Abstract
Purpose
CA-125 has been a valuable marker for detecting ovarian cancer, however, not sensitive enough to detect early stage disease and not specific for ovarian cancer. The purpose of our study was to identify autoantibody markers that are specific for ovarian cancer regardless of CA-125 levels.
Methods
Top-down and iTRAQ quantitative proteomics methods were used to identify high frequency autoantibodies in ovarian cancer. Protein microarrays comprising the recombinant autoantigens were screened using serum samples from various stages of ovarian cancer with diverse levels of CA-125 as well as benign and healthy controls. ROC curve and dot blot analyses were performed to validate the sensitivity and specificity of the autoantibody markers.
Results
The proteomics methodologies identified >60 potential high frequency autoantibodies in ovarian cancer. Individual serum samples from ovarian cancer stages I-IV compared to control samples that were screened on a microarray containing native recombinant autoantigens revealed a panel of stage I high frequency autoantibodies. Preliminary ROC curve and dot blot analyses performed with the ovarian cancer samples showed higher specificity and sensitivity as compared to CA-125. Three autoantibody markers exhibited higher specificity in various stages of ovarian cancer with low and normal CA-125 levels.
Conclusions
Proteomics technologies are suitable for the identification of protein biomarkers and also the identification of autoantibody biomarkers when combined with protein microarray screening. Using native recombinant autoantigen arrays to screen autoantibody markers, it is possible to identify markers with higher sensitivity and specificity than CA-125 that are relevant for early detection of ovarian cancer.
Keywords: Proteomics, iTRAQ, autoantibodies, biomarkers, microarray, dot blot
Introduction
The American Cancer Society estimates that in 2013 about 22,240 new cases of ovariancancer will be diagnosed and 14,030 women will die of ovarian cancer in the United States (ovariancancer.org) (Zhang et al. 2004; Hays et al. 2010; Jemal et al. 2010). Ovarian cancer is typically asymptomatic until well advanced; however, when ovarian cancer is detected early the five year survival rate is over 90% (Piver and Wong 1998). Although used for screening, serum CA-125 has only a modest ability to detect early stage ovarian cancer. In 20% of advanced stage ovarian cancer cases and 50% of early stage ovarian cancer cases the CA-125 level is not elevated despite the presence of ovarian cancer (Partridge et al. 2009; Mor et al. 2005). CA-125 screening is also not reliable enough for routine monitoring and is only administered to high risk patients. Additionally, these tests have a very low predictive value and a high false positive rate (Partridge et al. 2009). Accurate and routine screening tests are urgently needed for early diagnosis of ovarian cancer. Advanced stage cancer patients also need a reliable monitoring test because the majority of these patients relapse and require further aggressive treatments. Development of improved diagnostic tools for the early detection of ovarian cancer, including the discovery of new ovarian cancer biomarkers, has the potential to significantly improve the survival rate.
The human immune system aptly called the ‘ultimate biosensor’ (Chatterjee et al. 2006), is considered the most sensitive and specific sentinel early warning system for cancer, eliciting a clear amplified response in the form of ‘autoantibodies’ that can be detected even in the presence of very few tumor cells (Burnham 1972; Tan 1991; Tan et al. 1988; Vogelstein and Kinzler 1993; Levine 1997; Hanahan and Weinberg 2000; Robinson et al. 2000). Detection of circulating tumor associated antigen (i.e. CA-125) relies on aberrant expression of specific proteins and their release from tumor cells to achieve a steady state, which normally occurs well after the tumor is established (Taylor and Gercel-Taylor 1998). However, aberrant expression of tumor associated proteins at the initial transformation event may trigger an autoantibody response (Gagnon et al. 2008; Gercel-Taylor et al. 2001). The appearance of autoantibodies in the patient's blood may signal the earliest presence of abnormal tumor proteins up to two years before a tumor is visible by conventional methods, such as imaging, and prior to detection of circulating antigens (Anderson and LaBaer 2005; Nesterova et al. 2006). Autoantibody biomarkers are also very sensitive monitors of the effectiveness of cancer treatment because their re-appearance in the serum post-surgery and chemotherapy may be an early signal of cancer recurrence.
The sensitivity, specificity, and ultimate usefulness of autoantibodies to detect malignancy are highly dependent upon the target antigens selected. Many earlier studies of cancer autoantibodies used serum probing of recombinant proteins including cDNA libraries (Old and Chen 1998; Luo et al. 2002; Yamamoto et al. 1996; Jager et al. 1998; Gure et al. 1998), phage display (Chatterjee et al. 2006), or denatured proteins resolved by 2-D electrophoresis as the target antigens (Caron et al. 2007). These methods have significant limitations, notably the absence of proper conformation and post-translational modifications on recombinant proteins used in antibody recognition (Fossa et al. 2004; Suzuki et al. 2004; Le Naour et al. 2002; Zhang et al. 2003) and the generation of ‘mis-sense’ or ‘nonsense’ products from cDNA libraries and phage displays that do not occur in vivo (Wang et al. 2005). Thus, these approaches have the potential to overlook proteins relevant to cancer specificity because they do not evaluate native proteins that are known serum components, and may possess protein conformational and post-translational modifications that can profoundly influence antibody specificity (Fossa et al. 2004; Suzuki et al. 2004; Zhang et al. 2003) (Brichory et al. 2001; von Mensdorff-Pouilly et al. 2000).
In the last decade, with the evolution of various proteomics technologies, the potential for the identification of biomarkers has increased tremendously despite the extreme complexity of the serum with a dynamic range in concentration of several orders of magnitude (Anderson and Anderson 2002). More recently, proteomics, using a combination of sophisticated methods, provided new opportunities for screening and identifying autoantigens (Caron et al. 2007). Current proteomics based discovery approaches include “top-down” proteomics which utilizes intact protein analysis using the combination of 2-D gel and 2-D Western blotting, or “bottom-up” proteomics which utilizes multiple affinity protein profiling using combined ion exchange, reverse phase, and affinity chromatography for the purification of putative autoantigens followed by nano-LC-MS/MS analysis (Caron 2005). In addition, recent developments in multiplex quantitative proteomics, such as iTRAQ (Ross et al. 2004; Aggarwal et al. 2005; Shetty et al. 2012), have been useful in biomarker discovery but have yet to be utilized in the discovery of autoantibody biomarkers.
In the current study we compared two different but complementary proteomics technologies, including protein microarray screening and iTRAQ multiplex quantitative proteomics methods, to identify a panel of autoantigens targeting ovarian cancer. Protein microarrays comprised of the recombinant autoantigens were screened using serum samples from donors at various stages of ovarian cancer with diverse levels of CA-125, as well as benign and healthy controls. In this preliminary study, we report a panel of highly sensitive and specific autoantibodies that distinguishes early stage ovarian cancer with normal or low levels of CA-125.
Materials and Methods
Human serum samples
Twenty serum samples for autoantibody discovery (Table1A) were collected from each group: confirmed stage I ovarian cancer, stage II-IV ovariancancer and benign pelvic mass or endometriosis. Ovarian cancer serum samples were purchased from Proteogenex (Culver City, CA). Age matched healthy female serum samples were purchased from Research Blood Components, LLC (Brighton, MA). Samples with various CA-125 levels (Table 1C) were selected from Duke University and Proteogenex cohorts. Samples were obtained under IRB approved protocols from patients with ovarian cancer undergoing treatment at Duke University Medical Center. Serum samples were collected and stored at −80 °C.
Table 1. Serum samples used in the study.
1A: Sample groups used for autoantibody discovery. 1B: Sample groups used for autoantibody pre-validation studies. 1C: Sample groups used for autoantibody prevalidation studies.
| Table 1A. Sample groups used for autoantibody discovery | ||||||
|---|---|---|---|---|---|---|
| Group | Clinical Status | Number | TNM | Stage | CA125 | Age |
| 1 | Healthy female subjects | 20 | NA | NA | NA | 45-75 |
| 2 | Benign pelvic mass or endometriosis | 20 | NA | NA | NA | 45-75 |
| 3 | Stage l-IV ovarian cancer: | 20 | NA | I-IV | 3.924≤x≤3924 | 36≤x≤78 |
| 1. Clear cell carcinoma and endometrioid adenocarcinoma | T1cN0M0 | I | 10.31 | 78 | ||
| 2. Endometrioid adenxarcinoma | T1cN0M0 | I | 96 | 51 | ||
| 3. Serous adenocarcinoma | T1cN0M0 | I | 439.6 | 71 | ||
| 4. Serous adenocarcinoma | T1aN0M0 | I | 165 | 57 | ||
| 5. Mucinous adenocarcinoma | T1cN0M0 | I | 49 | 62 | ||
| 6. Serous adenocarcinoma | T1cN0M0 | I | 312 | 47 | ||
| 7. Serous papillary adenocarcinoma | T1aN0M0 | I | 92 | 54 | ||
| 8. Serous adenocarcinoma | T1cN0M0 | I | 218 | 48 | ||
| 9. Endometrioid adenocarcinoma | T1cN0M0 | I | 144 | 44 | ||
| 10. Serous adenocarcinoma | T1cN0M0 | I | 165 | 66 | ||
| 11. Endometrioid cystadenocarcinoma | T2bN0M0 | II | 112 | 66 | ||
| 12. Endometrioid adenocarcinoma | T2aN0M0 | II | 166 | 41 | ||
| 13. Serous adenocarcinoma | T2bN0M0 | II | 88 | 68 | ||
| 14. Endometrioid adenocarcinoma | T2aN0M0 | II | 480 | 56 | ||
| 15. Serous adenocarcinoma | T3cN0M0 | III | 605 | 72 | ||
| 16. Serous adenocarcinoma | T3aN0MO | III | 384 | 74 | ||
| 17. Clear cell carcinoma | T3cNIM0 | III | 524 | 68 | ||
| 18. Serous adenocarcinoma | T3cN0M1 | IV | 3924 | 64 | ||
| 19. Clear cell cystadenocarcinoma | T3cN0M1 | IV | 3.924 | 36 | ||
| 20. Endometrioid adenocarcinoma | T3cN1M1 | IV | 46 | 63 | ||
| Table 1B. Sample groups used for autoantibody pre-validation studies | ||
|---|---|---|
| Group | Clinical Status | Number |
| 1 | Healthy female subjects | 20 |
| 2 | Benign pelvic mass | 20 |
| 3 | Stage I ovarian cancer with various levels of CA-125 (10-439) | 20 |
| 4 | Stage II-IV ovarian cancer with various levels of CA-125 (46-3,924) | 20 |
| Table 1C. Sample groups used for autoantibody pre-validation | |||
|---|---|---|---|
| Group | Clinical status | CA-125 levels | Number |
| 1 | Ovarian cancer | Normal (<5) | 5 |
| 2 | Ovarian cancer | Low (10.31-49) | 10 |
| 3 | Ovarian cancer | Medium (55.8-96) | 20 |
| 4 | Ovarian cancer | High (112-480) | 20 |
| 5 | Ovarian cancer | Very-high (524-23422) | 20 |
| 6 | Non-ovarian cancer | (17.3-640) | 5 |
Protein microarray analysis
Ovarian cancer cell lysates were prepared by combining 5×108 SK-OV3 and 5×108 OVCAR3 cells by homogenization and freeze/thaw cycles in lysis buffer containing 1% NP40 (IBI-Scientific), 150mM NaCl, 10mM Na2HPO4, 1mM EDTA and a cocktail of protease inhibitors (all reagents from Calbiochem). For protein microarray preparation (work flow in Figure 1), 1 mL of the lysate was diluted to 2.5 mL total volume with ProteoSep Start Buffer (SB) and then buffer exchanged and desalted using a PD-10 Column (GE Healthcare). The eluent was diluted to 5.5 mL with SB and the contents injected onto the PF2D fractionation system (Beckman-Coulter, Inc) for 2D fractionation using the standard protocol (Caiazzo et al. 2011(). Glycerol was added to each of the fractions collected and the plates were dried down using a SpeedVac. Microarray printing buffer was added and the contents were transferred to 384 well plates (Genetix) for printing onto PATH slides (Gentel Biosciences) for serological assays. 2D Microarrays of the 2D fractions and controls were printed using 150 um pins and a Genetix printer.
Figure 1. Proteomics work flow used for autoantibody discovery.
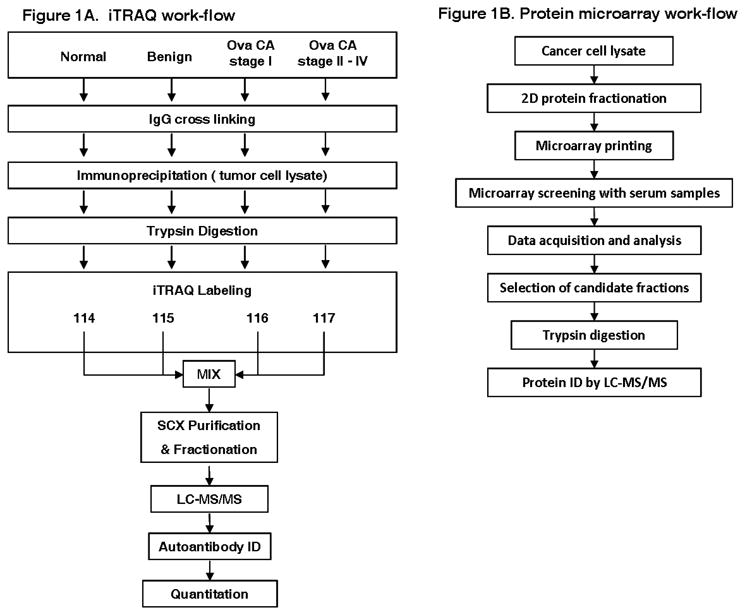
1A: iTRAQ 4plex quantitative proteomics method using pooled serum samples (Table 1A). 1B: Top down proteomics in combination with protein microarray screening method. Protein microarray was generated from 2D separation of ovarian tumor lysate samples and individual serum samples were screened on the protein microarray.
iTRAQ proteomics analysis
Individual serum samples from ovarian cancer and control cohorts were processed for iTRAQ proteomics analysis (Ross et al. 2004; Aggarwal et al. 2005) following the work flow shown in Figure 1. For the iTRAQ analysis serum samples were pooled from healthy normal, benign, stage I and stage II-IV ovarian cancer individuals and applied to protein A/G column to capture and cross link the IgG fraction from the serum. Serum IgG's were purified from 10μl of serum and cross-linked using an immunoprecipitation kit (Pierce-Thermo scientific) according to the manufacturer's instructions. Sera were incubated overnight at room temperature (RT) with 50 uL of protein A/G ultralink resin (Pierce-Thermo). The beads were then washed three times with coupling buffer. Cross-linking was achieved by incubating the protein A/G–immobilized antibody beads with 0.5 mM disuccinimidyl suberate for one hour. Beads were then washed three times with elution buffer and twice with cold IP lysis/wash buffer. Protein lysate prepared from ovarian cancer cell lines was added to the protein A/G–immobilized IgG beads prepared from each control and cancer serum samples and incubated overnight at 4°C on a rocking platform. The suspensions were then washed three times with IP lysis/wash buffer and three times with PBS. The immunoprecipitated proteins were recovered by eluting with 10 mL of elution buffer. The eluted samples were reduced with dithiothretol (DTT) (5μg/μL in 50mM AB) by incubating the mixture at 65°C for 45 minutes, alkylated with iodoacetamide (15μg/μL in 50mM AB) and digested following Waters’ protocol using RapiGest as a detergent. Briefly, digestion was carried out by Trypsin (Promega) (5 ng/μL in 50 mM AB) overnight at 37°C in a water bath. The tryptic peptide mixture was purified by sep-pak (C-18) columns (Waters). The columns were then washed thoroughly with 0.1% TFA and the tryptic peptides were eluted in 1 mL of 80% acetonitrile and concentrated under vacuum. Peptide mixtures were labeled using 4-plex iTRAQ labeling kit (Applied Biosystems) following the vendor's recommended protocol. The four iTRAQ labeled peptide solutions were mixed and purified using cation exchange (SCX) chromatography as described elsewhere (Shetty et al. 2012). The SCX purified iTRAQ labeled peptide mixture was fractionated by a Dionex C-18 RP column (4.6 mm diameter × 150 mm length) using an offline ultimate 3000 HPLC (Dionex, Sunnyvale, CA). A total of 35 fractions were collected and each fraction was concentrated to 6 μL under vacuum. The purified and fractionated iTRAQ labeled tryptic peptides were analyzed by data dependent nano LC-MS/MS experiments on an Velos LTQ-Orbitrap mass spectrometer (Thermo Fisher) interfaced with a nano ultimate HPLC (Dionex). The sample was loaded onto a trap column of 100 μm ID X 2 cm (L) packed with 5-μm Magic C18 AQ (200 A, 3 μm, Michrom) and washed using 97.95% H2O, 2% ACN, 0.05% TFA buffer at a flow rate of 10 μL/min for 5 minutes. The peptides were then separated by a self-packed 75μm ID X 50 cm (L) fused cilia column packed with 3-μm Magic C18 AQ (200 A, 3 μm, Michrom) using a linear gradient of Buffer B from 4% to 55% in 50 minutes at a flow rate of 300 nl/min. Buffer A was constituted of 0.1% formic acid in water, and buffer B was constituted of 0.1% formic acid, 80% acetonitrile in water. The analytical column was coupled to the mass spectrometer via a nanospray ion source (Proxeon) with a metal emitter. The peptides were analyzed in the Orbitrap operated at 60,000 resolution in full scan (300–2000m/z) followed by 10 Data-Dependent HCD MS/MS scans (100–2000m/z) with 7,500 resolution and normalized collision energy of 45%. Survey scans were acquired in profile mode and MS/MS scans were acquired in centroid mode. Maximum injection times for MS and MS/MS were set to 500 and 1000ms, respectively. The precursor isolation width was set at ±1.2 Da and monoisotopic precursor selection was enabled and singly charged ions are excluded from MS/MS. The minimum intensity threshold for MS/MS fragmentation in the orbitrap analyzer was 5000 counts and the dynamic exclusion was set to 60 seconds with repeat count as one. Proteins were identified by searching the LC-MS/MS raw data in Swissprot human database using Proteome Discoverer software (2.0) with Sequest search algorithm (Thermo). The search results were also verified manually to identify the correct peptide sequence. Only unique peptides were considered for the calculation of protein ratios and these ratios resulted from the median ratios. Peptides/proteins quantified with ≥ 1.5 fold increase in concentration were considered as up-regulated in the current analysis.
Serum sample screening on the microarray
Individual serum samples from different cohorts were diluted and added each to microarray slide. Briefly, the slides were rinsed with 1×PBS+0.5% Tween 20 (0.5%PBST) and then blocked for 1 hour at RT in 0.5%PBST+1% IgG free BSA. After drying, a 1:75 dilution of serum samples was made in 0.5%PBST+1% IgG free BSA and 500 uL of this solution was incubated on the slide for 1 hour at RT. The slides underwent three 5 minute washes in 0.5%PBST and dried by centrifugation. Next, the slides were incubated with 5μg/mL anti-human IgG-Biotin in 0.5%PBST+1% IgG free BSA for 1 hour at RT. The slides were washed three timess, and dried by centrifugation before being incubated with 1μg/mL Strepavidin 647 in 0.5%PBST+1% IgG free BSA for1 hour at RT. Finally, the slides were washed three times for minutes and then rinsed briefly in dH2O, dried by centrifugation, scanned on a ScanArray Lite (Perkin Elmer) and analyzed by Genepix software.
Statistical analysis and selection of fractions
The resulting data set was analyzed using M2 statistics and a PAMR statistical package and compared for overlap to identify the fractions that would distinguish one patient subgroup from another as determined by the cohort. For the PAMR analysis the data was normalized (Log10 then quartile normalized) and appropriate thresholds were used to “pick” the top 30-40 discriminating fractions of interest. These fractions were then compared to the M2 statistical analysis and the raw data to generate a set of statistically significant fractions for further analysis by mass spectrometry for the identification of autoantibody reactive proteins.
Mass spectrometry analysis of the fractions
Selected protein fractions were subjected to Trypsin digestion. Briefly, the protein solution was mixed with 30 μL of 50 mM AB containing 0.1% of Rapigest SF and reduced with DTT (5 μm/μlL in 50 mM AB) at 65°C for 45 minutes and alkylated with IA (15 μM in 50 mM AB) in the dark for 30 minutes. Then the alkylated proteins were digested by trypsin (5 ng/μL in 50 mM AB) overnight in a 37°C water bath. RapiGest was removed according to the vendor's recommended procedure. The digested peptides were purified by C-18 chromatography. Each fraction was then concentrated to 6 μl under vacuum and subjected to mass spectrometry. A 3000 Nano Ultimate HPLC (Dionex, Sunnyvale, CA) was coupled with LTQ mass spectrometer (Thermo Electron, San Jose, CA) equipped with advanced nanospray source to analyze peptides The peptides were analyzed by normal data dependent mode method in which the instrument was set to acquire fragment ion (MS/MS) spectra on the 4 most abundant precursor ions from each MS scan with a repeat count set of 1 and duration of 30 seconds. The raw data from LTQ mass spectrometer was searched against Swissprot human database with Proteome Discoverer algorithm. Results were filtered using Xcorr threshold for +1 of 1.8, +2 of 2.2 and +3 of 2.5. Proteins identified with at least 2 unique tryptic peptides were considered true positives. The search results were further verified manually.
Protein Preparation and Microarray Printing
Sixty high frequency ovarian cancer specific autoantibody reactive proteins identified by protein microarray and iTRAQ analysis were selected (Table S1) for the generation of recombinant proteins. Recombinant protein generation was outsourced to Origene (Rockville, MD), where protein production was performed in CHO cells and the proteins were purified to 80-90% purity. After determining the volume of each protein received, the volume was brought to 100 μL with dH2O and 40 μL was used to print the high density protein microarray. Proteins were diluted in PBS pH 7.4 at 0.1mg/mL in a 384 well source plate (Genetix), and loaded onto a Scienion sciFLEXARRAYER S3 ultra low volume non- contact microarrayer. Microarray slides coated with ultrathin nitrocellulose (PATH, Gentel Biosciences) were pre-gridded into 52 identical sub-arrays by using a wax imprinter (Gel Company). 30 autoantigens and 4 control proteins were spotted onto each subarray in triplicate, resulting in a 10×10 protein array within each subarray. The controls were influenza HA protein, purified human IgG, alpha gal antibody and biotinylated anti-human IgG. The microarray slides were stored in a vacuum sealed cassette with desiccant at 4°C before use.
Detection of autoantibody in patients’ serum samples
Pre-validation serum samples (Table 1B and 1C) were diluted 10 to 100 times by using PBS+ 0.1% Tween 20 (0.1%PBST). Microarray slides were pre-equilibrated to RT, and blocked in 0.5%PBST+1% IgG free BSA for 1 hour, and dried by centrifugation. 6 μL of each diluted serum sample was applied onto each sub-array on the microarray slide, and incubated in a humidified chamber with gentle shaking at RT for 1 hour. After three five-minute washes with 0.5%PBST and drying by centrifigation, 6 μL of 1μg/mL of biotinylated goat F(ab’)2 anti-human IgG (Jackson Immuno Research) was applied onto each sub-array and the slides were incubated in a humidified chamber at RT for 1 hour. After repeating the wash/dry cycle again, the microarray slides were probed with 1μg/mL of Strepavidin-647 (Invitrogen #S20992) at RT for 1 hour. Following three five-minute washes with wash buffer, the slides were washed briefly with dH2O, and then dried and scanned on a ScanArray Lite (Perkin Elmer) at a resolution of 10μm. The data was extracted by using Genepix 6.0 (Figure 2). T tests were performed and P-values were generated using a one tail, two sample equal variance T test and those proteins with a P-value of less than 0.1 were flagged for further analysis. In order to determine the sensitivity and specificity of the tested autoantibody markers capable of distinguishing benign disease versus cancer, nonparametric receiver-operating curves (ROCs) were performed, in which the value for sensitivity was plotted against false-positive rate (1-specificity), In addition, an area under the ROC curve (AUC) with 95% confidence intervals (CI) was calculated for each marker. In all tests, a P-value of ≤ 0.05 was considered to be statistically significant. The average values for each patient were used and ROC curves were generated using GraphPad Prism 5.
Figure 2. Recombinant protein microarray analysis.
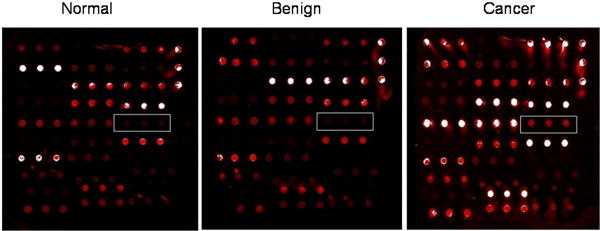
Individual serum samples from ovarian cancer, benign ovarian disease and normal control donors were screened on a microarray containing recombinant autoantigens. Positive image for one of the autoantigen is shown.
Dot Blot analysis
Dot blot analysis was performed using pooled cancer, benignand ovarian cancer samples with various CA-125 levels. The PVDF membrane (Immobilon, EMD Millipore) was designated into 0.5”×1.0” areas using a pencil and wet with 100% methanol (MeOH) for fifteen seconds at RT. Then the membrane was rinsed in dH2O with gentle agitation for 2 minutes at RT. Finally, the PVDF membrane was equilibrated in blotting buffer (1×Tris-Glycine-SDS buffer with 20% MeOH) for 5 minutes with gentle agitation at RT. The proteins were spotted in duplicate into a designated grid-like section at a volume of 3 μL per spot. Once all the proteins were spotted, the membrane was allowed to dry at RT for about 10 minutes (or until the protein spots were absorbed into the membrane). The PVDF membrane was then blocked [1:1 PBS + Odyssey Blocking Buffer (PBS+OBB); LiCOR Biosciences] for 1 hour with gentle agitation at RT. Dilutions of pooled serum samples (1:100) were prepared using 1:1 0.1%PBST + 1×Odyssey Blocking Buffer (0.1%PBST+OBB). After blocking, the membrane was cut along the 0.5” × 1.0” designations and each piece of the membrane was placed into a 60mm dish with a different pool of serum. The membrane was probed with the serum sample for 2 hours with gentle agitation at RT. The membranes then received three five-minute washes in 0.1%PBST with agitation at RT. Next, the membranes were probed with an IRDye 800CW goat anti-mouse antibody (1:5000) (LiCOR Biosciences) in 0.1%PBST+OBB for one hour with gentle agitation at RT. Again, the membranes were washed three times for five minutes in 0.1%PBST with agitation at RT. Finally the membranes were imaged using a LiCOR Odyssey Imaging System to acquire quantitative data.
Results
Proteomics methods to identify high frequency autoantibodies in ovarian cancer
To identify high frequency autoantibodies in ovarian cancer, we investigated twocomplementary proteomics methodologies. Protein microarrays (Figure 1) were generated from cell lysates by combining two well characterized ovarian cancer cell lines. The combined lysate was 2D fractionated to yield over 1000 fractions. Microarrays printed using these fractions were screened using serum samples from patients with ovarian cancer and benign gynecologic conditions (endometriosis and benign pelvic masses), as well as age-matched healthy females (Table 1A). Over 50 fractions were selected based on cancer specific autoantibody reactivity. Autoantigens present in the selected fractions were identified by LC-MS/MS analysis. Since we applied mass spectrometry analysis of the active and adjacent fractions, the first criterion for the selection of targets was the presence of tryptic peptides from the parent protein only in the active fractions. The next criterion was a greater number of tryptic peptides present in the active fraction as compared to the adjacent fractions. Based on these criteria, we selected a panel of autoantibody targets for ovarian cancer. We identified over 100 proteins with various specificities, i.e. highly reactive with cancer serum samples as compared to either benign or healthy control samples. The iTRAQ quantitative analysis (Figure 1) was performed using the same cell lysate and serum samples from the same cohorts. iTRAQ analysis yielded over 100 proteins that showed >1.5 fold higher reactivity with the autoantibodies in cancer serum samples as opposed to benign and healthy controls. The protein data between these two proteomics methods were compared to select 60 autoantigen proteins for recombinant protein generation (Table S1). There was ∼20% overlap between the autoantigens identified using these two methods. Also, a few autoantigens we identified were reported by others in ovarian and other cancers (Table S1) including p53 (Taylor et al. 2009; Abendstein et al. 2000), NY-ESO-1 (Sahin et al. 1995), HOXA7 (Naora et al. 2001), and plakoglobin (Gagnon et al. 2008). Benign ovarian disease had very few to no measurable autoantibodies. However, high levels of autoantibodies were identified in ovarian cancer samples. Interestingly, using ovarian cancer and benign patients’ serum samples as well as normal and cancer ovarian tissue lysates in western blot analysis showed that benign serum samples had very few autoantibodies as opposed to elevated diverse autoantibody levels in ovarian cancer patients that recognized multiple antigens from ovarian cancer tissue.
Custom autoantigen microarray to identify high frequency ovarian cancer autoantibodies
Native recombinant proteins were generated for 60 proteins selected from high frequencyautoantibodies identified by iTRAQ quantitative proteomics and by protein microarray proteomics inovarian cancer as compared to benign ovarian disease and healthy individuals (Table S1). These recombinant proteins, in addition tocontrols, were printed on microarray slides for pre-validation screening of serum samples toidentify the most high frequency autoantibodies in ovarian cancer for further analysis (Table 1B). Each of the ovarian cohorts of samples were applied tothe custom microarray and analyzed for autoantibodies present in the cancer samples as compared tothe benign and healthy serum samples. A typical positive image for cancer is shown in Figure 2. Data analysis for significant levels of autoantibodies in ovarian cancer generated 11 specific autoantibodies compared to benign and healthy samples (Table 2A). Interestingly, we also found there were 7 autoantibodies that were significantly higher in stage I cancer samples as compared to stage II-IV samples, which can form the foundation for potential early stage diagnostics (Table 2B). Although all the autoantibody targets are implicated in cancer initiation and development, Ezrin (Song et al. 2005), Cofilin-1 (Nishimura et al. 2011; Li et al. 2013) and PDZ domain-containing protein (Chaib et al. 2001) are implicated as prognostic markers in ovarian and other cancers.
Table 2. High frequency ovarian cancer autoantibody targets.
High frequency autoantibody targets identified by top-down proteomics microarray screening and iTRAQ quantitative proteomics analysis. (A) Autoantibodies found in ovarian cancer as compared to benign ovarian disease and age matched healthy female samples. (B) Autoantibodies that were significantly higher in stage I cancer samples as compared to stage II-IV samples.
| 2A. OVCA Specific as compared to benign and normal | |||
|---|---|---|---|
| Protein Name | Symbol | Uniprot | P-Value |
| Cofilin-1 | CFL1 | P23528 | 0.095925132 |
| Ezrin | EZR | P15311 | 0.086806974 |
| HistoneH1.2 | HIST1H1C | P16403 | 0.034922637 |
| Heterogeneous nuclear ribonucleoprotein A/B | HNRNPAB | Q99729 | 0.079023531 |
| Stress-70 protein, mitochondrial | HSPA9 | P38646 | 0.00865349 |
| PDZ domain-containing protein 11 | PDZD11 | Q5EBL8 | 0.000796403 |
| Profilin-1 | PFN1 | P07737 | 0.000864174 |
| Peptidyl-prolyl cis-1rans isomer ase A | PPIA | P62937 | 0.000187017 |
| Small EDRK-rich factor 2 | SERF2 | P84101 | 0.043832054 |
| Tubulin alpha-1C chain | TUBA1C | Q9BQE3 | 0.001168379 |
| Junction plakoglobin | JUP | P14923 | 0.051950189 |
| 2B. OVCA stage I specific as compared to stage II-IV | |||
|---|---|---|---|
| Protein Name | Symbol | Uniprot | P-Value |
| Cofilin-1 | CFL1 | P23528 | 0.042964969 |
| Ezrin | EZR | P15311 | 0.040955999 |
| HistoneH1.2 | HIST1H1C | P16403 | 0.002287586 |
| Heat schock-related 70 kDa protein 2 | HSPA2 | P54652 | 0.000998804 |
| PDZ domain-containing protein 11 | PDZD11 | Q5EBL8 | 0.042129066 |
| Tubulin alpha-1C chain | TUBA1C | Q9BQE3 | 0.01001449 |
| Junction plakoglobin | JUP | P14923 | 0.067963873 |
Specificity and sensitivity analysis of autoantibody markers
To assess the sensitivity and specificity of the autoantibody markers, we performedreceiver operating characteristic (ROC) curves. The microarray data for the selected four markersthat were present in high frequency in stage I ovarian cancer as compared to stage II-IV and benign samples are shown in figure 3. The cut-off value of 2000 spot density units (SDU), as identified from the microarray data (Figure 3), was used to generate ROC curves (Figure 4) that would distinguish stage I ovarian cancer from control samples (Table 1B). As shown in Figure 4, the selected autoantibodies showed higher specificity in distinguishing cancer from benign samples. Since we did not have a cohort of benign disease with CA-125 levels, we could not directly compare the autoantibodies markers with CA-125.
Figure 3. Microarray analysis of high frequency autoantibodies.
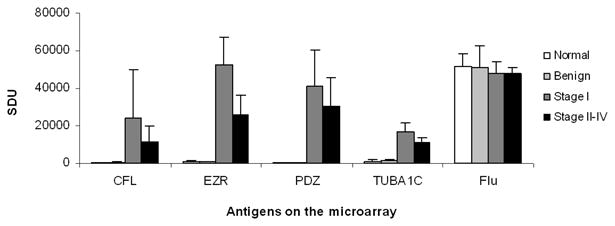
Individual serum samples from ovarian cancer, benign ovarian disease and healthy individuals were screened on a microarray containing recombinant CFL1, EZR, PDZ and TUBA1C autoantigens and influenza HA protein as controls. The average spot density generated by GenPix microarray reader is plotted for each of the autoantigens.
Figure 4. Receiver operating characteristic (ROC) curves discriminating ovarian cancer from benign controls.
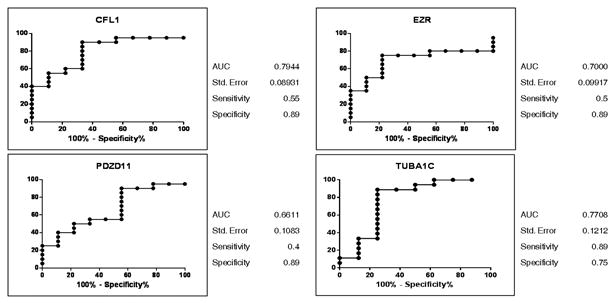
The ROC curves for selected autoantibody markers for ovarian cancer were generated using GraphPad Prism 5. Area under the ROC curve (AUC) with 95% confidence intervals (CI) was calculated and a P-value of ≤ 0.05 was considered to be statistically significant.
In addition to ROC curve analysis, we performed dot blot analysis using the native recombinant proteins (CFL1, EZR, PDZ and TUBA1C) and pooled cancer and benign serum samples. Cancer samples were pooled as three groups (Stage I, Stage II-IV and Stage I-IV). The sample pools were prepared based on the stage of the ovarian cancer regardless of their CA-125 levels. As shown in Figure 5, all the stages of ovarian cancer showed high immunoreactivity with CFL1, EZR and PDZ proteins and, interestingly, stage Iexhibited higher immunoreactivity with CFL1 and PDZ proteins.
Figure 5. Dot blot analysis with pooled cancer and benign samples.
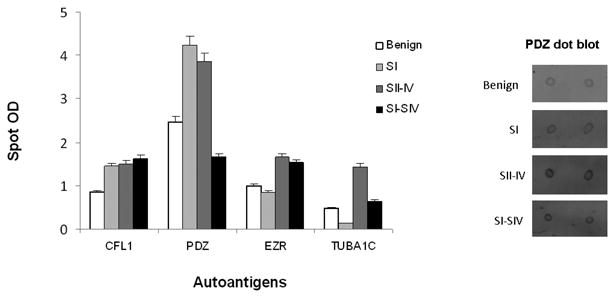
Dot blot analysis was performed using recombinant autoantigens and pooled serum samples from benign ovarian disease, stage I, stage II-IV, and stage I-IV ovarian cancer (Table 1B). An example (PDZ) of the dot blot image is given on the right.
Autoantibody targets distinguishes different stages of ovarian cancer with various CA-125 levels
To further validate the candidate markers, we performed dot blots with CFL1, EZR and PDZrecombinant proteins and serum samples pooled based on normal to very high levels of CA-125regardless of the stage of the ovarian cancer and samples from non-ovarian cancer (Non-OV) with various CA-125 levels (Table 1C). The immunoreactivities for non-ovarian cancer samples were negative to all the antigens tested. The mean for all the ovarian sample cohorts were statistically different from non-ovarian cancer samples. For most of the antigens tested, the mean reactivity was greater for samples with normal and low CA-125 levels than medium through very high levels of CA-125 (Figure 6). Specifically, the reactivity of samples with normal and low CA-125 levels with PDZ antigen was significantly greater than CFL1 and EZR antigens. All the dot blot analyses were performed with pooled samples. Although not significant, when the samples from stage I-IV were pooled, the immunoreactivity was diminished (Figure 5) perhaps due to the pooling effect. Pooled samples, although normalizing a less diverse group, may mask the effect of the true marker if more diverse samples are pooled.
Figure 6. Dot blot analysis of samples with various CA-125 levels.
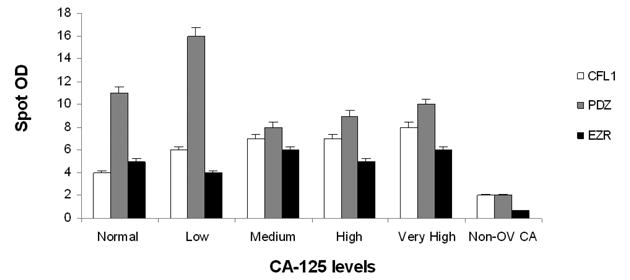
Dot blot analysis was performed with recombinant autoantigens and serum samples from ovariancancer and non-ovarian cancer patients with various levels of CA-125 (Table 1C).
Discussion
Circulating autoantibodies against tumor proteins have are present at the very earlieststages of ovarian (Luborsky et al. 2005) and prostate cancer(Suzuki et al. 2004) along with a variety of other cancers (Chen 2004) including leukemia, malignant melanoma, lung, breast, gastrointestinal, nasopharyngeal, and hepatocellular carcinomas (Burnham 1972). Production of autoantibodies occurs when certain intracellular proteins become immunogenic due to dysregulation of their function or alteration in molecular structure or localization, leading to the elicitation of a tumor-specific immune response (Tan 1991; Tan et al. 1988). These antibodies react to a variety of tumor-associated antigens (Mueller-Pillasch et al. 1997; Vogelstein and Kinzler 1993; Levine 1997; Hanahan and Weinberg 2000). The detection of anti-p53 antibodies can predate the diagnosis of cancer (Soussi 2000; Vogl et al. 2000; Vogl et al. 1999), strongly suggesting the potential to use autoantibodies as a means for early detection of the presence of tumors (Haab 2005). Various approaches have been used in the past to discover autoantibodies of diagnostic value. Although cDNA and phage display library screening yielded autoantibody targets for various cancers, the data on characterization and large scale screening of individual targets resulted in limited diagnostic significance. However, emerging proteomics methods have shown promising utility in the discovery of biomarkers, including cancer specific autoantibodies (Caron et al. 2007). In this report, using two different proteomics methods in combination with protein microarray screening, we have identified and characterized a panel of autoantigens in ovarian cancer. Different proteomics methods resulted in a group of ovarian cancer specific high frequency autoantibodies as compared to benign samples. Among the two proteomics methods used, iTRAQ method gave the best results and all the 11 validated markers were identified by iTRAQ method with six markers overlapping with the protein microarray methodology (Table S1). Furthermore, screening of serum samples using protein array containing native recombinant high frequency autoantibody targets (autoantigens) identified a panel of stage I ovarian cancer specific autoantibody markers (Table 2).
In our analysis, significant levels of autoantibodies were not detected in the serum ofthe patients with benign ovarian diseases, consistent with previous reports (Old and Chen 1998; Taylor et al. 2009). Specifically, Taylor et al. demonstrated that high levels of autoantibodies directed against multiple antigens in ovarian cancer were detectable in cancerous, but not benign, serum samples (Taylor et al. 2009). We also identified a few autoantigens that were reported by others using various methods, such as p53 (Abendstein et al. 2000; Sahin et al. 1995), NY-ESO-1(Sahin et al. 1995), HOXA7 (Naora et al. 2001) and plakoglobin (Gagnon et al. 2008). However, in our screening of various stages of ovarian cancer serum samples using an individual protein microarray designed to include high frequency autoantibody targets only resulted in identification of one previously reported prostate cancer autoantigen (Junction plakaglobin) (Ehrlich et al. 2006). Interestingly, using the native recombinant protein microarray as the final screen, we identified several novel autoantibodies in ovarian cancer; specifically in stage I ovarian cancer samples that could potentially be useful in the development of early stage diagnostics (Table 2).
We screened the stage I and stage II-IV ovarian cancer samples using dot blot analysisand ROC sensitivity/specificity analysis to further select the autoantibody targets specific for stage I ovarian cancer (Figures 2 and 3). Immunoreactivity with CFL1, EZR and PDZ autoantigens were significant in all stages of ovarian cancer indicating sustained autoantibody responses to these tumor associated antigens (Figure 5). We further demonstrated that these autoantibodies were not only present in all stages of ovarian cancer but were also present in normal to low level CA-125 positive ovarian cancer regardless of staging (Table 1C and Figure 6). Although our patient sample numbers were low, it is a significant observation that autoantibodies can distinguish ovarian cancer with normal levels of CA-125 from non-ovarian cancer with high levels of CA-125. CA-125 testing is not sensitive for detecting early stage ovarian cancer with normal levels of CA-125 (Ferrini 1997). For example, 79% of all ovarian cancers are positive for CA-125, whereas the remainder does not express this antigen at all (Rosen et al.). Also, only about 50% of patients with early stage ovarian cancer have elevated CA-125 levels, meaning that CA-125 has particularly poor sensitivity for ovarian cancer before the onset of symptoms (Sasaroli D; Nossov et al.). We report here that the sensitivity and specificity of the autoantibody markers are not significantly different from CA-125 in detecting stage I-IV ovarian cancer. However, the key finding of our work is twofold. First, the autoantibodies have a high rate of sensitivity and specificity for correctly classifying ovarian cancer with normal CA-125 levels (<35U/ml). This finding is important because it could help clarify the primary origin of the tumor in women with peritoneal metastases of unknown primary when the CA-125 is normal. Second, the autoantibodies are able to distinguish ovarian cancer, regardless of the stage of the disease, from non-ovarian cancer when the CA-125 levels are >35U/ml. This finding is important because it could help clarify the primary origin of the tumor in women with peritoneal metastases of unknown primary and an elevated CA-125 levels (>35U/ml). In some instances, tumors other than ovarian cancer that involve the peritoneum can cause elevated CA-125 levels.
Another important aspect of our study was the identification of previously unreported autoantibodies against CFL1, EZR AND PDZ. Our results demonstrate that the presence of autoantibodies against CFL1, EZR and PDZ antigens can not only differentiate between benign ovarian masses and stage I-IV ovarian cancer but can also distinguish ovarian cancer with normal to low levels of CA-125 from non-ovarian cancer with significant CA-125 levels. The proteins ezrin (EZR) (Song et al. 2005), cofilin (CFL-1) (Nishimura et al. 2011; Li et al. 2013) and PDZ (Chaib et al. 2001) are implicated as prognostic markers in ovarian and other cancers. InhibitingCFL-1, a protein directly involved in cell migration (Zhu et al. 2006), affects cell motility in carcinoma cells (Hotulainen et al. 2005), reduces the invasiveness of carcinoma cells by reducing the assembly and stability of invadopodia (Yamaguchi et al. 2005). When over-expressed, CFL-1 increases the velocity of cell migration (Yap et al. 2005). In addition, Nishimura et al demonstrated that the progression-free survival was longer in CFL-1 protein-negative cases; therefore, CFL-1 may predict the progression-free survival of patients with advanced epithelial ovarian cancer receiving standard therapy (Nishimura et al. 2011). Similar to CFL-1, the protein EZR is also a membrane cytoskeleton-linking protein that regulates cell motility, signal transduction, cell-to-cell and cell-to matrix recognition, cell growth, and invasion (Ohtani et al. 1999; Chen et al. 2001; Polesello et al. 2002; Gautreau et al. 2002). Proliferation of cancer cells and the metastatic phenotype of ovarian carcinoma depends on the expression of EZR protein (Ohtani et al. 1999; Chen et al. 2001). Furthermore, Song et al. has shown that estrogen induces over-expression of EZR, Matrigel membrane invasion and proliferation of ovarian cancer cells (Song et al. 2005). Unlike the CFL-1 and EZR, very little is known about PDZ in ovarian cancer. However, PDZ motifs are likely involved in protein clustering and scaffolding and studies suggest that the inappropriate up-regulation of these proteins s may contribute to tumorigenesis in prostate cancer (Chaib et al. 2001).
Overall, it is intriguing to note that in our study we found antibodies to CFL-1, EZR and PDZ motif containing proteins in all stages of ovarian cancer. This response needs to be investigated further since it may be involved in the suppression of the effect of these proteins and could contribute to the progression-free survival at all stages of ovarian cancer. The majority of tumor-derived antigens that have been identified as eliciting a humoral response in cancer patients are not the products of mutated genes. They include differentiation antigens and other proteins that are over-expressed in tumors (Old and Chen 1998; Luo et al. 2002). There is some evidence that the occurrence of autoantibodies in cancer is of prognostic relevance as well (Maddison et al. 1999; Blaes et al. 2000; Hirasawa et al. 2000). Individual or combinations of these three autoantibodies in ovarian cancer may have utility as early diagnostic markers. Since multiple antibody measurements may improve the predictive accuracy it will be necessary to test the clinical usefulness of these autoantibodies in large and independent patient cohort to improve the statistical power and validate as a clinically reliable approach.
Supplementary Material
Acknowledgments
This work was supported by USAMRAA grant W81XWH-10-1-0307
Footnotes
Conflict of interest statement: No actual or potential conflict of interest in relation to this article exists
References
- Abendstein B, Marth C, Muller-Holzner E, Widschwendter M, Daxenbichler G, Zeimet AG. Clinical significance of serum and ascitic p53 autoantibodies in epithelial ovarian carcinoma. Cancer. 2000;88(6):1432–1437. doi: 10.1002/(sici)1097-0142(20000315)88:6<1432::aid-cncr22>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Aggarwal K, Choe LH, Lee KH. Quantitative analysis of protein expression using amine-specific isobaric tags in Escherichia coli cells expressing rhsA elements. Proteomics. 2005;5(9):2297–2308. doi: 10.1002/pmic.200401231. [DOI] [PubMed] [Google Scholar]
- Anderson KS, LaBaer J. The sentinel within: exploiting the immune system for cancer biomarkers. Journal of proteome research. 2005;4(4):1123–1133. doi: 10.1021/pr0500814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1(11):845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- Blaes F, Klotz M, Huwer H, Straub U, Kalweit G, Schimrigk K, Schafers HJ. Antineural and antinuclear autoantibodies are of prognostic relevance in non-small cell lung cancer. Ann Thorac Surg. 2000;69(1):254–258. doi: 10.1016/s0003-4975(99)01198-4. [DOI] [PubMed] [Google Scholar]
- Brichory FM, Misek DE, Yim AM, Krause MC, Giordano TJ, Beer DG, Hanash SM. An immune response manifested by the common occurrence of annexins I and II autoantibodies and high circulating levels of IL-6 in lung cancer. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(17):9824–9829. doi: 10.1073/pnas.171320598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnham TK. Antinuclear antibodies in patients with malignancies. Lancet. 1972;2(7774):436. doi: 10.1016/s0140-6736(72)91842-9. [DOI] [PubMed] [Google Scholar]
- Caiazzo RJ, Jr, O'Rourke DJ, Barder TJ, Nelson BP, Liu BC. Native antigen fractionation protein microarrays for biomarker discovery. Methods in molecular biology (Clifton, NJ) 2011;723:129–148. doi: 10.1007/978-1-61779-043-0_9. [DOI] [PubMed] [Google Scholar]
- Caron M, Choquet-Kastylevsky G, Joubert-Caron R. Cancer Immunomics Using Autoantibody Signatures for Biomarker Discovery. Molecular & Cellular Proteomics. 2007;6(7):1115–1122. doi: 10.1074/mcp.R600016-MCP200. [DOI] [PubMed] [Google Scholar]
- Caron MJCR, Canelle L, Hardouin J. Serological proteome analysis (SERPA) and multiple affinity protein profiling (MAPPING) to discover cancer biomarkers. Molecular & Cellular Proteomics. 2005;4(S142) [Google Scholar]
- Chaib H, Rubin MA, Mucci NR, Li L, Taylor JMG, Day ML, Rhim JS, Macoska JA. Activated in prostate cancer: a PDZ domain-containing protein highly expressed in human primary prostate tumors. Cancer research. 2001;61(6):2390–2394. [PubMed] [Google Scholar]
- Chatterjee M, Mohapatra S, Ionan A, Bawa G, Ali-Fehmi R, Wang X, Nowak J, Ye B, Nahhas FA, Lu K, Witkin SS, Fishman D, Munkarah A, Morris R, Levin NK, Shirley NN, Tromp G, Abrams J, Draghici S, Tainsky MA. Diagnostic markers of ovarian cancer by high-throughput antigen cloning and detection on arrays. Cancer research. 2006;66(2):1181–1190. doi: 10.1158/0008-5472.CAN-04-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Fadiel A, Feng Y, Ohtani K, Rutherford T, Naftolin F. Ovarian epithelial carcinoma tyrosine phosphorylation, cell proliferation, and ezrin translocation are stimulated by interleukin 1alpha and epidermal growth factor. Cancer. 2001;92(12):3068–3075. doi: 10.1002/1097-0142(20011215)92:12<3068::aid-cncr10149>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Ehrlich JR, Qin S, Liu BC. The ‘reverse capture’ autoantibody microarray: a native antigen-based platform for autoantibody profiling. Nature protocols. 2006;1(1):452–460. doi: 10.1038/nprot.2006.66. [DOI] [PubMed] [Google Scholar]
- Ferrini R. Screening asymptomatic women for ovarian cancer: American College of Preventive Medicine practice policy. American journal of preventive medicine. 1997;13(6):444–446. [PubMed] [Google Scholar]
- Fossa A, Alsoe L, Crameri R, Funderud S, Gaudernack G, Smeland EB. Serological cloning of cancer/testis antigens expressed in prostate cancer using cDNA phage surface display. Cancer Immunol Immunother. 2004;53(5):431–438. doi: 10.1007/s00262-003-0458-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon A, Kim JH, Schorge JO, Ye B, Liu B, Hasselblatt K, Welch WR, Bandera CA, Mok SC. Use of a combination of approaches to identify and validate relevant tumor-associated antigens and their corresponding autoantibodies in ovarian cancer patients. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14(3):764–771. doi: 10.1158/1078-0432.CCR-07-0856. [DOI] [PubMed] [Google Scholar]
- Gautreau A, Louvard D, Arpin M. ERM proteins and NF2 tumor suppressor: the Yin and Yang of cortical actin organization and cell growth signaling. Current opinion in cell biology. 2002;14(1):104–109. doi: 10.1016/s0955-0674(01)00300-3. [DOI] [PubMed] [Google Scholar]
- Gercel-Taylor C, Bazzett LB, Taylor DD. Presence of aberrant tumor-reactive immunoglobulins in the circulation of patients with ovarian cancer. Gynecologic oncology. 2001;81(1):71–76. doi: 10.1006/gyno.2000.6102. [DOI] [PubMed] [Google Scholar]
- Gure AO, Altorki NK, Stockert E, Scanlan MJ, Old LJ, Chen YT. Human lung cancer antigens recognized by autologous antibodies: definition of a novel cDNA derived from the tumor suppressor gene locus on chromosome 3p21.3. Cancer research. 1998;58(5):1034–1041. [PubMed] [Google Scholar]
- Haab BB. Antibody arrays in cancer research. Mol Cell Proteomics. 2005;4(4):377–383. doi: 10.1074/mcp.M500010-MCP200. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hays JL, Kim G, Giuroiu I, Kohn EC. Proteomics and ovarian cancer: integrating proteomics information into clinical care. J Proteomics. 2010;73(10):1864–1872. doi: 10.1016/j.jprot.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirasawa Y, Kohno N, Yokoyama A, Kondo K, Hiwada K, Miyake M. Natural autoantibody to MUC1 is a prognostic indicator for non-small cell lung cancer. Am J Respir Crit Care Med. 2000;161(2 Pt 1):589–594. doi: 10.1164/ajrccm.161.2.9905028. [DOI] [PubMed] [Google Scholar]
- Hotulainen P, Paunola E, Vartiainen MK, Lappalainen P. Actin-depolymerizing factor and cofilin-1 play overlapping roles in promoting rapid F-actin depolymerization in mammalian nonmuscle cells. Molecular biology of the cell. 2005;16(2):649–664. doi: 10.1091/mbc.E04-07-0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager E, Chen YT, Drijfhout JW, Karbach J, Ringhoffer M, Jager D, Arand M, Wada H, Noguchi Y, Stockert E, Old LJ, Knuth A. Simultaneous humoral and cellular immune response against cancer-testis antigen NY-ESO-1: definition of human histocompatibility leukocyte antigen (HLA)-A2-binding peptide epitopes. J Exp Med. 1998;187(2):265–270. doi: 10.1084/jem.187.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60(5):277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- Le Naour F, Brichory F, Misek DE, Brechot C, Hanash SM, Beretta L. A distinct repertoire of autoantibodies in hepatocellular carcinoma identified by proteomic analysis. Mol Cell Proteomics. 2002;1(3):197–203. doi: 10.1074/mcp.m100029-mcp200. [DOI] [PubMed] [Google Scholar]
- Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88(3):323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- Li M, Yin J, Mao N, Pan L. Upregulation of phosphorylated cofilin 1 correlates with taxol resistance in human ovarian cancer in vitro and in vivo. Oncology reports. 2013;29(1):58–66. doi: 10.3892/or.2012.2078. [DOI] [PubMed] [Google Scholar]
- Luborsky JL, Barua A, Shatavi SV, Kebede T, Abramowicz J, Rotmensch J. Anti-tumor antibodies in ovarian cancer. Am J Reprod Immunol. 2005;54(2):55–62. doi: 10.1111/j.1600-0897.2005.00287.x. [DOI] [PubMed] [Google Scholar]
- Luo LY, Herrera I, Soosaipillai A, Diamandis EP. Identification of heat shock protein 90 and other proteins as tumour antigens by serological screening of an ovarian carcinoma expression library. Br J Cancer. 2002;87(3):339–343. doi: 10.1038/sj.bjc.6600439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison P, Newsom-Davis J, Mills KR, Souhami RL. Favourable prognosis in Lambert-Eaton myasthenic syndrome and small-cell lung carcinoma. Lancet. 1999;353(9147):117–118. doi: 10.1016/S0140-6736(05)76153-5. [DOI] [PubMed] [Google Scholar]
- Mor G, Visintin I, Lai Y, Zhao H, Schwartz P, Rutherford T, Yue L, Bray-Ward P, Ward DC. Serum protein markers for early detection of ovarian cancer. Proc Natl Acad Sci U S A. 2005;102(21):7677–7682. doi: 10.1073/pnas.0502178102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller-Pillasch F, Lacher U, Wallrapp C, Micha A, Zimmerhackl F, Hameister H, Varga G, Friess H, Buchler M, Beger HG, Vila MR, Adler G, Gress TM. Cloning of a gene highly over-expressed in cancer coding for a novel KH-domain containing protein. Oncogene. 1997;14(22):2729–2733. doi: 10.1038/sj.onc.1201110. [DOI] [PubMed] [Google Scholar]
- Naora H, Montz FJ, Chai CY, Roden RB. Aberrant expression of homeobox gene HOXA7 is associated with mullerian-like differentiation of epithelial ovarian tumors and the generation of a specific autologous antibody response. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(26):15209–15214. doi: 10.1073/pnas.011503998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesterova M, Johnson N, Cheadle C, Cho-Chung YS. Autoantibody biomarker opens a new gateway for cancer diagnosis. Biochimica et biophysica acta. 2006;1762(4):398–403. doi: 10.1016/j.bbadis.2005.12.010. [DOI] [PubMed] [Google Scholar]
- Nishimura S, Tsuda H, Kataoka F, Arao T, Nomura H, Chiyoda T, Susumu N, Nishio K, Aoki D. Over-expression of cofilin 1 can predict progression-free survival in patients with epithelial ovarian cancer receiving standard therapy. Human pathology. 2011;42(4):516–521. doi: 10.1016/j.humpath.2010.07.019. [DOI] [PubMed] [Google Scholar]
- Nossov V, Amneus M, Su F, Lang J, Janco JM, Reddy ST, Farias-Eisner R. The early detection of ovarian cancer: from traditional methods to proteomics. Can we really do better than serum CA-125? American journal of obstetrics and gynecology. 2008;199(3):215–223. doi: 10.1016/j.ajog.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Ohtani K, Sakamoto H, Rutherford T, Chen Z, Satoh K, Naftolin F. Ezrin, a membrane-cytoskeletal linking protein, is involved in the process of invasion of endometrial cancer cells. Cancer Lett. 1999;147(1-2):31–38. doi: 10.1016/s0304-3835(99)00272-4. [DOI] [PubMed] [Google Scholar]
- Old LJ, Chen YT. New paths in human cancer serology. J Exp Med. 1998;187(8):1163–1167. doi: 10.1084/jem.187.8.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge E, Kreimer AR, Greenlee RT, Williams C, Xu JL, Church TR, Kessel B, Johnson CC, Weissfeld JL, Isaacs C, Andriole GL, Ogden S, Ragard LR, Buys SS. Results from four rounds of ovarian cancer screening in a randomized trial. Obstet Gynecol. 2009;113(4):775–782. doi: 10.1097/AOG.0b013e31819cda77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piver MS, Wong C. Role of prophylactic surgery for women with genetic predisposition to cancer. Clin Obstet Gynecol. 1998;41(1):215–224. doi: 10.1097/00003081-199803000-00026. [DOI] [PubMed] [Google Scholar]
- Polesello C, Delon I, Valenti P, Ferrer P, Payre F. Dmoesin controls actin-based cell shape and polarity during Drosophila melanogaster oogenesis. Nature cell biology. 2002;4(10):782–789. doi: 10.1038/ncb856. [DOI] [PubMed] [Google Scholar]
- Robinson C, Callow M, Stevenson S, Scott B, Robinson BW, Lake RA. Serologic responses in patients with malignant mesothelioma: evidence for both public and private specificities. Am J Respir Cell Mol Biol. 2000;22(5):550–556. doi: 10.1165/ajrcmb.22.5.3930. [DOI] [PubMed] [Google Scholar]
- Rosen DG, Wang L, Atkinson JN, Yu Y, Lu KH, Diamandis EP, Hellstrom I, Mok SC, Liu J, Bast RC., Jr Potential markers that complement expression of CA-125 in epithelial ovarian cancer. Gynecologic oncology. 2005;99(2):267–277. doi: 10.1016/j.ygyno.2005.06.040. [DOI] [PubMed] [Google Scholar]
- Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3(12):1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- Sahin U, Tureci O, Schmitt H, Cochlovius B, Johannes T, Schmits R, Stenner F, Luo G, Schobert I, Pfreundschuh M. Human neoplasms elicit multiple specific immune responses in the autologous host. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(25):11810–11813. doi: 10.1073/pnas.92.25.11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaroli DCG, Scholler N. Beyond CA-125: the coming of age of ovarian cancer biomarkers. Are we there yet? Biomark Med. 2009;3:275–288. doi: 10.2217/bmm.09.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty V, Hafner J, Shah P, Nickens Z, Philip R. Investigation of ovarian cancer associated sialylation changes in N-linked glycopeptides by quantitative proteomics. Clin Proteomics. 2012;9(1):10. doi: 10.1186/1559-0275-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Fadiel A, Edusa V, Chen Z, So J, Sakamoto H, Fishman DA, Naftolin F. Estradiol-induced ezrin over-expression in ovarian cancer: a new signaling domain for estrogen. Cancer Letters. 2005;220(1):57–65. doi: 10.1016/j.canlet.2004.04.024. [DOI] [PubMed] [Google Scholar]
- Soussi T. p53 Antibodies in the sera of patients with various types of cancer: a review. Cancer research. 2000;60(7):1777–1788. [PubMed] [Google Scholar]
- Suzuki H, Akakura K, Igarashi T, Ueda T, Ito H, Watanabe M, Nomura F, Ochiai T, Shimada H. Clinical usefulness of serum antip53 antibodies for prostate cancer detection: a comparative study with prostate specific antigen parameters. J Urol. 2004;171(1):182–186. doi: 10.1097/01.ju.0000101501.54931.4a. [DOI] [PubMed] [Google Scholar]
- Tan EM. Autoantibodies in pathology and cell biology. Cell. 1991;67(5):841–842. doi: 10.1016/0092-8674(91)90356-4. [DOI] [PubMed] [Google Scholar]
- Tan EM, Chan EK, Sullivan KF, Rubin RL. Antinuclear antibodies (ANAs): diagnostically specific immune markers and clues toward the understanding of systemic autoimmunity. Clin Immunol Immunopathol. 1988;47(2):121–141. doi: 10.1016/0090-1229(88)90066-9. [DOI] [PubMed] [Google Scholar]
- Taylor DD, Gercel-Taylor C. Tumor-reactive immunoglobulins in ovarian cancer: diagnostic and therapeutic significance? (review) Oncology reports. 1998;5(6):1519–1524. doi: 10.3892/or.5.6.1519. [DOI] [PubMed] [Google Scholar]
- Taylor DD, Gercel-Taylor C, Parker LP. Patient-derived tumor-reactive antibodies as diagnostic markers for ovarian cancer. Gynecologic oncology. 2009;115(1):112–120. doi: 10.1016/j.ygyno.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Vogelstein B, Kinzler KW. The multistep nature of cancer. Trends Genet. 1993;9(4):138–141. doi: 10.1016/0168-9525(93)90209-z. [DOI] [PubMed] [Google Scholar]
- Vogl FD, Frey M, Kreienberg R, Runnebaum IB. Autoimmunity against p53 predicts invasive cancer with poor survival in patients with an ovarian mass. Br J Cancer. 2000;83(10):1338–1343. doi: 10.1054/bjoc.2000.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogl FD, Stickeler E, Weyermann M, Kohler T, Grill HJ, Negri G, Kreienberg R, Runnebaum IB. p53 autoantibodies in patients with primary ovarian cancer are associated with higher age, advanced stage and a higher proportion of p53-positive tumor cells. Oncology. 1999;57(4):324–329. doi: 10.1159/000012069. [DOI] [PubMed] [Google Scholar]
- von Mensdorff-Pouilly S, Petrakou E, Kenemans P, van Uffelen K, Verstraeten AA, Snijdewint FG, van Kamp GJ, Schol DJ, Reis CA, Price MR, Livingston PO, Hilgers J. Reactivity of natural and induced human antibodies to MUC1 mucin with MUC1 peptides and n-acetylgalactosamine (GalNAc) peptides. Int J Cancer. 2000;86(5):702–712. doi: 10.1002/(sici)1097-0215(20000601)86:5<702::aid-ijc16>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- Wang X, Yu J, Sreekumar A, Varambally S, Shen R, Giacherio D, Mehra R, Montie JE, Pienta KJ, Sanda MG, Kantoff PW, Rubin MA, Wei JT, Ghosh D, Chinnaiyan AM. Autoantibody signatures in prostate cancer. N Engl J Med. 2005;353(12):1224–1235. doi: 10.1056/NEJMoa051931. [DOI] [PubMed] [Google Scholar]
- Yamaguchi H, Lorenz M, Kempiak S, Sarmiento C, Coniglio S, Symons M, Segall J, Eddy R, Miki H, Takenawa T, Condeelis J. Molecular mechanisms of invadopodium formation: the role of the N-WASP-Arp2/3 complex pathway and cofilin. The Journal of cell biology. 2005;168(3):441–452. doi: 10.1083/jcb.200407076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Shimizu E, Ogura T, Sone S. Detection of auto-antibodies against L-myc oncogene products in sera from lung cancer patients. Int J Cancer. 1996;69(4):283–289. doi: 10.1002/(SICI)1097-0215(19960822)69:4<283::AID-IJC8>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Yap CT, Simpson TI, Pratt T, Price DJ, Maciver SK. The motility of glioblastoma tumour cells is modulated by intracellular cofilin expression in a concentration-dependent manner. Cell motility and the cytoskeleton. 2005;60(3):153–165. doi: 10.1002/cm.20053. [DOI] [PubMed] [Google Scholar]
- Zhang JY, Casiano CA, Peng XX, Koziol JA, Chan EK, Tan EM. Enhancement of antibody detection in cancer using panel of recombinant tumor-associated antigens. Cancer Epidemiol Biomarkers Prev. 2003;12(2):136–143. [PubMed] [Google Scholar]
- Zhang Z, Bast RC, Jr, Yu Y, Li J, Sokoll LJ, Rai AJ, Rosenzweig JM, Cameron B, Wang YY, Meng XY, Berchuck A, Van Haaften-Day C, Hacker NF, de Bruijn HW, van der Zee AG, Jacobs IJ, Fung ET, Chan DW. Three biomarkers identified from serum proteomic analysis for the detection of early stage ovarian cancer. Cancer Res. 2004;64(16):5882–5890. doi: 10.1158/0008-5472.CAN-04-0746. [DOI] [PubMed] [Google Scholar]
- Zhu B, Fukada K, Zhu H, Kyprianou N. Prohibitin and cofilin are intracellular effectors of transforming growth factor beta signaling in human prostate cancer cells. Cancer research. 2006;66(17):8640–8647. doi: 10.1158/0008-5472.CAN-06-1443. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


