The diuretic drug spironolactone up-regulates NKG2D ligand expression in colon cancer cells via activation of the ATM–Chk2–mediated checkpoint pathway to enhance the antitumor function of NK cells.
Abstract
Tumor metastasis and lack of NKG2D ligand (NKG2DL) expression are associated with poor prognosis in patients with colon cancer. Here, we found that spironolactone (SPIR), an FDA-approved diuretic drug with a long-term safety profile, can up-regulate NKG2DL expression in multiple colon cancer cell lines by activating the ATM–Chk2-mediated checkpoint pathway, which in turn enhances tumor elimination by natural killer cells. SPIR can also up-regulate the expression of metastasis-suppressor genes TIMP2 and TIMP3, thereby reducing tumor cell invasiveness. Although SPIR is an aldosterone antagonist, its antitumor effects are independent of the mineralocorticoid receptor pathway. By screening the human nuclear hormone receptor siRNA library, we identified retinoid X receptor γ (RXRγ) instead as being indispensable for the antitumor functions of SPIR. Collectively, our results strongly support the use of SPIR or other RXRγ agonists with minimal side effects for colon cancer prevention and therapy.
Colorectal cancer afflicts ∼150,000 Americans, and one million people worldwide, annually (Cappell, 2008). Significant progress has been made in understanding its molecular carcinogenesis and in early detection. Surgery, chemotherapy, and radiation have remained the mainstays of therapy. Unfortunately, colorectal cancer remains the second leading cause of death from cancer in the United States, and patient survival has improved only modestly during the past two decades. Therefore, further investigations in novel therapies such as immunotherapy are warranted.
Both NK and cytotoxic T cells express the NKG2D receptor that can recognize NKG2D ligands (NKG2DLs; MICA/B and ULBP1-6) on tumor cells (González et al., 2008; Champsaur and Lanier, 2010). Although the molecular mechanism that regulates NKG2DL expression is not clear, a variety of stimuli, such as oxidation, heat shock, and DNA damage, which induce cell stress and tumor transformation, have been shown to selectively up-regulate NKG2DL expression (Groh et al., 1996; Yamamoto et al., 2001; Gasser et al., 2005). During tumor progression, however, most tumor cells eventually acquire various mechanisms, such as ligand shedding and microRNA expression, which down-regulate the surface expression of NKG2DLs, resulting in immunosurveillance escape (Salih et al., 2002; Stern-Ginossar et al., 2008). In colorectal carcinoma, high expression of NKG2DLs is frequently observed in early stages of tumor development. The ligand expression, however, progressively decreases in later stages of the disease, and the level of expression correlates with patient survival (McGilvray et al., 2009). Studies are needed to elucidate the mechanism of NKG2DL regulation and to identify therapeutic drugs that can up-regulate their surface expression (González et al., 2008).
To this aim, we established a high-throughput compound library screening to identify drug candidates that can up-regulate NKG2DL expression. We found that the aldosterone antagonist spironolactone (SPIR) could up-regulate NKG2DL surface expression on multiple colorectal cancer cells through the activation of ATM–Chk2–mediated checkpoint pathway that greatly enhanced cancer cell susceptibility to NK cell cytolysis. We also found that SPIR enhanced the expression of metastasis-suppressor genes TIMP2 and TIMP3, resulting in a significant reduction of cancer metastasis in animal models. The drug effects were independent of mineralocorticoid receptor (MR) but required the activation of retinoid X receptor γ (RXRγ). Collectively, our findings support the potential use of SPIR or other RXRγ agonists in colorectal cancer.
RESULTS
SPIR up-regulates NKG2DL expression in multiple colorectal carcinoma cell lines
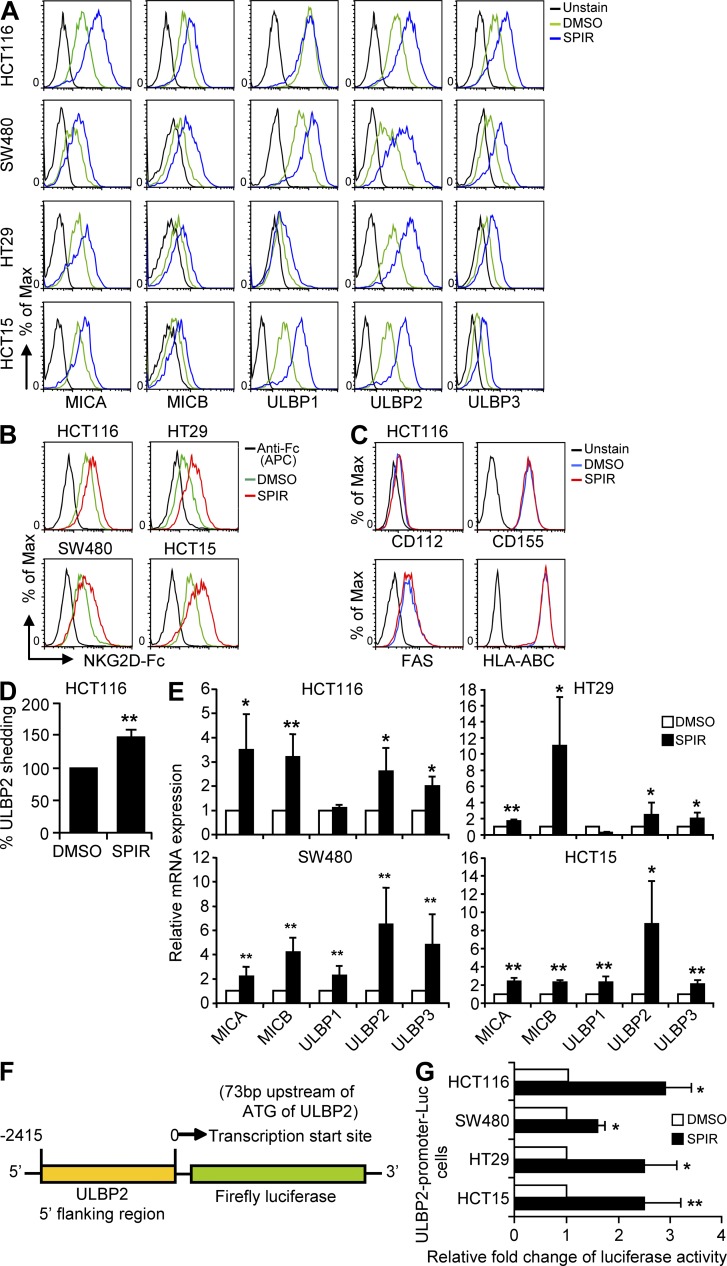
In our high-throughput screening of 5,600 bioactive compounds, the surface expression of an NKG2DL, ULBP2, was significantly increased in 293T cells in the presence of the aldosterone antagonist SPIR (unpublished data). We therefore investigated whether SPIR could increase NKG2DL expression on four representative colorectal carcinoma cell lines (HCT116, HT29, SW480, and HCT15) covering the most common tumorigenic mutations (Table S1). Consistent with the screening results in 293T cells, SPIR treatment significantly increased ULBP2 expression in all cancer cell lines (Fig. 1 A) in a dose-dependent manner without affecting the viability of the cells (not depicted). We also observed a moderate enhancement of MICA/B and ULBP3 expression in all SPIR-treated cells and a selective increase of ULBP1 expression in SW480 and HCT15 cells. Consistent NKG2DL up-regulation was also observed by staining the SPIR-treated colon cancer cell lines with soluble NKG2D-Fc molecules (Fig. 1 B). However, there was no increase in expression of other NK cell ligands such as DNAM-1 ligands (CD112 and CD155), Fas, and HLAs in SPIR-treated HCT116 cells (Fig. 1 C), indicating that the up-regulation of NKG2DLs by SPIR was specific. A similar increase in NKG2DL expression was observed upon SPIR treatment in two other colorectal carcinoma cell lines (COLO201 and LS174T) and in other types of human cancer cells, including head and neck squamous cell carcinoma (CAL27), prostate cancer (DU145), hepatocellular carcinoma (HepG2), and rhabdomyosarcoma (RD), but not in normal cells such as peripheral blood mononuclear cells (unpublished data).
Figure 1.
SPIR up-regulates NKG2DL expression. (A) Cell surface expression of MICA-B and ULBP1-3 was analyzed by flow cytometry in multiple colon cancer cell lines treated with DMSO (solvent control) or SPIR (56 µM) for 3 d. Data are representative of five independent experiments. (B) NKG2D-Fc staining was performed to evaluate the change of total NKG2DL expression in the colon cancer cell lines upon SPIR treatment. Controls were stained with APC-conjugated anti–human Fc antibody alone without preincubation with NKG2D-Fc. Data are representative of three independent experiments. (C) DNAM1 ligands (CD112 and CD155), Fas, and HLA-ABC surface expressions on HCT16 cells treated with DMSO or SPIR (56 µM) for 3 d were analyzed by flow cytometry. Results are representative of two independent experiments. (D) Shedding of ULBP2 to the culture supernatant from SPIR-treated (56 µM for 3 d) or untreated HCT116 cells was detected by ELISA (n = 3). (E) NKG2DL mRNA expression in the colon cancer cell lines was analyzed by qRT-PCR analysis. Data are normalized to GAPDH mRNA levels and are presented as fold change relative to the expression in DMSO-treated cells (n = 5). (F) Lentiviral-based luciferase reporter construct containing the putative ULBP2 promoter (–2,415 to 0 bp from the transcription start site: 73 bp upstream of ATG) was transduced into the colon cancer cell lines. Positively transduced cells were selected by puromycin treatment (2 µg/ml) for 2 wk. (G) Luciferase activities of the colon cancer cell lines transfected with the putative ULBP2 promoter construct were determined 3 d after DMSO or SPIR treatment. Data shown are expressed as fold change relative to the luciferase activity observed in DMSO-treated cells (n = 3). *, P < 0.05; **, P < 0.01.
Ligand shedding mediated by metalloproteinases has been observed in various types of cancer (Waldhauer and Steinle, 2006; Waldhauer et al., 2008). We compared the amount of soluble ULBP2 in the culture supernatant of SPIR-treated or untreated HCT116 cells by ELISA. As shown in Fig. 1 D, SPIR treatment did not reduce but rather moderately increased the amount of soluble ULBP2 from HCT116 cells. Quantitative real-time polymerase chain reaction (qRT-PCR) assays showed an increase in mRNA levels (Fig. 1 E) corresponding to the enhanced surface expression of NKG2DLs. We also observed a significant increase in luciferase activity (∼1.5-fold to 3-fold over the solvent control, DMSO treatment) in all SPIR-treated colon cancer cell lines bearing a luciferase reporter construct driven by a putative ULBP2 promoter (Fig. 1, F and G). Collectively, these data suggest that SPIR up-regulates NKG2DL expression by promoting gene transcription and protein production rather than by inhibiting shedding.
SPIR enhances tumor cell sensitivity to NK cell–mediated cytolysis
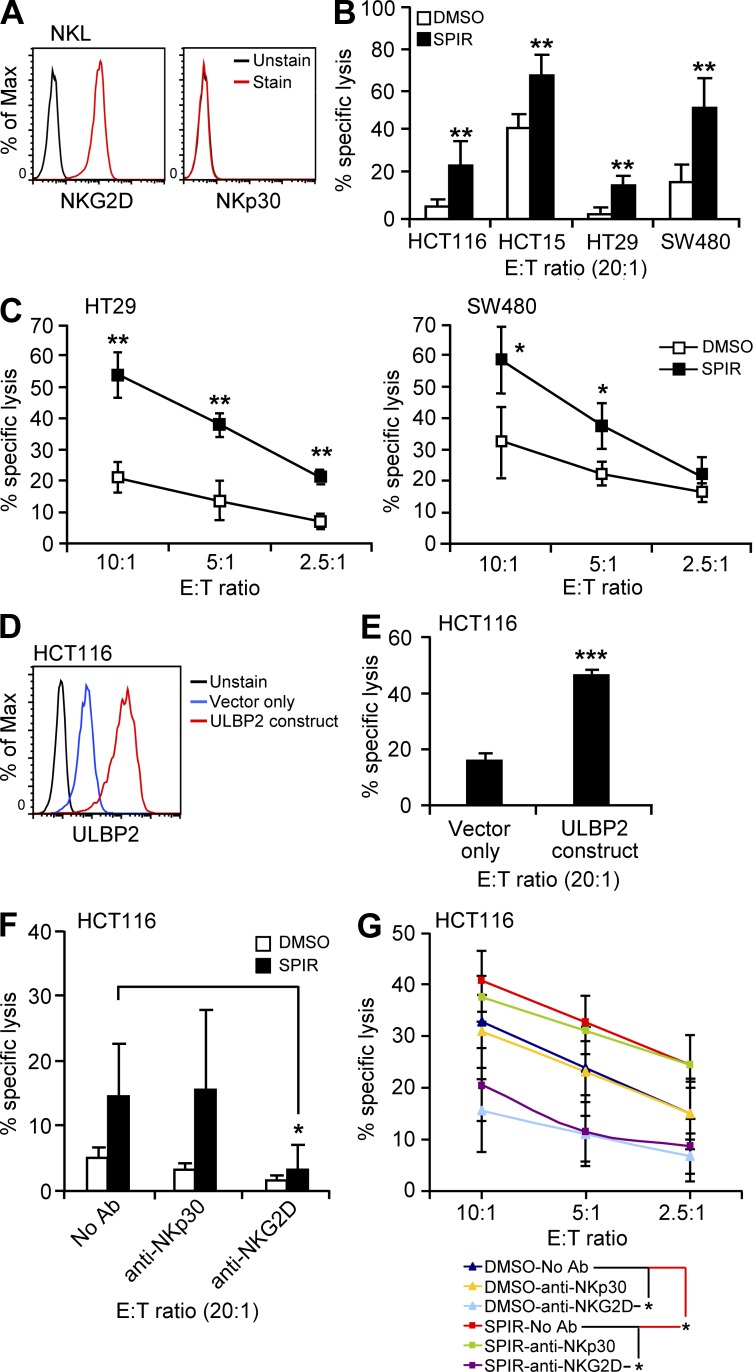
To determine whether the increased expression of NKG2DLs induced by SPIR enhanced tumor cell lysis by NK cells, we evaluated NK cell cytotoxicity to the drug-treated or untreated cells by using the NKG2D-expressing NK cell line NKL (Fig. 2, A and B) and interleukin-2–activated primary NK cells (Fig. 2 C). Upon SPIR treatment, the susceptibility of all cell lines to NKL lysis was significantly increased. Similarly, treatment of the HT29 and SW480 cells with SPIR markedly enhanced their susceptibility to primary NK cell–mediated lysis.
Figure 2.
SPIR enhances tumor cell sensitivity to NK cell killing. (A) NKL cells express high level of NKG2D but not NKp30 on their cell surface as determined by flow cytometry. Results are representative of two independent experiments. Therefore, the use of anti-NKp30 in NKL killing assay (described in panel F) was considered as a nonspecific IgG blockade relative to anti-NKG2D. (B) NK cell cytotoxicity on the colon cancer cell lines treated with DMSO or SPIR (56 µM) for 3 d was determined by a BATDA release assay using NKL cells as effector cells (n = 3). (C) NK cell cytotoxicity on HT29 and SW480 cells treated with DMSO or SPIR (56 µM) for 3 d was determined by a BATDA release assay using IL-2 (10 U/ml)–primed primary NK cells isolated from healthy donors at various E:T ratios (n = 3). (D) HCT116 cells transduced with control or a ULBP2-overexpressing lentiviral construct were analyzed by flow cytometry for the surface expression of ULBP2. Results are representative of two independent experiments. (E) NK cell cytotoxicity on the ULBP2-transduced HCT116 cells was determined by a BATDA release assay using NKL cells as effector cells (n = 3). (F) DMSO- or SPIR-treated (3 d) HCT116 cells were subjected to a BATDA release assay using NKL cells in the presence or absence of anti-NKp30 or anti-NKG2D antibodies (10 µg/ml; n = 3). (G) DMSO- or SPIR-treated (3 d) HCT116 cells were subjected to a BATDA release assay using IL-2 (10 U/ml)-primed primary NK cells isolated from healthy donors at various E:T ratios in the presence or absence of anti-NKp30 or anti-NKG2D antibodies (10 µg/ml; n = 3). *, P < 0.05; **, P < 0.005; ***, P < 0.0001.
To verify that the enhancement of tumor cell lysis directly correlated with increased NKG2DL expression, we first overexpressed ULBP2 in HCT116 cells by lentiviral transduction (Fig. 2 D). As shown in Fig. 2 E, increased expression of ULBP2 clearly rendered HCT116 cells more susceptible to NKL–mediated lysis. Additionally, the enhancement of NK cell cytotoxicity against SPIR-treated HCT116 cells was completely abolished in the presence of blocking antibodies against NKG2D but not with those against NKp30 (an activating receptor that induces NK cell cytotoxicity independent of NKG2D–NKG2DL interactions (André et al., 2004; Fig. 2, F and G). This result confirmed the direct involvement of NKG2D–NKG2DL interaction in NK-mediated lysis of the SPIR-treated cells.
SPIR exerts antitumor activities in vivo
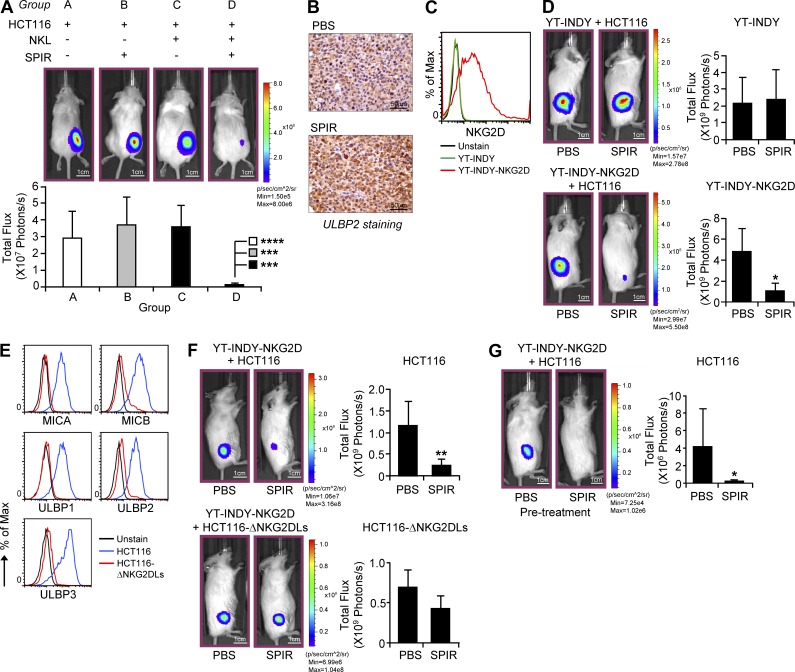
To evaluate the in vivo antitumor efficacy of SPIR, we established a xenograft mouse model (luciferase-expressing HCT116 cells in NOD-SCID γ [NSG] mice) and performed bioluminescence imaging to monitor tumor growth. HCT116 cells were injected with or without NKL cells subcutaneously into NSG mice. The mice were untreated or treated with SPIR 2 d after the tumor implantation (twice a week for 2 wk before tumor imaging). As shown in Fig. 3 A, the treatment with SPIR alone (group B) or NKL alone (group C) did not affect tumor growth in the mice. However, injecting SPIR into the mice bearing NKL cells (group D) suppressed tumor growth considerably. In line with our in vitro data, SPIR injection significantly enhanced ULBP2 expression in the HCT116 xenografts (Fig. 3 B).
Figure 3.
SPIR exerts antitumor activities in vivo. (A) 106 luciferase-expressing HCT116 cells, alone or with 2 × 107 NKL cells, were subcutaneously injected into 8–10-wk-old NSG mice (n = 4). 2 d after the cell injection, SPIR (1.25 mg/mouse) or PBS was intraperitoneally injected into the mice twice a week for 2 wk. HCT116 xenograft growth was analyzed by performing bioluminescence imaging on day 15. (B) 106 HCT116 cells were subcutaneously injected into 8–10-wk-old NSG mice. 2 d after the cell injection, SPIR (1.25 mg/mouse) or PBS was intraperitoneally injected into the mice twice a week for 2 wk (n = 3 per group). Tumor sections were prepared for ULBP2 immunohistochemical staining on day 15. (C) NKG2D expression on the cell surface of YT-INDY and YT-INDY-NKG2D cells was analyzed by flow cytometry. Results are representative of two independent experiments. (D) 106 luciferase-expressing HCT116 cells were subcutaneously injected into 8–10-wk-old NSG mice with either 106 YT-INDY or YT-INDY-NKG2D cells (n = 8 per group). 2 d after the cell injection, SPIR (1.25 mg/mouse) or PBS was intraperitoneally injected into the mice (n = 4 per group) twice a week for 2 wk. HCT116 xenograft growth was analyzed by performing bioluminescence imaging on day 15. (E) MICA-B and ULBP1-3 expressions on the cell surface of HCT116 or HCT116-ΔNKG2DLs cells were analyzed by flow cytometry. Results are representative of two independent experiments. (F) 106 luciferase-expressing HCT116 or HCT116-ΔNKG2DLs cells were subcutaneously injected into 8–10-wk-old NSG mice with 106 YT-INDY-NKG2D cells (n = 8 per group). 2 d after the cell injection, SPIR (1.25 mg/mouse) or PBS was intraperitoneally injected into the mice (n = 4 per group) every other day for three times. HCT116 xenograft growth was analyzed by performing bioluminescence imaging on day 10. (G) 8–10-wk-old NSG mice were intraperitoneally injected with PBS or SPIR (1.25 mg per mouse) twice a week for two weeks (n = 10 per group). 1 × 106 each of luciferase-expressing HCT116 cells and YT-INDY-NKG2D were injected subcutaneously into the mice on day 0. HCT116 xenograft growth was analyzed by performing bioluminescence imaging on day 3. *, P < 0.05; **, P < 0.01; ***, P < 0.005; ****, P < 0.0005.
To confirm that the in vivo antitumor effect of SPIR depends on the expression of NKG2D in NK cells, we established another HCT116 xenograft model with either NKG2D-negative NK cell line YT-INDY or YT-INDY stably expressing NKG2D (YT-INDY-NKG2D; Fig. 3 C). As shown in Fig. 3 D, SPIR treatment did not reduce the HCT116 xenograft growth in YT-INDY–bearing mice. The drug treatment, however, strongly suppressed the tumor growth in the presence of YT-INDY-NKG2D. To further confirm that the in vivo antitumor effect of SPIR also requires the increased surface expression of NKG2DLs in tumor cells, we generated NKG2DL–deficient HCT116 cells (HCT116-ΔNKG2DLs; Fig. 3 E). As shown in Fig. 3 F, decreased surface expression of NKG2DLs in HCT116 cells significantly reduced tumor susceptibility to NK cell killing triggered by SPIR in vivo. Collectively, these data suggest that SPIR increases NKG2DL expression in tumor cells in vivo, thereby enhancing NKG2D-dependent tumor control by NK cells. Moreover, pretreating NSG mice with SPIR twice a week for 2 wk before HCT116 and YT-INDY-NKG2D implantation greatly inhibited tumor development (Fig. 3 G), indicating that SPIR may also serve as a cancer prevention drug.
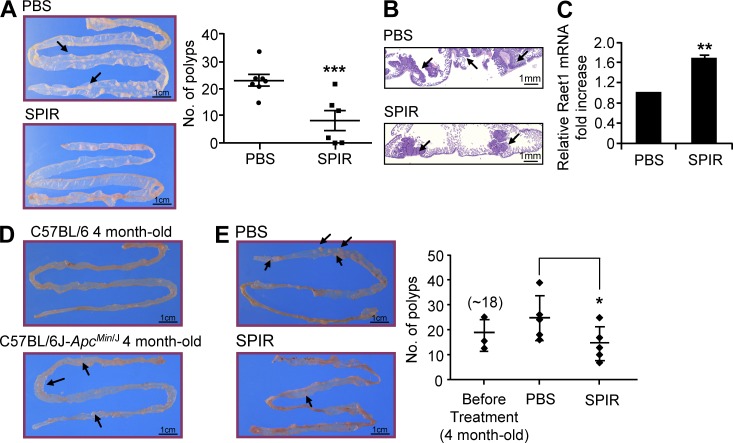
To further explore the potential of SPIR in colon cancer prevention and therapy in vivo, we studied the C57BL/6J-ApcMin/J mice, an APC-mutated strain that is highly susceptible to spontaneous intestinal neoplasia formation (Su et al., 1992). For cancer prevention, we treated 8-wk-old C57BL/6J-ApcMin/J mice, in which polyp development has not yet started (unpublished data) with SPIR twice a week for 3 mo. Intestines were collected from the mice for polyp counting and immunohistochemistry (Fig. 4, A and B). The number of polyps formed in SPIR-treated mice was significantly less than that in untreated mice. We then collected the polyp samples from SPIR-treated or untreated mice to prepare cDNA for qRT-PCR. As shown in Fig. 4 C, SPIR treatment significantly increased the expression of the Raet1 ligand in the polyps (either Raet1d or Raet1e in the B6 mouse background; Nausch and Cerwenka, 2008), showing SPIR-mediated NKG2DL up-regulation similar to that observed in the previous human xenograft models. To further explore the therapeutic potential of SPIR in this model, 4-mo-old C57BL/6J-ApcMin/J mice that had already developed intestinal polyps (Fig. 4 D) were treated with SPIR twice a week for 3 wk. A significant reduction of the number of polyps was observed in SPIR-treated mice compared with the untreated controls (Fig. 4 E). These findings support the use of SPIR in both colon cancer prevention and treatment.
Figure 4.
SPIR treatment demonstrates both preventive and therapeutic potential in C57BL/6J-ApcMin/J Mouse model. (A) 8-wk-old C57BL/6J-ApcMin/J mice were given intraperitoneal injections of PBS (n = 7) or SPIR (n = 6) twice a week for 3 mo. The number of spontaneous intestinal polyps (arrows) was counted and compared between the two groups of mice at the end of the 3 mo period. (B) The difference in adenoma growth (arrows) between the PBS- and SPIR-treated mouse groups was confirmed in intestine tissue sections. (C) Polyp samples were collected from C57BL/6J-ApcMin/J mice given intraperitoneal injections of PBS or SPIR (n = 3 per group) twice a week for two months. Total RNA were collected for cDNA preparation and the expressions of the mouse NKG2D ligands Raet1 family were analyzed by qRT-PCR. (D) Representative appearances of small intestine of 4-mo old C57BL/6 (no intestinal polyp) and C57BL/6J-ApcMin/J (displayed obvious polyps - arrows) mice were shown (n = 2). (E) Four month-old C57BL/6J-ApcMin/J mice were given intraperitoneal injections of PBS or SPIR (1.25 mg per mouse; n = 5 per group) twice a week for 3 wk. The number of spontaneous intestinal polyps (arrows) was counted and compared between the two groups of mice. *, P ≤ 0.05, **, P ≤ 0.01, ***, P < 0.005.
The up-regulation of NKG2DL expression by SPIR is independent of the MR pathway
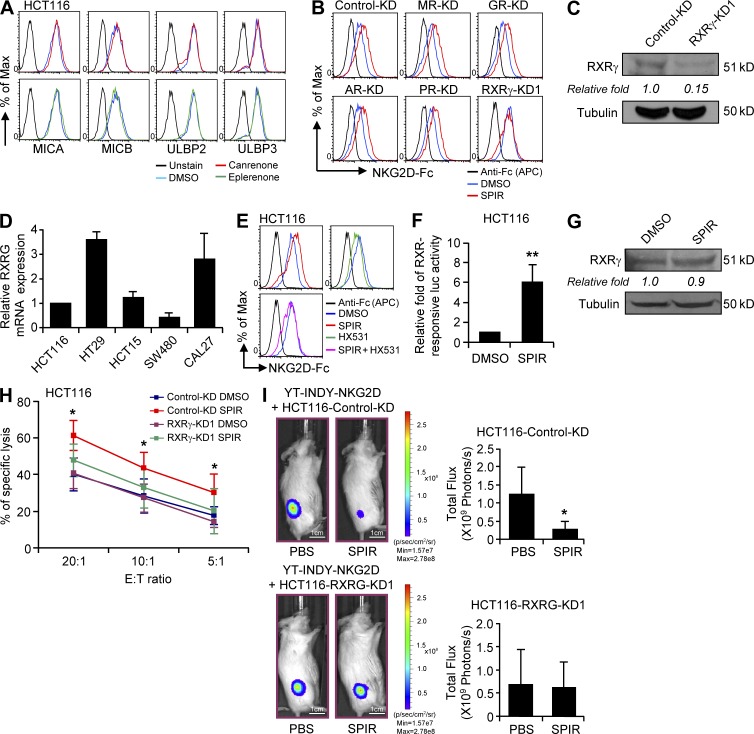
SPIR has long been clinically used as an aldosterone antagonist, competing with aldosterone for interaction with mineralocorticoid receptor (MR; Struthers et al., 2008). Using MR expression in 293T cells as a relative standard, we found that both mRNA and protein levels of MR were significantly lower in all the colon cancer cell lines and in CAL27 cells (Fig. S1, A and B). Additionally, shRNA knockdown of MR in 293T cells did not affect the up-regulation of ULBP2 upon SPIR treatment (Fig. S1, C and D). Treating HCT116 cells with two other potent MR antagonists, canrenone and eplerenone, did not reproduce the phenotypes of enhanced expression of NKG2DL upon SPIR treatment (Fig. 5 A), confirming that MR is not involved in SPIR’s mechanism of action.
Figure 5.
SPIR exerts its effects on colon cancer through the activation of RXRγ but not MR. (A) NKG2DL surface expression was analyzed in HCT116 cells cultured with DMSO, canrenone, or eplerenone for 3 d. Data shown are representative of three independent experiments. (B) NKG2DL expression in control, MR, GR, AR, PR and RXRγ knockdown HCT116 cells was compared after DMSO or SPIR treatment (3 d) by NKG2D-Fc staining. Data shown are representative of three independent experiments. (C) The protein level of RXRγ was compared between control and RXRγ-KD1 HCT116 cells by Western blot analysis. Data shown are representative of two independent experiments. (D) The mRNA expression of RXRγ in the carcinoma cell lines was confirmed by using qRT-PCR assays. Data are presented as fold differences relative to the mRNA expression in HCT116 cells (n = 2). (E) NKG2DL expression was analyzed in HCT116 cells treated for 3 d with SPIR (56 µM), HX531 (10 µM), or both. Data shown are representative of three independent experiments. (F) Luciferase activity was measured in HCT116 cells transfected with an RXRγ-responsive luciferase construct after DMSO or SPIR treatment for 24 h (n = 3). (G) The protein expression of RXRγ was compared between DMSO- and SPIR-treated HCT116 cells. Data shown are representative of three independent experiments. (H) HCT116 cells were transfected with control KD or RXRγ-KD1 siRNA on day 0. The cells were treated with DMSO or SPIR (56 µM) on day 2. NK cell cytotoxicity on the tumor cells was determined by a BATDA release assay on day 5 using IL-2 (10 U/ml)–primed primary NK cells isolated from healthy donors at various E:T ratios (n = 3). (I) Luciferase-expressing HCT116 cells were transfected with either control-KD or RXRγ-KD1 siRNA on day 0. The transfected cells were collected on the next day to inject (106 cells/mouse) subcutaneously into 8–10-wk-old NSG mice with 106 YT-INDY-NKG2D cells (n = 8 per group). 2 d after the cell injection, SPIR (1.25 mg/mouse) or PBS was intraperitoneally injected into the mice (n = 4 per group) every other day, three times. HCT116 xenograft growth was analyzed by performing bioluminescence imaging on day 10. *, P < 0.05; **, P < 0.0001.
SPIR acts as an RXRγ agonist for the up-regulation of NKG2DL expression and enhancement of tumor susceptibility to NK cytolysis
To identify the nuclear hormone receptor (NHR) responsible for the effect of SPIR, we used a siRNA library (3 specific siRNAs per target gene) to target 47 NHRs in HCT116 cells. Consistent with our previous results (Fig. S1, C and D), specific knockdown of MR did not affect NKG2DL up-regulation in HCT116 cells by SPIR (Fig. 5 B). Although SPIR is also known to interact with glucocorticoid receptor (GR), androgen receptor (AR), and progesterone receptor (PR; Williams et al., 2006), none of the siRNAs targeting these NHRs affected the SPIR-mediated NKG2DL up-regulation. In contrast, five different siRNAs (three from the library and two added for validation) specifically knocked down the expression of RXRγ (Dawson and Xia, 2012; Fig. 5 C) and significantly reduced SPIR’s up-regulation of NKG2DLs (Fig. 5 B and not depicted). Consistent with previous studies, RXRγ was constitutively expressed in HCT116 cells (Fig. 5 C; Papi et al., 2010). We also observed expression of RXRγ mRNA in all other cell lines tested (Fig. 5 D).
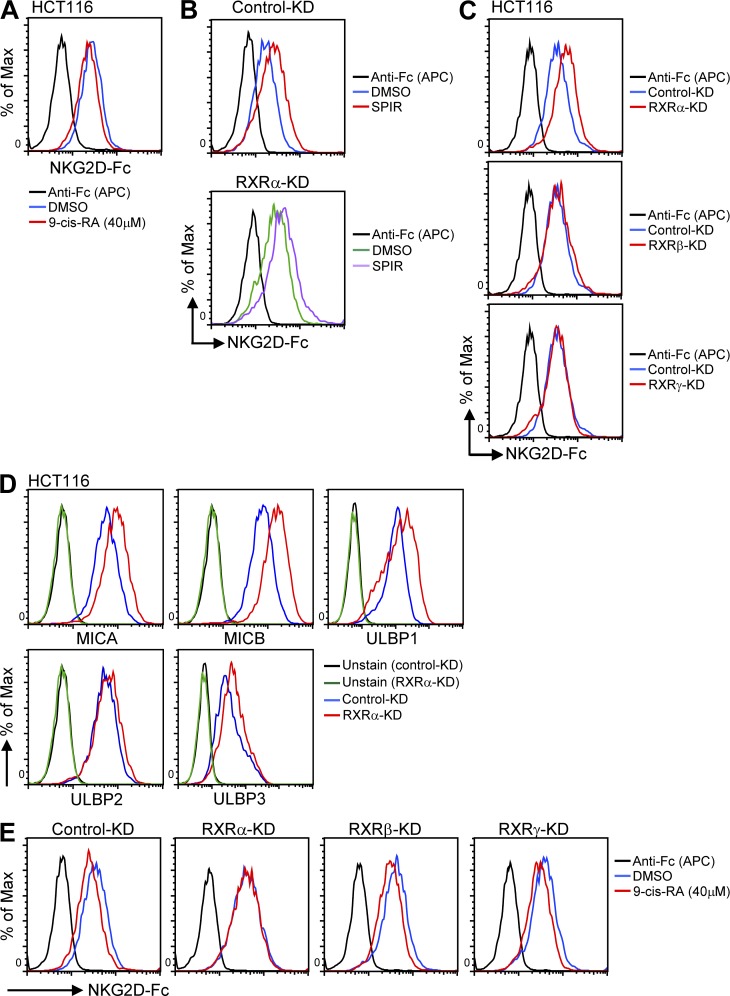
To further support the hypothesis that RXRγ is directly responsible for SPIR’s effect, we co-treated HCT116 cells with SPIR and HX531, a specific RXR antagonist (Huang et al., 2011). In line with the siRNA-knockdown results, HX531 treatment significantly reduced SPIR’s up-regulation of NKG2DLs (Fig. 5 E). We next transfected HCT116 cells with an RXRγ-responsive luciferase construct. Upon SPIR treatment, luciferase activity was significantly increased (Fig. 5 F), but the RXRγ protein level did not change (Fig. 5 G). In addition, specific knockdown of RXRγ in HCT116 cells significantly reduced the SPIR-mediated enhancement of tumor susceptibility to NK cytolysis both in vitro (Fig. 5 H) and in vivo (Fig. 5 I). These findings suggest that SPIR functions as an RXRγ agonist and confirm the absolute requirement of RXRγ for the antitumor effect of SPIR. Surprisingly, by treating HCT116 cells with 9-cis-retinoic acid (9-cis-RA), a high-affinity ligand of RXRs, we observed a decrease rather than an increase in NKG2DL expression (Fig. 6 A). In fact, from the results of our NHR siRNA library screening, we found that, although specific knockdown of another RXR family member, RXRα, did not affect the potential of SPIR in up-regulating NKG2DL expression in HCT116 cells (Fig. 6 B), the gene knockdown by itself without any drug treatment resulted in an increase of NKG2DL expression (Fig. 6, C and D). Moreover, with the silencing of RXRα (but not RXRβ or RXRγ), the effect of 9-cis-RA in reducing NKG2DL expression in HCT116 cells was abolished (Fig. 6 E). These results implicated that the activation of RXRα conversely inhibits NKG2DL expression, underscoring the complexity of the RXR family in the regulation of NKG2DLs.
Figure 6.
9-cis-RA treatment reduces NKG2DL expression, whereas specific siRNA knockdown of RXRα up-regulates NKG2DL expression. (A) Total NKG2DL expression was analyzed in HCT116 cells treated with DMSO or 9-cis-RA (40 µM) for 3 d by flow cytometry. Results are representative of three independent experiments. (B) HCT116 cells were transfected with control-KD or RXRα-KD siRNAs 48 h before treatment with DMSO or SPIR (56 µM). The expression of total NKG2DLs was analyzed by flow cytometry 3 d after the drug treatment. Results are representative of three independent experiments. (C) Total NKG2DL expression was analyzed in HCT116 cells 48 h after transfected with specific siRNAs targeting RXRα, RXRβ, or RXRγ. Results are representative of three independent experiments. (D) MICA-B and ULBP1-3 expression were analyzed in HCT116 cells transfected with control-KD or RXRα-KD siRNAs after DMSO or SPIR treatment for 3 d. Results are representative of three independent experiments. (E) HCT116 cells were transfected with control, RXRα-, RXRβ-, or RXRγ-specific siRNAs 48 h before treatment with DMSO or 9-cis-RA (40 µM). The expression of total NKG2DLs was analyzed by flow cytometry 3 d after the drug treatment. Results are representative of three independent experiments.
SPIR-augmented NKG2DL expression depends on the ATM–Chk2 checkpoint pathway
We next investigated how SPIR–RXRγ signaling regulated the transcription of NKG2DLs. Previous studies have shown that histone deacetylase (HDAC) inhibitors such as sodium valproate can enhance the transcription of MICA/B in various cell lines (Armeanu et al., 2005; Diermayr et al., 2008), and HDAC3 specifically represses ULBP1-3 expression in epithelial tumor cells (López-Soto et al., 2009). However, SPIR treatment did not inhibit HDAC activities in SW480 or HCT116 cells, and there was no significant change in the mRNA expression of HDAC3 after SPIR treatment (unpublished data), suggesting that SPIR-RXRγ induction of NKG2DL transcription is independent of HDAC.
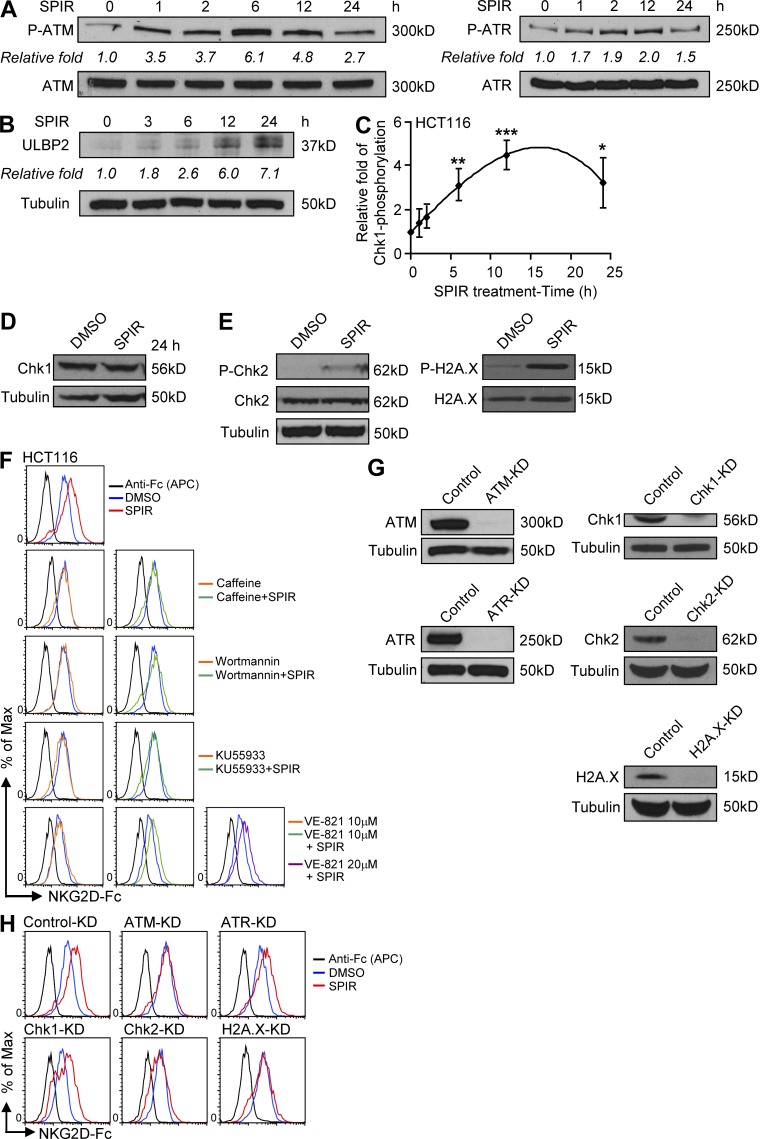
The DNA damage checkpoint pathway initiated by ATM–ATR kinases may regulate NKG2DL expression (Gasser et al., 2005; Soriani et al., 2009). Here, we observed that SPIR treatment induced ATM and ATR phosphorylation (Fig. 7 A) in a time-dependent manner that preceded the increase in ULBP2 protein expression (Fig. 7 B). To further examine the downstream signaling through ATM–ATR kinases, we performed ELISAs and found that SPIR-treated HCT116 cells showed a time-dependent increase of Ser-317 phosphorylation of Chk1 (P-Chk1; Fig. 7 C) but not the total protein level (Fig. 7 D). By Western blot analysis, we also detected an increase in the amount of phosphorylated Chk2 and H2A.X downstream of the ATM–ATR pathway upon SPIR treatment of HCT116 cells (Fig. 7 E). Moreover, in the presence of ATM–ATR inhibitors such as caffeine or wortmannin, SPIR’s effect on NKG2DL expression was completely abolished in HCT116 (Fig. 7 F) and SW480 cells (not depicted). To further confirm that ATM and ATR played a role in SPIR-mediated NKG2DL up-regulation, we treated HCT116 cells with SPIR together with either an ATM-specific inhibitor, KU55933, or a selective ATR inhibitor, VE-821. Consistent with the aforementioned ATM–ATR inhibitor treatments, treatment with 10 µM KU55933 completely abolished the up-regulation of NKG2DLs induced by SPIR. On the contrary, treatment with 10 µM VE-821 only partially reduced the effect of SPIR in NKG2DL expression. Increasing the drug concentration to 20 µM did not further affect NKG2DL expression (Fig. 7 F). Furthermore, specific siRNA knockdown of ATM, Chk2, or H2A.X, but not of ATR or Chk1 (Fig. 7 G), completely abolished up-regulation of NKG2DL in SPIR-treated HCT116 cells (Fig. 7 H). Collectively, these results suggest that although SPIR activates both ATM and ATR, the ATM–Chk2, but not the ATR–Chk1 pathway was primarily responsible for the up-regulation of NKG2DLs.
Figure 7.
The up-regulation of NKG2DLs by SPIR requires activation of the ATM–Chk2 signaling pathway. HCT116 cells were treated with SPIR (56 µM). Total cell lysates were collected at various time points for Western blot analysis to detect ATM (Ser1981) and ATR (Ser428) phosphorylation (A) and ULBP2 expression (B). Data shown are representative of three independent experiments. (C) HCT116 cells were cultured with SPIR (56 µM). Total cell lysates were prepared at various time points (with the same total protein concentration as determined by BCA protein assay; 0–24 h) for phospho-Chk1 by ELISA. Data were expressed as fold change relative to the degree of Chk1 phosphorylation at 0 h (n = 3). (D) The protein level of Chk1 upon SPIR treatment for 24 h was determined by Western blot analysis. Data are representative of three independent experiments. (E) Phosphorylation of Chk2 (Thr68) and histone H2A.X (Ser139) in HCT116 cells treated with DMSO or SPIR for 24 h were determined by Western blot analysis. Data are representative of three independent experiments. (F) NKG2DL expression was analyzed in HCT116 cells treated with SPIR (56 µM), with or without caffeine (1.5 mM), wortmannin (10 µM), KU55933 (10 µM), or VE-821 (10 and 20 µM) for 3 d. Data shown are representative of three independent experiments. (G) The knockdown efficiencies of various siRNAs in HCT116 cells were evaluated by Western blot analysis. Results are representative of three independent experiments. (H) NKG2DL expression was analyzed in HCT116 cells transfected with various siRNAs after DMSO or SPIR treatment for 3 d. Data shown are representative of three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
SPIR triggers the ATM–ATR checkpoint pathway without causing DNA damage but requires the activation of RXRγ
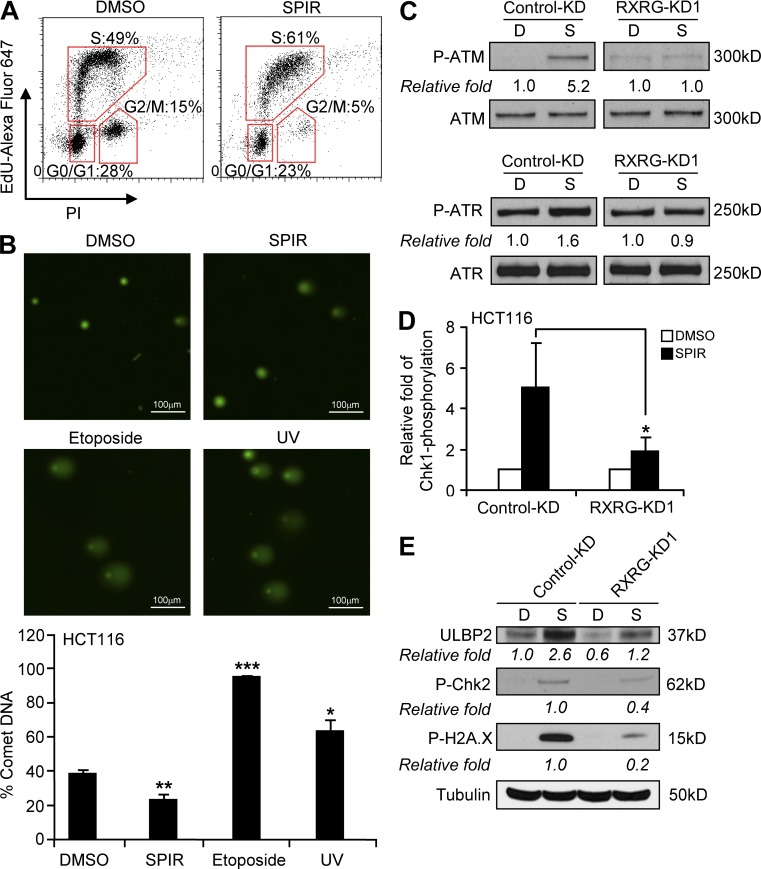
Because SPIR has been used widely in the clinic without evidence of inducing DNA breaks or stalled DNA replication, we hypothesized that its induction of ATM–ATR activity was not caused by a DNA stress response. Consistent with the enhanced phosphorylation of multiple signaling molecules in the ATM–ATR checkpoint pathways, we observed S-phase cell cycle arrest in SPIR-treated HCT116 cells (Fig. 8 A). However, this cell cycle arrest was associated with a moderate decrease in DNA breakage rather than an increase, as shown in comet assays (Fig. 8 B), indicating that SPIR is not genotoxic. The induction of the ATM–ATR pathways and NKG2DL expression by SPIR requires RXRγ activation, as knockdown of RXRγ in HCT116 cells diminished the SPIR-induced phosphorylation of ATM, ATR (Fig. 8 C), Chk1 (Fig. 8 D), Chk2, and H2A.X (Fig. 8 E), resulting in less ULBP2 expression.
Figure 8.
SPIR does not induce DNA damage, and its activation of the ATM–ATR pathway requires the activation of RXRγ. (A) Cell cycle analysis of HCT116 cells after DMSO or SPIR treatment for 24 h was performed by staining the cells with propidium iodide and EdU-Alexa Fluor 647. Data shown are representative of three independent experiments. (B) DNA damage in HCT116 cells after treatment with SPIR (56 µM for 24 h), etoposide (20 µM for 24 h), or UV exposure (4 h) was analyzed by performing comet assays (n = 3). (C) HCT116 cells were transfected with control-KD or RXRγ-KD1 siRNA on day 0. Transfected cells were treated with DMSO or SPIR (56 µM) on day 2. Total cell lysates were collected 6 h after the drug treatment for Western blot analysis to detect phosphorylated ATM and ATR, or 24 h after the drug treatment for ELISA to detect phosphorylated Chk1 (D), and for Western blot analysis to detect ULBP2 expression and phosphorylated Chk2 and H2A.X (E). Data shown are representative of three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.0001.
SPIR suppresses motility and invasiveness of tumor cells
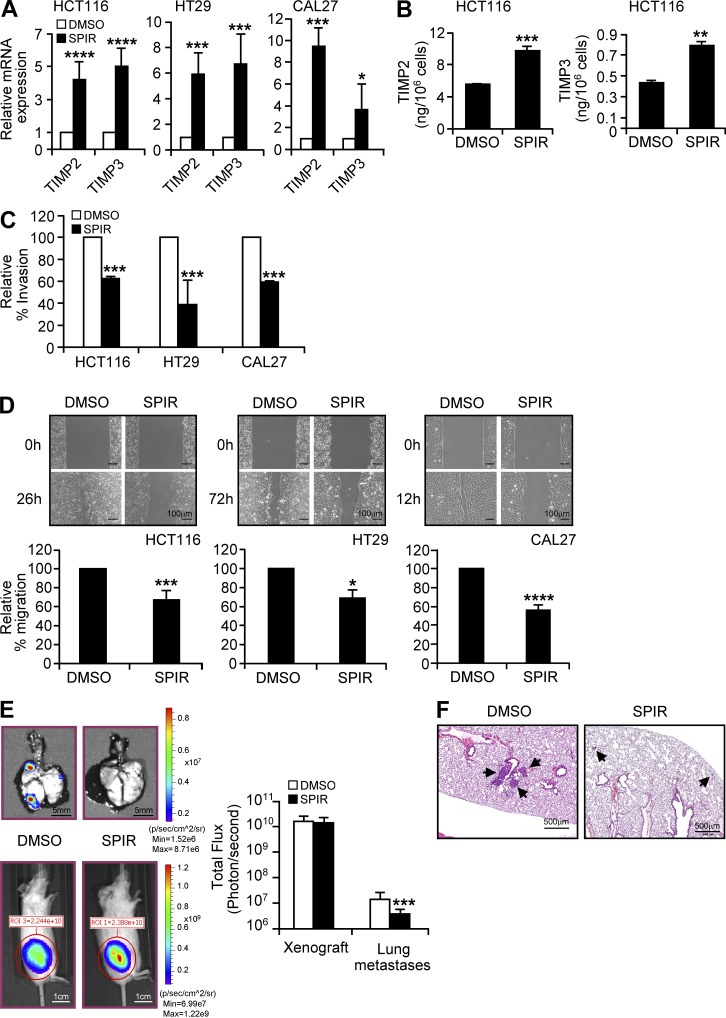
We next attempted to identify other genes that may be regulated by SPIR in tumor cells. We performed a microarray analysis to compare gene expression between treated and untreated HCT116 and CAL27 cells. Selected gene candidates were then confirmed by qRT-PCR analysis. SPIR significantly up-regulated mRNA expression of two metastasis-suppressor genes, TIMP2 and TIMP3 (Albini et al., 1991; Loging and Reisman, 1999) in CAL27 and all the colon cancer cell lines (Fig. 9 A and not depicted). ELISAs revealed corresponding increases in TIMP2 and TIMP3 in HCT116 culture supernatant (Fig. 9 B). The SPIR-induced expression of TIMPs, however, was independent of the ATM–ATR checkpoint pathways. We performed gene knockdown of ATM or ATR in HCT116 cells and confirmed that the reduction of ATM or ATR expression did not affect the SPIR up-regulation of TIMP2 and TIMP3 as determined by qRT-PCR analysis (unpublished data).
Figure 9.
SPIR inhibits tumor metastasis. (A) qRT-PCR was performed to analyze the mRNA expression of TIMP2 and TIMP3 in HCT116, HT29, and CAL27 cells treated with DMSO or SPIR (56 µM) for 3 d (n = 3). (B) The protein level of TIMP2 and TIMP3 in the culture supernatant of HCT116 cells treated for 3 d with DMSO or SPIR (56 µM) was detected by ELISA (n = 3). (C) ECM invasion assays were performed by culturing HCT116, HT29, and CAL27 cells on ECM-coated Transwell membranes with DMSO or SPIR. The number of cells that invaded the ECM was determined after 24 h. Data are expressed as percentage change in tumor invasion relative to that of DMSO-treated cells (n = 3). (D) Wound healing by DMSO- and SPIR-treated HCT116, HT29, and CAL27 cells was compared. Data are calculated as the percentage of change in distance of migration relative to that of DMSO-treated cells (n = 3). (E) NSG mice (n = 8) given subcutaneous injections (day 0) of luciferase-expressing HT29 cells (107) were treated with PBS or SPIR (1.25 mg; intraperitoneally) twice a week for 3 wk. Tumor xenografts and lung metastases were detected by bioluminescence imaging on day 25. Data are shown as the mean total flux (photons/second) ± SD (n = 8). (F) The lung metastases (arrows) were confirmed by examining lung tissue sections. *, P ≤ 0.05; **, P < 0.01; ***, P ≤ 0.005; ****, P < 0.0001.
We then examined the antimetastatic potential of SPIR with HCT116, HT29, and CAL27 cells. We observed that SPIR significantly suppressed invasion of the cell lines through extracellular matrix (ECM)-coated membranes in transwell assays (Fig. 9 C) and delayed cell migration in wound-healing assays (Fig. 9 D).
HT29 cells injected subcutaneously into mice are known to metastasize naturally to the lungs (Jojovic and Schumacher, 2000). In an in vivo metastasis model, bioluminescence imaging (Fig. 9 E) and lung tissue sectioning (Fig. 9 F) revealed a significant reduction of lung metastasis in NSG mice bearing HT29 xenografts and receiving intraperitoneal injections of SPIR twice a week for 3 wk.
The antimetastatic effect of SPIR is independent of MR but requires the activation of RXRγ
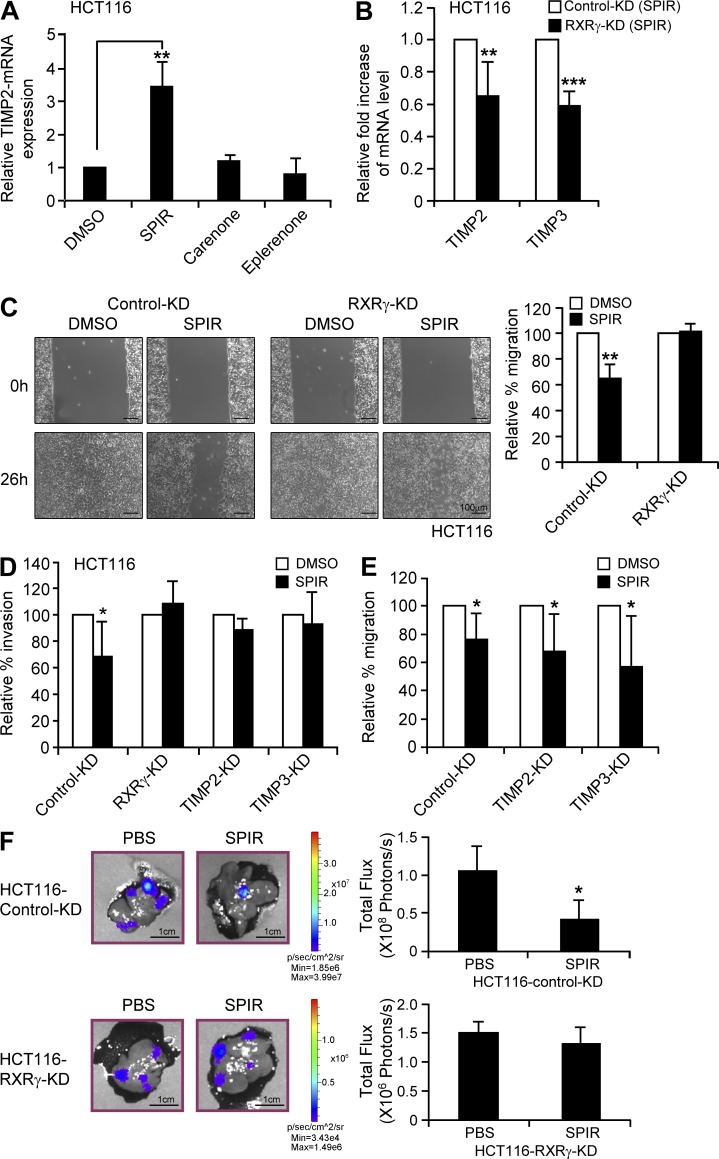
To test whether the antimetastatic effect of SPIR depended on the MR pathway, we compared the expression of TIMP2 in HCT116 cells treated with SPIR, canrenone, or eplerenone. As shown in Fig. 10 A, neither canrenone nor eplerenone up-regulated TIMP2 expression. Moreover, treating HCT116 cells with these alternative MR antagonists did not reproduce the suppressive effect of SPIR on cell invasion in ECM-coated trans-well assays and cell migration in wound-healing assays (unpublished data), suggesting that SPIR did not modulate tumor metastasis through MR signaling.
Figure 10.
The antimetastatic effect of SPIR is independent of MR but requires the activation of RXRγ. (A) TIMP2 mRNA expression was compared by qRT-PCR on HCT116 cells treated with DMSO, SPIR, canrenone, or eplerenone for 3 d (n = 3). (B) HCT116 cells were transfected with control KD or RXRγ-KD1 siRNA on day 0. The cells were treated with DMSO or SPIR (56 µM) on day 2. The mRNA expression of TIMP2 and TIMP3 was analyzed by qRT-PCR on day 5. Data are shown as relative mRNA fold change of SPIR-treated RXRγ-KD1 cells to control KD cells (n = 3). (C) HCT116 cells were transfected with control KD or RXRγ-KD1 siRNA on day 0. Wound healing assay on the cells treated with DMSO or SPIR (56 µM) was performed on day 2. Data are calculated as the percentage change in distance of migration relative to that of DMSO-treated cells (n = 3). (D) HCT116 cells were transfected with control KD, RXRγ-KD1, TIMP2-KD or TIMP3-KD siRNA on day 0. Transfected cells were cultured on ECM-coated Transwell membranes with DMSO or SPIR on day 2. The number of cells that invaded the ECM was determined after 36 h. Data are expressed as percentage change in tumor invasion relative to that of DMSO-treated cells (n = 3). (E) HCT116 cells were transfected with control-KD, TIMP2-KD, or TIMP3-KD siRNA on day 0. Wound healing assay on the cells treated with DMSO or SPIR (56 µM) was performed on day 2 (n = 3). (F) Luciferase-expressing HCT116 cells were transfected with either control-KD or RXRγ-KD1 siRNA on Day 0. The transfected cells were collected on the next day to inject (5 × 105 cells/mouse) intrasplenically into 8–10-wk-old NSG mice (n = 8 per group). SPIR (1.25 mg/mouse) or PBS was intraperitoneally injected into the mice (n = 4 per group) every other day for three times. Hepatic metastasis was analyzed by performing bioluminescence imaging on day 10. *, P < 0.05; **, P < 0.005; ***, P < 0.0001.
In contrast, specific knockdown of RXRγ in HCT116 cells not only significantly reduced the effect of SPIR on the up-regulation of TIMP2 and TIMP3 (Fig. 10 B), but also completely abolished the drug effect on suppressing cell migration in wound-healing assays (Fig. 10 C) and cell invasion in ECM-coated trans-well assays (Fig. 10 D). Knockdown of TIMP2 or TIMP3 directly produced effects similar to those of RXRγ-KD on cell invasion (Fig. 10 D) but had no effects on cell migration (Fig. 10 E). These results suggest that RXRγ is essential in both SPIR-mediated suppression of cell invasion and migration and that its role in SPIR-mediated up-regulation of TIMP2 and TIMP3 correlated with the inhibition of cell invasion.
In a liver metastasis model (Hamada et al., 2008) in which HCT116 cells were implanted into the spleen of 8-wk-old NSG mice, SPIR treatment (intraperitoneal injection every other day for 10 d) significantly suppressed tumor metastasis (Fig. 10 F). The antimetastatic effect of SPIR was dramatically reduced when RXRγ was knocked down in HCT116 cells before intrasplenic injection. These results suggest that RXRγ is essential for the antimetastatic effect of SPIR.
DISCUSSION
NK cells are being increasingly considered in immunotherapy for cancers, including colorectal cancer (Ljunggren and Malmberg, 2007). Although most studies have focused on extrinsic conditions to optimize missing self-recognition and to activate NK cells (Fujisaki et al., 2009; Chan et al., 2012; Rujkijyanont et al., 2013), great efforts are underway to identify new drug candidates that enhance intrinsic tumor sensitivity to immunotherapy with minimal side effects (Bae et al., 2010). To this end, we performed a high-throughput screening of a bioactive compound library and found that SPIR could up-regulate NKG2DL expression in multiple colorectal carcinoma cell lines, thereby enhancing tumor susceptibility to NK cell cytotoxicity.
Although our findings showed a general increase in both mRNA and protein levels of NKG2DLs in SPIR-treated cells, the level of mRNA transcript increase (e.g., MICB in some cell lines such as HT29) may not always correlate with an increase in NKG2DL protein expression on the cell surface. The reason is that cell surface expression of NKG2DLs has been shown to be regulated by various posttranscriptional controls such as ligand shedding and intracellular retention (Waldhauer and Steinle, 2006; Fuertes et al., 2008). In this regard, we have observed that NKG2DLs may undergo diverse protein maturation processes in different cell lines that significantly affect protein stability and cell surface expression independent of gene transcription (unpublished data).
Gasser et al. (2005) found that genotoxic stress such as ionizing radiation and treatment with inhibitors of DNA replication could up-regulate various NKG2DL expressions in both mouse and human tumor cells through the activation of ATM–ATR signaling pathways. In response to DNA damage, ATM primarily activates Chk2 (when there are DNA double-strand breaks), whereas ATR mainly activates Chk1 (responding to stalled DNA replication fork caused by bulky DNA lesions; Kastan and Lim, 2000; Bartek and Lukas, 2003; Shiloh, 2003). Depending on the nature of treatment, NKG2DL up-regulation can be solely dependent on ATM or ATR, or it can require both (Gasser et al., 2005; Valés-Gómez et al., 2008). In our colon cancer experiments, SPIR activated both ATM–Chk2 and ATR–Chk1 pathways, but its specific effect in NKG2DLs was primarily through the activation of the ATM–Chk2 pathway.
The SPIR effect is independent of DNA damage, as SPIR has been used widely in the clinic without evidence of inducing DNA breaks or stalled DNA replication. In fact, recent studies have shown the potential of SPIR to prevent DNA and chromosomal damage caused by aldosterone (Schupp et al., 2010a,b). Furthermore, there are studies showing that ATM activation can be induced by nongenotoxic events, such as oxidative stress, heat shock, and microtubule depolymerization (Shen et al., 2006; Hunt et al., 2007; Guo et al., 2010), or by chromatin modification independent of DNA damage (Bakkenist and Kastan, 2003; Gupta et al., 2005; Lai et al., 2005; Schou et al., 2008). In this regard, RXRs have been shown to play an essential role in chromatin remodeling, i.e., chromatin decondensation in association with other early transcription regulators such as Stat5a and C/EBPβ to establish transcription factor “hotspots,” which in turn recruit other transcription factors to promote gene transcription (Dilworth and Chambon, 2001; Siersbæk et al., 2011; Eeckhoute et al., 2012). Moreover, treatments such as sodium valproate that modify chromatin to a more open structure have been shown to enhance NKG2DL expression (Armeanu et al., 2005). Accordingly, we hypothesize that SPIR activates RXRγ, which initiates chromatin remodeling, resulting in DNA damage–independent activation of the ATM–Chk2 DNA repair checkpoint pathway that enhances NKG2DL expression. The chromatin decondensation and the establishment of transcription factor hotspots induced by activated RXRγ may facilitate not only NKG2DLs, but also the expression of metastasis suppressor genes such as TIMP2 and TIMP3, which ultimately enhance tumor susceptibility to NK cell cytolysis and inhibit tumor metastasis. Validation of this hypothesis is required to make a more definitive conclusion.
The RXR family consists of three members: RXRα, RXRβ, and RXRγ (Dawson and Xia, 2012). Various naturally occurring and synthetic retinoids have been identified as RXR ligands. However, due to the observation of numerous side effects upon short- and long-term administration (partly because of their ability to activate multiple RXR members), the systemic use of these agents has been limited to cancer treatment (Boehm et al., 1994). Therefore, identification and development of novel, receptor subtype–specific compounds are critical to improve the therapeutic potential of RXR agonists. In this study, we showed that SPIR can up-regulate NKG2DLs specifically via RXRγ. Alternatively, specific siRNA knockdown of RXRα (but not RXRβ or RXRγ) can also enhanced the expression of NKG2DLs without any drug treatment (Fig. 6, C and D). Treating HCT116 cells with 9-cis-RA, an agonist that can broadly bind to all members of RXR families, could not induce the up-regulation of NKG2DL expression, suggesting that the negative signaling through RXRα override the positive effect via RXRγ. Collectively, these data underscore the importance of further investigations into the complex interplay among RXR family members in the regulation of NKG2DL expression.
SPIR has been commonly used to treat hypertension and edema and has a good long-term safety record. In the screening phase, we confirmed that it was safe in mice and observed no detrimental effect on the viability or NKG2DL expression of normal human lymphocytes (unpublished data), suggesting that SPIR could act as a safe and effective RXRγ-specific modulator in colon cancer therapy. Furthermore, we showed the potential of SPIR in colon cancer prevention in vivo (Fig. 3 G and Fig. 4 A). Treatment with SPIR for 2–12 wk successfully achieved a drug level that could prevent colon cancer development in both the NSG and APC mouse model.
Tumor metastasis has been associated with poor prognosis and a low survival rate in colorectal cancer patients (Murray et al., 1996; Cappell, 2008). In this study, we provide strong evidence that administration of SPIR significantly inhibited colon cancer metastasis that was associated with increased expression of TIMP2 and TIMP3. TIMPs are endogenous protein inhibitors that suppress the activities of matrix metalloproteinases (MMPs; Brew and Nagase, 2010), and therefore play a pivotal role in regulating ECM degradation, an essential step in tumor invasion (Visse and Nagase, 2003). The clinical significance of TIMP expression in suppressing metastasis of various types of cancer has been widely reported (Ring et al., 1997; Song et al., 2010). Recent studies of gene polymorphisms in TIMP2 and TIMP3 further revealed their associations with tumorigenesis and the cancer survival rate (Messerli, 2004; Malemud, 2006a,b). It has been reported that increased expression of MMP-1 correlates with poor prognosis and metastasis in colon cancer (Murray et al., 1996; Sunami et al., 2000). Blockade of its enzyme activity or gene expression has been shown to inhibit tumor development and metastasis in mouse models (Pulukuri and Rao, 2008). In line with these results, we observed a dramatic decrease of MMP-1 activity (unpublished data) in the culture supernatant from HT29 and CAL27 cells upon SPIR treatment, possibly because of the increased expression of TIMP2 and TIMP3. Papi et al. (2010) reported that activation of RXRγ together with PPAR inhibits tumor invasiveness in association with an increase of TIMP2 expression in colon cancer cells. These results corroborate our observation that SPIR suppresses tumor metastasis by increasing the expression of TIMP2 and TIMP3 through the activation of RXRγ.
In summary, this is the first study showing a novel RXRγ pathway linking tumor cell–intrinsic NK cell immunosurveillance and metastasis control. Specifically, SPIR is a potent anticancer agent that favorably regulates NKG2DL and metastasis-suppressor gene expression, enhancing tumor susceptibility to NK cell cytotoxicity and inhibiting metastasis. The drug’s effects were not restricted to colon cancer, being observed also in head and neck, prostate, liver, and smooth muscle cancer cells. Our data provide strong evidence to support further investigations of SPIR and other RXRγ agonists in cancer treatment and prevention, and encourage the development of novel SPIR analogues that possess excellent anticancer activity without the side effect of MR inhibition.
MATERIALS AND METHODS
Statement.
The Institutional Animal Care and Use Committee of St. Jude Children’s Research Hospital approved all animal protocols.
Cell lines, mouse strains, antibodies, and reagents.
CAL27, HCT116, HT29, HCT15, SW480, COLO201, LS174T, HepG2, DU145, and RD cell lines were obtained from American Type Culture Collection (ATCC). The NKL cell line was a gift from J.L. Strominger (Harvard University, Cambridge, MA); the YT-INDY cell line was a generous gift from Z. Brahmi (Indiana University, Indianapolis, IN), and the anti-ULBP3 (M550) antibody was a gift from Amgen. NSG and C57BL/6J-ApcMin/J mice were obtained from The Jackson Laboratory. Anti-ULBP1 (AUMO2) and anti-ULBP2 (BUMO1; as capture antibody for ELISA) antibodies were purchased from BAMOMAB; allophycocyanin-conjugated anti-ULBP2, anti-MICA, and anti-MICB, blocking antibodies anti-NKG2D (clone 149810) and anti-NKp30, anti-ULBP2 (IgG2a as detection antibody for ELISA), anti-Chk1 and goat anti-ULBP2 (for Western blot), and human NKG2D-Fc (for flow cytometry) were purchased from R&D Systems; allophycocyanin-conjugated anti–mouse IgG antibody and HRP-conjugate goat anti–mouse IgG2a were from Jackson ImmunoResearch Laboratories; allophycocyanin-conjugated mouse anti–human IgG (Fc) were purchased from Southern Biotech; anti-MR (N-17) antibody were obtained from Santa Cruz Biotechnology, Inc.; anti-ATM, anti-ATR, anti–P-ATR, anti–P-Chk2, and anti–γ-H2A.X antibodies were purchased from Cell Signaling Technology; anti-tubulin (DM1A+DM1B), anti-H2A.X, anti-Chk2, and anti-ATM(Phospho-S1981; EP1890Y) antibodies were purchased from Abcam; SPIR, eplerenone, and caffeine were obtained from Sigma-Aldrich; canrenone, HX531, and KU55933 from TOCRIS Bioscience; wortmannin from InvivoGen; VE-821 from Selleckchem; human nuclear hormone receptor siRNA library and siRNAs targeting RXRγ (siRNA-ID: s200454, s200455, s200456, 251285, and 251287) were purchased from Life Technologies; the control knockdown and prevalidated siRNAs targeting Chk1 and Chk2 was purchased from Cell Signaling Technology; siRNA targeting histone H2A.X from QIAGEN; siRNA (siGENOME) targeting ATM, ATR, TIMP2, TIMP3, MICA, and ULBP3 were obtained from Thermo Fisher Scientific. qRT-PCR primers for mouse Raet1 family (QT01782564: pan-Raet1 primers detecting all members including Raet1a, Raet1b, Raet1c, Raet1d, and Raet1e) were purchased from QIAGEN.
Flow cytometry.
Cell lines were stained with allophycocyanin-conjugated anti-MICA, anti-MICB, or anti-ULBP2 antibodies for 20 min at 4°C. For ULBP1 and ULBP3 staining, cells were first incubated with anti-ULBP1 and anti-ULBP3 antibodies, and then with allophycocyanin-conjugated goat anti–mouse IgG antibody. To analyze total NKG2DL cell surface expression, cells were first incubated with human NKG2D-Fc for 20 min at 4°C, and then with allophycocyanin-conjugated mouse anti–human IgG (Fc) antibody. Cells were washed and analyzed on a C6 flow cytometer (Accuri). Data were analyzed using FlowJo software (Tree Star).
ULBP2 ELISA.
ULBP2 shed from cell surface to the culture supernatant was detected using ELISA, as previously described (Waldhauer and Steinle, 2006). In brief, 100 µl (triplicate) of cultured supernatant was added into a 96-well plate precoated with 1 µg/ml of mouse anti-ULBP2 (BUMO1) and incubated at 4°C overnight. The plate was washed, and then incubated with mouse anti-ULBP2 (IgG2a; 100 µl/well at 1 µg/ml) for 1 h. followed by incubation with HRP-conjugated goat anti–mouse IgG2a for 1 h at room temperature. TMB substrate was added, and the plate was read using Victor2 plate reader (PerkinElmer).
Cytotoxicity assay.
Cell lines treated for 5 d with DMSO, SPIR (56 µM), or other agents as indicated were subjected to BATDA release assays (PerkinElmer) according to the manufacturer’s instructions. The results were calculated as follows: percentage of specific lysis = (experimental release − spontaneous release)/(maximum release − spontaneous release) × 100.
Quantitative real-time PCR (qRT-PCR) assay and microarray analysis.
For qRT-PCR, total RNA was extracted from untreated or drug-treated tumor cells by using the RNA Clean & Concentrator (Zymo Research). All cDNA was generated by using the SuperScript VILO cDNA Synthesis kit (Life Technologies) and diluted 10-fold for analysis using the 7900HT Fast Real-time PCR System (Applied Biosystems). The data were calculated as the CT of target genes normalized to the CT of GAPDH of each sample. Primer sequences of the target genes are: MICA-F, 5′-CCTTGGCCATGAACGTCAGG-3′; MICA-R, 5’-CCTCTGAGGCCTCGCTGCG-3′; MICB-F, 5′-ACCTTGGCTATGAACGTCACA-3′; MICB-R, 5′-CCCTCTGAGACCTCGCTGCA-3′; ULBP1-F, 5′-ATCAGCGCCTCCTGTCCAC-3′; ULBP1-R, 5′-AAAGACAGTGTGTGTCGACCCAT-3′; ULBP2-F, 5′-AAATGTCACAACGGCCTG-3′; ULBP2-R, 5′-TGAGGGGTTCCTTGGG-3′; ULBP3-F, 5′-CGATTCTTCCGTACCTGCTATTCG-3′; ULBP3-R, 5′-ATTCTTCTGATCCACCTGGCTCT-3′; TIMP2-F, 5′-TCGGTCTGAAAGGTGTGGCCTTTA-3′; TIMP2-R, 5′-AGGTATTTGAGCGGCTTCCTCTGT-3′; TIMP3-F, 5′-TCCTGCTACTACCTGCCTTGCTTT-3′; TIMP3-R, 5′-AGCCAGGGTAACCGAAATTGGAGA-3′; GAPDH-F, 5′-ATGGGGAAGGTGAAGGTCG-3′; GAPDH-R, 5′-GGGGTCATTGATGGCAACAATA-3′. Microarray analysis was performed using the GeneChip U133 Plus 2.0 expression microarray (Affymetrix) on SPIR (56 µM) treated or untreated (DMSO) HCT116 and CAL27 cells (n = 2; Gene Expression Omnibus accession no. GSE50010). Differentially expressed transcripts were analyzed by ANOVA on Spotfire program.
Generation of YT-INDY-NKG2D cells.
YT-INDY cells were transduced with lentiviral vector MSCV-DAP10-IRES-YFP followed by MSCV-NKG2D-IRES-GFP. GFP and YFP double-positive cells were sorted and the surface expression of NKG2D was analyzed by flow cytometry. In vitro validation had confirmed that YT-INDY-NKG2D could kill NKG2DL-expressing cells such as K562, Jurkat, and HCT116 cells, whereas the parental YT-INDY cells did not (unpublished data).
Generation of HCT116-ΔNKG2DLs cells.
HCT116 cells were transduced with a CMV-driven puromycin-selective lentiviral vector derived from pLL3.7 that contained UL16 sequence (human herpesvirus 5 strain Towne; gene ID, 3077464). Previous studies have shown that UL16 viral proteins interact with various NKG2DLs that interrupt their surface expression (Dunn et al., 2003). We performed flow cytometry on the puromycin selected cells and we found that the surface expression of MICB, ULBP1, and ULBP2 was dramatically reduced. We next transfected the HCT116-UL16 cells with siRNAs targeting MICA and ULBP3 to lower the surface level of MICA and ULBP3 also before injecting the cells into NSG mice for in vivo studies.
In vivo xenograft and metastasis model.
Luciferase-expressing HCT116 and HT29 cells were generated by lentiviral transduction of the cells with a CMV promoter–driven luciferase expression construct engineered from the pLL3.7 vector. For the HCT116 xenograft model, 106 cells were subcutaneously injected into 8–10-wk-old NSG mice with or without effector cells (NKL, YT-INDY or YT-INDY-NKG2D cells) followed by the treatment of SPIR as described in Fig. 10 F. Bioluminescence imaging was performed in a Xenogen imaging system to monitor tumor growth, and the data were analyzed using Living Image software (PerkinElmer). For the lung metastasis model using HT29 cells, lungs were harvested for bioluminescence imaging, fixed in 10% neutral buffered formalin, paraffin embedded, sectioned, and stained with hematoxylin and eosin. A total of 6 lung sections at 100-µm intervals were collected for the observation of pulmonary nodules. For the hepatic metastasis model, 0.5 × 106 of luciferase-expressing HCT116 cells were intrasplenically implanted into 6–8-wk-old NSG mice. Livers were harvested for bioluminescence imaging after SPIR treatment as described in Fig. 10 F.
C57BL/6J-ApcMin/J mouse model.
By examining the intestines collected from different age groups of C57BL/6J-APCMin/J mice (n = 2–3 per group), we found that none of the 8-wk-old mice developed intestinal polyps. The mice started to develop polyps when they were 10–12 wk of age, and there were 15–25 intestinal adenomas established in the intestinal tracts of all the 4-mo-old mice. Most of the mice died at 5–6 mo, with 25–30 adenomas in their intestines. SPIR (1.25 mg/mouse) or PBS was intraperitoneally injected into 8-wk-old C57BL/6J-ApcMin/J mice twice a week for 3 mo. The number of intestinal polyps was counted, and the intestine was fixed in 10% neutral buffered formalin, paraffin embedded, sectioned, and stained with hematoxylin and eosin. Intestine sections at 100-µm intervals were collected for the observation of neoplasia without knowledge of treatment assignment.
TIMP2 and TIMP3 ELISA.
Culture supernatant from 2 × 106 cells treated for 3 d with DMSO or SPIR (56 µM) were collected to determine the protein level of secreted TIMP2 and TIMP3 by ELISA according to the manufacturer’s instructions (R&D Systems).
siRNA transfection.
For the transfection study, 2 × 105 cells were transfected with 60 pmol of siRNA molecules using Lipofectamine RNAiMAX according to the manufacturer’s instructions (Invitrogen).
In vitro cell invasion assay.
The invasiveness of the tumor cells was determined by using the CytoSelect 96-well Cell Invasion Assay with Basement Membrane (Cell Biolabs) according to the manufacturer’s instructions.
Wound-healing assay.
Confluent cell layers of CAL27 and HT29 cells with a 500 ± 50 µm gap were formed by using Ibidi culture inserts (RPI Corp.). Gap distances were measured at various times using an Eclipse Ti inverted microscope and NIS-Elements software (Nikon).
RXRγ reporter assay.
The Cignal RXR reporter assay system containing an RXRγ-responsive luciferase construct, which encodes the firefly luciferase reporter gene under the control of a minimal CMV promoter and tandem repeats of the RXRγ transcriptional response element, was purchased from QIAGEN. The experiment was performed in HCT116 cells according to the manufacturer’s instructions.
Phospho-Chk1 ELISA.
Ser-317 phosphorylation of Chk1 was determined by using the PathScan phosphor-Chk1 (Ser317) Sandwich ELISA kit (Cell Signaling Technology). Data were normalized to the total Chk1 levels obtained from an ELISA with anti-Chk1 (total) used as the detection antibody.
Comet assay.
HCT116 cells treated with SPIR (56 µM) or etoposide (20 µM) for 24 h or exposed to UV irradiation for 2 h were assayed by using the OxiSelect comet assay kit (Cell Biolabs, Inc.). Experiments were performed according to the manufacturer’s instructions.
Online supplemental material.
Fig. S1 shows that SPIR-mediated NKG2DL up-regulation was independent of MR pathways. Table S1 summarizes the information of the colon cancer cell lines that we used in this study. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20122292/DC1.
Supplementary Material
Acknowledgments
We thank Amgen for providing the anti-ULBP3 antibody. The NKL cell line was a kind gift from Professor Jack Strominger. The YT-INDY cell line was a generous gift from Zacharie Brahmi. We thank David Galloway and Cherise Guess for editing the manuscript.
This study was supported by the Assisi Foundation of Memphis and the American Lebanese Syrian Associated Charities.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- 9-cis-RA
- 9-cis-retinoic acid
- AR
- androgen receptor
- GR
- glucocorticoid receptor
- MR
- mineralocorticoid receptor
- NHR
- nuclear hormone receptor
- NKG2DL
- NKG2D ligand
- PR
- progesterone receptor
- RXRγ
- retinoid X receptor γ
- SPIR
- spironolactone
References
- Albini A., Melchiori A., Santi L., Liotta L.A., Brown P.D., Stetler-Stevenson W.G. 1991. Tumor cell invasion inhibited by TIMP-2. J. Natl. Cancer Inst. 83:775–779 10.1093/jnci/83.11.775 [DOI] [PubMed] [Google Scholar]
- André P., Castriconi R., Espéli M., Anfossi N., Juarez T., Hue S., Conway H., Romagné F., Dondero A., Nanni M., et al. 2004. Comparative analysis of human NK cell activation induced by NKG2D and natural cytotoxicity receptors. Eur. J. Immunol. 34:961–971 10.1002/eji.200324705 [DOI] [PubMed] [Google Scholar]
- Armeanu S., Bitzer M., Lauer U.M., Venturelli S., Pathil A., Krusch M., Kaiser S., Jobst J., Smirnow I., Wagner A., et al. 2005. Natural killer cell-mediated lysis of hepatoma cells via specific induction of NKG2D ligands by the histone deacetylase inhibitor sodium valproate. Cancer Res. 65:6321–6329 10.1158/0008-5472.CAN-04-4252 [DOI] [PubMed] [Google Scholar]
- Bae J.H., Kim J.Y., Kim M.J., Chang S.H., Park Y.S., Son C.H., Park S.J., Chung J.S., Lee E.Y., Kim S.H., Kang C.D. 2010. Quercetin enhances susceptibility to NK cell-mediated lysis of tumor cells through induction of NKG2D ligands and suppression of HSP70. J. Immunother. 33:391–401 10.1097/CJI.0b013e3181d32f22 [DOI] [PubMed] [Google Scholar]
- Bakkenist C.J., Kastan M.B. 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 421:499–506 10.1038/nature01368 [DOI] [PubMed] [Google Scholar]
- Bartek J., Lukas J. 2003. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell. 3:421–429 10.1016/S1535-6108(03)00110-7 [DOI] [PubMed] [Google Scholar]
- Boehm M.F., Zhang L., Badea B.A., White S.K., Mais D.E., Berger E., Suto C.M., Goldman M.E., Heyman R.A. 1994. Synthesis and structure-activity relationships of novel retinoid X receptor-selective retinoids. J. Med. Chem. 37:2930–2941 10.1021/jm00044a014 [DOI] [PubMed] [Google Scholar]
- Brew K., Nagase H. 2010. The tissue inhibitors of metalloproteinases (TIMPs): an ancient family with structural and functional diversity. Biochim. Biophys. Acta. 1803:55–71 10.1016/j.bbamcr.2010.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappell M.S. 2008. Pathophysiology, clinical presentation, and management of colon cancer. Gastroenterol. Clin. North Am. 37:1–24: v (v.) 10.1016/j.gtc.2007.12.002 [DOI] [PubMed] [Google Scholar]
- Champsaur M., Lanier L.L. 2010. Effect of NKG2D ligand expression on host immune responses. Immunol. Rev. 235:267–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan W.K., Kung Sutherland M., Li Y., Zalevsky J., Schell S., Leung W. 2012. Antibody-dependent cell-mediated cytotoxicity overcomes NK cell resistance in MLL-rearranged leukemia expressing inhibitory KIR ligands but not activating ligands. Clin. Cancer Res. 18:6296–6305 10.1158/1078-0432.CCR-12-0668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson M.I., Xia Z. 2012. The retinoid X receptors and their ligands. Biochim. Biophys. Acta. 1821:21–56 10.1016/j.bbalip.2011.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diermayr S., Himmelreich H., Durovic B., Mathys-Schneeberger A., Siegler U., Langenkamp U., Hofsteenge J., Gratwohl A., Tichelli A., Paluszewska M., et al. 2008. NKG2D ligand expression in AML increases in response to HDAC inhibitor valproic acid and contributes to allorecognition by NK-cell lines with single KIR-HLA class I specificities. Blood. 111:1428–1436 10.1182/blood-2007-07-101311 [DOI] [PubMed] [Google Scholar]
- Dilworth F.J., Chambon P. 2001. Nuclear receptors coordinate the activities of chromatin remodeling complexes and coactivators to facilitate initiation of transcription. Oncogene. 20:3047–3054 10.1038/sj.onc.1204329 [DOI] [PubMed] [Google Scholar]
- Dunn C., Chalupny N.J., Sutherland C.L., Dosch S., Sivakumar P.V., Johnson D.C., Cosman D. 2003. Human cytomegalovirus glycoprotein UL16 causes intracellular sequestration of NKG2D ligands, protecting against natural killer cell cytotoxicity. J. Exp. Med. 197:1427–1439 10.1084/jem.20022059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eeckhoute J., Oger F., Staels B., Lefebvre P. 2012. Coordinated Regulation of PPARγ Expression and Activity through Control of Chromatin Structure in Adipogenesis and Obesity. PPAR Res. 2012:164140 10.1155/2012/164140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuertes M.B., Girart M.V., Molinero L.L., Domaica C.I., Rossi L.E., Barrio M.M., Mordoh J., Rabinovich G.A., Zwirner N.W. 2008. Intracellular retention of the NKG2D ligand MHC class I chain-related gene A in human melanomas confers immune privilege and prevents NK cell-mediated cytotoxicity. J. Immunol. 180:4606–4614 [DOI] [PubMed] [Google Scholar]
- Fujisaki H., Kakuda H., Shimasaki N., Imai C., Ma J., Lockey T., Eldridge P., Leung W.H., Campana D. 2009. Expansion of highly cytotoxic human natural killer cells for cancer cell therapy. Cancer Res. 69:4010–4017 10.1158/0008-5472.CAN-08-3712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser S., Orsulic S., Brown E.J., Raulet D.H. 2005. The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature. 436:1186–1190 10.1038/nature03884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González S., López-Soto A., Suarez-Alvarez B., López-Vázquez A., López-Larrea C. 2008. NKG2D ligands: key targets of the immune response. Trends Immunol. 29:397–403 10.1016/j.it.2008.04.007 [DOI] [PubMed] [Google Scholar]
- Groh V., Bahram S., Bauer S., Herman A., Beauchamp M., Spies T. 1996. Cell stress-regulated human major histocompatibility complex class I gene expressed in gastrointestinal epithelium. Proc. Natl. Acad. Sci. USA. 93:12445–12450 10.1073/pnas.93.22.12445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Kozlov S., Lavin M.F., Person M.D., Paull T.T. 2010. ATM activation by oxidative stress. Science. 330:517–521 10.1126/science.1192912 [DOI] [PubMed] [Google Scholar]
- Gupta A., Sharma G.G., Young C.S., Agarwal M., Smith E.R., Paull T.T., Lucchesi J.C., Khanna K.K., Ludwig T., Pandita T.K. 2005. Involvement of human MOF in ATM function. Mol. Cell. Biol. 25:5292–5305 10.1128/MCB.25.12.5292-5305.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada K., Monnai M., Kawai K., Nishime C., Kito C., Miyazaki N., Ohnishi Y., Nakamura M., Suemizu H. 2008. Liver metastasis models of colon cancer for evaluation of drug efficacy using NOD/Shi-scid IL2Rgammanull (NOG) mice. Int. J. Oncol. 32:153–159 [PubMed] [Google Scholar]
- Huang J.K., Jarjour A.A., Nait Oumesmar B., Kerninon C., Williams A., Krezel W., Kagechika H., Bauer J., Zhao C., Baron-Van Evercooren A., et al. 2011. Retinoid X receptor gamma signaling accelerates CNS remyelination. Nat. Neurosci. 14:45–53 10.1038/nn.2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt C.R., Pandita R.K., Laszlo A., Higashikubo R., Agarwal M., Kitamura T., Gupta A., Rief N., Horikoshi N., Baskaran R., et al. 2007. Hyperthermia activates a subset of ataxia-telangiectasia mutated effectors independent of DNA strand breaks and heat shock protein 70 status. Cancer Res. 67:3010–3017 10.1158/0008-5472.CAN-06-4328 [DOI] [PubMed] [Google Scholar]
- Jojovic M., Schumacher U. 2000. Quantitative assessment of spontaneous lung metastases of human HT29 colon cancer cells transplanted into SCID mice. Cancer Lett. 152:151–156 10.1016/S0304-3835(99)00443-7 [DOI] [PubMed] [Google Scholar]
- Kastan M.B., Lim D.S. 2000. The many substrates and functions of ATM. Nat. Rev. Mol. Cell Biol. 1:179–186 10.1038/35043058 [DOI] [PubMed] [Google Scholar]
- Lai M., Zimmerman E.S., Planelles V., Chen J. 2005. Activation of the ATR pathway by human immunodeficiency virus type 1 Vpr involves its direct binding to chromatin in vivo. J. Virol. 79:15443–15451 10.1128/JVI.79.24.15443-15451.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljunggren H.G., Malmberg K.J. 2007. Prospects for the use of NK cells in immunotherapy of human cancer. Nat. Rev. Immunol. 7:329–339 10.1038/nri2073 [DOI] [PubMed] [Google Scholar]
- Loging W.T., Reisman D. 1999. Inhibition of the putative tumor suppressor gene TIMP-3 by tumor-derived p53 mutants and wild type p53. Oncogene. 18:7608–7615 10.1038/sj.onc.1203135 [DOI] [PubMed] [Google Scholar]
- López-Soto A., Folgueras A.R., Seto E., Gonzalez S. 2009. HDAC3 represses the expression of NKG2D ligands ULBPs in epithelial tumour cells: potential implications for the immunosurveillance of cancer. Oncogene. 28:2370–2382 10.1038/onc.2009.117 [DOI] [PubMed] [Google Scholar]
- Malemud C.J. 2006a. Matrix metalloproteinases (MMPs) in health and disease: an overview. Front. Biosci. 11:1696–1701 10.2741/1915 [DOI] [PubMed] [Google Scholar]
- Malemud C.J. 2006b. Matrix metalloproteinases: role in skeletal development and growth plate disorders. Front. Biosci. 11:1702–1715 10.2741/1916 [DOI] [PubMed] [Google Scholar]
- McGilvray R.W., Eagle R.A., Watson N.F., Al-Attar A., Ball G., Jafferji I., Trowsdale J., Durrant L.G. 2009. NKG2D ligand expression in human colorectal cancer reveals associations with prognosis and evidence for immunoediting. Clin. Cancer Res. 15:6993–7002 10.1158/1078-0432.CCR-09-0991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerli F.H. 2004. TIMPs, MMPs and cardiovascular disease. Eur. Heart J. 25:1475–1476 10.1016/j.ehj.2004.07.015 [DOI] [PubMed] [Google Scholar]
- Murray G.I., Duncan M.E., O’Neil P., Melvin W.T., Fothergill J.E. 1996. Matrix metalloproteinase-1 is associated with poor prognosis in colorectal cancer. Nat. Med. 2:461–462 10.1038/nm0496-461 [DOI] [PubMed] [Google Scholar]
- Nausch N., Cerwenka A. 2008. NKG2D ligands in tumor immunity. Oncogene. 27:5944–5958 10.1038/onc.2008.272 [DOI] [PubMed] [Google Scholar]
- Papi A., Rocchi P., Ferreri A.M., Orlandi M. 2010. RXRgamma and PPARgamma ligands in combination to inhibit proliferation and invasiveness in colon cancer cells. Cancer Lett. 297:65–74 10.1016/j.canlet.2010.04.026 [DOI] [PubMed] [Google Scholar]
- Pulukuri S.M., Rao J.S. 2008. Matrix metalloproteinase-1 promotes prostate tumor growth and metastasis. Int. J. Oncol. 32:757–765 [PMC free article] [PubMed] [Google Scholar]
- Ring P., Johansson K., Höyhtyä M., Rubin K., Lindmark G. 1997. Expression of tissue inhibitor of metalloproteinases TIMP-2 in human colorectal cancer—a predictor of tumour stage. Br. J. Cancer. 76:805–811 10.1038/bjc.1997.466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rujkijyanont P., Chan W.K., Eldridge P.W., Lockey T., Holladay M., Rooney B., Davidoff A.M., Leung W., Vong Q. 2013. Ex vivo activation of CD56(+) immune cells that eradicate neuroblastoma. Cancer Res. 73:2608–2618 10.1158/0008-5472.CAN-12-3322 [DOI] [PubMed] [Google Scholar]
- Salih H.R., Rammensee H.G., Steinle A. 2002. Cutting edge: down-regulation of MICA on human tumors by proteolytic shedding. J. Immunol. 169:4098–4102 [DOI] [PubMed] [Google Scholar]
- Schou K.B., Schneider L., Christensen S.T., Hoffmann E.K. 2008. Early-stage apoptosis is associated with DNA-damage-independent ATM phosphorylation and chromatin decondensation in NIH3T3 fibroblasts. Cell Biol. Int. 32:107–113 10.1016/j.cellbi.2007.08.019 [DOI] [PubMed] [Google Scholar]
- Schupp N., Kolkhof P., Queisser N., Gärtner S., Schmid U., Kretschmer A., Hartmann E., Oli R.G., Schäfer S., Stopper H. 2011a. Mineralocorticoid receptor-mediated DNA damage in kidneys of DOCA-salt hypertensive rats. FASEB J. 25:968–978 10.1096/fj.10-173286 [DOI] [PubMed] [Google Scholar]
- Schupp N., Queisser N., Wolf M., Kolkhof P., Bärfacker L., Schäfer S., Heidland A., Stopper H. 2010b. Aldosterone causes DNA strand breaks and chromosomal damage in renal cells, which are prevented by mineralocorticoid receptor antagonists. Horm. Metab. Res. 42:458–465 10.1055/s-0029-1243253 [DOI] [PubMed] [Google Scholar]
- Shen K., Wang Y., Brooks S.C., Raz A., Wang Y.A. 2006. ATM is activated by mitotic stress and suppresses centrosome amplification in primary but not in tumor cells. J. Cell. Biochem. 99:1267–1274 10.1002/jcb.20848 [DOI] [PubMed] [Google Scholar]
- Shiloh Y. 2003. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer. 3:155–168 10.1038/nrc1011 [DOI] [PubMed] [Google Scholar]
- Siersbæk R., Nielsen R., John S., Sung M.H., Baek S., Loft A., Hager G.L., Mandrup S. 2011. Extensive chromatin remodelling and establishment of transcription factor ‘hotspots’ during early adipogenesis. EMBO J. 30:1459–1472 10.1038/emboj.2011.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B., Wang C., Liu J., Wang X., Lv L., Wei L., Xie L., Zheng Y., Song X. 2010. MicroRNA-21 regulates breast cancer invasion partly by targeting tissue inhibitor of metalloproteinase 3 expression. J. Exp. Clin. Cancer Res. 29:29 10.1186/1756-9966-29-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriani A., Zingoni A., Cerboni C., Iannitto M.L., Ricciardi M.R., Di Gialleonardo V., Cippitelli M., Fionda C., Petrucci M.T., Guarini A., et al. 2009. ATM-ATR-dependent up-regulation of DNAM-1 and NKG2D ligands on multiple myeloma cells by therapeutic agents results in enhanced NK-cell susceptibility and is associated with a senescent phenotype. Blood. 113:3503–3511 10.1182/blood-2008-08-173914 [DOI] [PubMed] [Google Scholar]
- Stern-Ginossar N., Gur C., Biton M., Horwitz E., Elboim M., Stanietsky N., Mandelboim M., Mandelboim O. 2008. Human microRNAs regulate stress-induced immune responses mediated by the receptor NKG2D. Nat. Immunol. 9:1065–1073 10.1038/ni.1642 [DOI] [PubMed] [Google Scholar]
- Struthers A., Krum H., Williams G.H. 2008. A comparison of the aldosterone-blocking agents eplerenone and spironolactone. Clin. Cardiol. 31:153–158 10.1002/clc.20324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L.K., Kinzler K.W., Vogelstein B., Preisinger A.C., Moser A.R., Luongo C., Gould K.A., Dove W.F. 1992. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science. 256:668–670 10.1126/science.1350108 [DOI] [PubMed] [Google Scholar]
- Sunami E., Tsuno N., Osada T., Saito S., Kitayama J., Tomozawa S., Tsuruo T., Shibata Y., Muto T., Nagawa H. 2000. MMP-1 is a prognostic marker for hematogenous metastasis of colorectal cancer. Oncologist. 5:108–114 10.1634/theoncologist.5-2-108 [DOI] [PubMed] [Google Scholar]
- Valés-Gómez M., Chisholm S.E., Cassady-Cain R.L., Roda-Navarro P., Reyburn H.T. 2008. Selective induction of expression of a ligand for the NKG2D receptor by proteasome inhibitors. Cancer Res. 68:1546–1554 10.1158/0008-5472.CAN-07-2973 [DOI] [PubMed] [Google Scholar]
- Visse R., Nagase H. 2003. Matrix metalloproteinases and tissue inhibitors of metalloproteinases: structure, function, and biochemistry. Circ. Res. 92:827–839 10.1161/01.RES.0000070112.80711.3D [DOI] [PubMed] [Google Scholar]
- Waldhauer I., Steinle A. 2006. Proteolytic release of soluble UL16-binding protein 2 from tumor cells. Cancer Res. 66:2520–2526 10.1158/0008-5472.CAN-05-2520 [DOI] [PubMed] [Google Scholar]
- Waldhauer I., Goehlsdorf D., Gieseke F., Weinschenk T., Wittenbrink M., Ludwig A., Stevanovic S., Rammensee H.G., Steinle A. 2008. Tumor-associated MICA is shed by ADAM proteases. Cancer Res. 68:6368–6376 10.1158/0008-5472.CAN-07-6768 [DOI] [PubMed] [Google Scholar]
- Williams T.A., Verhovez A., Milan A., Veglio F., Mulatero P. 2006. Protective effect of spironolactone on endothelial cell apoptosis. Endocrinology. 147:2496–2505 10.1210/en.2005-1318 [DOI] [PubMed] [Google Scholar]
- Yamamoto K., Fujiyama Y., Andoh A., Bamba T., Okabe H. 2001. Oxidative stress increases MICA and MICB gene expression in the human colon carcinoma cell line (CaCo-2). Biochim. Biophys. Acta. 1526:10–12 10.1016/S0304-4165(01)00099-X [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.