Abstract
We assessed the content, structure, and distribution of segmental duplications (≥90% sequence identity, ≥5 kb length) within the published version of the Rattus norvegicus genome assembly (v.3.1). The overall fraction of duplicated sequence within the rat assembly (2.92%) is greater than that of the mouse (1%–1.2%) but significantly less than that of human (∼5%). Duplications were nonuniformly distributed, occurring predominantly as tandem and tightly clustered intrachromosomal duplications. Regions containing extensive interchromosomal duplications were observed, particularly within subtelomeric and pericentromeric regions. We identified 41 discrete genomic regions greater than 1 Mb in size, termed “duplication blocks.” These appear to have been the target of extensive duplication over millions of years of evolution. Gene content within duplicated regions (∼1%) was lower than expected based on the genome representation. Interestingly, sequence contigs lacking chromosome assignment (“the unplaced chromosome”) showed a marked enrichment for segmental duplication (45% of 75.2 Mb), indicating that segmental duplications have been problematic for sequence and assembly of the rat genome. Further targeted efforts are required to resolve the organization and complexity of these regions.
Segmental duplications have long been recognized as important mediators of both gene and genome evolution (Muller 1936; Ohno 1970). From the genic perspective, such duplications often encode protein products which, although not essential for viability of the organism, are important for the adaptation of the species to specific ecological niches (Duda and Palumbi 1999). Among mammalian species, commonly duplicated genes include those associated with the recognition of environmental molecules and include genes associated with innate immunity, drug detoxification, olfaction, and sperm competition. From the perspective of genome structure, lineage-specific segmental duplications or large repeats often delineate regions of recurrent evolutionary lability (Eichler and Sankoff 2003). Recent comparative sequencing efforts among closely related eukaryotes, for example, shows that highly homologous repetitive sequence frequently associate with the breakpoints of large-scale chromosomal rearrangement (Dehal et al. 2001; Kellis et al. 2003). Understanding the nature and pattern of segmental duplications provides fundamental insight into functional redundancy, adaptive evolution, and the structural dynamics of chromosomal evolution.
One of the surprising findings from the analysis of the Human Genome Project data was the relevant abundance of large blocks of sequence with a high degree of sequence identity (Bailey et al. 2001; International Human Genome Sequencing Consortium [IHGSC] 2001). A variety of computational and experimental methods (Bailey et al. 2001, 2002; Cheung et al. 2001) now estimate 5%–6% of the human as duplicated (≥1kb and ≥90%). Compared to other sequenced organisms such as fly and worm, the human genome is enriched for recent segmental duplications, particularly interspersed duplications (Bailey et al. 2002). Such comparisons, however, typically assess duplication content with lower-bound estimates of length. For example, these cross-species comparisons rarely characterize duplications less than 500 bp in length. This may introduce an ascertainment bias, particularly among invertebrates, whose genomes can be orders of magnitude smaller compared to human. Larger genomes may simply harbor larger segmental duplications. The purported “unique” properties of the human genome can only be assessed by detailed comparison with other mammalian genomes where genome sizes are equivalent. With the whole-genome shotgun sequence assembly of the rat genome, we can now assess the nature and pattern of segmental duplication of a third mammalian genome (Rat Genome Sequencing Project Consortium [RGSPC] 2004; Waterston et al. 2002).
We present a preliminary, genome-wide analysis of the segmental duplication content of the rat (Rattus norvegicus). Any assessment of segmental duplication content is highly dependent on the methodology and quality of the sequence assembly (Bailey et al. 2001; Eichler 2001; Cheung et al. 2003a). Discrimination between highly paralogous copies and allelic regions that have not been properly assembled requires an estimate of the levels of both allelic variation and sequencing error. For most regions, paralogous sequences are more divergent than allelic copies. Another consideration is the method of genome assembly. Assembly algorithms based on sequence overlap from working-draft BAC clones were shown to overestimate the frequency of segmental duplication, due to a failure to properly merge allelic overlaps (Bailey et al. 2001; IHGSC 2001). Alternatively, assembly strictly from whole-genome shotgun sequence reads tends to over-collapse and therefore underrepresent such regions due to the recruitment of both paralogous and allelic sequence reads (Eichler 1998; Bailey et al. 2002; Estivill et al. 2002). Interestingly, the assembly algorithm of the rat genome represents a hybrid of whole-genome- and clone-ordered-based approaches. The ability of this approach to resolve segmental duplications has not been tested previously. In light of these inherent difficulties associated with the assembly of highly similar duplications, the analysis should be considered a first approximation of the recent duplication properties of the rat genome. Such initial analyses, however, are essential in providing a more accurate and robust “final” version of the rat genome as well as insight into genome-assembly approaches. The results of the present study have been made publicly available through the UCSC genome browser as well as through our own local database (http://ratparalogy.cwru.edu), providing a resource for the rat sequencing and genetics community.
RESULTS
We initially examined the entire draft genome of the rat using a previously described BLAST-based whole-genome sequence comparison method (see Methods; Bailey et al. 2001). Assembled draft sequence of any genome may be operationally divided into two categories: sequence which can be mapped to a chromosome, and that which cannot. In the case of the rat genome, 75 Mb of assembled sequence was ambiguous in its placement. Because segmental duplications are particularly enriched in this category, we separately considered this category throughout our analysis. In order to detect segmental duplications specific to the rat lineage, we examined all duplications that showed <10% sequence divergence. Based on sequence divergence between mouse and rat (0.175–0.195; RGSPC 2004), such regions likely represent either lineage-specific duplications or large-scale gene conversion events. During the initial phases of this analysis, we discovered an overabundance of pairwise alignments <5 kb in size (Table 1). Their high-copy number, relatively small size, well defined borders, and their highly interspersed nature both within and between chromosomes suggested contamination by high-copy repeats, despite the removal of rodent retroelements using the latest curated version of Repeatmasker (June 2003). Such contamination could be due to either incomplete masking of unknown repeat elements or transduction of flanking sequence (Goodier et al. 2000; Pickeral et al. 2000). As our goal was to identify genomic sequence that arose as a consequence of duplication (not retrotransposition or retroposon-induced transduction), we raised our threshold for seeding alignments to 5 kb—the effective insertion length of most retroelements is <5 kb in length, whereas most transduced sequences are less than 1 kb in length. For comparisons we considered additional alignment length thresholds (5, 10, 20 and 50 kb; Fig. 1) which were certain to exclude all transposable elements, including full-length retroviral repeats (Table 2).
Table 1.
Length vs. Number of Pairwise Alignments (Rat v. 3.1)
| Seed length (bp) | # Pairwise |
|---|---|
| >250 | 1,283,258 |
| >1000 | 532,720 |
| >5000 | 45,835 |
| >10,000 | 4798 |
| >20,000 | 171 |
Seed length determined after masking of common rat repeats (Repeatmasker June 2003 version).
Figure 1.
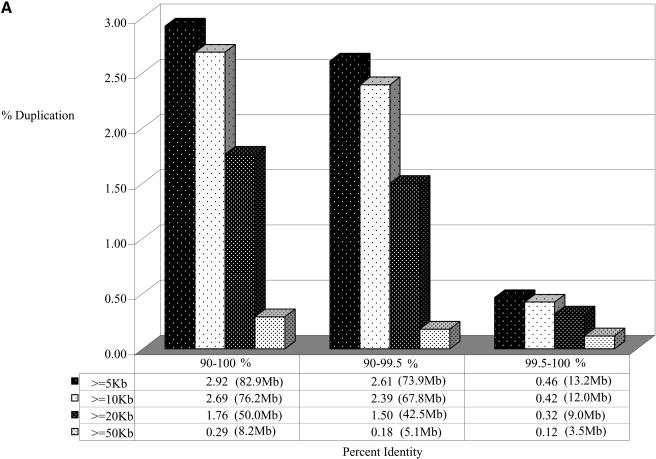
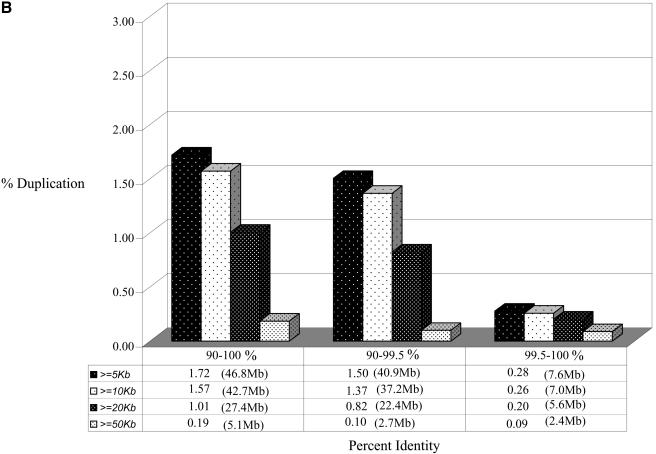
Duplicated fraction in the rat genome. The figure depicts the proportion of the genome that shows duplication (A) when all genomic sequence was compared, and (B) for the rat genome excluding random, unassigned sequence contigs. Various lengths and % identity thresholds are shown. A very small portion of the rat genome shows segmental duplications with ≥99.5% sequence identity. This suggests that the majority of segmental duplications are bona fide and are not the result of missed allelic overlaps during genome assembly.
Table 2.
Rat Segmental Duplication Sequence Alignment Statistics
|
Number of alignments
|
Average sequence identity
|
Average length
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Size (kb) | Inter | Intra | All | Inter | Intra | All | Inter | Intra | All |
| 5 | 568 | 1265 | 1833 | 0.940 | 0.952 | 0.948 | 5631 | 5630 | 5631 |
| 6 | 1255 | 3769 | 5024 | 0.941 | 0.947 | 0.945 | 6528 | 6529 | 6529 |
| 7 | 1532 | 3968 | 5500 | 0.936 | 0.944 | 0.942 | 7496 | 7495 | 7495 |
| 8 | 1329 | 4501 | 5830 | 0.934 | 0.942 | 0.940 | 8506 | 8472 | 8480 |
| 9 | 1145 | 3487 | 4632 | 0.938 | 0.943 | 0.942 | 9533 | 9463 | 9481 |
| 10-19 | 4367 | 13550 | 17917 | 0.944 | 0.946 | 0.945 | 13376 | 13280 | 13303 |
| 20-29 | 673 | 1571 | 2244 | 0.950 | 0.953 | 0.952 | 23894 | 23601 | 23689 |
| 30-39 | 134 | 277 | 411 | 0.956 | 0.960 | 0.959 | 33183 | 33858 | 33638 |
| 40-49 | 33 | 70 | 103 | 0.965 | 0.964 | 0.964 | 44493 | 44211 | 44301 |
| 50+ | 34 | 69 | 103 | 0.979 | 0.967 | 0.971 | 59546 | 56850 | 57740 |
| Total | 11070 | 32527 | 43597 | 0.941 | 0.946 | 0.944 | 11520 | 11253 | 11321 |
Alignments were binned into groups based on 1-kb increments (i.e., 5, 6 kb, etc) and 10 kb increments (i.e., 10-19.9 kb), the absolute number of alignments, average sequence identity, and average length for interchromosomal, intrachromosomal, and all alignments are shown after seed alignments were joined (see Methods). Consequently, the total number of joined alignments is less than the number of seed alignments (Table 1).
Sequence Properties of Rat Segmental Duplications
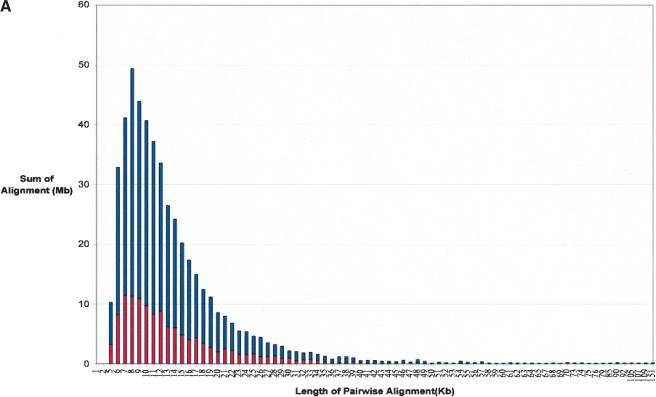
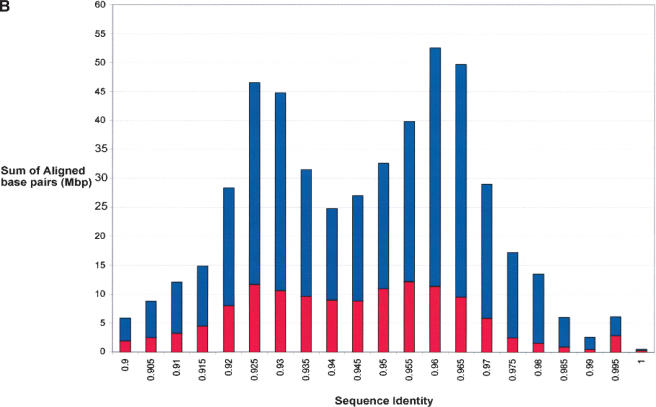
We calculate a total of 2.92% (82.8 Mb/2835 Mb) of the rat genome as duplicated (≥90% sequence identity, ≥5 kb; Figs. 1, 2). These correspond to 43,597 pairwise alignments and represent 3237 distinct regions of the rat genome (Table 2). Pairwise alignments may be redundant in nature, as the same sequence may be duplicated to multiple locations in the genome. Therefore, the number of distinct, nonoverlapping regions is substantially fewer. Figure 1 depicts the duplication content of the rat genome as a function of the length of alignment and the degree of sequence identity. As described above, we included and excluded the unplaced sequence contigs to show the disproportionate representation of duplicated sequence in this category. Based on our analysis of the entire genome, the median length of alignments (9749 bp) is not significantly different between interchromosomal and intrachromosomal duplications. The largest alignment detected is 104 kb. The average degree of sequence identity among all alignments is 94.4%. Interestingly, when we considered the percent nucleotide sequence identity for segmental duplications as a function of the number of aligned base pairs, we observed a distinct bimodal distribution (Fig. 2B). Two peaks were observed corresponding to 95.5% and 92.5% sequence identity (0.045 substitutions per site and 0.075 substitutions per site). This bimodal distribution was consistently observed whether unmapped genomic sequence was excluded or included in the analysis.
Figure 2.
Sequence properties of rat segmental duplications. Distributions of the (A) length and (B) percent nucleotide sequence identity for segmental duplications are shown as a function of the number of aligned bp. Interchromosomal duplications (red); intrachromosomal duplications (blue).
Estimates of segmental duplication from the human genome working draft sequence assembly initially overestimated the fraction of duplicated bases (10.8% of the genome). This was the result of a failure to merge allelic overlaps during the genome assembly process. Subsequent analysis of finished genome sequence showed that such alignments showed an extraordinary degree of sequence identity consistent with missed allelic overlaps (Bailey et al. 2001). To eliminate such potential artifacts in the rat genome assembly, we separately considered all alignments where the degree of sequence identity is less than 99.5%. We derive a conservative estimate of the duplication content of the rat genome to be 2.61% (73.9 Mb/2835.2 Mb, 2928 distinct genomic regions; Fig. 1). It is unlikely, therefore, that the majority of rat segmental duplications identified in this study arise as a consequence of a failure to merge overlaps during assembly.
Rat segmental duplications show a bias toward intrachromosomal alignments (68.1 Mb) compared to interchromosomal duplications (48.2 Mb; Figs. 2, 3, Table 2). Interestingly, the number of intrachromosomal and interchromosomal pairwise alignments differs more dramatically. By this measure, intrachromosomal duplications are three times more frequent than interchromosomal duplications (32,527 intrachromosomal alignments vs. 11,070 interchromosomal alignments; Table 2). It should be noted, however, that a significant fraction of the rat genome sequence (115.2 Mb) has not been assigned to a chromosome (unplaced chromosome), nor has it been assigned specifically within a chromosomal region (random chromosome bins). The above calculations treat the unmapped sequence as a separate chromosome when classifying duplications as inter- or intrachromosomal. We estimate that 45% (36.1 Mb/82.8 Mb) of the duplications are mapped to these intractable regions of the rat genome. Their map locations are ambiguous, and intra/interchromosomal distribution is technically unknown. If we exclude these two categories of sequence, a total of 1911 (46.7 Mb, 1.72% of the genome) regions of duplication are identified which have been unambiguously mapped within the rat genome. Again, a stronger preference for intrachromosomal duplications (38.8 Mb) was observed compared to interchromosomal duplications (17.7 Mb). With few exceptions, most intrachromosomal duplications are organized as clusters of tandem or inverted duplications within close proximity. Using these conservative criteria, ∼21% of the duplicated bases (8.8 Mb) were part of interchromosomal and intrachromosomal duplication alignments.
Figure 3.
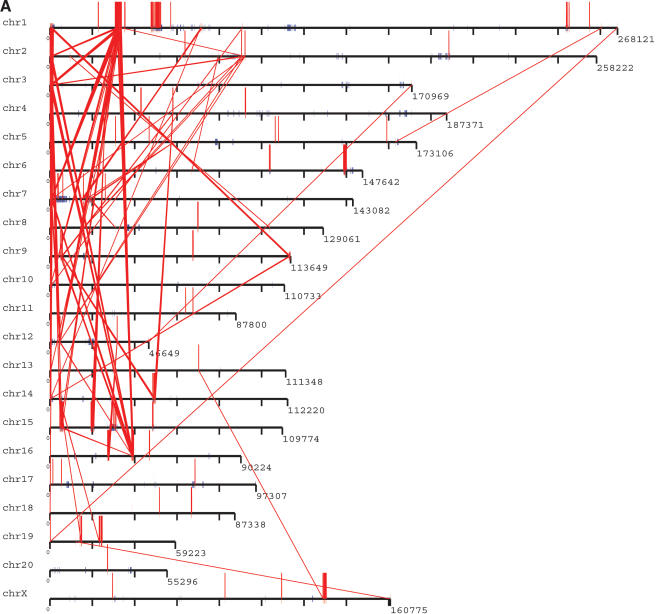
Distribution of segmental duplications (≥90% and ≥10 kb) in the rat genome. The pattern of (A) interchromosomal duplications (red) and (B) intrachromosomal duplications (blue) are depicted for all duplications ≥90% sequence identity and ≥10 kb in length. For clarity, interchromosomal distribution patterns with the random, unassigned sequence contigs (chrUn) are not shown for (A). For more detail, including % identity and pairwise relationships of all duplications and alignments, see http://ratparalogy.cwru.edu.
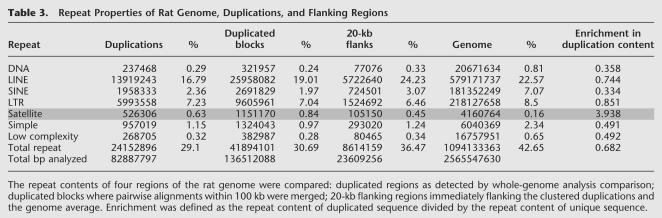
As a final analysis of the sequence properties of rat segmental duplications, we compared the repeat content of duplicated sequence, flanking sequence and the whole genome (Table 3, Methods). Unlike human segmental duplications, which are enriched for SINE repeats (Bailey et al. 2003), no SINE enrichment (nor any other retroelement) was associated with rat segmental duplications. The working draft nature of the rat genome sequence prevents a detailed analysis of the sequence structure at the transition regions between unique and duplicated sequence. Nevertheless, two clear patterns emerge regarding repeat content. Although the common repeat content of most duplications appears to be reduced, SINE content shows the greatest reduction compared to the genome average (1.97% vs. 7.1%). This gradually increases to the genome average as sequences flanking the duplications are considered (Table 3). An opposite trend is observed with respect to centromeric satellite repeat sequences. Rat segmental duplications show a fourfold enrichment for satellite repeat content compared to the genome average. When individual repeat subfamilies are considered, satellite repeat classes 91ES8_RN and RNSAT1 show the greatest enrichment (10-fold and sevenfold, respectively). This association is most pronounced among blocks of interchromosomal duplication (see below).
Table 3.
Repeat Properties of Rat Genome, Duplications, and Flanking Regions
The repeat contents of four regions of the rat genome were compared: duplicated regions as detected by whole-genome analysis comparison; duplicated blocks where pairwise alignments within 100 kb were merged; 20-kb flanking regions immediately flanking the clustered duplications and the genome average. Enrichment was defined as the repeat content of duplicated sequence divided by the repeat content of unique sequence.
Organization of Rat Segmental Duplications
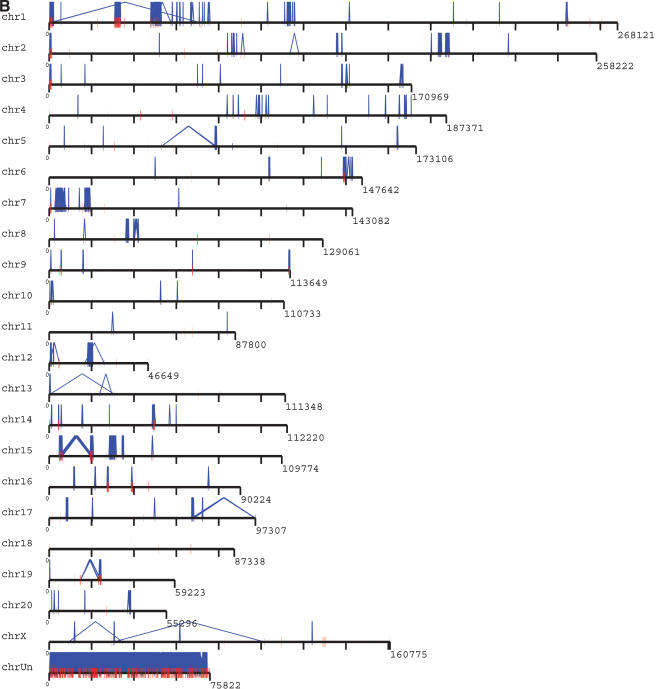
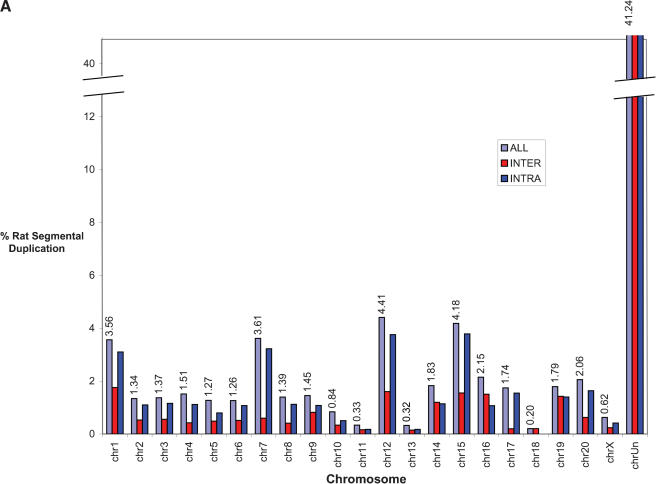
The recent segmental duplications of the rat genome are distributed in a nonrandom fashion at two different levels. First, duplication content varies significantly among different chromosomes. Chromosomes 12, 7, 15, and 1 show the greatest enrichment for segmental duplication (Fig. 4A) with twofold the duplication content of the genome average (excluding unplaced sequence contigs). Most of this effect is due to an increase in intrachromosomal duplication content localized as specific clusters. During the analysis of segmental duplications, large tracts were identified which were populated by a high density of segmental duplications. These tracts, termed “duplication blocks” (Bailey et al. 2001) ranged from 500 kb to as large as 3 Mb in size (Table 4), were generally gene-poor, and were characterized by assembly inconsistencies. A total of 41 discrete duplication blocks were identified which exceeded 1 Mb in length (Table 4). Typical block structures for chromosomes 1 and 7 are depicted (Fig. 4B). Analysis of the pairwise alignments underlying these block structures showed considerable variation in sequence identity (90%–99% identity), often within the same block. Two types of duplication block structures were distinguished: chromosome-specific blocks which consisted largely of interspersed segmental duplications (Table 5), and clustered interchromosomal pairwise alignments with considerable range in sequence identity. Interestingly, within a specific duplication block, multiple pairwise alignments among specific subsets of chromosomes could be identified (Table 5).
Figure 4.
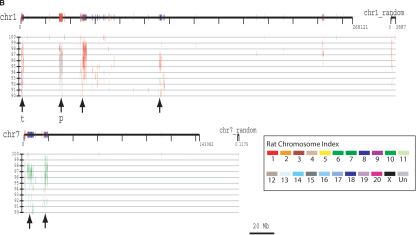
(A) Segmental duplication content per chromosome. The relative proportion of intrachromosomal and interchromosomal duplications for each chromosome is shown. The above calculations treat the unmapped sequence as a separate chromosome when classifying duplications as inter- or intrachromosomal. Forty-five percent of the unplaced chromosome is made up almost entirely of duplicated sequence. (B) Duplication blocks. Rat segmental duplications clustered into larger regions ranging from 100 to 3000 kb in length. We termed these structures “duplication blocks.” Examples of duplication blocks on chromosomes 1 and 7 are presented (arrows) with the underlying degree of sequence identity for each pairwise depicted below the graph. Chromosome 1, green; chromosome 7, red. A subtelomeric (t) and pericentromeric (p) block are indicated. The regions of the rat genome are typified by low gene density (RefSeq/EST/mRNA), a high frequency of gaps within the assembly, and an excess of pairwise alignments.
Table 4.
Block Structure of Rat Segmental Duplications
| Block Size | Assigned | Unknown | Total |
|---|---|---|---|
| ≥2 Mb | 12 | 5 | 17 |
| ≥1 Mb | 19 | 22 | 41 |
| ≥500 kb | 25 | 49 | 76 |
Unknown refers to unplaced sequence either random or “unknown” chromosome.
Table 5.
Largest Blocks of Segmental Duplication in the Rat Genome
| Chromosome | v.3.1begin | v.3.1end | Block size | Homology | Content |
|---|---|---|---|---|---|
| chr15 | 28153750 | 31785691 | 3631941 | chr15 | T-cell receptor |
| chr1 | 49778386 | 53005805 | 3227419 | chr1 | noRefseq, noRatmRNA, noRatEst, but homology to non-rat mRNA |
| chr1 | 30806167 | 33768308 | 2962141 | chr1, 3, 7, 12, 16 | noRefseq, noRatmRNA, limited RatEst |
| chr7 | 4210045 | 6569239 | 2359194 | chr7 | noRefseq, noRatmRNA, noRatEst, but homology to non-rat mRNA |
| chr1 | 5500 | 2196674 | 2191174 | chr1, 7, 9, 14 | noRefseq, noRatmRNA, limitedRatEst, subtelomeric homology |
| chr14 | 48526685 | 50326618 | 1799933 | chr14, 9 | noRefseq, noRatmRNA, RatEST |
| chr15 | 19229668 | 20983782 | 1754114 | chr15, 3, 12 | prostaglandin D2 receptor |
| chr15 | 4606290 | 6304612 | 1698322 | chr15 | MIC2 like 1/Rhombex40 |
| chr7 | 2608728 | 4103847 | 1495119 | chr7 | noRefseq, noRatmRNA, noRatEst, but homology to human mRNA |
| chr19 | 23253330 | 24673455 | 1420125 | chr14, 19 | noRefseq, noRatmRNA, noRatEst, but homology to non-rat mRNA |
| chr12 | 18092439 | 19509542 | 1417103 | chr12 | noRefseq, noRatmRNA, limited RatEst |
| chr6 | 138660326 | 140060338 | 1400012 | chr6 | Immunglobulin heavy chain variable region (IGHV) |
| chr17 | 67155883 | 68547346 | 1391463 | chr17 | noRefseq, noRatmRNA, RatEst |
| chr2 | 14691 | 1387166 | 1372475 | chr1, 2, 3, 7, 16 | noRefseq, limited RatmRNA and RatEST, subtelomeric |
| chr7 | 18156845 | 19521121 | 1364276 | chr7(prim), 8, 9 | noRefseq, noRatmRNA, RatEST |
| chr7 | 16446057 | 17651248 | 1205191 | chr7 | noRefseq, noRatmRNA, RatEST (conserved) |
| chr3 | 20974 | 1139288 | 1118314 | chr1, 2, 3, 7, 12, 15, 16 | noRefseq, noRatmRNA, RatEST |
| chr17 | 7851444 | 8937241 | 1085797 | chr17 | Cathepsin Ma |
| chr8 | 36551927 | 37612862 | 1060935 | chr8 | Atpase inhibitor |
| chr1 | 112988982 | 114038449 | 1049467 | chr1 | noRefseq, noRatmRNA, RatEST |
| chr5 | 78092980 | 79128821 | 1035841 | chr5 | alpha 2μ globulin PGCL3b Zfp37 |
| chr9 | 4870998 | 5902959 | 1031961 | chr9, 14, 16 | noRefseq, noRatmrna, RatEST |
Blocks were defined as clusters of segmental duplication with <100 kb of intervening sequence between duplicons. The largest 22 blocks which were assigned to a chromosome are shown. For a complete listing of all blocks, see http://ratparalogy.gene.cwru.edu. Begin and end coordinates within build 3.1, block size, and homologous regions are shown. Content was based on assigned Refseq, Rat mRNA, and ESTs within intron/exon structure within the UCSC browser. aBest sequence match of Cathepsin M is on chromosome 5. bEstimated 20 copies, but only five copies can be distinguished within the assembly.
In humans (Eichler et al. 1996; Jackson et al. 1999; Horvath et al. 2000) and to a lesser known extent in mouse (Thomas et al. 2003), segmental duplications show particular biases for pericentromeric and subtelomeric regions of the genome. Based on the current rat genome assembly, regions of segmental duplications (100 kb–1.5 Mb in size) were observed for 13 of the 40 possible most distal sequence contigs, suggesting a subtelomeric preponderance. Most of these subtelomeric blocks showed complex patterns of interchromosomal duplication among specific subsets of rat chromosomes. Characterization of a pericentromeric bias for segmental duplication is more difficult to determine, because the location of rat centromeres are generally not as well identified as mouse and human. We attempted to approximate the centromere position within the rat genome assembly using two independent methods (RGSPC 2004). The first approach mapped the most proximal STS/gene marker to the p and q arm of each rat chromosome by FISH, and considered the interval between these markers within the assembly as a possible centromere location. The second approach identified dense clusters of classic rat satellite repeats (particularly SATI_RN and ISAT_RN) within the assembly. Six rat chromosomes showed a correlation by these two different methods and allowed a likely assignment of the centromere region. Of these four chromosomes, large blocks of interchromosomal segmental duplication were identified ranging in size from 300 kb–3 Mb. Once again, analysis of underlying pairwise alignments identified sequence homology among specific sets of rat chromosomes (Table 5). A dearth of RefSeq genes or spliced rat mRNA within these regions was noted.
Gene Analysis of Recent Rat Segmental Duplications
We considered the genomic duplication content of all RefSeq mRNA aligned to the rat genome. To eliminate potential false positives, we limited our analysis to duplications showing ≥1% sequence divergence, well below the polymorphic level of variation for this inbred strain. The duplications therefore likely represent bona fide recent gene duplication or gene conversion events within the rat lineage. A total of 45/4250 rat RefSeq genes were identified that were embedded within the segmental duplications detected by whole-genome analysis comparison (Table 6). Even though interchromosomal duplications constitute ∼one-third of all pairwise alignments and 40% of all duplicated bases, genes are largely biased to intrachromosomal duplications (41/45 or 91%). Of these, almost all pairwise were <1 Mb apart, indicating that most “functional” duplicates within the rat genome are tandem gene clusters, as opposed to widely interspersed duplications. Indeed, in our analysis of the RefSeq genes alone, 19/45 genes belonged to known clusters of tandem gene families. Due to the limited number of characterized RefSeq genes, we broadened our analysis to consider known rat mRNA which possessed two or more exons. Although a few putative novel gene families were identified (e.g., α-latroxin G-coupled protein receptor, low-voltage activated calcium channel gene family, and a dynein-like protein subfamily; Supplemental Table 1), most mRNA corresponded to additional members of previously characterized genes (RGSPC 2004).
Table 6.
Genes Within Rat Segmental Duplications
| Accession | Gene name | Gene product | Chrom | txStart | txEnd | Exon count | Exons hit | Gene size | # Dupbp |
|---|---|---|---|---|---|---|---|---|---|
| NM_181693 | Adam28 | A disintegrin and metalloprotease domain 28 | chr15 | 49036202 | 49100982 | 22 | 12 | 2357 | 1200 |
| NM_023103 | Mug1 | Alpha(1)-inhibitor 3, variant I | chr4 | 158528836 | 158582575 | 36 | 21 | 4656 | 2829 |
| NM_147214 | LOC259246 | Alpha-2μ globulin PGCL1 | chr5 | 78712436 | 78715858 | 7 | 7 | 878 | 878 |
| NM_147212 | LOC259244 | Alpha-2μ globulin PGCL3 | chr5 | 78094054 | 78171960 | 7 | 7 | 807 | 807 |
| NM_147215 | LOC259247 | Alpha-2μ globulin PGCL4 | chr5 | 78093991 | 78097430 | 7 | 7 | 1010 | 1010 |
| NM_147213 | LOC259245 | Alpha-2μ globulin PGCL5 | chr5 | 78686553 | 78689873 | 7 | 7 | 733 | 733 |
| NM_012718 | Andpro | Androgen regulated 20 kDa protein | chr3 | 137991232 | 137997597 | 4 | 4 | 828 | 828 |
| NM_032072 | Appbpl | APP-binding protein 1 | chr19 | 382555 | 408667 | 20 | 7 | 1780 | 692 |
| NM_012915 | Atpi | ATPase inhibitor | chr8 | 36902394 | 36905032 | 3 | 3 | 415 | 415 |
| NM_022281 | Abccl | ATP-binding cassette, sub-family C (CFTR/MRP) | chr10 | 452238 | 575705 | 31 | 13 | 4998 | 2360 |
| NM_031565 | Ces1 | Carboxylesterase 1 | chr19 | 14892725 | 14929187 | 14 | 11 | 1936 | 1389 |
| NM_133295 | Ces3 | Carboxylesterase 3 | chr19 | 14933410 | 14971894 | 14 | 14 | 1892 | 1892 |
| NM_144743 | LOC246252 | Carboxylesterase isoenzyme gene | chr19 | 37685 | 44736 | 12 | 12 | 1872 | 1872 |
| NM_181378 | Ctsm | Cathepsin M | chr17 | 8931467 | 8936876 | 8 | 8 | 1355 | 1355 |
| NM_031561 | Cd36 | CD36 antigen | chr4 | 13472462 | 13554416 | 13 | 12 | 2447 | 2285 |
| NM_153313 | CYP2D1 | Cytochrome P450 2D1 | chr7 | 120803930 | 120808335 | 9 | 9 | 1632 | 1632 |
| NM_017158 | Cyp2c39 | Cytochrome P450, 2c39 | chr1 | 243799046 | 244827001 | 9 | 3 | 1591 | 754 |
| NM_017158 | Cyp2c39 | Cytochrome P450, 2c39 | chr1 | 243935780 | 244719024 | 12 | 10 | 1737 | 1518 |
| NM_173304 | Cyp2d5 | Cytochrome P450CMF1b | chr7 | 120794663 | 120799170 | 9 | 9 | 1599 | 1599 |
| NM_022849 | Dmbt1 | Deleted in malignant brain tumors 1 | chr1 | 190539537 | 190620410 | 33 | 7 | 4360 | 1635 |
| NM_138902 | Loc192264 | Eosinophil cationic protein | chr15 | 27287836 | 27288707 | 2 | 2 | 713 | 713 |
| NM_053689 | Crfg | G protein-binding protein CRFG | chr17 | 72111731 | 72131468 | 17 | 11 | 1927 | 1267 |
| NM_181440 | Grp-Ca | Glutamine/glutamic acid-rich protein GRP-Ca | chr4 | 170757736 | 170828910 | 5 | 5 | 876 | 876 |
| NM_138517 | Gzmb | Granzyme B | chr15 | 35211149 | 35214166 | 5 | 5 | 1035 | 1035 |
| NM_019261 | Klrc2 | Killer cell lectin-like receptor subfamily C | chr4 | 167201553 | 167212693 | 7 | 6 | 1309 | 751 |
| NM_133421 | Lkap | Limkain b1 | chr10 | 826802 | 872214 | 28 | 27 | 7645 | 7519 |
| NM_152848 | Ly49i2 | Ly49 inhibitory receptor 2 | chr4 | 168584327 | 168608948 | 7 | 7 | 1522 | 1522 |
| NM_153726 | Ly49s3 | Ly-49 stimulatory receptor 3 | chr4 | 168268533 | 168454309 | 7 | 4 | 1401 | 932 |
| NM_173291 | Ly49 | Lymphocyte antigen 49 complex | chr4 | 168135002 | 168354511 | 9 | 9 | 1145 | 1145 |
| NM_134459 | Mic211 | MIC2 like 1 | chr15 | 5673663 | 5719158 | 11 | 4 | 4177 | 359 |
| NM_022247 | Pdcl | Phosducin-like protein | chr3 | 17003800 | 17012826 | 4 | 1 | 2303 | 1825 |
| NM_022241 | Ptgdr2 | Prostaglandin D2 receptor | chr15 | 19345554 | 19352929 | 2 | 2 | 1317 | 1317 |
| NM_080770 | Psbp1 | Prostatic steroid-binding protein 1 | chr1 | 212320755 | 212323535 | 3 | 3 | 518 | 518 |
| NM_173315 | LOC286981 | Putative pheromone receptor (Go-VN2) | chr1 | 71238505 | 71258976 | 6 | 2 | 3572 | 512 |
| NM-173318 | LOC286984 | Putative pheromone receptor (Go-VN4) | chr1 | 61650206 | 61748606 | 8 | 8 | 3650 | 3650 |
| NM_173320 | LOC286986 | Putative pheromone receptor Go-VN13C | chr18 | 34192 | 61266 | 6 | 5 | 3346 | 3111 |
| NM_173113 | VN1 | Putative pheromone receptor VN1 | chr4 | 124479450 | 124720930 | 2 | 1 | 1378 | 251 |
| NM_173298 | VN2 | Putative pheromone receptor VN2 | chr4 | 124479445 | 124487113 | 3 | 3 | 1663 | 1663 |
| NM_012646 | RT1-N1 | RT1 class 1b gene, H2-TL-like, grc region | chr20 | 278581 | 2789031 | 8 | 8 | 1186 | 1186 |
| NM_176076 | S100RVP | S100 calcium-binding protein | chr2 | 183684144 | 183687494 | 4 | 4 | 831 | 831 |
| NM_012657 | Spin2b | Serine protease inhibitor 2b | chr6 | 128383141 | 128390546 | 5 | 5 | 1669 | 1669 |
| NM_031664 | Slc28a2 | Solute carrier family 2, member 2 | chr3 | 109256103 | 109276805 | 17 | 17 | 2644 | 2644 |
| NM_053752 | Suclg1 | Succinate-CoA ligase, GDP-forming, alpha | chr8 | 36133697 | 36137263 | 4 | 4 | 476 | 476 |
| NM_133547 | Sult1c2 | Sulfotransferase family, cytosolic, 1C, member | chr9 | 1030810 | 1160809 | 7 | 4 | 2432 | 2029 |
| NM_058209 | Zfp37 | Zinc finger protein 37 | chr5 | 78484370 | 79157374 | 6 | 3 | 2492 | 2167 |
aOnly genes within segmental duplications where the alignments were between 90%—99% identical are shown. A total of 63 genes were detected within duplications 90—100% identical. The number of exons (exons hit) and genic bases within the duplicated region (Dupbp) are indicated. Gene size is the sum of exon lengths from the rat genome assembly.
The genes identified in our analysis fall into three categories. These include genes associated with foreign compound detoxification (cytochrome P450 and carboxylesterase genes), environmental signal recognition (α-2 globulin and pheromone receptors), and innate immune response (rat serine protease inhibitors, natural killer cell receptors, T-cell receptor, major histocompatibility locus, and immunoglobulin variable heavy chain locus, etc.; RGSPC 2004). Despite the abundance of rat segmental duplications on the “unknown” chromosome, only eight duplicate genes with two or more exons are identified within this 45 Mb of duplicated sequence. This included caveolin-2 (AF439788), a vacuolar protein sorting homolog (U35244), two copies of a carboxylesterase E gene (D00362), and various gene fragments/orphons of immunoglobulin γ and ε variable chain, T-cell receptor, and cytochrome P450 genes. It is likely that these sequences represent displaced members of tandem gene clusters which proved difficult to integrate into the genome assembly due to the high degree of sequence identity.
DISCUSSION
We present a preliminary analysis of recent segmental duplication content of the rat genome. In order to avoid some of the difficulties and artifacts associated with detection and characterization of low-copy repeat sequence (Bailey et al. 2001), we implemented several precautions during our in silico analysis of the draft sequence. First, to avoid overestimating segmental duplication content due to common repeats, we purposefully set our alignment length criteria to exclude uncharacterized retroelements considering thresholds at both 5 and 10 kb. Second, we considered separately the proportion of duplications with near perfect sequence identity (Fig. 1; Table 1). Initial analyses of the Human Genome Project overestimated the amount of segmental duplication as much as threefold due to a failure to merge allelic overlap during the clone-ordered assembly process (IHGSC 2001). Subsequent analyses showed that such artifactual duplications were readily distinguished by an unusually high degree of sequence identity consistent with allelic levels of variation (≥99.8%). Our analysis of the rat genome indicates that the vast majority (≥91%) of the pairwise alignments show sequence identity <99.5%—far below the estimated levels of sequence variation due to error and/or allelic variation. In fact, the individual rat used for genome sequencing was highly inbred, with virtually no allelic variation—the product of more than 50 brother–sister matings (Methods). The fact that the majority of duplication alignments were <99.5% identical suggests relatively few false positives during this analysis.
There are some limitations of this analysis that should be noted. Regions of extremely high sequence identity may have been collapsed during assembly. Thus, the relative small fraction of the genome that shows duplication ≥99.5% (Fig. 1) may be an underestimate. The fact that the α-2-globulin cluster shows only five duplicated genes as opposed to the estimated 15–20 copies (McFadyen et al. 1999; McFadyen and Locke 2000) may be a consequence of such an effect. Although many of the expected rat gene duplications and highly homologous gene families (i.e., carboxylesterases, α-2-globulin, cytochrome P450 genes, serine protease inhibitor, T-cell receptor, MHC genes, etc.; Atchison and Adesnik 1986; Pages et al. 1990; Yan et al. 1995; McFadyen et al. 1999; Ioannidu et al. 2001; Oldfield et al. 2001; Rolstad et al. 2001) were validated during our analysis, not all were detected. For example, the pancreatic type ribonuclease I represents a single-copy gene within most mammalian lineages that has expanded specifically within the genus Rattus (Dubois et al. 2002). It was not detected as a duplicated gene within rat genome assembly v. 3.1 by our criteria. A more detailed analysis of the gene showed that segmental duplications were indeed present, but the effective length of these alignments was less than 5 kb (below our detection threshold). Surprisingly, duplications of this gene were detected within a ≥5-kb pairwise alignment within a previous rat genome assembly (v.2.1; http://ratparalogy.gene.cwru.edu). The presence of sequence gaps, changes in sequence contig orientation, and our length threshold prevented its detection within the newer assembly. It is clear that duplications have been problematic during sequence and assembly. The analysis of the unplaced chromosome and random chromosomal sequence provides the best testament to this effect. The “unplaced” chromosome showed a marked enrichment for blocks of segmental duplication, with almost half (36.1/82.8 Mb) of the duplications assigned to this category. Further targeted efforts are required to resolve the true location, organization, and complexity of these regions.
Despite these methodological and assembly limitations, some important trends regarding rat segmental duplications emerged during our study. The overall content of highly homologous duplications as determined by the sequence assembly is greater within the rat (2.92%) than the mouse (1%–1.2%; Cheung et al. 2003b). Both are significantly reduced for segmental duplications compared to human (4.78%) for similar length thresholds (>5 kb; Bailey et al. 2002). The threefold difference between rat and mouse is surprising and may reflect biological differences or differences in the strategy for genome assembly. The mouse genome assembly strategy depended almost solely upon whole-genome shotgun (WGS) assembly, which has been predicted to overcollapse segmental duplication (Eichler 1998, 2001; Waterston et al. 2002). In contrast, the rat genome was assembled using a hybrid strategy, termed “BAC-enrichment.” The BAC-enrichment hybrid strategy entailed low-pass sequencing of 20,987 individual rat BAC clones, followed by an enrichment phase where individual WGS reads were mapped to specific BAC projects based on sequence overlap (RGSPC 2004). In such a scenario, paralogous regions within BACs would compete to optimally place WGS mate pairs and in so doing prevent overcollapse of duplicated regions.
Based on the current assembly, recent duplications are distributed in a nonuniform fashion across the genome. In addition to chromosome differences, we identified 41 duplication blocks (Fig. 3B) over 1 Mb in size. The extreme variation in sequence identity underlying the pairwise alignments (http://ratparalogy.gene.cwru.edu) within these blocks suggests that these areas have been the target of recurrent duplication over millions of years of evolution. The majority of duplications are organized as clusters of tandem or inverted intrachromosomal duplications. A similar bias toward clustered duplications was observed in the mouse genome assembly (Cheung et al. 2003b). Regions of extensive interchromosomal duplication were observed, particularly near the subtelomeric and pericentromeric regions. In the absence of detailed mapping information regarding the precise positions of centromeres and telomeres, it is difficult to assess these properties for all rat chromosomes. Our preliminary analyses of the rat genome, however, clearly shows a pericentromeric and subtelomeric bias for segmental duplications, suggesting that these may be general properties of mammalian chromosomal architecture. An analysis of the evolutionary genetic distance of all segmental duplications as a function of the sum of aligned base pairs (43,597 alignments) showed a bimodal distribution, particularly for intrachromosomal segmental duplications. Two peaks were observed, at 0.045 substitutions per site and 0.075 substitutions per site. Assuming that the rat and mouse lineages diverged 16–23 million years ago (Springer et al. 2003) and a neutral sequence divergence range of 0.173–0.195 years (RGSPC 2004), this bimodal distribution may correspond to bursts of segmental duplication that occurred approximately four and eight million years ago, respectively.
An analysis of the RefSeq genes (Methods) showed that segmental duplications are generally gene-poor based on their genomic representation (∼1.3% vs. 2.9%). Of the 63 genes that were identified within duplicated sequence, 33 were part of alignments which contained a complete complement of exons. Most of the duplications that contained genes were part of intrachromosomal alignments. A similar effect was observed when assigned rat mRNA was considered. This suggests that regions containing interchromosomal duplications are conspicuously transcriptionally silent. Our analysis was designed to recover genes that had emerged specifically within the rat lineage, because aligned genomic sequence between rat and mouse shows on average 0.175–0.195 substitutions per site and our study was limited to alignments showing less than 0.10 substitutions/site. Many of the rat duplication gene clusters recovered during this analysis (natural-killer cell receptor, serine protease inhibitor, carboxylesterase, cytochrome P450 gene families, etc.; Atchison and Adesnik 1986; Pages et al. 1990; Yan et al. 1995; McFadyen et al. 1999; Ioannidu et al. 2001; Oldfield et al. 2001; Rolstad et al. 2001) were also detected during an analysis of “recent” segmental duplication within the mouse lineage (Cheung et al. 2003b). The fact that such tandem duplications with extensive sequence identity exist within both lineages argues for active gene conversion (Atchison and Adesnik 1986) to maintain such homologous structures within each species.
Since the original analyses of working draft sequences of human and mouse (IHGSC 2001; Waterston et al. 2002), global studies of segmental duplication content have become an effective measure to assess one aspect of the quality of whole-genome sequence assemblies (Bailey et al. 2001, 2002; Cheung et al. 2001). Regions of recent segmental duplication remain one of the greatest challenges to finishing a genome sequence. Within the “finished” human genome assembly, for example, there is a striking correspondence between the position of sequence “gaps” among finished chromosomes and regions of large highly homologous duplications. Such areas have proven problematic for both clone-based methods and whole-genome shotgun sequence (Bailey et al. 2001, 2002; Cheung et al. 2001). In general, it is well recognized that the greater the proportion of large, highly homologous repeats, the more difficult a genome is to finish. Among certain genomes such as the human and mouse, high-quality ordered and oriented finished genome sequence is the stated goal. Concomitantly, it is expected that the structure and organization of such regions will ultimately be resolved—albeit with considerable effort and expenditure (Eichler 2001). Among other genomes such as the rat, finished genome sequence is not the stated goal. An initial assessment of segmental duplication content therefore provides an important level of annotation for the user of genome sequence information in the design and interpretation of experiments. Moreover, we argue that these initial analyses precisely delineate potential regions where whole-genome shotgun or a BAC-enrichment strategy will provide insufficient information for the biologist. In this study for example, we have identified <100 regions where the segmental duplications and bona fide gene families intersect. These regions include gene families important in drug detoxification, chemotaxis, and immunity. The content and structure of these regions will be pivotal to the full realization of the rat as a physiological model of pharmacology and complex genetic diseases (Jacob and Kwitek 2002). We therefore propose that such highly duplicated, generich regions be uncoupled from WGS sequencing strategies and be targeted for high-quality BAC-based finishing. The analysis presented here should provide a framework for the prioritization of such regions.
METHODS
Genome Resources
All reported analyses were performed on the June 2003 rat genome assembly (version 3.1). A complete segmental duplication analysis was also performed on an earlier assembly (version 2.1). The results of both analyses including pairwise sequence alignment locations, statistics, and gene content are available at http://ratparalogy.gene.cwru.edu. Segmental duplication analyses for version 3.1 have been added as a segmental duplication browser track as part of the UCSC browser (http://genome.ucsc.edu). Both rat genome assemblies were constructed using the BAC-enrichment strategy, which represents a hybrid between whole-genome shotgun sequence and clone-ordered approaches (see http://www.hgsc.bcm.tmc.edu/ for details). Genome sequences used in this study were derived from an inbred strain (BN/SsNHsd/MCW) of the brown Norway rat (Rattus norvegicus). The original inbred founder pair Harlan Sprague Dawley showed limited allelic variation; 6 of ∼4338 microsatellite loci (http://rgd.mcw.edu/). Brother–sister matings were performed for an additional 13 and 14 generations. A mother–daughter pair were the source of the whole-genome shotgun sequence library and the large-insert BAC library (CHORI-230).
Rat Segmental Duplications Detection
To analyze rat segmental duplications, we applied a BLAST-based whole-genome assembly comparison (Bailey et al. 2001). This BLAST-based method was designed to detect highly similar (≥90% identity) lineage-specific segmental duplications (≥1kb) after extracting common repeat sequences. We applied this method to the rat but detected an excess of smaller putative segmental duplications (Table 1) after using an updated Repeatmasker library database (June 2003). Upon inspection, many of the shorter alignments corresponded to incompletely masked high-copy repeats (LTR elements) or composite repeat elements (LTR/LINE hybrids). Because our detection algorithm extends seeding alignments into adjacent high-copy repeats, partially masked repeats will be lengthened to include the entire element. To circumvent the high-copy repeat overabundance, we selected a higher length threshold (≥5000 bp of seeding sequence). At this threshold, most uncharacterized transposable element alignments were eliminated. These seeding alignments were then trimmed to better define their end points, and optimal global alignments were performed to generate accurate alignment statistics. Alignments were then joined for gaps up to 10 kb in size. To avoid the potential of larger transposable elements as well as composite repeats, we considered various length thresholds (5, 10, and 20 kb). Sequence alignment statistics were calculated from optimal global alignments as described (Bailey et al. 2001), and paralogous sequence relationships were generated using Parasight graphical visualization software (J. Bailey, unpubl.).
Block-Size Delineation
We clustered duplications into larger blocks by examining the proximity of flanking sequences. A “weld” was performed if another pairwise alignment was identified within 100 kb from the coordinates of a pairwise alignment (Table 3). Gaps were not included in this calculation. Clustering proceeded in both directions from the seed pairwise alignment until a unique region (no duplications) of at least 100 kb was encountered per each cluster. (Table 3). Analysis of flanking sequences was performed based on these “weld” coordinates.
Gene Analysis
Gene content of rat segmental duplications was assessed using two differences sources of data: LocusLink RefSeq gene annotations and rat mRNAs in GenBank. All mRNAs were aligned using BLAT as described (Kent et al. 2002), and intersections between segmental duplication coordinates and exon positions were compared using mySQL queries of the UCSC browser database. During our analysis, a total of 63 RefSeq genes (from a genome total of 4532) and 945 rat mRNAs (from a genome total of 11,560) were identified that had been assigned to duplicated regions. Of these, 716 mRNAs were identified that did not overlap with Ref-Seq gene coordinates. In addition, 61/63 of the RefSeq genes contained two or more exons.
Acknowledgments
We thank members of the Rat Genome Sequencing Project for open discussion and access to unpublished data during the preparation of this manuscript. We are particularly grateful to Norbert Huebner, Michael Jensen-Seaman, and Arian Smit for information regarding the putative centromere positions within the rat genome assembly. This work was supported in part by NIH grants GM58815 and HG002318 and U.S. Department of Energy grant ER62862 to E.E.E., an NIH Career Development Program in Genomic Epidemiology of Cancer (CA094816) to J.A.B., the W.M. Keck Foundation, and the Charles B. Wang Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1907504.
References
- Atchison, M. and Adesnik, M. 1986. Gene conversion in a cytochrome P-450 gene family. Proc. Natl. Acad. Sci. 83: 2300-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, J.A., Yavor, A.M., Massa, H.F., Trask, B.J., and Eichler, E.E. 2001. Segmental duplications: Organization and impact within the current human genome project assembly. Genome Res. 11: 1005-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, J.A., Gu, Z., Clark, R.A., Reinert, K., Samonte, R.V., Schwartz, S., Adams, M.D., Myers, E.W., Li, P.W., and Eichler, E.E. 2002. Recent segmental duplications in the human genome. Science 297: 1003-1007. [DOI] [PubMed] [Google Scholar]
- Bailey, J.A., Giu, L., and Eichler, E.E. 2003. An Alu transposition model for the origin and expansion of human segmental duplications. Am. J. Hum. Genet. 73: 823-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, J., Estivill, X., Khaja, R., MacDonald, J.R., Lau, K., Tsui, L.C., and Scherer, S.W. 2003a. Genome-wide detection of segmental duplications and potential assembly errors in the human genome sequence. Genome Biol. 4: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, J., Wilson, M.D., Zhang, J., Khaja, R., MacDonald, J.R., Heng, H.H., Koop, B.F., and Scherer, S.W. 2003b. Recent segmental and gene duplications in the mouse genome. Genome Biol. 4: R47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, V.G., Nowak, N., Jang, W., Kirsch, I.R., Zhao, S., Chen, X.N., Furey, T.S., Kim, U.J., Kuo, W.L., Olivier, M., et al. 2001. Integration of cytogenetic landmarks into the draft sequence of the human genome. The BAC Resource Consortium. Nature 409: 953-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal, P., Predki, P., Olsen, A.S., Kobayashi, A., Folta, P., Lucas, S., Land, M., Terry, A., Zhou, C.L.E., Rash, S., et al. 2001. Human chromosome 19 and related regions in mouse: Conservative and lineage specific evolution. Science 293: 104-111. [DOI] [PubMed] [Google Scholar]
- Dubois, J.Y., Jekel, P.A., Mulder, P.P., Bussink, A.P., Catzeflis, F.M., Carsana, A., and Beintema, J.J. 2002. Pancreatic-type ribonuclease 1 gene duplications in rat species. J. Mol. Evol. 55: 522-533. [DOI] [PubMed] [Google Scholar]
- Duda, T.F. and Palumbi, S.R. 1999. Molecular genetics of ecological diversification: Duplication and rapid evolution of toxin genes of the venomous gastropod Conus. Proc. Natl. Acad. Sci. 96: 6820-6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler, E.E. 1998. Masquerading repeats: Paralogous pitfalls of the human genome. Genome Res. 8: 758-762. [DOI] [PubMed] [Google Scholar]
- Eichler, E.E. 2001. Segmental duplications: What's missing, misassigned, and misassembled—And should we care? Genome Res. 11: 653-656. [DOI] [PubMed] [Google Scholar]
- Eichler, E.E. and Sankoff, D. 2003. Structural dynamics of eukaryotic chromosome evolution. Science 301: 793-797. [DOI] [PubMed] [Google Scholar]
- Eichler, E.E., Lu, F., Shen, Y., Antonacci, R., Jurecic, V., Doggett, N.A., Moyzis, R.K., Baldini, A., Gibbs, R.A., and Nelson, D.L. 1996. Duplication of a gene-rich cluster between 16p11.1 and Xq28: A novel pericentromeric-directed mechanism for paralogous genome evolution. Hum. Mol. Genet. 5: 899-912. [DOI] [PubMed] [Google Scholar]
- Estivill, X., Cheung, J., Pujana, M.A., Nakabayashi, K., Scherer, S.W., and Tsui, L.C. 2002. Chromosomal regions containing high-density and ambiguously mapped putative single nucleotide polymorphisms (SNPs) correlate with segmental duplications in the human genome. Hum. Mol. Genet. 11: 1987-1995. [DOI] [PubMed] [Google Scholar]
- Goodier, J.L., Ostertag, E.M., and Kazazian Jr., H.H. 2000. Transduction of 3′-flanking sequences is common in L1 retrotransposition. Hum. Mol. Genet. 9: 653-657. [DOI] [PubMed] [Google Scholar]
- Horvath, J., Schwartz, S., and Eichler, E. 2000. The mosaic structure of a 2p11 pericentromeric segment: A strategy for characterizing complex regions of the human genome. Genome Res. 10: 839-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium (IHGSC). 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-920.11237011 [Google Scholar]
- Ioannidu, S., Walter, L., Dressel, R., and Gunther, E. 2001. Physical map and expression profile of genes of the telomeric class I gene region of the rat MHC. J. Immunol. 166: 3957-3965. [DOI] [PubMed] [Google Scholar]
- Jackson, M.S., Rocchi, M., Thompson, G., Hearn, T., Crosier, M., Guy, J., Kirk, D., Mulligan, L., Ricco, A., Piccininni, S., et al. 1999. Sequences flanking the centromere of human chromosome 10 are a complex patchwork of arm-specific sequences, stable duplications, and unstable sequences with homologies to telomeric and other centromeric locations. Hum. Mol. Genet. 8: 205-215. [DOI] [PubMed] [Google Scholar]
- Jacob, H.J. and Kwitek, A.E. 2002. Rat genetics: Attaching physiology and pharmacology to the genome. Nat. Rev. Genet. 3: 33-42. [DOI] [PubMed] [Google Scholar]
- Kellis, M., Patterson, N., Endrizzi, M., Birren, B., and Lander, E.S. 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423: 241-254. [DOI] [PubMed] [Google Scholar]
- Kent, W.J., Sugnet, C.W., Furey, T.S., Roskin, K.M., Pringle, T.H., Zahler, A.M., and Haussler, D. 2002. The human genome browser at UCSC. Genome Res. 12: 996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadyen, D.A. and Locke, J. 2000. High-resolution FISH mapping of the rat α2u-globulin multigene family. Mamm. Genome 11: 292-299. [DOI] [PubMed] [Google Scholar]
- McFadyen, D.A., Addison, W., and Locke, J. 1999. Genomic organization of the rat α2u-globulin gene cluster. Mamm. Genome 10: 463-470. [DOI] [PubMed] [Google Scholar]
- Muller, H.J. 1936. Bar duplication. Science 83: 528-530. [DOI] [PubMed] [Google Scholar]
- Ohno, S. 1970. Evolution by gene duplication. Springer Verlag, Berlin.
- Oldfield, S., Grubb, B.D., and Donaldson, L.F. 2001. Identification of a prostaglandin E2 receptor splice variant and its expression in rat tissues. Prostaglandins 63: 165-173. [DOI] [PubMed] [Google Scholar]
- Pages, G., Rouayrenc, J.F., Rossi, V., Le Cam, G., Mariller, M., Szpirer, J., Szpirer, C., Levan, G., and Le Cam, A. 1990. Primary structure and assignment to chromosome 6 of three related rat genes encoding liver serine protease inhibitors. Gene 94: 273-282. [DOI] [PubMed] [Google Scholar]
- Pickeral, O.K., Makalowski, W., Boguski, M.S., and Boeke, J.D. 2000. Frequent human genomic DNA transduction driven by LINE-1 retrotransposition. Genome Res. 10: 411-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rat Genome Sequencing Project Consortium (RGSPC). 2004. Genome sequence of the Brown Norway Rat yields insights into mammalian evolution. Nature (in press). [DOI] [PubMed]
- Rolstad, B., Naper, C., Lovik, G., Vaage, J.T., Ryan, J.C., Backman-Petersson, E., Kirsch, R.D., and Butcher, G.W. 2001. Rat natural killer cell receptor systems and recognition of MHC class I molecules. Immunol. Rev. 181: 149-157. [DOI] [PubMed] [Google Scholar]
- Springer, M.S., Murphy, W.J., Eizirik, E., and O'Brien, S.J. 2003. Placental mammal diversification and the Cretaceous-Tertiary boundary. Proc. Natl. Acad. Sci. 100: 1056-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, J.W., Schueler, M.G., Summers, T.J., Blakesley, R.W., McDowell, J.C., Thomas, P.J., Idol, J.R., Maduro, V.V., Lee-Lin, S.Q., Touchman, J.W., et al. 2003. Pericentromeric duplications in the laboratory mouse. Genome Res. 13: 55-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- Yan, B., Yang, D., and Parkinson, A. 1995. Cloning and expression of hydrolase C, a member of the rat carboxylesterase family. Arch. Biochem. Biophys. 317: 222-234. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://ratparalogy.cwru.edu; Segmental Duplication Database for Rat at CWRU.
- http://genome.ucsc.edu; Genome browser at Univ. California–Santa Cruz.
- http://www.hgsc.bcm.tmc.edu/; Human Genome Sequencing Center at Baylor College of Medicine. [DOI] [PubMed]
- http://rgd.mcw.edu/; Rat Genome Database at Medical College of Wisconsin.