deardorff M.A., Clark D., Kaur M., Tandy S., Kondoh T., Rappaport E., et al. 2009. Transcriptional dysregulation in NIPBL and cohesin mutant human cells. PLoS Biol. 7:e1000119 10.1371/journal.pbio.1000119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manis J.P., Morales J.C., Xia Z., Kutok J.L., Alt F.W., Carpenter P.B. 2004. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat. Immunol. 5:481–487 10.1038/ni1067 [DOI] [PubMed] [Google Scholar]
- Michaelis C., Ciosk R., Nasmyth K. 1997. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 91:35–45 10.1016/S0092-8674(01)80007-6 [DOI] [PubMed] [Google Scholar]
- Moore J.K., Haber J.E. 1996. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:2164–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K., Haering C.H. 2009. Cohesin: its roles and mechanisms. Annu. Rev. Genet. 43:525–558 10.1146/annurev-genet-102108-134233 [DOI] [PubMed] [Google Scholar]
- Noordzij J.G., Verkaik N.S., van der Burg M., van Veelen L.R., de Bruin-Versteeg S., Wiegant W., Vossen J.M., Weemaes C.M., de Groot R., Zdzienicka M.Z., et al. 2003. Radiosensitive SCID patients with Artemis gene mutations show a complete B-cell differentiation arrest at the pre-B-cell receptor checkpoint in bone marrow. Blood. 101:1446–1452 10.1182/blood-2002-01-0187 [DOI] [PubMed] [Google Scholar]
- O’Reilly N., Charbin A., Lopez-Serra L., Uhlmann F. 2012. Facile synthesis of budding yeast a-factor and its use to synchronize cells of α mating type. Yeast. 29:233–240 10.1002/yea.2906 [DOI] [PubMed] [Google Scholar]
- Oka Y., Suzuki K., Yamauchi M., Mitsutake N., Yamashita S. 2011. Recruitment of the cohesin loading factor NIPBL to DNA double-strand breaks depends on MDC1, RNF168 and HP1γ in human cells. Biochem. Biophys. Res. Commun. 411:762–767 10.1016/j.bbrc.2011.07.021 [DOI] [PubMed] [Google Scholar]
- Pan-Hammarström Q., Jones A.M., Lähdesmäki A., Zhou W., Gatti R.A., Hammarström L., Gennery A.R., Ehrenstein M.R. 2005. Impact of DNA ligase IV on nonhomologous end joining pathways during class switch recombination in human cells. J. Exp. Med. 201:189–194 10.1084/jem.20040772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pié J., Gil-Rodríguez M.C., Ciero M., López-Viñas E., Ribate M.P., Arnedo M., Deardorff M.A., Puisac B., Legarreta J., de Karam J.C., et al. 2010. Mutations and variants in the cohesion factor genes NIPBL, SMC1A, and SMC3 in a cohort of 30 unrelated patients with Cornelia de Lange syndrome. Am. J. Med. Genet. A. 152A:924–929 10.1002/ajmg.a.33348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina-San-Martin B., Chen J., Nussenzweig A., Nussenzweig M.C. 2007. Enhanced intra-switch region recombination during immunoglobulin class switch recombination in 53BP1−/− B cells. Eur. J. Immunol. 37:235–239 10.1002/eji.200636789 [DOI] [PubMed] [Google Scholar]
- Schär P., Fäsi M., Jessberger R. 2004. SMC1 coordinates DNA double-strand break repair pathways. Nucleic Acids Res. 32:3921–3929 10.1093/nar/gkh716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoumans J., Wincent J., Barbaro M., Djureinovic T., Maguire P., Forsberg L., Staaf J., Thuresson A.C., Borg A., Nordgren A., et al. 2007. Comprehensive mutational analysis of a cohort of Swedish Cornelia de Lange syndrome patients. Eur. J. Hum. Genet. 15:143–149 10.1038/sj.ejhg.5201737 [DOI] [PubMed] [Google Scholar]
- Seitan V.C., Hao B., Tachibana-Konwalski K., Lavagnolli T., Mira-Bontenbal H., Brown K.E., Teng G., Carroll T., Terry A., Horan K., et al. 2011. A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature. 476:467–471 10.1038/nature10312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharbeen G., Yee C.W., Smith A.L., Jolly C.J. 2012. Ectopic restriction of DNA repair reveals that UNG2 excises AID-induced uracils predominantly or exclusively during G1 phase. J. Exp. Med. 209:965–974 10.1084/jem.20112379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjögren C., Nasmyth K. 2001. Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr. Biol. 11:991–995 10.1016/S0960-9822(01)00271-8 [DOI] [PubMed] [Google Scholar]
- Stavnezer J., Björkman A., Du L., Cagigi A., Pan-Hammarström Q. 2010. Mapping of switch recombination junctions, a tool for studying DNA repair pathways during immunoglobulin class switching. Adv. Immunol. 108:45–109 10.1016/B978-0-12-380995-7.00003-3 [DOI] [PubMed] [Google Scholar]
- Stewart G.S., Stankovic T., Byrd P.J., Wechsler T., Miller E.S., Huissoon A., Drayson M.T., West S.C., Elledge S.J., Taylor A.M. 2007. RIDDLE immunodeficiency syndrome is linked to defects in 53BP1-mediated DNA damage signaling. Proc. Natl. Acad. Sci. USA. 104:16910–16915 10.1073/pnas.0708408104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkaik N.S., Esveldt-van Lange R.E., van Heemst D., Brüggenwirth H.T., Hoeijmakers J.H., Zdzienicka M.Z., van Gent D.C. 2002. Different types of V(D)J recombination and end-joining defects in DNA double-strand break repair mutant mammalian cells. Eur. J. Immunol. 32:701–709 [DOI] [PubMed] [Google Scholar]
- Vrouwe M.G., Elghalbzouri-Maghrani E., Meijers M., Schouten P., Godthelp B.C., Bhuiyan Z.A., Redeker E.J., Mannens M.M., Mullenders L.H., Pastink A., Darroudi F. 2007. Increased DNA damage sensitivity of Cornelia de Lange syndrome cells: evidence for impaired recombinational repair. Hum. Mol. Genet. 16:1478–1487 10.1093/hmg/ddm098 [DOI] [PubMed] [Google Scholar]
- Watrin E., Peters J.M. 2009. The cohesin complex is required for the DNA damage-induced G2/M checkpoint in mammalian cells. EMBO J. 28:2625–2635 10.1038/emboj.2009.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C.T., Boboila C., Souza E.K., Franco S., Hickernell T.R., Murphy M., Gumaste S., Geyer M., Zarrin A.A., Manis J.P., et al. 2007. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 449:478–482 10.1038/nature06020 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.