Loss of Asxl1 results in myelodysplastic syndrome, whereas concomitant deletion of Tet2 restores HSC self-renewal and triggers a more severe disease phenotype distinct from that seen in single-gene knockout mice.
Abstract
Somatic Addition of Sex Combs Like 1 (ASXL1) mutations occur in 10–30% of patients with myeloid malignancies, most commonly in myelodysplastic syndromes (MDSs), and are associated with adverse outcome. Germline ASXL1 mutations occur in patients with Bohring-Opitz syndrome. Here, we show that constitutive loss of Asxl1 results in developmental abnormalities, including anophthalmia, microcephaly, cleft palates, and mandibular malformations. In contrast, hematopoietic-specific deletion of Asxl1 results in progressive, multilineage cytopenias and dysplasia in the context of increased numbers of hematopoietic stem/progenitor cells, characteristic features of human MDS. Serial transplantation of Asxl1-null hematopoietic cells results in a lethal myeloid disorder at a shorter latency than primary Asxl1 knockout (KO) mice. Asxl1 deletion reduces hematopoietic stem cell self-renewal, which is restored by concomitant deletion of Tet2, a gene commonly co-mutated with ASXL1 in MDS patients. Moreover, compound Asxl1/Tet2 deletion results in an MDS phenotype with hastened death compared with single-gene KO mice. Asxl1 loss results in a global reduction of H3K27 trimethylation and dysregulated expression of known regulators of hematopoiesis. RNA-Seq/ChIP-Seq analyses of Asxl1 in hematopoietic cells identify a subset of differentially expressed genes as direct targets of Asxl1. These findings underscore the importance of Asxl1 in Polycomb group function, development, and hematopoiesis.
Candidate gene and genome-wide discovery studies have identified a set of novel disease alleles in patients with myelodysplastic syndromes (MDSs), acute myeloid leukemia (AML), and myeloproliferative neoplasms (MPNs). These include somatic mutations in genes with a known or putative role in the epigenetic regulation of gene expression (Shih et al., 2012). Addition of Sex Combs Like 1 (ASXL1) is a Polycomb-associated protein that has been shown to be an essential cofactor for the nuclear deubiquitinase BAP1 (Dey et al., 2012), as well as a critical mediator of the function of the Polycomb repressive complex 2 (PRC2; Abdel-Wahab et al., 2012). Recurrent somatic loss-of-function mutations and deletions in ASXL1 are observed in MDS, MPN, and AML patients (Gelsi-Boyer et al., 2009). ASXL1 mutations are most common in MDS patients (Bejar et al., 2011, 2012; Thol et al., 2011; Sanada and Ogawa, 2012), including in 15–20% of MDS patients and in 40–60% in patients with MDS/MPN overlap syndromes (Gelsi-Boyer et al., 2009; Boultwood et al., 2010; Jankowska et al., 2011). ASXL1 mutations are associated with adverse overall survival in MDS, chronic myelomonocytic leukemia, AML, and MPN (Bejar et al., 2011, 2012; Metzeler et al., 2011; Patel et al., 2012; Itzykson et al., 2013; Vannucchi et al., 2013), highlighting the relevance of ASXL1 mutations to myeloid transformation and clinical outcome.
More recently, de novo constitutive ASXL1 mutations were identified in children with the developmental disorder Bohring-Opitz syndrome (Hoischen et al., 2011; Magini et al., 2012). Although these genetic data strongly implicate ASXL1 mutations in myeloid malignancies and in developmental defects, our understanding of the role of Asxl1 in steady-state hematopoiesis, hematopoietic stem/progenitor function, and myeloid malignancies has been limited by the lack of a mouse model for conditional and tissue-specific deletion of Asxl1. Fisher et al. (2010a,b) investigated the role of Asxl1 in hematopoiesis through the creation and analysis of a model of constitutive Asxl1 deletion with targeted insertion of a neo cassette into the Asxl1 locus. Disruption of Asxl1 expression in this manner resulted in partial perinatal lethality. Analysis of the remaining aged (beyond 15 wk of age) Asxl1 mutant mice revealed impairment of B and T cell lymphopoiesis and myeloid differentiation. However, constitutive Asxl1 loss did not alter long-term reconstitution in competitive repopulation studies using Asxl1-null fetal liver cells (Fisher et al., 2010a,b). These results suggested that Asxl1 has an important role in normal hematopoiesis; however, the effects of somatic loss of Asxl1 in hematopoietic cells were not evaluated. Here we investigate the effects of Asxl1 loss in a time- and tissue-dependent manner through the generation of a mouse model for conditional deletion of Asxl1. We also characterized the effect of Asxl1 loss on transcriptional output and gene regulation using epigenomic and transcriptomic analysis of hematopoietic stem/progenitor cells (HSPCs) from WT and Asxl1-deficient mice.
RESULTS
Development of a conditional Asxl1 KO allele
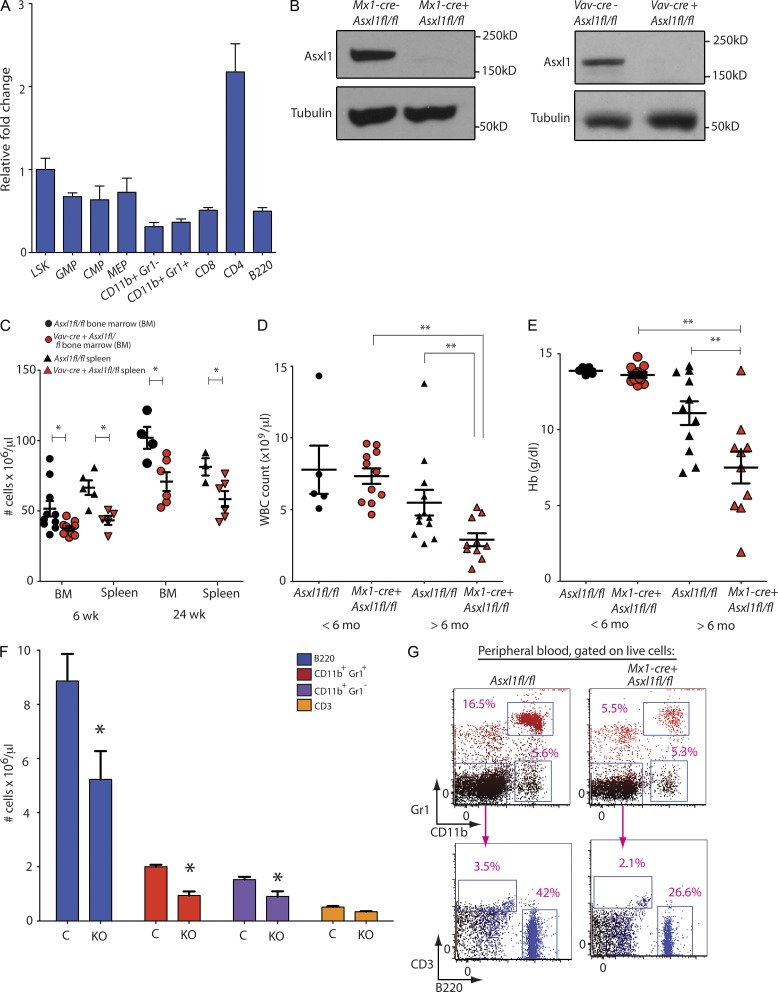
To delineate the role of Asxl1 in development and in hematopoiesis, we generated a conditional allele targeting Asxl1 in vivo (Fig. 1, A and B). We used embryonic stem (ES) cell targeting to insert two LoxP sites flanking exons 5–10 of Asxl1, as well as an Frt-flanked neomycin selection cassette in the upstream intron (Fig. 1, A and B). The generated mice (Asxl1fl/fl) were initially crossed to a germline Flp-deleter mouse line to eliminate the neomycin cassette and then subsequently crossed to germline EIIa-cre mice, IFN-α–inducible Mx1-cre, and hematopoietic-specific Vav-cre (all as described below). Asxl1 protein expression was not detectable in hematopoietic tissue from Vav-cre and Mx1-cre mice (Fig. 2 B), consistent with generation of a KO allele.
Figure 1.
Generation of a conditional Asxl1 allele and characterization of mice with constitutive Asxl1 loss. (A) Schematic depiction of the targeted Asxl1 allele. Exons 5–10 are targeted and flanked by LoxP sites upon Frt-mediated deletion of the Neo cassette. (B) Verification of correct homologous recombination of Asxl1-targeted allele using Southern blots on targeted ES cells. (C) Enumeration of offspring derived from mating EIIa-cre Asxl1+/Δ parents. (D and E) Gross pathology (D) and tissue sections (E) of Asxl1Δ/Δ mice at 14.5 and 18.5 d postcoitus (dpc). (F) Analysis of skeletal preparations from germline Asxl1-null mice surviving to E20.5 including hypoplastic mandibles (asterisk), lack of hyoid bone (arrowhead), and lower lumbar/sacral posterior homeotic transformations (arrow). (G) Gross phenotype of EIIa-cre Asxl1+/Δ and littermate control mice on bilateral microphthalmia. Bars: (D) 2 mm; (E and F [top]) 1 mm; (F, bottom) 2.5 mm; (G) 0.5 cm. (H) Immunophenotyping of fetal liver at 14.5 dpc on relative frequency of LSK cells, MPP cells (LSK, CD48+, CD150− cells), and LT-HSCs (LSK, CD48−, CD150+ cells) between mice with germline loss of 0, 1, or 2 copies of Asxl1. FACS analysis was performed with three to five independent fetal liver samples per genotype. (I) FACS analysis of fetal liver at 14.5 dpc reveals relative frequency of CD71+ single-positive, CD71/Ter119 double-positive, or Ter119 single-positive cells with constitutive loss of Asxl1. Antibody stainings are as indicated, and cells were gated on live cells in the parent gate. Error bars represent ±SD.
Figure 2.
Conditional deletion of Asxl1 results in age-dependent leukopenia and anemia. (A) qRT-PCR showing relative expression level of Asxl1 in purified progenitor and mature mouse hematopoietic stem and progenitor subsets. (B) Verification of Mx1-cre– and Vav-cre–mediated deletion of Asxl1 at the level of protein expression in Western blot of splenocytes. (C) Enumeration of nucleated cells in bilateral femurs and tibiae or whole spleens of control (Asxl1fl/fl) and Asxl1 hematopoietic-specific KO mice (Vav-cre Asxl1fl/fl) at 6 as well as 24 wk of age (n = 6–10 mice per genotype at each time point examined). (D and E) Enumeration of peripheral WBCs (D) and Hb (E) with postnatal deletion of Asxl1 (performed using Mx1-cre Asxl1fl/fl mice or Cre− Asxl1fl/fl controls). Counts in aged Asxl1 KO mice are compared with age-matched controls as well as younger KO and control mice (n = 6–12 mice per genotype at each time point examined). (F and G) Flow cytometric enumeration of B220+, CD11b+/Gr1+, CD3+, and CD11b+/Gr1− cells in the peripheral blood of >6-mo-old Mx1-cre Asxl1fl/fl (KO) and Asxl1fl/fl (C) mice (n = 5 mice per genotype were used for FACS analysis of peripheral blood). The right panel reveals peripheral blood FACS analysis. Antibody stainings are as indicated, and cells were gated on live cells in the parent gate. (A and C–F) Error bars represent ±SD (A and F); mean ± SEM is shown (C–E); *, P < 0.05; **, P < 0.001 (Mann–Whitney U test).
Germline Asx1 loss results in embryonic lethality and craniofacial abnormalities
We characterized the effects of constitutive deletion of Asxl1 by crossing mice bearing floxed Asxl1 alleles with germline EIIa-cre mice. The Asxl1 floxed allele was completely recombined in EIIa-cre Asxl1fl/fl mice (not depicted). We observed 100% embryonic lethality in mice with germline complete deletion of Asxl1 (Asxl1Δ/Δ), whereas mice with heterozygous germline deletion of Asxl1 (Asxl1+/Δ) were born at expected Mendelian ratios (Fig. 1 C). Asxl1Δ/Δ mice were no longer viable by embryonic day (E) 19.5 and were characterized by microphthalmia/anophthalmia (seen in 12/12 of homozygous Asxl1-null embryos examined; Fig. 1, D and E), frequent cleft palates (seen in 5/12 of homozygous Asxl1-null embryos examined; Fig. 1 E), and multiple skeletal abnormalities (mandibular hypoplasia, loss of hyoid bone formation, and posterior homeotic transformations; seen in 4/12 of homozygous Asxl1-null embryos examined; Fig. 1 F). Asxl1+/Δ were viable but exhibited craniofacial dysmorphism in 35% (14/40) of adult Asxl1+/Δ mice examined (Fig. 1 G). Immunophenotypic analysis of HSPCs and erythroid precursor cells in fetal liver from control, Asxl1+/Δ, and Asxl1Δ/Δ mice at E14.5 did not reveal differences among the genotypes (Fig. 1, H and I).
Hematopoietic-specific deletion of Asxl1 results in MDS
Asxl1 is expressed throughout the adult hematopoietic compartment (Fig. 2 A). To elucidate the effects of Asxl1 loss on postnatal hematopoiesis, Asxl1fl/fl mice were crossed to Vav-cre and IFN-α–inducible Mx1-cre transgenic mice for conditional deletion of Asxl1 in the hematopoietic compartment (termed as Asxl1 KO hereafter). In both cases, Asxl1 protein expression was not detectable in hematopoietic tissue (Fig. 2 B). Mice with hematopoietic-specific deletion of Asxl1 (Vav-cre Asxl1fl/fl) developed progressive BM and splenic hypocellularity relative to littermate controls (Cre− Asxl1fl/fl) beginning at 6 wk of age and likewise evident at 24 wk of age (n = 6–10 mice per genotype at each time point examined; Fig. 2 C). Asxl1 KO mice, but not littermate controls, developed progressive leukopenia (Fig. 2 D) and anemia (Fig. 2 E) that was most apparent at 6–12 mo of age. Although the hemoglobin (Hb) in Asxl1 KO mice was within normal limits (median of 13.4 g/dl, range 12.9–14.8 g/dl) in mice <6 mo of age, between 6 and 12 mo the Hb was a median of 8 g/dl (range 1.94–13.9 g/dl) in KO mice relative to a median of 11.4 g/dl in age-matched littermate control mice (range 7.17–14.2 g/dl; n = 6–12 mice with each genotype at each time point examined; Fig. 2 E). Similarly, the WBC count was within normal limits in Asxl1-null mice at <6 mo of age (median of 7.64 × 109 cells/µl, range 4.66–9.6 × 109 cells/µl), the WBC count fell to a median of 2.51 × 109 cells/µl (range 0.88–5.18 × 109 cells/µl) in Asxl1 KO mice between the ages of 6 and 12 mo compared with age-matched littermate Asxl1 WT mice (median WBC count of 4.51 × 109 cells/µl, range 2.62–13.8 × 109 cells/µl; Fig. 2 D). The leukopenia was predominantly caused by decrements in B220+ mature B cells, CD11b+ Gr1+ neutrophils, and CD11b+ Gr1− monocytes as indicated by flow cytometric and morphological analysis of peripheral blood (Fig. 2, F and G). The age-dependent anemia observed in Asxl1 KO mice was accompanied by an increase (median 1.4- to 2-fold) in CD71+/Ter119− erythroid precursor cells in both the BM and spleen, consistent with impaired erythroid differentiation (Fig. 3 A).
Figure 3.
Deletion of Asxl1 results in myeloid and erythroid dysplasia and impaired progenitor differentiation consistent with myelodysplasia. (A) Relative frequency of CD71+/Ter119− erythroid precursors in BM and spleen of 6.5-mo-old Mx1-cre Asxl1fl/fl (KO) and Cre− Asxl1fl/fl control mice (expressed as percentage of live cells; n = 3–5 mice per genotype in each tissue type examined by FACS analysis). (B) Histological (H&E) analysis of Mx1-cre Asxl1fl/fl and Cre− Asxl1fl/fl control BM from 6-mo-old littermate mice. (C) BM cytospins (Wright-Giemsa) from the same mice (arrows indicate erythroid precursors with prominent irregular nuclear contours). (D) Representative morphology of peripheral blood myeloid cells (top) and nucleated RBCs (bottom) in KO mice (Wright-Giemsa stain). (E) Number of colonies formed 7 d after plating of 1,000 CMP, GMP, or MEP cells into methylcellulose from 6-wk-old Vav-cre Asxl1fl/fl and littermate control (Cre− Asxl11fl/fl control) mice. The experiment was performed in biological duplicate. (F) Photograph of methylcellulose colony plate 7 d after plating of MEP cells from 6-wk-old KO and control mice. (G) Histological analysis by H&E staining of liver from 72-wk-old Mx1-cre Asxl1fl/fl mice and littermates. (H) Number of colonies formed 7 d after plating of 200,000 nucleated cells harvested from the liver of 72-wk-old Mx1-cre Asxl1fl/fl or littermate control mice in methylcellulose containing rmIL-3, rm-SCF, rh-IL6, rh-EPO (liver cells from n = 5 mice per genotype plated in methylcellulose). (I) Photomicrograph of colonies grown from cells taken from the liver and plated in methylcellulose is shown on right. Bars: (B) 50 µm; (C) 10 µm; (D) 5 µm; (G) 100 µm; (I) 200 µm. Error bars represent ±SD; *, P < 0.05 (Mann–Whitney U test).
Pathological analysis of Asxl1 KO hematopoietic tissues at 6 mo of age revealed morphological dysplasia of circulating myeloid cells (Fig. 3 D), frequent circulating nucleated red cells (Fig. 3 D), hypocellular marrow (Fig. 3 B), and dysplasia of erythroid precursors (Fig. 3 C). Previous characterization of HSPCs from patients with MDS (Sawada et al., 1993, 1995) and mouse models of MDS (such as the NUP98-HOXD13 transgenic mouse model [Choi et al., 2008]) have identified an impairment of sorted hematopoietic progenitors to form colonies ex vivo in methylcellulose containing myeloid and erythroid cytokines. Consistent with these prior observations and the impairment in mature myeloid and erythroid differentiation seen in Asxl1-deficient mice, in vitro analysis of sorted myeloid progenitor (MP) cells from 6-wk-old Asxl1 KO and control mice revealed a clear decrease in colony output of sorted common MPs (CMPs), granulocyte/macrophage progenitors (GMPs), and megakaryocyte/erythroid progenitors (MEPs) in KO versus control mice (Fig. 3, E and F).
Consistent with the age-dependent development of impaired myeloid and erythroid output in Asxl1 KO mice compared with age-matched littermate controls, Asxl1 KO (Mx1-cre Asxl1fl/fl) mice were found to have infiltration of liver with hematopoietic cells consistent with extramedullary hematopoiesis (EMH) with Asxl1 deletion (Fig. 3 G). To ascertain whether this hematopoietic infiltrate represented inflammatory infiltration of hematopoietic cells versus EMH, we plated 200,000 cells harvested from the liver of Asxl1 KO mice and littermate controls in methylcellulose semisolid media containing myeloid-erythroid cytokines (rmIL-3, rmSCF, rh-EPO, and rh-IL6). Colonies plated with cells derived from Asxl1 KO mice alone yielded abundant colonies (Fig. 3, H and I), demonstrating EMH.
Cell-autonomous effects of Asxl1 loss
Transplantation of whole BM from Asxl1 KO (Vav-cre Asxl1fl/fl) mice into lethally irradiated recipients resulted in a penetrant, lethal hematopoietic disorder (Fig. 4 A), indicating that the phenotype induced by Asxl1 loss was cell autonomous. For example, transplantation of whole BM from 70-wk-old primary Asxl1 KO mice into lethally irradiated recipients resulted in the death of recipient mice at a median of 50 wk after transplant (range 41–74 wk), whereas no mice transplanted with Asxl1 WT BM died during this period of observation. Furthermore, serial transplantation into tertiary recipients resulted in shorter latency of disease, with mice dying 24–42 wk after transplant (median of 28 wk). Transplantation of purified lineage− Sca-1+ c-KIT+ (LSK) cells from the BM of secondary recipients led to a lethal myeloid disease in all tertiary transplant recipients with more rapid onset (median of 10.3 wk, range 5.1–10.6 wk) than transplantation of unfractionated BM cells from the same secondary recipients (Fig. 4 A). Disease in transplanted mice was characterized by progressive anemia and cachexia (Fig. 4, B and C), BM hypocellularity (Fig. 4 D), and an increase in the relative frequency of HSPCs in both the BM and the spleen (Fig. 4, E and F). This was accompanied by splenomegaly caused by EMH and effacement of splenic architecture (Fig. 4, G–I). Anemia in the KO-transplanted recipient mice was evident even with gross inspection of bones (Fig. 4 J). As in primary Asxl1 KO mice, this anemia occurred despite an increase in erythroid precursors in both the BM and spleen (Fig. 4, K and L), consistent with a block in erythroid differentiation with Asxl1 loss. This block in erythroid differentiation was characterized by a significant increase in CD71/Ter119 double-positive erythroid precursors in the spleen (Fig. 4 L).
Figure 4.
Serial noncompetitive transplantation of Asxl1-null cells results in lethal myelodysplastic disorder. (A) Kaplan-Meier survival curve of recipient mice transplanted with 70-wk-old Vav-cre Asxl1fl/fl or Cre− Asxl1fl/fl littermate control whole BM after secondary and tertiary transplantation. Also shown is the survival of mice transplanted with purified LSK cells in tertiary transplantation (tertiary transplant of Asxl1fl/fl control LSK cells is not shown; no recipient mice from this group died by 40 wk [n = 5]). Cre− Asxl1fl/fl littermate controls were similarly transplanted in parallel in each experiment. Four to six recipient mice were transplanted in each experiment. (B) Hematocrit over time of secondary recipient mice transplanted with Asxl1-null or littermate control whole BM in a noncompetitive manner. The dashed line represents the lower limit of normal hematocrit for C57BL/6 mice (n = 4–6 mice per genotype at each time point). (C) Body weight of secondarily transplanted mice at 50 wk after transplantation (n = 4 mice per genotype). (D) BM histopathology of secondary recipient mice transplanted with Asxl1-null or littermate control whole BM at 50 wk. (E) Relative frequency of LSK cells, MPP cells (LSK+, CD150−, CD48+), and LT-HSCs (LSK+ CD150+ CD48−) in BM and spleen at 50 wk after noncompetitive secondary transplantation. Frequencies are expressed as frequency of live cells (n = 4 mice per genotype examined for FACS experiments). (F) Relative frequency of MP (lineage− c-Kit+ Sca-1−), CMP (lineage−, c-Kit+, Sca-1−, FcγR−, CD34+), GMP (lineage− c-Kit+ Sca-1−, FcγR+ CD34+), and MEP (lineage− c-Kit+ Sca-1− FcγR− CD34−) cells at 50 wk after noncompetitive secondary transplantation. Frequencies are expressed as a frequency of live cells. (G) Photographs of spleens from secondary recipient mice transplanted with Vav-cre Asxl1fl/fl or Cre− Asxl1fl/fl littermate control whole BM 50 wk after lethal irradiation. (H) Weight of the same spleens as shown in G (n = 4 mice per genotype). (I) Histopathology of spleens from secondary recipient mice transplanted with Asxl1-null or WT littermate control whole BM 50 wk after noncompetitive secondary transplantation revealing loss of normal splenic architecture. (J) Photographs of representative femur (top) and tibia (bottom) from secondary recipient mice transplanted with Vav-cre Asxl1fl/fl or Cre− Asxl1fl/fl littermate control whole BM 50 wk after noncompetitive secondary transplantation. Bars: (D and I) 50 µm; (J) 2 mm. (K) Relative quantification of CD71+/Ter119− and CD71/Ter119 double-positive cells from BM and spleen of secondary recipient mice transplanted with Vav-cre Asxl1fl/fl or Cre− Asxl1fl/fl littermate control whole BM 50 wk after noncompetitive secondary transplantation. Frequencies are expressed as a percentage of live cells (n = 4 mice per genotype examined by FACS analysis). (L) Representative FACS plots of data shown in K from splenocytes. Staining is as shown, and live cells were gated in parent gate. Error bars represent ±SD; *, P < 0.05 (Mann–Whitney U test).
Impaired self-renewal of Asxl1-deficient cells is rescued by concomitant Tet2 loss
We next assessed the effects of Asxl1 loss on hematopoietic stem cell (HSC) frequency and function. We observed an increase in the absolute number of immunophenotypically defined HSPCs in Asxl1 KO (Vav-cre Asxl1fl/fl) mice at 6 wk of age, including long-term HSCs (LT-HSC; CD150+ CD48− Lin− Sca-1+ c-Kit+; Fig. 5, A and B; quantified as the total number of live cells per femur). Although the number of immunophenotypic stem/progenitor cells was increased, we observed a decrease in serial plating in vitro in Asxl1 KO cells (Fig. 6 A), suggesting a potential defect in self-renewal. To assess the effects of Asxl1 deletion in vivo, 500,000 whole BM nucleated cells from 6-wk-old CD45.2 Vav-cre Asxl1fl/fl mice or Asxl1fl/fl littermate controls were transplanted in competition with an equal number of 6-wk-old CD45.1 competitor BM cells into lethally irradiated CD45.1 recipient mice (Fig. 5 C). Chimerism was assessed based on evaluation of the ratio of CD45.1 to CD45.2 peripheral blood mononuclear cells beginning 2 wk after transplantation and then monitored on a monthly basis until 16 wk thereafter. Consistent with the in vitro data, we observed a clear reduction in self-renewal in vivo. Asxl1 KO HSPCs had a significant disadvantage in competitive transplantation that was further accentuated with serial transplantation (Fig. 5, C and D).
Figure 5.
Asxl1−/− mice have increased stem/progenitor cells but impaired self-renewal. (A) Flow cytometric enumeration of BM LSK cells, LT-HSCs (LSK CD150+ CD48−), and MPP cells (LSK CD150− CD48+) in WT (Asxl1fl/fl) and KO (Vav-cre Asxl1fl/fl) mice at 6 wk of age (n = 4–6 mice per genotype as indicated). Data are expressed as total number of live cells per femur. (B) Representative FACS analysis of BM stem cell populations of Asxl1−/− (Vav-cre Asxl1fl/fl) and WT (Asxl1fl/fl) at 6 wk. Antibody stains are as indicated and parent gate is live, lineage− cells. (C) Schematic depiction of the competitive transplantation assay. Asxl1fl/fl and Vav-cre Asxl1fl/fl are positive for CD45.2, whereas WT competitor cells are positive for CD45.1. Recipient mice are also CD45.1. Representative FACS plots of the percentage of CD45.1 versus CD45.2 total chimerism in the peripheral blood of recipient animals at 16 wk after competitive transplantation is shown. (D) Percentage of CD45.1 versus CD45.2 total chimerism in the peripheral blood of recipient animals at 4 and 16 wk in primary competitive transplant and serial secondary competitive transplants are shown (n = 5 recipient mice for each genotype; C, control; KO, Asxl1 KO). The experiment was performed in biological duplicate. Error bars represent ±SD; *, P < 0.05 (Mann–Whitney U test).
Figure 6.
Combined loss of Asxl1 and Tet2 rescues the impaired self-renewal of Asxl1-deficient HSCs. (A) Enumeration of colonies and serial replating capacity of 20,000 whole BM cells from 6-wk-old littermate mice with hematopoietic-specific deletion of Asxl1 (Vav-cre Asxl1fl/fl), Tet2 (Vav-cre Tet2fl/fl), or both (Vav-cre Asxl1fl/fl Tet2fl/fl). (B) Schematic depiction of the competitive transplantation experiment. Control, Vav-cre Asxl1fl/fl, Vav-cre Tet2fl/fl, and Vav-cre Asxl1fl/flTet2fl/fl cells are positive for CD45.2, whereas WT competitor cells are positive for CD45.1. On the right, monthly assessment of donor chimerism in the peripheral blood of recipient animals is shown up to 16 wk after transplant (n = 5 recipient mice were used for each genotype and experiment was performed in biological duplicate). 16-wk chimerism was significantly higher in Tet2−/− transplanted mice compared with all other genotypes. (C) Representative FACS analysis of peripheral blood of mice transplanted with each genotype at 16 wk. Staining schemes are as indicated and parental gate was live cells. (D) Proportion of CD45.2+ peripheral blood cells of each lineage at 16 wk in mice transplanted with each genotype (n = 5 mice analyzed for each genotype) as determined by FACS analysis. Each competitive transplantation experiment was performed in biological duplicate with five recipient mice per genotype in each experiment. Error bars represent ±SD; *, P < 0.05 (Mann–Whitney U test).
After the final assessment of chimerism in the primary competitive transplantation experiments, primary recipient mice were sacrificed and a serial competitive transplantation experiment was performed by transplanting 1 million whole BM cells from primary recipient mice into lethally irradiated CD45.1 secondary recipient mice (Fig. 5 D). Serial competitive transplantation revealed an even further competitive disadvantage in Asxl1-deficient HSPCs (Fig. 5 D).
Given that MDS is characterized by impaired myeloid differentiation, multilineage cytopenias, and clonal dominance over time, we hypothesized that mutations that occur in concert with ASXL1 deletion/mutation in MDS might compensate for the impaired self-renewal observed with Asxl1 loss. Previous studies have shown that mutations in TET2 are most commonly observed with mutations in ASXL1 in MDS (Bejar et al., 2011, 2012). We and others previously demonstrated increased hematopoietic self-renewal in Tet2-deficient mice (Ko et al., 2011; Li et al., 2011; Moran-Crusio et al., 2011; Quivoron et al., 2011). We analyzed the in vitro and in vivo phenotype of Vav-cre Asxl1fl/fl Tet2fl/fl hematopoietic cells compared with control, Vav-cre Asxl1fl/fl, and Vav-cre Tet2fl/fl mice (Fig. 6 A). Colony assays of whole BM cells from the same mice revealed reduced serial replating activity of Asxl1 KO cells but restored serial-replating capacity of cells with compound Asxl1/Tet2 loss (Fig. 6 A). More importantly, competitive transplantation experiments revealed a competitive advantage for Vav-cre Asxl1fl/fl Tet2fl/fl whole BM compared with matched CD45.1 competitor BM (Figs. 6, B–D). These data demonstrate that concurrent Tet2 loss restores the self-renewal defect induced by Asxl1 loss.
Concomitant deletion of Asxl1 and Tet2 in vivo results in MDS
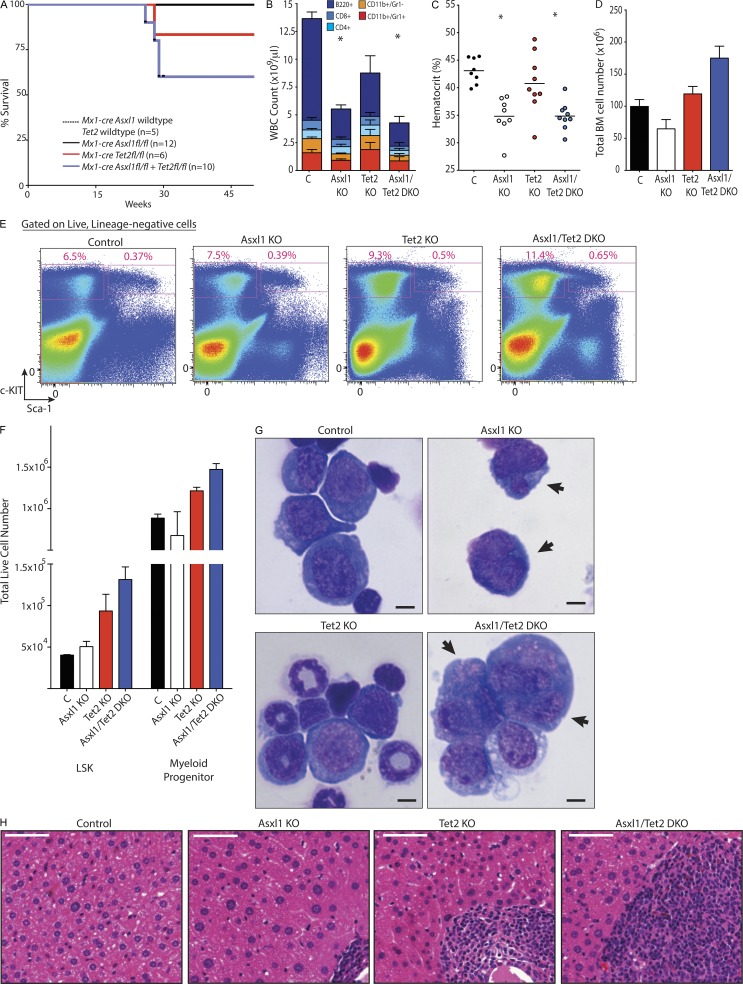
Given the restoration of self-renewal noted in mice with concomitant deletion of Tet2 and Asxl1 in the context of a competitive transplantation experiment, we investigated the phenotype of mice with compound deletion of Tet2 and Asxl1 compared with mice with deletion of each gene alone. A cohort of primary Cre− Asxl1 WT Tet2 WT, Mx1-cre Asxl1fl/fl (Asxl1 KO), Mx1-cre Tet2fl/fl (Tet2 KO), and Mx1-cre Asxl1fl/fl Tet2fl/fl (double KO [DKO]) mice were treated with polyinosinic-polycytidylic acid (polyI:polyC) at 4 wk of life and followed up to 50 wk after birth (46 wk after polyI:polyC administration). At the end of this observation period, 40% of DKO mice (4/10) and 17.7% of Tet2 KO died (1/6), whereas no Asxl1 KO (0/12) or control mice (0/5) died (Fig. 7 A).
Figure 7.
Concomitant deletion of Asxl1 and Tet2 results in myelodysplasia in mice. (A) Kaplan-Meier survival curve of primary Cre− Asxl1fl/fl (n = 5), Mx1-cre Asxl1fl/fl (n = 12), Mx1-cre Tet2fl/fl (n = 6), Mx1-cre Asxl1fl/fl Tet2fl/fl (n = 10 mice per genotype). Mice were treated with polyI:polyC at 4 wk after birth and then followed for 50 wk. (B) Peripheral WBC count and differential of recipient mice transplanted with BM from 6-wk-old Mx1-cre Asxl1 WT Tet2 WT (control; C), Mx1-cre Asxl1fl/fl (Asxl1 KO), Mx1-cre Tet2fl/fl (Tet2 KO), and Mx1-cre Asxl1fl/fl Tet2fl/fl (Asxl1/Tet2 DKO) mice 66 wk after transplantation (68 wk after polyI:polyC administration to recipient mice; n = 10 mice per genotype). Differential was determined by flow cytometric analysis of peripheral blood. (C and D) Hematocrit (C) and total number (D) of nucleated BM cells of same mice as shown in B. Horizontal lines indicate the mean. (E) Representative flow cytometric assessment of relative frequencies of MP and LSK cells in 72-wk-old mice. Parent population was live, lineage− cells. (F) Total numbers of LSK and MP cells (lineage− Sca-1− c-Kit+) in mice from each genotype at 72 wk of age. This was determined by flow cytometric quantification of living LSK and MP cells from c-KIT–enriched BM cells harvested from spine plus bilateral femurs, tibiae, and humeri of each mouse from each genotype at 72 wk of age (n = 3 mice per group). (G) Wright-Giemsa stain of BM representative erythroid precursor from cytospins of 72-wk-old control, Asxl1 KO, Tet2 KO, or Asx1/Tet2 DKO mice. Arrows indicate multinuclearity and nuclear fragmentation in erythroid precursors. (H) Representation histological sections of liver from 72-wk-old control, Asxl1 KO, Tet2 KO, or Asx1/Tet2 DKO mice Bars: (G) 5 µm; (H) 50 µm. For A and B, n = 10 mice per group; for C–H, n = 3 mice per group. Error bars represent ±SD; *, P < 0.05 (Mann–Whitney U test).
For further analyses, to obtain a sufficiently large number of mice for each genotype, BM from 6-wk-old CD45.2 Mx1-cre Asxl1 WT Tet2 WT, Mx1-cre Asxl1fl/fl (Asxl1 KO), Mx1-cre Tet2fl/fl (Tet2 KO), and Mx1-cre Asxl1fl/fl Tet2fl/fl (DKO) mice were transplanted into lethally irradiated CD45.1 recipient mice (10 recipient mice per genotype). All recipient mice (including those transplanted with Mx1-cre Asxl1 WT Tet2 WT BM) were then treated with polyI:polyC 2 wk after transplantation to delete Tet2 and/or Asxl1. At 72 wk after transplant, Asxl1-null and/or Asxl1/Tet2 compound–null mice had significantly lower WBC counts and hematocrit compared with WT or Tet2 single KO control mice (Fig. 7, B and C). As seen in mice with primary deletion of Asxl1, Asxl1 KO mice here had reduced BM cellularity compared with control or Tet2 KO mice (Fig. 7 D). However, despite the similar blood counts between Asxl1 KO and DKO mice, the DKO had greater BM cellularity than mice with deletion of just Asxl1 or even Tet2 (Fig. 7 D). Examination of the HSPC compartment across mice with the four genotypes at 72 wk indicated a greater total number as well as relative frequency of LSK and MP cells in the BM of DKO mice compared with other groups (Fig. 7, E and F). Morphologically, BM of Asxl1 KO mice was characterized by the presence of dysplastic erythroid precursor cells as seen in primary Asxl1 KO mice earlier (Fig. 7 G). The BM of DKO mice likewise was characterized by a similar presence of dysplastic erythroid precursors as well as dysplastic myeloid cells (Fig. 7 G) but lacked the hypocellularity seen in the BM of mice with Asxl1 deletion alone (Fig. 7 D). Histological analysis of liver tissue revealed increased hematopoietic cell infiltration in DKO mice compared with the other groups (Fig. 7 H). The presence of morphological dysplasia in precursor cells, decrease in peripheral circulating mature cells, and concurrent increased total BM cells and HSPCs were suggestive of the presence of MDS in the Asxl1/Tet2 compound–deficient mice.
Transcriptional effects of Asxl1 loss
To understand the basis for the impaired myeloid differentiation and self-renewal observed with Asxl1 loss, we performed expression analysis of sorted LSK and MP cells from cytopenic 1-yr-old Asxl1 KO mice and age-matched littermate controls (Fig. 8, A and B; and Tables S1 and S2). Analysis of RNA-Seq data identified a set of differentially expressed genes in Asxl1 KO LSK cells (797 genes) and MP cells (1,095 genes). Integrative analysis identified a set of 75 genes that were differentially expressed in Asxl1 KO LSK and MP cells, including 41 up-regulated and 34 down-regulated genes in Asxl1 KO mice relative to controls (Fig. 8 B).
Figure 8.
Identification of genes significantly dysregulated with deletion of Asxl1 alone and in concert with deletion of Tet2 and their functional impact. (A) Volcano plot of differentially expressed transcripts from RNA-Seq data of 1-yr-old control versus littermate Asxl1 KO (Mx1-cre Asxl1fl/fl) LSK and MP (lineage− Sca-1− c-Kit+) cells (experiment included cells from two individual mice per genotype). (B) Venn diagrams of genes significantly up- and down-regulated with Asxl1 loss in LSK and MP (lineage−, Sca-1−, cKit+) cells from 1-yr-old Mx1-cre Asxl1fl/fl mice and littermate Cre− controls as identified in A. (C) qRT-PCR analysis of HoxA and Hox-associated transcription factor genes in LSK cells of 1-yr-old Cre− Asxl1fl/fl control versus littermate Vav-cre Asxl1fl/fl. (D) qRT-PCR analysis of p16INK4a in LT-HSCs (lineage−, Sca-1+, c-Kit+, CD150+, CD48−) and MPP cells (lineage−, Sca-1+, c-Kit+, CD150−, CD48+) from 6-wk- and 6-mo-old control (C) versus littermate Vav-cre Asxl1fl/fl (KO) mice. (E) Cell cycle analysis of MPPs from 72-wk-old Vav-cre Asxl1fl/fl or littermate Cre− Asxl1fl/fl control mice with in vivo BrdU administration. Representative FACS plot analysis showing gating on MPP cells followed by BrdU versus DAPI stain is shown on the left (parent gate is LSK cells). Relative quantification of the percentage of MPP cells in S, G2/M, and G0/1 phase is shown on the right (n = 5 mice per group). (F) Assessment of the proportion of HSPCs undergoing apoptosis was performed by Annexin V/DAPI stain of LSK cells from 72-wk-old Vav-cre Asxl1fl/fl mice or Cre− Asxl1fl/fl littermate controls. Representative FACS plot analysis showing gating on LSK cells followed by Annexin V versus DAPI stain is shown on the left (parent gate is lineage− cells). Relative quantification of the percentage of Annexin V+/DAPI− and Annexin V+/DAPI+ LSK cells is shown on the right (n = 5 mice per group). (G) Comparison of significant differentially expressed genes in LSK cells from 6-wk-old Mx1-cre Asxl1fl/fl, Mx1-cre Tet2fl/fl, or Mx1-cre Asxl1fl/fl Tet2fl/fl relative to controls (or Mx1-cre Asxl1 WT Tet2 WT). 99 genes are uniquely down-regulated in Asxl1/Tet2 DKO mice relative to all other genotypes (left), whereas 49 genes are significantly up-regulated (right). (H) GSEA of overlapping and statistically significant gene sets enriched in the LSK cells of mice with deletion of Asxl1 alone or with combined Asxl1 and Tet2 deletion. (I) Gene sets uniquely enriched in mice with concomitant deletion of Asxl1 and Tet2 relative to all other genotypes as determined by GSEA. Error bars represent ±SD; *, P < 0.05 (Mann–Whitney U test).
Consistent with previous in vitro data (Abdel-Wahab et al., 2012), we observed increased expression of posterior HoxA genes in Asxl1 KO LSK cells, including HoxA7 and HoxA9 as well as the Hox-associated transcription factors Hes5 and Gdf11 (Fig. 8 C). Meis1 was not up-regulated, with Asxl1 loss consistent with a previous study in in vitro systems (Fig. 8 C; Abdel-Wahab et al., 2012). We also noted a progressive, age-dependent increase p16INK4a expression in LT-HSCs and multipotent progenitor (MPP) cells (LSK CD48+ CD150−) of Asxl1 KO mice compared with age-matched controls (Fig. 8 D). The p16INK4a locus is a known PRC2 target (Jacobs et al., 1999; Bracken et al., 2007; Hidalgo et al., 2012; Tanaka et al., 2012), and in vitro loss of Asxl1 has been linked to defective Polycomb repression and reduced H3K27 methylation (Abdel-Wahab et al., 2012). Given the increase in p16INK4a expression in KO MPP cells from Asxl1 KO mice, we examined the in vivo cell proliferation of MPP cells from 72-wk-old Asxl1 KO (Vav-cre Asxl1fl/fl) versus control mice (Cre− Asxl1fl/fl) via an in vivo BrdU incorporation assay. The MPP cells of Asxl1 KO mice showed a significant decrease in S-phase compared with littermate control cells (Fig. 8 E). Flow cytometric quantitative assessment of apoptosis in LSK cells of the same mice revealed a significant increase in Annexin-V+/DAPI− and Annexin-V+/DAPI+ LSK cells (Fig. 8 F; n = 5 mice per group) consistent with cell cycle exit and an increase in apoptosis in vivo.
To understand what transcriptional differences might exist between mice with compound deletion of Asxl1 and Tet2 relative to mice with deletion of Asxl1, Tet2, or neither gene, we performed RNA-Seq on LSK cells sorted from the BM of 6-wk-old Mx1-cre Asxl1 WT Tet2 WT, Mx1-cre Asxl1fl/fl, Mx1-cre Tet2fl/fl, and Mx1-cre Asxl1fl/fl Tet2fl/fl mice (Fig. 8 G and Table S3). Of the 1,744 genes significantly up-regulated in any KO mice relative to controls, the majority of these genes were shared between Asxl1 KO and Tet2 KO LSKs but not DKO LSKs (32.6% of up-regulated genes [569/1,744 genes]), followed by genes shared between Asxl1 KO, Tet2 KO, and DKO mice (29.2% of up-regulated genes [510/1,744 genes]). Likewise, for the 1,363 significantly down-regulated genes, the majority of these were shared between Asxl1 KO and Tet2 KO LSKs but not DKO LSKs (37.6% of up-regulated genes [513/1,363 genes]), followed by genes shared between Asxl1 KO, Tet2 KO, and DKO mice (28.0% of up-regulated genes [382/1,363 genes]).
We next performed gene set enrichment analysis (GSEA) to identify gene sets enriched in HSPCs from Asxl1 KO mice or Asxl1/Tet2 DKO mice compared with other groups (Subramanian et al., 2005). We identified gene sets enriched in HSCs (Ramalho-Santos et al., 2002) and apoptosis (http://www.genome.jp/kegg/pathway/hsa/hsa04210.html) in mice with Asxl1 deletion and with concomitant Asxl1/Tet2 deletion (Fig. 8 H). We also identified gene sets that were uniquely enriched in Tet2/Asxl1 DKO mice and not seen in the other groups. This prominently included gene sets characteristic of apoptosis signatures, purified HSPCs (Ivanova et al., 2002), cell cycle regulators (http://www.genome.jp/kegg/pathway/hsa/hsa04110.html), and signatures from MLL-rearranged primary leukemias (Fig. 8 I; Ross et al., 2003; Mullighan et al., 2007).
Genome-wide binding of Asxl1 and global effects of Asxl1 loss on the epigenome
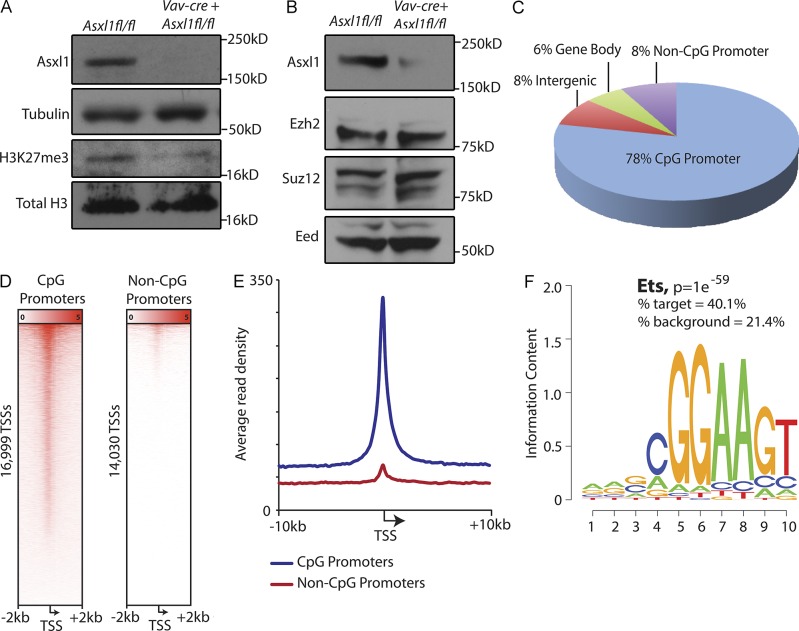
Asxl1 has been shown to interact with epigenetic modifiers known to impact transcription (Abdel-Wahab et al., 2012; Dey et al., 2012). This includes binding to the core members of the PRC2, where loss of ASXL1 has previously been found to result in global down-regulation of histone H3 lysine 27 (H3K27) methylation (Abdel-Wahab et al., 2012) in vitro and in ex vivo ASXL1 mutant primary patient samples. Consistent with this, H3K27 trimethyl (H3K27me3) levels were significantly reduced after Asxl1 deletion (Fig. 9 A) despite sustained expression of the core PRC2 components (Fig. 9 B).
Figure 9.
Effect of Asxl1 loss in vivo on H3K27me3 and identification of Asxl1-regulated genes ChIP-Seq. (A) Western blot analysis of H3K27me3 and total histone H3 in splenocytes of 6-wk-old Vav-cre Asxl1fl/fl mice relative to littermate control. (B) Levels of core PRC2 members Ezh2, Suz12, and Eed in splenocytes of same mice as shown in A. (C) Characterization of Asxl1-binding sites identified by anti-Asxl1 ChIP-Seq analysis in mouse WT BMDMs. (D) Heat map representation of Asxl1 ChIP-Seq signal centered around TSSs (±2 kb) of CpG (left) and non-CpG (right) promoters. (E) Mean Asxl1 ChIP-Seq signal density of CpG and non-CpG promoters centered around the TSS ± 10 kb. (F) Motif enrichment analysis of Asxl1-binding sites identified significant enrichment of Ets transcription factor binding sites (P = 1 ×10−59, percent target = 40.1%, and percent background = 21.4%).
Although the effects of Asxl1 loss on transcription caused by alterations in histone posttranslational modifications have previously been described (Abdel-Wahab et al., 2012), direct transcriptional targets of Asxl1 through characterization of Asxl1 binding throughout the genome have never previously been assessed. We therefore performed chromatin immunoprecipitation (ChIP) for Asxl1 followed by DNA sequencing (ChIP-Seq) in purified mouse myeloid hematopoietic cells. Asxl1 was found to bind to many sites throughout the genome with the majority of significantly enriched Asxl1 peaks (78%) located at CpG-rich transcription start sites (TSSs; Fig. 9, C–E; and Tables S4 and S5). Motif enrichment analysis of the Asxl1-binding sites revealed that the top occurring motifs are most similar to known binding sites of the Ets family of transcription factors (Fig. 9 F; P = 1 × 10−59, percent target = 40.1%, and percent background = 21.4%). A significant subset of genes with dysregulated expression in Asxl1 KO LSK/MP cells were confirmed as direct targets of Asxl1 in our ChIP-Seq analysis (14 up-regulated and 9 down-regulated genes; Table 1).
Table 1.
Genes directly regulated by Asxl1 as determined by differential expression in HSPCs with Asxl1 deletion and directly bound by Asxl1
| Gene | MPs | LSK cells | Anti-Asxl1 ChIP-Seq | ||||
| log2 (fold change) | P-value | log2 (fold change) | P-value | Peak p-value | Fold enrichment | Peak FDR | |
| Tmem87a | 3.91 | 9.60 × 10−6 | 3.11 | 3.68 × 10−6 | 2.78 × 10−7 | 4.21 | 0.29 |
| Pusl1 | 1.54 | 2.21 × 10−7 | 1.30 | 1.29 × 10−3 | 3.04 × 10−8 | 5.24 | 0.15 |
| Ddx23 | 1.45 | 1.15 × 10−10 | 1.09 | 1.79 × 10−3 | 7.45 × 10−15 | 6.93 | 0.09 |
| Pan3 | 1.01 | 1.36 × 10−5 | 0.79 | 8.21 × 10−4 | 4.49 × 10−29 | 9.3 | 0 |
| Srrm2 | 0.86 | 6.02 × 10−8 | 0.98 | 1.61 × 10−6 | 1.85 × 10−12 | 4.49 | 0.06 |
| Dusp6 | 0.80 | 4.11 × 10−4 | 0.63 | 2.45 × 10−3 | 4.48 × 10−30 | 8.43 | 0 |
| Cdca2 | 0.77 | 6.77 × 10−8 | 0.62 | 3.84 × 10−4 | 3.78 × 10−8 | 3.6 | 0.16 |
| Apobec3 | 0.54 | 2.25 × 10−10 | 0.36 | 8.29 × 10−4 | 4.48 × 10−10 | 6.18 | 0.09 |
| Cdc40 | 0.41 | 1.68 × 10−4 | 0.59 | 1.93 × 10−5 | 4.79 × 10−10 | 5.91 | 0.09 |
| Rpl30 | 0.39 | 1.73 × 10−10 | 0.31 | 5.74 × 10−5 | 1.57 × 10−22 | 6.99 | 0 |
| Rad51 | 0.39 | 3.17 × 10−3 | 0.51 | 3.48 × 10−3 | 2.28 × 10−10 | 7.16 | 0.1 |
| Nbr1 | 0.34 | 4.65 × 10−8 | 0.43 | 0.00 | 4.04 × 10−19 | 5.69 | 0 |
| Slc24a6 | 0.30 | 4.66 × 10−4 | 0.82 | 3.24 × 10−10 | 7.52 × 10−11 | 8.09 | 0.1 |
| Dhrs3 | 0.28 | 4.27 × 10−3 | 0.37 | 2.16 × 10−4 | 4.33 × 10−49 | 5.15 | 0 |
| Cdk2 | 0.23 | 1.51 × 10−4 | 0.21 | 3.94 × 10−4 | 1.06 × 10−16 | 7.89 | 0.11 |
| Ifngr1 | 0.18 | 4.59 × 10−3 | 0.38 | 1.99 × 10−4 | 3.02 × 10−20 | 11.89 | 0 |
| Hsp90ab1 | 0.16 | 1.08 × 10−3 | 0.24 | 9.11 × 10−5 | 1.41 × 10−45 | 12.71 | 0 |
| Mtmr9 | −0.44 | 4.53 × 10−4 | −0.47 | 3.62 × 10−4 | 4.07 × 10−11 | 7.19 | 0.11 |
| Ppp1cb | −0.48 | 1.35 × 10−8 | −0.64 | 2.14 × 10−7 | 5.79 × 10−12 | 4.77 | 0.09 |
| Mthfr | −0.51 | 1.65 × 10−3 | −0.47 | 1.21 × 10−3 | 2.47 × 10−9 | 4.82 | 0.11 |
| Ncf4 | −0.52 | 7.21 × 10−9 | −0.66 | 4.38 × 10−5 | 2.16 × 10−18 | 7.53 | 0.07 |
| Slc25a19 | −0.96 | 7.68 × 10−11 | −0.75 | 3.50 × 10−6 | 2.24 × 10−7 | 6.64 | 0.28 |
| Rpl29 | −1.18 | 0.00 | −0.76 | 0.00 | 2.47 × 10−9 | 5.96 | 0.11 |
| Mettl23 | −1.33 | 8.43 × 10−10 | −1.10 | 1.21 × 10−5 | 1.10 × 10−7 | 3.68 | 0.2 |
| Caskin2 | −1.54 | 3.42 × 10−5 | −2.45 | 1.53 × 10−4 | 4.97 × 10−13 | 5.6 | 0.07 |
| Thbs1 | −1.75 | 4.71 × 10−7 | −1.84 | 1.63 × 10−6 | 9.53 × 10−8 | 5.06 | 0.19 |
FDR, false discovery rate. Differentially expressed genes in RNA sequencing of MPs (lineage− Sca-1− c-KIT+ cells) from 1-yr-old Mx1-cre Asxl1fl/fl mice relative to Cre− Asxl1fl/fl littermate controls. Differentially expressed genes in RNA sequencing of LSK cells (lineage− Sca-1+ c-KIT+ cells) from 1-yr-old Mx1-cre Asxl1fl/fl mice relative to Cre− Asxl1fl/fl littermate controls. Data derived from anti-Asxl1 ChIP-Seq in WT C57BL/6H BMDMs.
DISCUSSION
Here we show that conditional deletion of Asxl1, a gene commonly mutated in human MDS, in hematopoietic cells resulted in the development of progressive anemia and leukopenia with concomitant multilineage myeloid dysplasia in vivo. Asxl1 deletion was associated with an increase in the frequency and total number of HSPCs, increased apoptosis, and altered cell cycle distribution of HSPCs in vivo. Asxl1 loss also led to a reduction in myeloid colony output. MDS is characterized by variable cytopenias caused by ineffective production of mature granulocyte, erythroid, and/or megakaryocyte populations and a risk of transformation to AML. Functional characterization of primary samples from patients with MDS has identified an expansion of the primitive HSC compartment (comprised of long-term and short-term HSCs; Will et al., 2012; Pang et al., 2013), the presence of specific genetic alterations throughout the diseased clone originating in the most immature HSCs (Nilsson et al., 2000, 2007; Tehranchi et al., 2010; Will et al., 2012; Pang et al., 2013), dysplastic clonogenic activity of HSCs with reduced in vitro colony formation from MDS-derived HSCs (Sawada et al., 1993, 1995; Will et al., 2012), and an increase in the frequency and absolute number of HSPCs in the setting of decreased mature circulating cells with a concomitant increase in apoptosis of HSPCs in MDS patients (Sawada et al., 1993, 1995; Pang et al., 2013). Collectively, the phenotype of Asxl1 loss recapitulates these central features of human MDS.
One aspect of the Asxl1 conditional KO mouse model that differs from human MDS is the BM hypocellularity observed in Asxl1 KO mice, in contrast to the increased BM cellularity in most MDS patients. Nevertheless, the impaired production of mature myeloid and erythroid cells in the context of an increased relative and total numbers of HSPCs in Asxl1 conditional KO mice does recapitulate key features of human MDS. Moreover, concomitant loss of Asxl1 and Tet2, which are commonly mutated in concert in human MDS, resulted in increased BM cellularity and disease severity with pathological evidence of multilineage dysplasia.
In contrast to MPN and AML, there are few previously described models of MDS, and to date no models of MDS based on mutations in recurrently mutated MDS disease alleles. The most widely used model of MDS is based on transgenic expression of a NUP98-HOXD13 fusion allele (Lin et al., 2005). Although this model has many of the characteristic features of human MDS, the NUP98-HOXD13 fusion was identified in a young patient with therapy-related AML (Raza-Egilmez et al., 1998) and has not been identified in MDS patients to date. In contrast, ASXL1 mutations occur in 15–20% of patients with MDS.
Prior studies of the effects of Asxl1 loss on development and hematopoiesis were performed using a constitutive Asxl1 KO mouse model (Fisher et al., 2010a,b). Our conditional model allows for evaluation of the effects of postnatal deletion of Asxl1 and obviates the problems associated with a high frequency of perinatal lethality in mice with constitutive Asxl1 deletion. Notably, Fisher et al. (2010a,b) identified a 72% reduction in the expected number of Asxl1 homozygous KO mice by postnatal day 21; when mice were backcrossed more than eight generations to a consistent genetic background, KO mice were 100% embryonic lethal, preventing analysis of adult constitutive Asxl1 KO mice. Similar to the germline model, we observed an age-dependent decrease in mature B lymphocytes, splenomegaly caused by EMH, and decreased formation of myeloid and erythroid colonies from Asxl1 KO cells (Fisher et al., 2010a,b). In addition, Fisher et al. (2010a,b) observed dysregulated expression of HoxA genes and homeotic transformation of homozygous Asxl1 mutant embryos, consistent with the current Asxl1 germline and conditional KO models described in this study.
Despite these similarities, several differences exist between the two models. First, no reproducible differences in peripheral blood counts, BM cellularity, or BM cell morphology were seen in Asxl1 constitutive KO mice. In contrast, mice with conditional, homozygous, postnatal deletion of Asxl1 developed leukopenia, anemia, myeloerythroid dysplasia, and BM hypocellularity starting at 6 mo of life. This could be caused by differences in the strain of the mice analyzed, the timing of Asxl1 loss, or cell-nonautonomous effects observed in the constitutive KO model.
Of note, the more profound hematologic abnormalities seen with serial competitive and noncompetitive transplantation of Asxl1 conditional KO hematopoietic cells here have no counterpart in the study of the prior constitutive KO model as serial transplantation was not performed in the prior study (Fisher et al., 2010b). The marked impairment in serial transplantability observed with Asxl1 loss is consistent with the progressive defects in HSC function observed in mice with other alterations in other Polycomb group functions (Ohta et al., 2002).
Although noncompetitive transplantation experiments demonstrated that the MDS phenotype was cell autonomous, Asxl1 deficiency was associated with a defect in HSC self-renewal in competitive transplantation assays and in in vitro colony formation. These data suggested that concurrent genetic or epigenetic alterations in ASXL1 mutant MDS cells must promote self-renewal to allow for clonal dominance of MDS cells. Indeed, concurrent Tet2 loss restored the self-renewal defect induced by Asxl1 loss. Moreover, mice with concomitant loss of Asxl1 and Tet2 developed a larger increase in HSPCs, increased BM cellularity, and decreased numbers of circulating mature cells compared with single-gene KO mice. This phenotype is consistent with more severe MDS and suggests a functional interdependency between these two disease alleles in MDS. Subsequent studies may identify additional disease alleles that can rescue the self-renewal defect seen in ASXL1-deficient stem cells and may lead to the identification of additional mutational interdependencies in MDS and in other malignant contexts.
We previously demonstrated that loss of ASXL1 in vitro results in global down-regulation in H3K27me3 (Abdel-Wahab et al., 2012), the repressive histone modification placed by the PRC2. Here we demonstrate that Asxl1 deletion in the hematopoietic compartment results in reduced H3K27me3 in vivo. We used ChIP-Seq for Asxl1 itself to show that Asxl1 is enriched at gene promoter regions throughout the genome, suggesting a potential role for Asxl1 in direct regulation of gene transcription. In addition, motif enrichment analysis of Asxl1-binding sites revealed enrichment in known binding sites of the Ets family of transcription factors. The significant overlap between genome-wide binding of Asxl1 and Ets family members is critically supportive of the importance of Asxl1 in hematopoiesis as the Ets family of transcription factors are very well understood to play a key role in the growth, survival, differentiation, and activation of hematopoietic cells (Mizuki et al., 2003; Vangala et al., 2003; Koschmieder et al., 2005; Steidl et al., 2006; Choi et al., 2008). Deletion, mutation, and translocation of ETS family members are well-described in myeloid malignancies (Gilliland, 2001), including ETV6 mutations/translocations in MDS and chronic myelomonocytic leukemia (Haferlach et al., 2012) and loss-of-function mutations of PU.1 in AML (Mueller et al., 2002). Moreover, common oncogenic events seen in patients with myeloid malignancies have been demonstrated to transform myeloid cells through suppression of expression of key Ets members. For example, FLT3-ITD mutations and the AML1-ETO (t(8;21)) fusion oncoprotein have been shown to suppress PU.1 expression and function (Mizuki et al., 2003; Vangala et al., 2003). In addition, down-regulation of PU.1 expression results in impaired myeloid differentiation (Rosenbauer et al., 2004; Steidl et al., 2006). Further work to understand the involvement of individual Ets family members and/or a shared transcriptional program between Asxl1 loss and Ets family member loss (Steidl et al., 2006) in the pathogenesis of ASXL1 mutant myeloid malignancy will be critical.
Collectively, our experiments reveal that deletion of Asxl1 results in craniofacial and skeletal developmental abnormalities and mice with hematopoietic-specific Asxl1 loss developed hallmark features of MDS, including progressive ineffective hematopoiesis, impaired myeloid differentiation, multilineage dysplasia, and increased apoptosis and altered cell cycle regulation of HSPCs. Given the paucity of mouse models of human MDS based on known, recurrent MDS disease alleles, we believe the development of a genetically accurate model of MDS will inform subsequent studies aimed to elucidate the molecular basis for MDS and to develop novel therapies for MDS patients.
MATERIALS AND METHODS
Animals.
All animals were housed at New York University School of Medicine or at Memorial Sloan-Kettering Cancer Center. All animal procedures were conducted in accordance with the Guidelines for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committees at New York University School of Medicine and Memorial Sloan-Kettering Cancer Center.
Generation of Asxl1-deficient mice.
The Asxl1 allele was deleted by targeting exons 5–10. Two LoxP sites flanking exon 5–10 and an Frt-flanked neomycin selection cassette were inserted in the upstream intron (Fig. 1 A). 10 µg of the targeting vector was linearized by NotI and then transfected by electroporation of BAC-BA1 (C57BL/6 × 129/SvEv) hybrid ES cells. After selection with G418 antibiotic, surviving clones were expanded for PCR analysis to identify recombinant ES clones. Secondary confirmation of positive clones identified by PCR was performed by Southern blotting analysis. DNA was digested with BamHI and electrophoretically separated on a 0.8% agarose gel. After transfer to a nylon membrane, the digested DNA was hybridized with a probe targeted against the 3′ or 5′ external region. DNA from C57BL/6 (B6), 129/SvEv (129), and BA1 (C57BL/6 × 129/SvEv; Hybrid) mouse strains was used as WT controls. Positive ES clones were expanded and injected into blastocysts.
The generated mice (Asxl1fl/fl) were initially crossed to a germline Flp-deleter (The Jackson Laboratory), to eliminate the neomycin cassette, and subsequently to the IFN-α–inducible Mx1-cre (The Jackson Laboratory), the hematopoietic-specific Vav-cre, and the germline EIIa-cre (Kühn et al., 1995; Lakso et al., 1996; Stadtfeld and Graf, 2005). Mice were backcrossed for six generations to C57BL/6 mice.
Asxl1fl/fl, Asxl1fl/+, and Asxl1+/+ littermate mice were genotyped by PCR with primers Asxl1-F3 (5′-CAGCCGTTTTACCACAGTTT-3′) and Asxl1-R3 (5′-AGGGAAAGGGACAGAATGAC-3′) using the following parameters: 95°C for 4 min, followed by 35 cycles of 95°C for 45 s, 56°C for 45 s, and 72°C for 1 min, and then 72°C for 5 min. The WT allele was detected as a band at 200 bp, whereas the floxed allele was detected as a band of 380 bp. Excision after Cre recombination was confirmed by PCR with primers to detect a floxed portion of the construct (Asxl1-RecF, 5′-ACGCCGGCTTAAGTGTACACG-3′; and Asxl1-RecR, 5′-GACTAAGTTGCCGTGGGTGCT-3′) using the same parameters as above.
In vivo experiments.
Mx1-cre Asxl1fl/fl conditional and Cre− Asxl1fl/fl control mice received five intraperitoneal injections of polyI:polyC every other day at a dose of 20 mg/kg of body weight starting at 2 wk after birth. For the hematopoietic-specific Vav-cre line, Asxl1fl/flVav-cre+, and Asxl1fl/flVav-cre− mice were analyzed between 3 and 60 wk of age. BM, spleen, and peripheral blood were analyzed by flow cytometry. Formalin-fixed paraffin-embedded tissue sections were stained with hematoxylin and eosin (H&E). Peripheral blood was smeared on a slide and stained using the Wright-Giemsa staining method. Tissue sections and blood smears were evaluated by a hematopathologist (C.Y. Park) Deletion of the Asxl1 allele and transcript was measured by genomic PCR and Western blot analysis.
BM transplantation.
Freshly dissected femurs and tibias were isolated from Asxl1fl/fl CD45.2+ or Vav-cre+Asxl1fl/fl CD45.2+ mice. BM was flushed with a 3-cc insulin syringe into PBS supplemented with 3% fetal bovine serum. The BM was spun at 0.5 g by centrifugation at 4°C, and RBCs were lysed in ammonium chloride-potassium bicarbonate lysis buffer for 5 min. After centrifugation, cells were resuspended in PBS plus 3% FBS, passed through a cell strainer, and counted. Finally, 0.5 × 106 total BM cells of Asxl1fl/fl CD45.2+ or Vav-cre+ Asxl1fl/fl CD45.2+ mice were mixed with 0.5 × 106 WT CD45.1+ support BM and transplanted via tail vein injection into lethally irradiated (two times 450 cGy) CD45.1+ host mice. Chimerism was measured by FACS in peripheral blood at 4 wk after transplant (week 0, pre-polyI:polyC). Chimerism was followed via FACS in the peripheral blood every 4 wk (week 0, 4, 6, 8, 12, and 16 after polyI:polyC injection). Additionally, for each bleeding, whole blood cell counts were measured on a blood analyzer, and peripheral blood smears were scored. Chimerism in the BM, spleen, and thymus was evaluated at 16 wk via animal sacrifice and subsequent FACS analysis. The above procedure was also repeated with Asxl1fl/fl CD45.2+, Vav-cre+ Asxl1fl/fl CD45.2+, Vav-cre+ Tet2fl/fl CD45.2+, and Vav-cre+ Asxl1fl/fl Tet2fl/fl CD45.2+ mice for competitive transplantation of mice with loss of Asxl1, Tet2, or both. For noncompetitive transplantation experiments, 106 total BM cells of Asxl1fl/fl CD45.2+, littermate Vav-cre+ Asxl1fl/fl CD45.2+, or littermate Mx1-cre+ Asxl1fl/fl CD45.2+ mice were injected into lethally irradiated (two times 450 cGy) CD45.1+ host mice. Similarly, for LSK transplants, 1,000 FACS-sorted LSK cells from secondarily transplanted Asxl1 KO or control mice were transplanted into lethally irradiated CD45.1 host mice. Recipient mice were then followed until moribund or 80 wk after transplantation.
In vitro colony-forming assays.
LSK, CMP, GMP, and MEP cells were sorted from the BM of Asxl1fl/fl and littermate Vav-cre+ Asxl1fl/fl mice and seeded at a density of 500 cells/replicate for LSK cells and 1,000 cells/replicate for CMP, GMP, and MEP subsets into cytokine-supplemented methylcellulose medium (Methocult M3434; STEMCELL Technologies). Colonies propagated in culture were scored at day 7. Representative colonies were isolated from the plate for cytospins. Remaining cells were resuspended and counted, and a portion was taken for replating (20,000 cells/replicate) for a total of seven platings. Cytospins were performed by resuspending in warm PBS and spun onto the slides at 350 g for 5 min. Slides were air-dried and stained using the Giemsa-Wright method.
Antibodies, FACS, and Western blot analysis.
Antibody staining and FACS analysis was performed as previously described (Klinakis et al., 2011). BM or spleen mononuclear cells were stained with a lineage cocktail comprised of antibodies targeting CD4, CD8, B220, NK1.1, Gr-1, CD11b, Ter119, and IL-7Rα. Cells were also stained with antibodies against c-Kit, Sca-1, FcγRII/III, and CD34. Cell populations were analyzed using a FACS-LSRII (BD) and sorted with a FACSAria instrument (BD). All antibodies were purchased from BD or eBioscience. We used the following antibodies: c-Kit (2B8), Sca-1 (D7), Mac-1/CD11b (M1/70), Gr-1 (RB6-8C5), NK1.1 (PK136), Ter-119, IL7-Rα (A7R34), CD34 (RAM34), FcγRII/III (2.4G2), CD4 (RM4-5), CD4 (H129.19), CD8 (53-6.7), CD45.1 (A20), CD45.2 (104), CD150 (9D1), and CD48 (HM48-1). The following antibodies were used for Western blot analysis: Asxl1 (clone N-13; Santa Cruz Biotechnology, Inc.), Ezh2 (EMD Millipore), Suz12 (Abcam), EED (Abcam), H3K27me3 (Abcam), total H3 (Abcam), and tubulin (Sigma-Aldrich).
Cell cycle and apoptosis analyses.
For cell cycle analysis, the BrdU-APC kit was used (BD) according to the manufacturer’s protocol. Mice were treated with 1 mg BrdU intraperitoneally, followed by harvest of BM cells 24 h later. For evaluation of apoptosis, the Annexin V–FITC apoptosis detection kit was used (BD) according to the manufacturer’s recommendations. DAPI was used as counterstain in both BrdU and annexin V experiments.
Histological analyses.
Mice were sacrificed and autopsied, and then dissected tissue samples were fixed for 24 h in 4% paraformaldehyde, dehydrated, and embedded in paraffin. Paraffin blocks were sectioned at 4 µm and stained with H&E. Images were acquired using an Axio Observer A1 microscope (Carl Zeiss).
Peripheral blood analysis.
Blood was collected by retroorbital bleeding using heparinized microhematocrit capillary tubes (Thermo Fisher Scientific). Automated peripheral blood counts were obtained using a HemaVet 950 (Drew Scientific) according to standard manufacturer’s instruction. Differential blood counts were realized on blood smears stained using Wright-Giemsa staining and visualized using an Axio Observer A1 microscope.
RNA-Seq and quantitative real-time PCR (qRT-PCR) analysis.
Total RNA was isolated using the RNeasy Plus Mini kit (QIAGEN), and cDNA was synthesized using the SuperScript First-Strand kit (Invitrogen). Quantitative PCR was performed using SYBR green iMaster and a LightCycler 480 (Roche). For RNA-Seq analysis, Fastq files were aligned to mm9 using TopHatV1.4 with default parameters. Differential expression tests were performed using the Cuffdiff module of Cufflinks with RefSeq genes provided as an annotation (-N, -u and -M options engaged). We considered genes that had a q < 0.05 to be significantly different between genotypes.
ChIP-Seq and analysis.
Because low chromatin yields from HSPC populations precluded ChIP-Seq experiments, BMDMs from WT C57BL/6 mice were used as a surrogate to identify genome-wide Asxl1 binding sites. The antibody used for Asxl1 ChIP-Seq experiments was obtained from Santa Cruz Biotechnology, Inc. ChIP was performed as described previously (Dey et al., 2012).
ChIP and input DNAs were prepared for amplification by converting overhangs into phosphorylated blunt ends and adding an adenine to the 3′ ends. Illumina adaptors were added and the library was size-selected (175–225 bp) on an agarose gel. The adaptor- ligated libraries were amplified for 18 cycles. The resulting DNA libraries were purified, quantified, and tested by qPCR at the same specific genomic regions as the original ChIP DNA to assess quality of the amplification reactions. DNA libraries then were sequenced on the Illumina Genome Analyzer II.
Sequenced reads were aligned to the reference genomes (mm9) using bowtie with maximum two mismatches, keeping only uniquely mapping reads. Peak calling was performed using MACS1.4 with the following options: -p 1e-7,–nomodel True,–shiftsize 100,–keep-dup 1. Peaks were assigned to genes using bedtools. We considered Asxl1-bound genes to be any mouse RefSeq entry containing a peak overlapping the gene or 2 kb upstream of the TSS. ChIP-Seq read profile and heat map densities were generated using genomic tools. Mouse RefSeq and CpG island annotations were downloaded from the UCSC Genome Bioinformatics Table Browser.
Skeletal preparations.
Skeletal preparations were performed as described previously (de Pontual et al., 2011).
Online supplemental material.
Table S1, included as a separate PDF file, shows differentially expressed transcripts between 1-yr-old Mx1-cre Asxl1fl/fl and Asxl1fl/fl LSK cells by RNA sequencing. Table S2, included as a separate PDF file, shows differentially expressed transcripts between 1-yr-old Mx1-cre Asxl1fl/fl and Asxl1fl/fl MPs by RNA sequencing. Table S3, included as a separate PDF file, shows differentially expressed genes in LSK cells from 6-wk-old Mx1-cre Asxl1fl/fl, Mx1-cre Tet2fl/fl, and Mx1-cre Asxl1fl/fl Tet2fl/fl. Table S4, included as a separate PDF file, shows regions with significant Asxl1 binding by anti-Asxl1 ChIP-Seq. Table S5, included as a separate PDF file, shows genes with significant Asxl1 binding ±2 kb from the TSS as determined by anti-Asxl1 ChIP-Seq. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20131141/DC1.
Supplementary Material
Acknowledgments
This work was supported by a grant from the Starr Cancer Consortium to R.L. Levine and B.E. Bernstein, by grants from Gabrielle’s Angel Fund to R.L. Levine, I. Aifantis, and O. Abdel-Wahab, by National Institutes of Health (NIH) grant 1R01CA138234-01 to R.L. Levine and 5RO1CA173636-01 to R.L. Levine and I. Aifantis, by a Leukemia and Lymphoma Society (LLS) Translational Research Program grant to R.L. Levine and I. Aifantis, and by National Heart, Lung, and Blood Institute grant 5U01HL100395 to B.E. Bernstein. I. Aifantis is supported by the NIH (grants RO1CA133379, RO1CA105129, 1RO1CA173636, RO1CA149655, and 5RO1CA173636), the LLS Chemotherapy Foundation, the William Lawrence Blanche Hughes Foundation, and the V Foundation for Cancer Research. J. Gao is supported by the New York University T32 CA009161 (Levy). I. Aifantis and B.E. Bernstein are Howard Hughes Medical Institute Early Career Scientists. O. Abdel-Wahab is supported by an NIH K08 Clinical Investigator Award (1K08CA160647-01), a US Department of Defense Postdoctoral Fellow Award in Bone Marrow Failure Research (W81XWH-12-1-0041), the Josie Roberston Investigator Program, and a Damon Runyon Clinical Investigator Award with support from the Evans Foundation. R.L. Levine is a Scholar of the LLS.
All authors declare no conflicts of interest.
Author contributions: O. Abdel-Wahab, J. Gao, I. Aifantis, and R.L. Levine designed the study. O. Abdel-Wahab, J. Gao, M. Adli, Y.R. Chung, J.Y. Shin, P.K. Bhatt, O.A. Guryanova, E. Kim, L. Telis, and S. Pandey performed the experiments. D. Ndiaye-Lobry, L.M. LaFave, A.H. Shih, and S. Pandey helped with animal generation, genotyping, and maintenance. S. Monette and C.Y. Park performed phenotypic and histological analysis of tissues. M. Adli, A. Dey, T. Trimarchi, R. Koche, C. Kuscu, J. Liu, and B.E. Bernstein performed ChIP and sequencing analysis. O. Abdel-Wahab, J. Gao, M. Adli, T. Trimarchi, T. Hricik, C. Kuscu, R. Koche, S. Li, X. Zhao, and C.E. Mason analyzed the data. O. Abdel-Wahab, J. Gao, I. Aifantis, and R.L. Levine prepared the manuscript with input from the other authors.
Footnotes
Abbreviations used:
- AML
- acute myeloid leukemia
- ChIP
- chromatin immunoprecipitation
- CMP
- common MP
- DKO
- double KO
- EMH
- extramedullary hematopoiesis
- ES
- embryonic stem
- GMP
- granulocyte/macrophage progenitor
- GSEA
- gene set enrichment analysis
- Hb
- hemoglobin
- HSC
- hematopoietic stem cell
- HSPC
- hematopoietic stem/progenitor cell
- LT-HSC
- long-term HSC
- MDS
- myelodysplastic syndrome
- MEP
- megakaryocyte/erythroid progenitor
- MP
- myeloid progenitor
- MPN
- myeloproliferative neoplasm
- MPP
- multipotent progenitor
- polyI:polyC
- polyinosinic-polycytidylic acid
- qRT-PCR
- quantitative real-time PCR
- TSS
- transcription start site
References
- Abdel-Wahab O., Adli M., LaFave L.M., Gao J., Hricik T., Shih A.H., Pandey S., Patel J.P., Chung Y.R., Koche R., et al. 2012. ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer Cell. 22:180–193 10.1016/j.ccr.2012.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejar R., Stevenson K., Abdel-Wahab O., Galili N., Nilsson B., Garcia-Manero G., Kantarjian H., Raza A., Levine R.L., Neuberg D., Ebert B.L. 2011. Clinical effect of point mutations in myelodysplastic syndromes. N. Engl. J. Med. 364:2496–2506 10.1056/NEJMoa1013343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejar R., Stevenson K.E., Caughey B.A., Abdel-Wahab O., Steensma D.P., Galili N., Raza A., Kantarjian H., Levine R.L., Neuberg D., et al. 2012. Validation of a prognostic model and the impact of mutations in patients with lower-risk myelodysplastic syndromes. J. Clin. Oncol. 30:3376–3382 10.1200/JCO.2011.40.7379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boultwood J., Perry J., Pellagatti A., Fernandez-Mercado M., Fernandez-Santamaria C., Calasanz M.J., Larrayoz M.J., Garcia-Delgado M., Giagounidis A., Malcovati L., et al. 2010. Frequent mutation of the polycomb-associated gene ASXL1 in the myelodysplastic syndromes and in acute myeloid leukemia. Leukemia. 24:1062–1065 10.1038/leu.2010.20 [DOI] [PubMed] [Google Scholar]
- Bracken A.P., Kleine-Kohlbrecher D., Dietrich N., Pasini D., Gargiulo G., Beekman C., Theilgaard-Mönch K., Minucci S., Porse B.T., Marine J.C., et al. 2007. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 21:525–530 10.1101/gad.415507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C.W., Chung Y.J., Slape C., Aplan P.D. 2008. Impaired differentiation and apoptosis of hematopoietic precursors in a mouse model of myelodysplastic syndrome. Haematologica. 93:1394–1397 10.3324/haematol.13042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pontual L., Yao E., Callier P., Faivre L., Drouin V., Cariou S., Van Haeringen A., Geneviève D., Goldenberg A., Oufadem M., et al. 2011. Germline deletion of the miR-17∼92 cluster causes skeletal and growth defects in humans. Nat. Genet. 43:1026–1030 10.1038/ng.915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A., Seshasayee D., Noubade R., French D.M., Liu J., Chaurushiya M.S., Kirkpatrick D.S., Pham V.C., Lill J.R., Bakalarski C.E., et al. 2012. Loss of the tumor suppressor BAP1 causes myeloid transformation. Science. 337:1541–1546 10.1126/science.1221711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher C.L., Lee I., Bloyer S., Bozza S., Chevalier J., Dahl A., Bodner C., Helgason C.D., Hess J.L., Humphries R.K., Brock H.W. 2010a. Additional sex combs-like 1 belongs to the enhancer of trithorax and polycomb group and genetically interacts with Cbx2 in mice. Dev. Biol. 337:9–15 10.1016/j.ydbio.2009.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher C.L., Pineault N., Brookes C., Helgason C.D., Ohta H., Bodner C., Hess J.L., Humphries R.K., Brock H.W. 2010b. Loss-of-function Additional sex combs like 1 mutations disrupt hematopoiesis but do not cause severe myelodysplasia or leukemia. Blood. 115:38–46 10.1182/blood-2009-07-230698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelsi-Boyer V., Trouplin V., Adélaïde J., Bonansea J., Cervera N., Carbuccia N., Lagarde A., Prebet T., Nezri M., Sainty D., et al. 2009. Mutations of polycomb-associated gene ASXL1 in myelodysplastic syndromes and chronic myelomonocytic leukaemia. Br. J. Haematol. 145:788–800 10.1111/j.1365-2141.2009.07697.x [DOI] [PubMed] [Google Scholar]
- Gilliland D.G. 2001. The diverse role of the ETS family of transcription factors in cancer. Clin. Cancer Res. 7:451–453 [PubMed] [Google Scholar]
- Haferlach C., Bacher U., Schnittger S., Alpermann T., Zenger M., Kern W., Haferlach T. 2012. ETV6 rearrangements are recurrent in myeloid malignancies and are frequently associated with other genetic events. Genes Chromosomes Cancer. 51:328–337 10.1002/gcc.21918 [DOI] [PubMed] [Google Scholar]
- Hidalgo I., Herrera-Merchan A., Ligos J.M., Carramolino L., Nuñez J., Martinez F., Dominguez O., Torres M., Gonzalez S. 2012. Ezh1 is required for hematopoietic stem cell maintenance and prevents senescence-like cell cycle arrest. Cell Stem Cell. 11:649–662 10.1016/j.stem.2012.08.001 [DOI] [PubMed] [Google Scholar]
- Hoischen A., van Bon B.W., Rodríguez-Santiago B., Gilissen C., Vissers L.E., de Vries P., Janssen I., van Lier B., Hastings R., Smithson S.F., et al. 2011. De novo nonsense mutations in ASXL1 cause Bohring-Opitz syndrome. Nat. Genet. 43:729–731 10.1038/ng.868 [DOI] [PubMed] [Google Scholar]
- Itzykson R., Kosmider O., Renneville A., Gelsi-Boyer V., Meggendorfer M., Morabito M., Berthon C., Adès L., Fenaux P., Beyne-Rauzy O., et al. 2013. Prognostic score including gene mutations in chronic myelomonocytic leukemia. J. Clin. Oncol. 31:2428–2436 10.1200/JCO.2012.47.3314 [DOI] [PubMed] [Google Scholar]
- Ivanova N.B., Dimos J.T., Schaniel C., Hackney J.A., Moore K.A., Lemischka I.R. 2002. A stem cell molecular signature. Science. 298:601–604 10.1126/science.1073823 [DOI] [PubMed] [Google Scholar]
- Jacobs J.J., Kieboom K., Marino S., DePinho R.A., van Lohuizen M. 1999. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 397:164–168 10.1038/16476 [DOI] [PubMed] [Google Scholar]
- Jankowska A.M., Makishima H., Tiu R.V., Szpurka H., Huang Y., Traina F., Visconte V., Sugimoto Y., Prince C., O’Keefe C., et al. 2011. Mutational spectrum analysis of chronic myelomonocytic leukemia includes genes associated with epigenetic regulation: UTX, EZH2, and DNMT3A. Blood. 118:3932–3941 10.1182/blood-2010-10-311019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinakis A., Lobry C., Abdel-Wahab O., Oh P., Haeno H., Buonamici S., van De Walle I., Cathelin S., Trimarchi T., Araldi E., et al. 2011. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature. 473:230–233 10.1038/nature09999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M., Bandukwala H.S., An J., Lamperti E.D., Thompson E.C., Hastie R., Tsangaratou A., Rajewsky K., Koralov S.B., Rao A. 2011. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc. Natl. Acad. Sci. USA. 108:14566–14571 10.1073/pnas.1112317108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koschmieder S., Rosenbauer F., Steidl U., Owens B.M., Tenen D.G. 2005. Role of transcription factors C/EBPalpha and PU.1 in normal hematopoiesis and leukemia. Int. J. Hematol. 81:368–377 10.1532/IJH97.05051 [DOI] [PubMed] [Google Scholar]
- Kühn R., Schwenk F., Aguet M., Rajewsky K. 1995. Inducible gene targeting in mice. Science. 269:1427–1429 10.1126/science.7660125 [DOI] [PubMed] [Google Scholar]
- Lakso M., Pichel J.G., Gorman J.R., Sauer B., Okamoto Y., Lee E., Alt F.W., Westphal H. 1996. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. USA. 93:5860–5865 10.1073/pnas.93.12.5860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Cai X., Cai C.L., Wang J., Zhang W., Petersen B.E., Yang F.C., Xu M. 2011. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 118:4509–4518 10.1182/blood-2010-12-325241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.W., Slape C., Zhang Z., Aplan P.D. 2005. NUP98-HOXD13 transgenic mice develop a highly penetrant, severe myelodysplastic syndrome that progresses to acute leukemia. Blood. 106:287–295 10.1182/blood-2004-12-4794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magini P., Della Monica M., Uzielli M.L., Mongelli P., Scarselli G., Gambineri E., Scarano G., Seri M. 2012. Two novel patients with Bohring-Opitz syndrome caused by de novo ASXL1 mutations. Am. J. Med. Genet. A. 158A:917–921 10.1002/ajmg.a.35265 [DOI] [PubMed] [Google Scholar]
- Metzeler K.H., Becker H., Maharry K., Radmacher M.D., Kohlschmidt J., Mrózek K., Nicolet D., Whitman S.P., Wu Y.Z., Schwind S., et al. 2011. ASXL1 mutations identify a high-risk subgroup of older patients with primary cytogenetically normal AML within the ELN Favorable genetic category. Blood. 118:6920–6929 10.1182/blood-2011-08-368225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuki M., Schwable J., Steur C., Choudhary C., Agrawal S., Sargin B., Steffen B., Matsumura I., Kanakura Y., Böhmer F.D., et al. 2003. Suppression of myeloid transcription factors and induction of STAT response genes by AML-specific Flt3 mutations. Blood. 101:3164–3173 10.1182/blood-2002-06-1677 [DOI] [PubMed] [Google Scholar]
- Moran-Crusio K., Reavie L., Shih A., Abdel-Wahab O., Ndiaye-Lobry D., Lobry C., Figueroa M.E., Vasanthakumar A., Patel J., Zhao X., et al. 2011. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 20:11–24 10.1016/j.ccr.2011.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller B.U., Pabst T., Osato M., Asou N., Johansen L.M., Minden M.D., Behre G., Hiddemann W., Ito Y., Tenen D.G. 2002. Heterozygous PU.1 mutations are associated with acute myeloid leukemia. Blood. 100:998–1007 10.1182/blood.V100.3.998 [DOI] [PubMed] [Google Scholar]
- Mullighan C.G., Kennedy A., Zhou X., Radtke I., Phillips L.A., Shurtleff S.A., Downing J.R. 2007. Pediatric acute myeloid leukemia with NPM1 mutations is characterized by a gene expression profile with dysregulated HOX gene expression distinct from MLL-rearranged leukemias. Leukemia. 21:2000–2009 10.1038/sj.leu.2404808 [DOI] [PubMed] [Google Scholar]
- Nilsson L., Astrand-Grundström I., Arvidsson I., Jacobsson B., Hellström-Lindberg E., Hast R., Jacobsen S.E. 2000. Isolation and characterization of hematopoietic progenitor/stem cells in 5q-deleted myelodysplastic syndromes: evidence for involvement at the hematopoietic stem cell level. Blood. 96:2012–2021 [PubMed] [Google Scholar]
- Nilsson L., Edén P., Olsson E., Månsson R., Astrand-Grundström I., Strömbeck B., Theilgaard-Mönch K., Anderson K., Hast R., Hellström-Lindberg E., et al. 2007. The molecular signature of MDS stem cells supports a stem-cell origin of 5q myelodysplastic syndromes. Blood. 110:3005–3014 10.1182/blood-2007-03-079368 [DOI] [PubMed] [Google Scholar]
- Ohta H., Sawada A., Kim J.Y., Tokimasa S., Nishiguchi S., Humphries R.K., Hara J., Takihara Y. 2002. Polycomb group gene rae28 is required for sustaining activity of hematopoietic stem cells. J. Exp. Med. 195:759–770 10.1084/jem.20011911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang W.W., Pluvinage J.V., Price E.A., Sridhar K., Arber D.A., Greenberg P.L., Schrier S.L., Park C.Y., Weissman I.L. 2013. Hematopoietic stem cell and progenitor cell mechanisms in myelodysplastic syndromes. Proc. Natl. Acad. Sci. USA. 110:3011–3016 10.1073/pnas.1222861110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel J.P., Gönen M., Figueroa M.E., Fernandez H., Sun Z., Racevskis J., Van Vlierberghe P., Dolgalev I., Thomas S., Aminova O., et al. 2012. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N. Engl. J. Med. 366:1079–1089 10.1056/NEJMoa1112304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivoron C., Couronné L., Della Valle V., Lopez C.K., Plo I., Wagner-Ballon O., Do Cruzeiro M., Delhommeau F., Arnulf B., Stern M.H., et al. 2011. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 20:25–38 10.1016/j.ccr.2011.06.003 [DOI] [PubMed] [Google Scholar]
- Ramalho-Santos M., Yoon S., Matsuzaki Y., Mulligan R.C., Melton D.A. 2002. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science. 298:597–600 10.1126/science.1072530 [DOI] [PubMed] [Google Scholar]
- Raza-Egilmez S.Z., Jani-Sait S.N., Grossi M., Higgins M.J., Shows T.B., Aplan P.D. 1998. NUP98-HOXD13 gene fusion in therapy-related acute myelogenous leukemia. Cancer Res. 58:4269–4273 [PubMed] [Google Scholar]
- Rosenbauer F., Wagner K., Kutok J.L., Iwasaki H., Le Beau M.M., Okuno Y., Akashi K., Fiering S., Tenen D.G. 2004. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat. Genet. 36:624–630 10.1038/ng1361 [DOI] [PubMed] [Google Scholar]
- Ross M.E., Zhou X., Song G., Shurtleff S.A., Girtman K., Williams W.K., Liu H.-C., Mahfouz R., Raimondi S.C., Lenny N., et al. 2003. Classification of pediatric acute lymphoblastic leukemia by gene expression profiling. Blood. 102:2951–2959 10.1182/blood-2003-01-0338 [DOI] [PubMed] [Google Scholar]
- Sanada M., Ogawa S. 2012. Genome-wide analysis of myelodysplastic syndromes. Curr. Pharm. Des. 18:3163–3169 10.2174/1381612811209023163 [DOI] [PubMed] [Google Scholar]
- Sawada K., Sato N., Tarumi T., Sakai N., Koizumi K., Sakurama S., Ieko M., Yasukouchi T., Koyanagawa Y., Yamaguchi M., et al. 1993. Proliferation and differentiation of myelodysplastic CD34+ cells in serum-free medium: response to individual colony-stimulating factors. Br. J. Haematol. 83:349–358 10.1111/j.1365-2141.1993.tb04656.x [DOI] [PubMed] [Google Scholar]
- Sawada K., Sato N., Notoya A., Tarumi T., Hirayama S., Takano H., Koizumi K., Yasukouchi T., Yamaguchi M., Koike T. 1995. Proliferation and differentiation of myelodysplastic CD34+ cells: phenotypic subpopulations of marrow CD34+ cells. Blood. 85:194–202 [PubMed] [Google Scholar]
- Shih A.H., Abdel-Wahab O., Patel J.P., Levine R.L. 2012. The role of mutations in epigenetic regulators in myeloid malignancies. Nat. Rev. Cancer. 12:599–612 10.1038/nrc3343 [DOI] [PubMed] [Google Scholar]
- Stadtfeld M., Graf T. 2005. Assessing the role of hematopoietic plasticity for endothelial and hepatocyte development by non-invasive lineage tracing. Development. 132:203–213 10.1242/dev.01558 [DOI] [PubMed] [Google Scholar]
- Steidl U., Rosenbauer F., Verhaak R.G., Gu X., Ebralidze A., Otu H.H., Klippel S., Steidl C., Bruns I., Costa D.B., et al. 2006. Essential role of Jun family transcription factors in PU.1 knockdown-induced leukemic stem cells. Nat. Genet. 38:1269–1277 10.1038/ng1898 [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Miyagi S., Sashida G., Chiba T., Yuan J., Mochizuki-Kashio M., Suzuki Y., Sugano S., Nakaseko C., Yokote K., et al. 2012. Ezh2 augments leukemogenicity by reinforcing differentiation blockage in acute myeloid leukemia. Blood. 120:1107–1117 10.1182/blood-2011-11-394932 [DOI] [PubMed] [Google Scholar]
- Tehranchi R., Woll P.S., Anderson K., Buza-Vidas N., Mizukami T., Mead A.J., Astrand-Grundström I., Strömbeck B., Horvat A., Ferry H., et al. 2010. Persistent malignant stem cells in del(5q) myelodysplasia in remission. N. Engl. J. Med. 363:1025–1037 10.1056/NEJMoa0912228 [DOI] [PubMed] [Google Scholar]
- Thol F., Friesen I., Damm F., Yun H., Weissinger E.M., Krauter J., Wagner K., Chaturvedi A., Sharma A., Wichmann M., et al. 2011. Prognostic significance of ASXL1 mutations in patients with myelodysplastic syndromes. J. Clin. Oncol. 29:2499–2506 10.1200/JCO.2010.33.4938 [DOI] [PubMed] [Google Scholar]
- Vangala R.K., Heiss-Neumann M.S., Rangatia J.S., Singh S.M., Schoch C., Tenen D.G., Hiddemann W., Behre G. 2003. The myeloid master regulator transcription factor PU.1 is inactivated by AML1-ETO in t(8;21) myeloid leukemia. Blood. 101:270–277 10.1182/blood-2002-04-1288 [DOI] [PubMed] [Google Scholar]
- Vannucchi A.M., Lasho T.L., Guglielmelli P., Biamonte F., Pardanani A., Pereira A., Finke C., Score J., Gangat N., Mannarelli C., et al. 2013. Mutations and prognosis in primary myelofibrosis. Leukemia. 27:1861–1869 10.1038/leu.2013.119 [DOI] [PubMed] [Google Scholar]
- Will B., Zhou L., Vogler T.O., Ben-Neriah S., Schinke C., Tamari R., Yu Y., Bhagat T.D., Bhattacharyya S., Barreyro L., et al. 2012. Stem and progenitor cells in myelodysplastic syndromes show aberrant stage-specific expansion and harbor genetic and epigenetic alterations. Blood. 120:2076–2086 10.1182/blood-2011-12-399683 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.