SUMMARY
Understanding the mechanism by which tumor cells influence osteoclast differentiation is crucial for improving treatment of osteolytic metastasis. Here, we report broad microRNA (miRNA) expression changes in differentiating osteoclasts after exposure to tumor-conditioned media, in part through activation of NFκB signaling by soluble intracellular adhesion molecule (sICAM1) secreted from bone-metastatic cancer cells. Ectopic expression of multiple miRNAs down-regulated during osteoclastogenesis suppresses osteoclast differentiation by targeting important osteoclast genes. Intravenous delivery of these miRNAs in vivo inhibits osteoclast activity and reduces osteolytic bone metastasis. Importantly, serum levels of sICAM1 and two osteoclast miRNAs, miR-16 and miR-378, which are elevated in osteoclast differentiation, correlate with bone metastasis burden. These findings establish miRNAs as potential therapeutic targets and clinical biomarkers of bone metastasis.
INTRODUCTION
Osteolytic bone metastasis is a frequent occurrence in late stage breast, lung, thyroid, bladder and many other cancers, leading to pathological fractures, pain, and hypercalcemia (Weilbaecher et al., 2011). The development of bone lesions depends upon the orchestrated interactions between tumor cells and functional cells within the bone, namely osteoblasts and osteoclasts (Ell and Kang, 2012; Weilbaecher et al., 2011). The bone resorbing osteoclasts play an important role in physiological bone remodeling (Boyle et al., 2003; Teitelbaum and Ross, 2003), while aberrant osteoclast activity can lead to pathological conditions including Paget’s disease and lytic bone metastasis (Weilbaecher et al., 2011). Osteoclast differentiation is canonically dependent on two essential molecules, macrophage colony-stimulating factor (M-CSF) and receptor activator of NF-κB ligand (RANKL) (Boyle et al., 2003; Teitelbaum and Ross, 2003), although a number of RANKL independent pathways have been described (Hemingway et al., 2011). Aberrant expression of these signaling molecules by bone-metastatic cancer cells has been shown to recruit pre-osteoclasts to the site of osteolytic metastasis and induce their differentiation, leading to degradation of the bone and the subsequent release of bone matrix-embedded tumor-promoting growth factors such as TGFβ (Ell and Kang, 2012; Korpal et al., 2009; Weilbaecher et al., 2011). The role of osteoclasts in bone metastasis is further underscored by the efficacy of treatments targeting osteoclast differentiation and activity (Clezardin, 2011).
MicroRNAs (miRNAs) are a class of short (~22nt) non-coding RNAs capable of repressing gene expression through complementary binding of the “seed sequence” of target mRNAs (Bartel, 2009). MiRNAs have been well recognized as playing vital roles in cellular processes such as differentiation and development (Kloosterman and Plasterk, 2006), and numerous studies have linked aberrant miRNA expression to pathological conditions such as cancer (Croce, 2009). While a number of miRNAs have been identified as regulators of metastasis (Le et al., 2010), the function of miRNAs in organ-specific metastasis to bone remains poorly understood. Furthermore, few studies to date investigate the role of stromal miRNAs as potential mediators and biomarkers of metastasis. Given the prominent role of osteoclasts in osteolytic bone metastasis, miRNAs that regulate osteoclastogenesis may play a key role in osteolytic bone metastasis. Recent findings have revealed a general necessity for miRNAs in osteoclastogenesis, as genetic or siRNA-mediated ablation of factors important for biogenesis of miRNAs, including Dicer1, Dgcr8 and Ago2, blocked osteoclast differentiation (Mizoguchi et al., 2010; Sugatani and Hruska, 2009). Additionally, ectopic expression of miR-155 (Mann et al., 2010; Mizoguchi et al., 2010; Zhang et al., 2012) or repression of miR-21 (Sugatani et al., 2011) inhibit osteoclast differentiation, while conflicting functions of miR-223 in osteoclastogenesis have also been reported (Sugatani and Hruska, 2007; Sugatani and Hruska, 2009). While these results indicate a crucial role for miRNAs in physiological osteoclast differentiation, a comprehensive understanding of miRNAs in pathological osteoclastogenesis is needed to evaluate their potential application in clinical management of bone metastasis. Here, we examine the miRNA expression changes during physiological and pathological osteoclastogenesis, and evaluate the application of osteoclast miRNAs as possible therapeutic targets and biomarkers for metastatic disease.
RESULTS
Conditioned media from bone-metastatic cancer cells induces osteoclast differentiation
As previously shown (Sethi et al., 2011), the murine pre-osteoclast cell lines RAW264.7 (Figure S1A, S1B) and MOCP-5 (data not shown) can be induced to differentiate into mature, multi-nucleated osteoclasts through the addition of 20–50 ng/ml RANKL. To examine the potential for tumor conditioned media (CM) to induce osteoclast differentiation, RAW264.7 or MOCP-5 pre-osteoclast cells were treated with CM from two pairs of cancer cell lines with differing bone metastasis capabilities: (1) the highly metastatic 4T1.2 mouse mammary tumor cell line and weakly metastatic 4T1 parental line (Lelekakis et al., 1999); and (2) the highly metastatic TSU-Pr1-B2 human bladder cancer cell line and the weakly metastatic TSU-Pr1 parental line (Chaffer et al., 2005). After 6 days of treatment with CM, TRAP staining revealed mature osteoclasts only in cells treated with CM from the highly bone metastatic 4T1.2 and TSU-Pr1-B2 (Figure 1A, 1B), although at significantly lower levels than treatment with 50 ng/ml RANKL (RANKLHigh). Combined treatment of RAW264.7 cells with CM and a limited level of RANKL (20 ng/ml, RANKLLow) significantly increased differentiation, approaching that seen by treatment with 50 ng/ml of RANKL (Figure 1A–C). Treatment with CM induced similar differentiation in mouse bone marrow-derived primary pre-osteoclasts (Figure S1C) and MOCP5 cells (Figure S1D–F). Importantly, qRT-PCR indicated that osteoclast genes were elevated in RAW264.7 cells upon treatment with RANKL and bone metastatic CM (Figure S1G, S1H). Taken together, these results indicate that bone-metastatic breast cancer cells are capable of secreting factors that induce differentiation and maturation of osteoclasts.
Figure 1. RANKL and conditioned media of highly bone metastatic cancer cell lines induce similar changes in miRNA expression in osteoclasts.
(A) Representative images of RAW264.7 cells stained for TRAP (red) after treatment for 6 days with conditioned media from indicated cell lines, or conditioned media with 20 ng/ml RANKL (RANKLLow), or 50 ng/ml RANKL (RANKLHigh). Scale Bar 200 μm. (B) Quantification of TRAP+ osteoclasts from experiments in (A). **p < 0.01, ***p < 0.001. (C) Quantification of nuclei in TRAP+ osteoclasts from experiments in (A). *p < 0.05, **p < 0.01. (D) Heat map depicting miRNA microarray expression profiling in RAW264.7 cells treated with conditioned media from 4T1.2 or TSU-Pr1-B2 cells as compared to corresponding weakly metastatic cells (4T1 and TSU-Pr1), or 50 ng/ml RANKL as compared to non-treatment control. (E) Correlation of miRNA expression changes in the indicated treatment pairs as in (D). Red dots represent miRNAs selected for further analysis. (F) qRT-PCR analysis of selected miRNAs in RAW264.7 or primary bone marrow derived osteoclasts after indicated length of treatment with 50ng/ml RANKL. *p < 0.05, ***p < 0.001. Data in the figure represent average ± SEM; p values were based on Student’s t-test. See also Figure S1 and Table S1.
MiRNA microarrays reveal differentially regulated miRNAs
MiRNA microarray profiling was performed to compare miRNA expression changes in RAW264.7 cells treated for seven days with CM from 4T1 versus 4T1.2 cells or CM from TSU-Pr1 versus TSU-Pr1-B2 cells, and with or without 50 ng/ml RANKL. A total of 334 mouse miRNAs were detected, revealing 42 up-regulated and 45 down-regulated miRNAs with a >2.2 fold change across treatment groups (Figure 1D, left panel). As an initial step toward identifying miRNAs that may inhibit osteoclast differentiation, we focused on 22 unique miRNAs consistently down-regulated across three sets of samples (Figure 1D, right panel). Notably, there were significant correlations of miRNA expression changes between the treatment groups (Figure 1E), indicating conservation in the miRNA regulatory network during osteoclastogenesis in physiological and pathological conditions.
Genes known to be important for osteoclast differentiation and function were examined for predicted miRNA binding sites based on TargetScan (Grimson et al., 2007) and PicTar (Krek et al., 2005) predictions (Table S1). Five miRNAs, miR-33a-5p, miR-133a, miR-141-3p, miR-190, and miR-219-5p (hereafter miR-33a, miR-141, and miR-219) were selected for further analysis based upon their significant down-regulation, multiple predicted osteoclast targets, and sequence conservation between human and mouse. Quantitative real-time PCR (qPCR) analysis confirmed that all five miRNAs examined decreased significantly after 6 days of RANKL treatment in RAW264.7, primary bone marrow-derived precursors (Figure 1F) and MOCP-5 cells (data not shown). Importantly, these miRNA changes were also observed during the differentiation of primary human osteoclasts (Figure S1I–K).
Ectopic expression of miRNAs inhibits osteoclast differentiation
To examine the functional role of the down-regulated miRNAs in osteoclastogenesis, we ectopically expressed the miRNAs in pre-osteoclast cells prior to RANKL-induced osteoclast differentiation. Overexpression of all five miRNAs resulted in a significant decrease in osteoclast differentiation in both RAW264.7 cells (Figure 2A) and primary pre-osteoclasts (Figure S2A). We observed no change in cell growth or apoptosis after any of the miRNA treatments (data not shown). The defect in osteoclastogenesis was confirmed by qRT-PCR analysis that found repression of multiple osteoclast marker genes after ectopic miRNA expression (Figure S2B). In vitro bone resorption assay was then used to measure the effect of ectopic miRNA expression on osteoclast activity. Interestingly, while miR-33a caused only a small decrease in bone resorption, the remaining four miRNAs dramatically limited osteoclast activity (Figure 2B).
Figure 2. Ectopic expression of miRNAs downregulated during osteoclastogenesis inhibits osteoclast differentiation.
(A) Quantification and representative images of TRAP+ osteoclasts in RAW264.7 cells transfected with indicated miRNAs followed by treatment with 50 ng/ml RANKL. Scale bar, 200 μm **p < 0.01. (B) Quantification and representative images of bone resorption by RAW264.7 cells after treatment in conditions as in (A), relative to control cells. Scale bar, 200 μm. **p < 0.01, ***p < 0.001. (C) Quantification of TRAP+ osteoclasts from RAW264.7 treated cells transfected with 1 pM indicated miRNAs, followed by treatment with 20 ng/ml RANKL. *p < 0.05. (D) Quantification of relative bone resorption in RAW264.7 cells transfected with1 pM indicated miRNA or 20 nM Zometa. *p < 0.05. Data in the figure represent average ± SEM. p values were based on Student’s t-test. See also Figure S2.
Since these miRNAs might inhibit osteoclast differentiation or function by targeting different mRNAs, we further examined the repression of osteoclast differentiation using combinations of inhibitory miRNAs. Given that transient transfection with 10 pM of miRNA precursors was sufficient to inhibit up to ~90% of osteoclast differentiation, we treated RAW264.7 cells with a combination of miRNAs at a reduced concentration of 1 pM total precursors. Combined ectopic expression of miR-141, miR-190, and miR-219 revealed a significant additive effect on osteoclast maturation (Figure 2C) and bone resorption (Figure 2D, Figure S2C) that is equivalent to the treatment with 20 nM zoledronic acid (Figure 2D, Figure S2C). Importantly, ectopic expression of the miRNAs combined with zoledronic acid revealed an additive effect, leading to almost complete ablation of osteoclast activity (Figure 2D, Figure S2C). These results reveal a necessity for repression of multiple miRNAs during osteoclast differentiation, with ectopic expression of miR-133a, miR-141, and miR-219 strongly inhibiting osteoclast maturation and activity in all models tested, and partial inhibition seen after ectopic expression of miR-33a and miR-190.
To confirm that miRNA mediated inhibition of osteoclast differentiation is not specific to the RAW264.7 model, we examined the effect of ectopic miRNA expression in additional models. MOCP5 cells transfected with pre-miRNAs revealed a similar pattern of inhibition (Figure S2D and S2E). A previous study revealed greatly enhanced osteoclast differentiation after activation of the Notch pathway by tumor-derived Jagged1 (Sethi et al., 2011). To examine the effect of miRNAs on Jagged1-enhanced osteoclast differentiation, RAW264.7 cells were plated on Jagged1 coated plates, followed by treatment with 50 ng/ml RANKL. Ectopic miRNA expression significantly inhibited Jagged1-dependent enhancement of osteoclast differentiation (Figure S2F–H). Taken together, these results illustrate the broad capacity for ectopic expression of these miRNAs to inhibit osteoclast differentiation.
Identification of miRNA targets involved in osteoclast differentiation and function
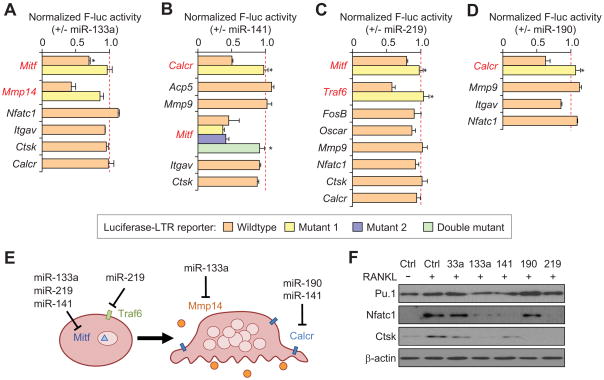
We sought to examine direct miRNA targeting of mRNAs related to osteoclastogenesis using luciferase reporters containing the 3′UTR of prospective target mRNAs (Table S2). Reporter constructs were co-transfected with pre-miRs, as well as a Renilla luciferase plasmid for normalization. Reporter assays revealed a number of direct targets for the candidate miRNAs. MiR-133a, miR-141, and miR-219 were all found to repress expression of Mitf, and specific targeting of Mitf by these miRNAs was confirmed by site-directed mutagenesis of the predicted miRNA binding sites (Figure 3A–C). Interestingly, miR-141 was found to contain two active binding sites in the Mitf 3′UTR, reducing expression by ~50% (Figure 3B), while a single site in miR-219 was able to repress Mitf expression by ~20% (Figure 3C). In addition, miR-133a was found to repress Mmp14, miR-141 repressed Calcr, miR-219 repressed Traf6, and miR-190 was able to repress Calcr (Figure 3A–D). These miRNA targets represent important factors involved in osteoclast differentiation and function, with Mitf functioning as an essential transcription factor, Calcr and Traf6 serving as important signal transducers, and Mmp14 functioning as a secreted matrix metalloproteinase during osteoclastogenesis (Figure 3E). To better examine the capacity of the miRNAs to inhibit osteoclast differentiation, osteoclast markers were examined in RAW264.7 cells transfected with the miRNA of interest, followed by RANKL treatment. Western blot analysis of Pu.1, Nfatc1, and Ctsk revealed that miR-133a, miR-141, and miR-219 seem to be inhibiting early osteoclast differentiation, with minimal expression of Nfatc1, which plays a role in early osteoclast commitment, or the mature osteoclast marker Ctsk (Teitelbaum and Ross, 2003) (Figure 3F). These results are in agreement with their targeting of Mitf or Traf6, either of which is predicted to inhibit the earliest stages of osteoclast commitment. In comparison, miR-190 transfected cells expressed Pu.1 and Nfatc1, but not Ctsk, indicating that miR-190 might be inhibiting osteoclast differentiation after commitment to an osteoclast fate. miR-33a transfected cells retained expression of all three osteoclast genes, consistent with its relatively weak effect on inhibiting osteoclastogenesis (Figure 3F). Taken together, these results indicate that the miRNAs are inhibiting osteoclast differentiation through the functional targeting of essential osteoclast genes functioning at different stages of osteoclast differentiation.
Figure 3. Validation of candidate miRNA genes.
(A–D) Normalized activity of luciferase reporter containing the 3′ UTR of candidate genes upon cotransfection with indicated miRNAs, relative to transfection with negative control miRNA. *p < 0.05. (E) Schematic indicating miRNA targets that are involved in osteoclast differentiation or function. (F) Western blot analysis for Pu.1, Nfatc1, or Ctsk in RAW264.7 cells after transient transfection with indicated miRNA and treatment with 50ng/ml RANKL. Data in the figure represent average ± SEM; p values were based on Student’s t-test. See also Table S2.
Systemic miRNA treatment inhibits osteoclasts in vivo
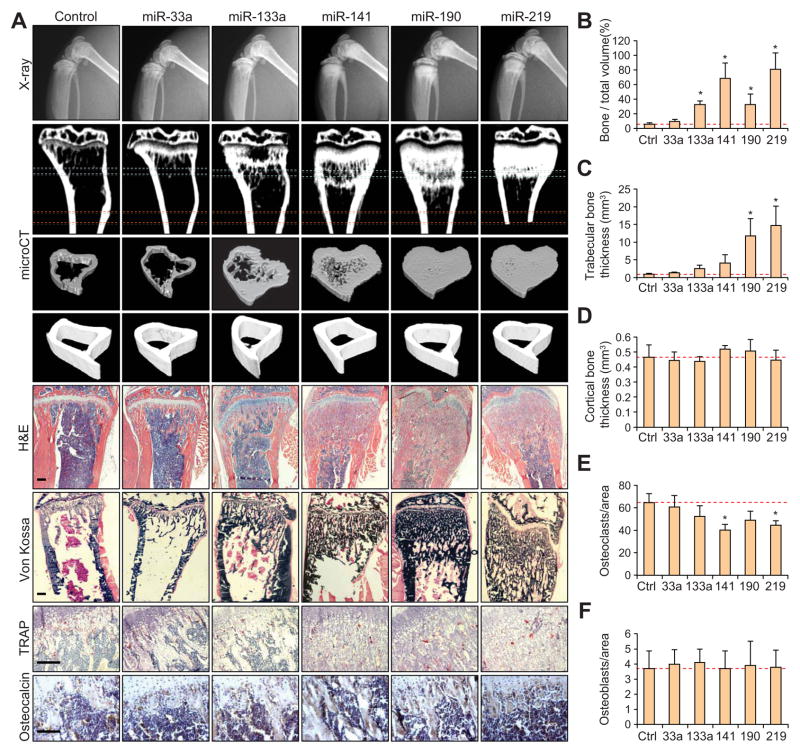
To evaluate the capacity for these miRNAs to inhibit mouse osteoclast function in vivo, we injected 10 μg of pre-miRNA into the lateral tail-vein of Balb/c mice weekly for four weeks. Mice were examined weekly by X-ray radiography, revealing increased bone density in the hind limbs, particularly in the distal femur and proximal tibia (data not shown). Examination by microCT revealed increased trabecular bone in both femur and tibia (Figure 4A). Further quantitative analyses confirmed a significant increase in bone volume (Figure 4B) and trabecular thickness (Figure 4C), while no significant differences in cortical bone thickness was found (Figure 4A, D).
Figure 4. Micro-computed tomography and histological analysis revealing alterations in bone homeostasis after treatment of miRNAs downregulated in osteoclastogenesis.
(A) Representative X-ray, μCT and histological images for TRAP, H&E, Von Kossa, or osteocalcin staining of bones from mice treated with indicated miRNAs. Regions of interest for trabecular and cortical bone scan and analysis are marked by blue and red dashed lines, respectively. Scale bar, 200 μm. n = 6. (B) Quantification of bone volume relative to total volume from representative μCT scans in (A). n = 3. *p<0.05. (C) Quantification of trabecular thickness from representative μCT scans of mice treated with miRNAs as in (B). *p<0.05. (D) Quantification of cortical bone thickness from representative μCT scans as in (B). (E) Quantification of TRAP+ osteoclasts from decalcificed histological sections of hindlinbs from mice in (A), n = 6. *p<0.05. (F) Quantification of osteocalsin-positive osteoblasts from bone sections from mice in (A). Data in the figure represent average ± SEM; p values were based on Student’s t-test.
Histological TRAP staining of decalcified bone sections revealed a significant decrease in the number of osteoclasts relative to bone surface area in mice treated with miR-141 and miR-219 (Figure 4A, E) and Von Kossa staining revealed the increase of calcified tissues (Figure 4A), confirming the significant expansion in trabecular bone that was seen in the microCT analysis. Osteocalcin immunohistochemical staining revealed no significant difference in osteoblast number (Figure 4A, F), consistent with a previous report that inhibition of miRNA biogenesis does not affect osteoblast differentiation (Mizoguchi et al., 2010). While these results indicated osteoclasts as a major bone cell type affected by systemic delivery of these miRNAs, it is important to note that the differentiation or activity of additional bone stromal cells might also be impacted.
MiR-141 and miR-219 inhibit experimental bone metastasis
Given the ability of multiple miRNAs to inhibit osteoclast differentiation in vitro and in vivo, we next examined the capacity of these miRNAs to inhibit breast cancer bone metastasis in a mouse model. We first examined the potential of CM from multiple SCP cell lines, clonal derivatives of the human MDA-MB-231 breast cancer line (Kang et al., 2003) with different bone metastatic capabilities, to regulate osteoclast differentiation. Similar to our previous results (Blanco et al., 2012), CM from highly bone metastatic SCP cell lines (SCP2, 28 and 46), but not from weakly metastatic lines (SCP4, SCP6 and parental MDA-MB-231), stimulated osteoclast differentiation in RAW264.7 cells (Figure S3A and S3B), increased the expression of osteoclast marker genes (Figure S3C), and induced miRNA expression changes consistent with our previous findings (Figure S3D). After further validating the ability of highly metastatic SCP28 to induce osteoclastogenesis in MOCP5 cells (Figure S3E–G), we used SCP28 for analyzing the effect of miRNAs on bone metastasis development in mice.
Nude mice were systemically treated with control or experimental pre-miRNAs immediately before, and once per week subsequently, after inoculation with SCP28 cells via intracardiac injection. Metastatic progression was monitored by weekly bioluminescence imaging (BLI) using a firefly luciferase reporter stably expressed in the cell line. Whereas treatment with miR-133a and miR-190 had no effect on metastatic progression by BLI, there was a significant decrease in hind-limb tumor burden after treatment with either miR-141 (p = 0.024) or miR-219 (p = 0.015) (Figure 5A and 5B). qPCR analysis of serum samples from mice inoculated with the pre-miRNAs revealed a substantial decrease in miR-190 and miR-133a within 6 hours of injection, with nearly full clearance within 24 hours (Figure 5C). In contrast, miR-141 and miR-219 took substantially longer to clear from the serum, maintaining ~20% of the normalized expression at 24 hours. X-ray imaging revealed decreased bone lesions in miR-141 and miR-219 treated mice while miR-190, miR-133a and control pre-miRNA injected mice exhibited significant bone degradation (Figure 5A and 5D). Histological analysis revealed a decrease in osteolysis by hematoxylin and eosin (H&E) staining and a decrease in TRAP+ osteoclast recruitment at the tumor-bone interface. Consistent with the results in miRNA-treated healthy mice (Figure 4), miR-190, miR-141, and miR-219 treated mice showed a significant decrease in TRAP+ osteoclast number (Figure 5E), while there was no noticeable change in osteoblast number from any of the treatments (Figure 5A and 5F). To rule out the potential influence of the miRNAs on the growth or survival of the SCP28 cells, we treated the cells with miRNAs in culture and found no significant difference in proliferation (data not shown). Taken together, these results reveal a significant decrease in metastatic burden after systemic treatment with miR-141 or miR-219, likely due to decreased osteoclast activity.
Figure 5. Systemic treatment with miRNAs downregulated in osteoclastogenesis inhibits bone metastasis.
(A) BLI, X-ray, Osteocalcin, H&E, and TRAP images from mice inoculated with SCP28 breast cancer cells and treated with indicated miRNAs. BLI images show three representative mice from each experimental group. White arrows indicate osteolytic bone lesions in the X-ray images. Scale bar, 200 μm. n = 10. (B) Normalized bone metastasis BLI signals from mice in (A). n = 10. **p < 0.01 by Mann-Whitney test. (C) qRT-PCR miRNA expression levels from serum samples of mice taken at indicated times after injection with miRNAs. **p < 0.01 (D) Quantification of osteolytic lesion area in hindlimbs from indicated experimental group. (n = 6) **p < 0.01. (E) Quantification of TRAP+ osteoclasts from histology in experiment from (A). **p < 0.01. (F) Quantification of osteocalcin+ osteoblasts from histology in experiment from (A). Data in the figure represent average ± SEM; p values were based on Student’s t-test unless otherwise indicated. See also Figure S3.
We further compared the therapeutic effect of miRNAs with treatment of 100μg/kg zoledronic acid (Zometa) (Figure S3H and S3I). As expected, mice treated with Zometa experienced a significant decrease in tumor burden, with an overall trend of better response than individual miR-141 or miR-219 treatment, although the difference did not reach statistical significance. These results indicate that systemic treatment with miR-141 and miR-219 inhibits bone metastasis burden to a similar extent as current therapeutics.
Serum miRNA levels correlate with bone metastasis
To further evaluate the function of miRNAs that change during osteoclast differentiation we next examined a subset of four miRNAs with significant up-regulation during osteoclastogenesis (miR-16, miR-211, miR-378 and Let-7a, Figure 1D). Interestingly, we did not observe significant differences after ectopic expression or repression of any of the candidate miRNAs tested, although this result did not rule out the functional importance of additional candidates that we have not tested so far. As miRNAs are often released from cells into circulation, miRNAs with increased expression during osteoclastogenesis may potentially serve as biomarkers for osteolytic bone metastasis. To evaluate the possibility of using osteoclast miRNAs as biomarkers, we examined the expression of miRNAs up-regulated during osteoclastogenesis (Figure 1D) in serum samples of mice with different bone metastasis burdens. Serum samples were collected at 0, 7, and 35 days from nude mice after intracardiac injection of highly metastatic SCP2 cells and miRNA expression was analyzed by qRT-PCR. Examination of multiple miRNAs with the most dramatic increase during differentiation revealed consistently elevated expression of miR-378 and miR-16 at 35 days post-injection (Figure 6A).
Figure 6. Serum levels of miRNAs upregulated during osteoclastogenesis correlate with bone metastasis.
(A) BLI and qRT-PCR expression analysis of miRNAs in mice after intravenous inoculation with SCP2 breast cancer cells. Left, BLI images of two representative mice at each time point. Right, qRT-PCR for miRNA expression levels of miRNAs derived from serum samples taken on indicated day post-injection. Data represent average ± SEM. n = 10. *p < 0.05 by Student’s t-test. (B) BLI and qRT-PCR expression analysis of miRNAs in mice 28 days after intravenous inoculation with TSU-Pr1 or TSU-Pr1-B2 bladder cancer cells. Data represent average ± SEM. n = 10. *p < 0.05 by Student’s t-test. (C) qRT-PCR miRNA expression analysis of matched micro-dissected primary breast tumor (Primary) or bone metastasis (BM) samples. n = 12. p values based on Mann-Whitney test. (D) qRT-PCR miRNA expression analysis of serum samples from healthy donors (HD) or breast cancer patients with bone metastasis (BM). HD, n = 21. BM, n = 38. p values based on Mann-Whitney test.
Similarly, only miR-378 and miR-16 were elevated when we compared mice with low of high bone metastatic tumor burden after inoculation with weakly metastatic TSU-Pr1 or highly metastatic TSU-Pr1-B2 cell lines, respectively (Figure 6B). Based on these findings, we examined the expression of miR-378 and miR-16 in matched primary (Primary) or bone-metastatic (BM) tumor samples from 12 breast cancer patients. Both miR-16 and miR-378 have elevated expression in bone metastases, reflecting the presence of osteoclasts (Figure 6C). Next, we analyzed the expression of these two miRNAs in serum samples of healthy female donors (HD) or breast cancer patients with bone metastasis (BM) and again observed significantly increased level of both miRNAs in patients with bone metastasis (Figure 6D). Taken together, these results indicated the potential for using these miRNAs as biomarkers for bone metastasis progression.
Soluble ICAM1 from metastatic cells enhances osteoclast differentiation
Since tumor CM induced osteoclastogenesis produced similar miRNA changes to RANKL-induced physiological osteoclast differentiation, we decided to identify soluble factor(s) in CM that promote osteoclast activation. First, we investigated whether RANKL present in CM is accountable for such activities. ELISA quantification of RANKL levels in CM samples revealed very low levels (4–6 ng/ml) of RANKL even in highly metastatic MDA-MB-231, 4T1 or TSU sublines, although these sublines produced significantly higher levels of RANKL than their weakly metastatic isogenic counterparts (Figure S4A). These levels of RANKL are insufficient to induce osteoclast differentiation (see Figure S1A, B). Furthermore, osteoclast differentiation induced by 30 ng/ml RANKL is inhibited by more than 80% after treatment with 200 ng/ml of OPG, a decoy receptor and inhibitor of RANKL. In contrast, such treatment resulted in only ~50% inhibition of CM induced osteoclast differentiation (Figure 7A, Figure S4B). These results indicated that additional factor(s), other than RANKL, in the tumor CM play an important role in enhancing osteoclastogenesis. A number of RANKL-independent osteoclast differentiation factors have been previously described, such as IL-6, MIF-1α, PTHrP, TNFα, GM-CSF and IGF (Weilbaecher et al., 2011). To examine whether any of these or other cytokines were secreted by the metastatic cells to induce osteoclastogenesis, we evaluated cytokine expression in CM from the three series of cell lines with differential bone metastatic abilities (Figure 7B). Interestingly, among the cytokines analyzed, only soluble ICAM1 (sICAM1), the cleaved extracellular domain of ICAM1, was consistently overexpressed in the highly metastatic cell lines. Cell surface ICAM1 has been previously implicated in osteoclast differentiation, and antibodies against ICAM1 reduce osteoclast maturation (Harada et al., 1998; Kurachi et al., 1993). However, the functional role and mechanism of tumor-derived sICAM1 in osteoclastogenesis is unknown. Treatment of RAW264.7 cells with 50 ng/ml sICAM1 and 50 ng/ml RANKL together prompted osteoclast differentiation within 2 days, while treatment with RANKL alone required approximately 4 days for the first mature osteoclasts to differentiate (Figure 7C). Analysis of osteoclast marker genes confirmed that sICAM1 treatment together with 50ng/ml RANKL reduced the time necessary for osteoclast differentiation compared to RANKL treatment alone, although there was no difference in endpoint expression levels for any of the mRNAs (Figure S4C). Although sICAM1 alone was not sufficient to induce differentiation, it exhibited an additive effect on differentiation when combined with limiting concentrations of RANKL (10 ng/ml RANKL, Figure 7D), and a significant inhibition in the expression of miRNAs downregulated during osteoclastogenesis, which was magnified after co-treatment with RANKL (Figure 7E).
Figure 7. Soluble ICAM1 in tumor conditioned media synergizes with RANKL to promote osteoclast differentiation.
(A) Quantification of RAW264.7 cells treated with 30 ng/ml RANKL or conditioned media ± 200 ng/ml OPG. **p < 0.01. (B) Heat map from cytokine array depicting relative protein expression in conditioned media from indicated cell lines as compared to their weakly metastatic counterparts (4T1 vs 4T1.2, TSU-Pr1 vs TSU-PR1-B2, and MDA-231 vs SCP28, respectively). (C) Quantification of TRAP+ osteoclasts from RAW264.7 cells treated with 50 ng/ml RANKL ± 50 ng/ml sICAM1 for indicated length of time. ***p < 0.001. (D) Quantification of TRAP+ osteoclasts from RAW264.7 cells treated with 10 ng/ml RANKL, 50 ng/ml sICAM1, or both for 6 days. *p < 0.05. (E) qRT-PCR analysis of selected miRNAs in RAW264.7 cells after indicated length of treatment with 10 ng/ml RANKL or/and 50 ng/ml sICAM1. *p < 0.05 by analysis of variance (ANOVA) for all miRNAs. (F) Quantification of osteoclasts/field and nuclei/osteoclast in RAW264.7 cells treated with 4 ng/ml RANKL plus indicated concentration of sICAM1 for 6 days. **p < 0.01, ***p < 0.001. (G) Quantification of TRAP+ osteoclasts from RAW264.7 cells treated with 30ng/ml RANKL or SCP28 conditioned media followed by treatment with indicated concentration of ICAM1 antibody (in μg/mL). *p < 0.05, **p < 0.01. Data in the figure represent average ± SEM; p values were based on Student’s t-test unless otherwise indicated. See also Figure S4.
Since the ELISA results from CM samples revealed very low RANKL concentrations that are insufficient to support osteoclast maturation, and treatment with sICAM1 and RANKL revealed an additive effect, we hypothesized that CM-induced differentiation was due to the combined effect of both secreted factors. To test this, RAW264.7 cells were treated with 4 ng/ml RANKL (equivalent to the level in tumor CM) and increasing concentrations of sICAM1. While low levels of sICAM1 had no effect on RAW264.7 cells in the presence of 4 ng/ml RANKL, we observed high levels of TRAP+ osteoclasts after treatment with 50 ng/ml sICAM1, approaching that of CM treatments (Figure 7F, Figure S4D). Furthermore, when RAW264.7 cells were cultured with a function blocking monoclonal antibody (mAb) against ICAM1, they exhibited a dose-dependent decrease in osteoclast differentiation (Figure 7G, Figure S4E) induced by SCP28 CM, while RANKL-induced osteoclastogenesis was unaffected. Additionally, treating SCP28 CM with sICAM1 mAb resulted in an additive inhibitory effect when combined with OPG (Figure S4F), and ICAM1 mAb was able to inhibit the bone resorbing activity of CM-treated RAW264.7 cells (Figure S4G). Finally, RAW264.7 cells treated with sICAM1 showed increased migration in a dose dependent manner, which was once again inhibited by the ICAM1 mAb (Figure S4H). Taken together, these results support a functional role for sICAM1 as a crucial component of tumor CM in stimulating osteoclastogenesis.
Previous studies have identified αLβ2 and αMβ2 integrins as receptors of sICAM1 (Carlos and Harlan, 1994). Indeed, a function blocking antibody against the β2 subunit inhibited SCP28 CM, but not RANKL, -induced differentiation (Figure 8A, Figure S4I). Analysis of RAW264.7 cells revealed strong expression of αM and β2 subunits on the cell surface, and lower expression of αL (Figure 8B). Signaling through β2 integrins has been previously shown to activate the NFκB pathway in neutrophils under stimulation by GM-CSF or IL-6 (Kettritz et al., 2004), although it is not known if sICAM1 can similarly activate NFκB. We examined activation of the pathway by monitoring the activation of an NFκB luciferase reporter (Figure 8C), degradation of IκBα (Figure 8D), or phosphorylation of p65 (Figure S5A) in RAW264.7 cells treated with TNFα, RANKL, or sICAM1 over time and observed similar responses in these three treatment conditions. SCP28 CM treatment also showed a decrease in IκBα, which was inhibited by the addition of an antibody against the β2 integrin or OPG (Figure 8E). Similarly, the β2 integrin antibody inhibited sICAM1, but not RANKL, induced activation of NFκB (Figure S5B). Interestingly, while sICAM1-induced miRNA changes were abrogated after RAW264.7 cells were treated with the IKK inhibitor PS1145 (Figure 8F), miRNA transfection did not affect NFκB activation induced by TNFα, RANKL, or sICAM1 (Figure S5C and S5D), suggesting that these miRNAs act downstream of NFκB activation to influence osteoclastogenesis. Together, these results implicate sICAM1 as a bone metastatic cell-secreted factor that functions through β2 integrin and NFκB signaling to enhance osteoclast differentiation in the presence of low levels of RANKL.
Figure 8. sICAM1 promotes osteoclastogenesis by activating NFκB signaling, and sICAM1 levels in patient serum samples are correlated with miR-16 and miR-378 levels, and associated with bone metastasis.
(A) Quantification of TRAP+ osteoclasts from RAW264.7 cells treated with RANKL or SCP28 conditioned media ± 20μg ICAM1 antibody or 20μg β2 integrin antibody. Data represent average ± SEM. **p<0.01 by Student’s t-test. (B) Expression of αL, αM, and β2 integrin subunits on RAW264.7 by IF (left) and flow cytometry (right). Scale bar, 25 μm. (C) Normalized activity of NFκB luciferase reporter in RAW264.7 cells after indicated treatment with TNFα, RANKL, or sICAM1 for indicated length of time. Data represent average ± SEM. (D) Western blot analysis of IκB in RAW264.7 cells after treatment with TNFα, RANKL, or sICAM1 for indicated length of time. (E) Western blot analysis of IκB in RAW264.7 cells after treatment with indicated concentration of RANKL or SCP28 CM ± 20 ng β2 antibody or 200 ng/ml OPG for 2 hours. (F) Relative qRT-PCR miRNA expression level in RAW264.7 cells treated with 50ng/ml sICAM1, with or without 20μM IKK inhibitor PS1145 for 6 days, after normalization to the expression level without sICAM1 treatment. Data represent average ± SEM. *p<0.05. (G) ELISA quantification of sICAM1 expression levels in serum samples collected from healthy female donors (HD, n = 41), disease-free breast cancer patients (DFP, n = 16), or breast cancer patients with bone metastasis (BM, n = 38). p values based on Mann-Whitney test. (H) Correlation of qRT-PCR miR-16 or miR-378 expression levels with ELISA sICAM1 protein expression levels in serum samples from healthy donors or bone metastasis patients from (G). See also Figure S5.
Correlation of serum sICAM1 and miRNA expression in patients with bone metastasis
To further investigate the clinical significance of sICAM1 as a tumor-derived factor in inducing osteoclastogenesis and associated miRNA changes during bone metastasis, we analyzed the expression levels of sICAM1 in serum samples collected from healthy female donors (HD), disease-free breast cancer patients showing no occurrence of bone metastasis (DFP, samples taken immediately following resection of the primary tumor), or breast cancer patients with bone metastasis (BM). Patients with bone metastases exhibited significantly increased serum sICAM1 level compared to healthy donors or disease free patients (Figure 8G). Importantly, a strong correlation was seen between serum expression of sICAM1 and miR-16 or miR-378 (Figure 8H) in bone metastasis patients, while little correlation was seen in samples from healthy donors.
To further investigate the diagnostic potential of miR-16 and miR-378 as secreted biomarkers for osteolytic bone metastasis, we compared the serum expression of the miRNAs against N-terminal telopeptide (NTX), a standard marker of bone turnover. The sensitivity and specificity of using serum levels of miR-16, miR-378, sICAM1 and NTX, alone or in combination, for detecting bone metastasis were determined (Figure S5E and S5F). The diagnostic values of miR-378 and sICAM1 were lower than that of either miR-16 or NTX as individual variables, and combining sICAM1 with NTX did not significantly increase the value. In contrast, miR-16 revealed increased specificity over NTX, while maintaining a similar sensitivity. Importantly, the combination of miR-16 with NTX produced increased sensitivity, with only a slight reduction in specificity (Figure S5F). These findings underscored the clinical significance of elevated sICAM1 and miR-16/-378 expression in the serum of bone metastasis, and suggest their potential use as biomarkers for diagnosis of bone metastasis or predictive marker for anti-ICAM1 therapeutics.
DISCUSSION
The present study provides experimental and clinical evidence to support the role of miRNAs in osteoclast activation during breast cancer bone metastasis. Genetic ablation of Dicer in the osteoclast lineage has clearly established the importance of miRNAs in physiological osteoclast development (Mizoguchi et al., 2010; Sugatani and Hruska, 2009). However, little is known about miRNA regulation during tumor induced osteoclastogenesis. In this study, we revealed consistent up- and down-regulated miRNAs during osteoclast differentiation in both physiological and pathological conditions. We demonstrated inhibition of osteoclast differentiation and osteolytic bone metastasis after ectopic expression of several miRNAs down-regulated during osteoclastogenesis. We further identified sICAM1 as an important tumor-secreted factor that enhances osteoclast differentiation and influences osteoclast miRNA expression via the NFκB pathway. Importantly, serum levels of both sICAM1 and miRNAs are indicators of bone metastasis burden in breast cancer patients. These results suggest multiple avenues for clinical translation of the findings.
A previous microarray-based analysis of miRNA expression changes in mouse primary osteoclasts was conducted 24h after M-CSF/RANKL treatment and revealed altered expression of dozens of miRNAs (Sugatani et al., 2011). In our current study, we focus on miRNA changes induced by both RANKL and tumor conditioned media, and samples were collected after 7 days of treatment with RANKL or CM, after we observed full osteoclast differentiation. Such analysis is likely to reveal not only miRNAs involved in regulating osteoclast differentiation, but also those that influence the function of mature osteoclasts. Notably, although our miRNA microarray analysis revealed similar trends of expression changed in miRNAs reported in the previous study (Sugatani et al., 2011) (Figure S5G), a distinct set of most differentially expressed miRNAs were identified in our current study. It is possible that the differences across the two studies can be explained by the differences in cell lines used, treatment conditions and time points analyzed. Importantly, we used three different human and mouse cancer cell lines from two different tumor types in the CM treatment experiments. Despite the fact that these three distinctively different tumor cells all secrete a sub-minimal amount of RANKL into their CM, miRNA expression changes stimulated by their CM showed high levels of consistency with RANKL-induced osteoclastogenesis. Moreover, ectopic overexpression of down-regulated miRNAs inhibited osteoclastogenesis induced by multiple conditions, including RANKL, tumor CM, and Jagged1, in multiple osteoclast cell lines or primary preosteoclasts. These findings support a highly conserved miRNA regulatory network that controls osteoclast differentiation in both pathological and physiological conditions.
Our investigation into the target genes of osteoclast-inhibiting miRNAs revealed direct targeting a number of known osteoclast genes, including Mitf, Calcr, Traf6, and Mmp14. MITF is a basic-helix-loop-helix-zipper transcription factor (Hodgknson et al., 1993) that is crucial for osteoclast development (Hershey and Fisher, 2004; Sharma et al., 2007; Weilbaecher et al., 2001). Traf6 encodes a member of the tumor necrosis factor receptor (TNFR)-associated factor family of cytokine receptor adaptor proteins that play an essential role in signaling transduction of the RANKL pathway (Lomaga et al., 1999). Calcitonin receptor (CALCR) is a well characterized cell-surface receptor capable of influencing osteoclast-mediated bone resorption in vitro and in vivo (Davey et al., 2008). Mmp14 knockout mice feature severe skeletal defects, including osteopenia and skeletal dysplasia (Holmbeck et al., 1999). Interestingly, additional studies have implicated Mmp14 in osteoclast fusion during maturation, and Mmp14-null osteoclasts had decreased activity in vitro (Gonzalo et al., 2010). Therefore, decreased expression of Mitf, Traf6, Calcr, or Mmp14 is likely to constitute a miRNA target gene network responsible for the defect we see in osteoclast differentiation after ectopic expression of these miRNAs. Our analysis of stage-specific markers of osteoclast differentiation revealed that miR-133a, miR-141, and miR-219 seem to be inhibiting early osteoclastogenesis, while miR-190 might inhibit osteoclast differentiation or function after commitment to an osteoclast fate.
CM from highly bone metastatic cells was able to induce osteoclast differentiation and miRNA expression changes despite the presence of very low levels of RANKL, which is insufficient to induce osteoclastogenesis. Importantly, we identified sICAM1 as a tumor-derived factor in the CM that enhances osteoclast activation at the minimal concentration of RANKL. ICAM1 has been previously observed to increase in expression level during osteoclast differentiation, while ablation of ICAM1 inhibited osteoclast differentiation from peripheral blood mononuclear cells (Nakano et al., 2004). However, it was previously unknown how tumor-derived sICAM1 influences osteoclast differentiation. Our discovery that sICAM1 is capable of increasing RANKL induced osteoclast differentiation reveals an additional mechanism for osteoclast regulation from bone metastatic cells and suggests a potential target for therapeutic intervention. Various tumor-derived factors have been previously shown to induce osteoclast differentiation, including cell surface ligand Jagged1 and secreted factors RANKL, GM-CSF, MIP-1α, PTHrP, IL-8 and IL-6 (Ell and Kang, 2012; Weilbaecher et al., 2011). Our findings indicate a similar role for tumor secreted ICAM1, which is insufficient for independently inducing osteoclastogenesis but capable of enhancing differentiation in the presence of low, but physiological, levels of RANKL. We found that sICAM1 binding to its cognate receptor β2 integrins activates NFκB signaling, which is essential for canonical, RANK-mediated osteoclast differentiation (Boyle et al., 2003; Teitelbaum and Ross, 2003), potentially explaining the mechanism by which sICAM1 enhances osteoclast differentiation. Although it is not known how β2 integrins activate the NFκB pathway, it has been proposed that they provide a co-stimulatory effect on NFκB signaling in neutrophils treated with GM-CSF or IL-8 (Kettritz et al., 2004). Furthermore, β2 integrin clustering can activate NFκB (Kim et al., 2004). Importantly, sICAM1-induced alterations in miRNA expression appear to occur downstream of NFκB, as inhibiting the pathway prevented miRNA changes, while none of the miRNAs examined in this study altered NFκB signaling. Additionally, sICAM1 increases the migration of pre-osteoclast cells, which may indicate a role for bone-metastasis derived sICAM1 in the recruitment of osteoclasts to the developing bone lesion. This hypothesis is in line with results from a previous study which identified soluble VCAM1 as an important bone metastasis factor in the recruitment of osteoclasts to the osteolytic lesion through the binding of α4β1 integrins on pre-osteoclast cells (Lu et al., 2011). Notably, VCAM1 and ICAM1 are both endothelial adhesion molecules of the Ig gene superfamily that bind leukocyte integrins. It is therefore not surprising that they have conserved functions in enhancing osteoclastogenesis in bone metastasis.
Our findings reveal several avenues for potential translational applications in the clinical management of bone metastasis. Intravenous injection of pre-miR-133a, -141, -190, or -219 significantly reduced osteoclast activity in vivo. These effects were seen most clearly in the trabecular region of the femur and tibia, while no significant difference was seen in cortical bone thickness. This finding is similar to the observation in animals treated with bisphosphonates (Quattrocchi et al., 2012), possibly because of the higher rates of turnover in the trabecular bone. Consistently, treatment of mice with the pre-miR-141 and -219 was capable of inhibiting osteolytic bone metastasis. It is curious that miR-133a and miR-190 had no measurable effect on bone metastasis, despite a substantial influence on normal bone remodeling. It seems unlikely that this is due to a tumor-intrinsic mechanism, since in vitro studies showed no effect on SCP28 growth or survival after miRNA treatment. Instead, is possible that this is explained by the measured differences in stability of the miRNAs in circulation, as inhibition of bone metastasis may require greater levels of miRNAs to reach the bone than inhibition of normal bone remodeling.
In our studies, systemic injection of unconjugated miRNAs was sufficient to induce broad changes in bone remodeling and appeared to be well tolerated by the mice. While our data from in vitro and in vivo experiments illustrate miRNA-mediated inhibition of osteoclasts, it is possible that these miRNAs might also target additional cells in vivo. Therefore, additional further analyses of other potential cellular and molecular targets of these miRNAs during long-term treatment in vivo should be conducted when developing potential miRNA-based therapeutic applications. It is possible that improved delivery methods might enhance the pharmacokinetics and efficacy of the miRNAs on bone metastasis and potentially reveal an effect from miR-133a and miR-190. The therapeutic effects of miR-141 and miR-219 mimic those seen in mice after treatment with Zometa. In addition, we noted an additive effect from combined treatment with miR-141/-190/-219 and Zometa on bone resorption in vitro. Thus, it is possible that combinatorial treatments including miRNAs and currently approved osteoclast-targeting agents such as biosphosphonates and denosumab (RANKL antibody) might provide enhanced clinical efficacy. Furthermore, the functional role of tumor-derived sICAM1 in pathological osteoclast differentiation suggests the potential for using ICAM1 blocking antibodies for targeted therapeutics. Therefore, osteoclast miRNAs and sICAM1 represent potential targets in the treatment of aberrant osteoclast activity, namely bone degenerative diseases such as osteolytic bone metastasis, osteoporosis and Paget’s disease. Finally, our discovery that miR-16 and miR-378 levels increase in bone lesions and serum samples of bone metastasis patients presents the potential for their use as metastasis biomarkers. In particular, combined miR-16 and NTX as biomarkers increased the sensitivity of bone metastasis diagnosis, although the potential clinical application of this combination still awaits further large scale prospective analysis.
EXPERIMENTAL PROCEDURES
Tumor xenografts and bioluminescence analysis
All procedures involving mice and experimental protocols were approved by Institutional Animal Care and Use Committee (IACUC) of Princeton University. For bone metastasis studies, 1×105 tumor cells were injected into the left cardiac ventricle of anesthetized female athymic Ncr-nu/nu. MiRNA precursors (10μg/mouse in 100μl PBS, Applied Biosystems) and Zometa (100μg/kg) were injected intravenously. Development of metastases was monitored by measuring photon flux of BLI signals in the hindlimbs of mice after retro-orbital injection of 75 mg/kg D-Luciferin and image acquisition using the Xenogen IVIS 200 Imaging System. Data were normalized to the signal on day 0. X-ray examination was performed as previously described (Kang et al., 2003).
Analysis of primary tumors and bone metastases
Women with resected breast cancer were selected from patients followed from 1995 to 2010 in IRCCS IRST, Meldola, Italy. Tumor specimens were de-identified and were considered exempt samples in accordance with the institutional review board of the Local Ethic Committee, Forlì, Italy. Tumor specimens were fixed in formalin and embedded in paraffin. Tissues collected were 13 matched primary breast tumors and 13 bone metastatic tissues. Total RNA was collected from 20 μm thick sections from formalin-fixed paraffin embedded (FFPE) tissue blocks using the FFPE RNA/DNA Purification kit (Norgen) according to the manufacturer’s instructions.
Serum case series and sample collection
Breast cancer patients with no evidence of disease for at least 5 years (16 patients) and patients at first diagnosis of bone metastases (38 patients, radiologically confirmed) were recruited by the Osteoncology and Rare Tumors Center of IRCCS IRST (Meldola, Italy) from January 2007 to December 2009. A group of 41 healthy donors was also enrolled in the study. Informed consent was obtained from all subjects in accordance with the protocol approved by the institutional review board of the Local Ethic Committee, Forlì, Italy. A 5 ml venous blood sample from donors and patients was collected in tubes without anticoagulant and centrifuged at 2,500 rpm for 15 minutes at room temperature then stored at −80°C until processing. Total RNA from 400 μl of serum was extracted using the miRVana miRNA isolation kit (Ambion) according to the manufacturer’s instructions.
Statistical Analysis
Results are presented as average ± standard deviation or as average ± standard error of the mean (SEM), as indicated in figure legends. BLI signals were analyzed by nonparametric Mann-Whitney test. For serum markers analysis, in the absence of internationally available cut off values for markers, the cut off values maximally discriminating between patients with no evidence of disease and BM patients were identified using receiver operating characteristic (ROC) curve analysis. Sensitivity and specificity were calculated and their statistical significance were analyzed by chi-square test comparing BM patients versus patients with no evidence of disease or versus healthy donors. The correlation between serum levels of the markers was assessed with the Spearman rank test. The diagnostic relevance of the combinations NTX-mir16 and NTX-sICAM1 considered as continuous variables (subjected to natural logarithmic transformation) was analyzed by the logistic regression model. The linear predictor or logit resulting from the model was used as a new diagnostic test on which the ROC curve was calculated (Flamini et al., 2006). Statistical analysis was done using SPSS software. All other comparisons were analyzed by unpaired, two-sided, independent Student’s t test without equal variance assumption, unless otherwise described in figure legends.
Supplementary Material
SIGNIFICANCE.
The activation and recruitment of osteoclasts is of vital importance for the development of osteolytic bone metastasis. Significant advances have been made in understanding tumor-stromal interactions that foster hyperactive osteoclast differentiation. However, the role of miRNAs in osteoclastogenesis during bone metastasis remains poorly understood. We show that tumor-conditioned media from multiple cancer types activates a consensus miRNA response signature in osteoclasts. This signature includes up-regulated miRNAs that correlate with bone metastasis burden and show potential as clinical biomarkers. Importantly, functional studies reveal impaired osteoclast development, and reduced bone metastasis burden, after ectopic expression of down-regulated miR-141 and miR-219. These findings establish miRNAs as essential regulators of osteoclastogenesis, and potential therapeutic targets and biomarkers during bone metastasis.
HIGHLIGHTS.
Consistent miRNA changes in physiological and pathological osteoclastogenesis
MiR-141 and miR-219 inhibit osteoclastogenesis by targeting osteoclast genes
sICAM1 enhances osteoclastogenesis via β2 intergrin-mediated NFκB signaling
Elevated serum miR-16, miR-378, and sICAM1 levels correlate with bone metastasis
Acknowledgments
We thank C. DeCoste for assistance with flow cytometry, E. Williams, F. Miller, and R. L. Anderson for the TSU-PR1, 4T1 cell lines series. This research was supported by grants from the Komen for the Cure (KG110464), Department of Defense (BC123187), Brewster Foundation, the National Institutes of Health (R01CA134519 and R01CA141062), and the Champalimaud Foundation to Y.K., and European Research Council (Advanced Investigator Grant “DISSECT”, No. 269081), Deutsche Forschungsgemeinschaft, and German Minister of Education and Research (BMBF) to K.P. This research was also supported by the Preclinical Imaging and Flow Cytometry Shared Resources of the Cancer Institute of New Jersey (P30CA072720).
Footnotes
ACCESSION NUMBERS
The raw and normalized microarray data have been deposited in the Gene Expression Ominbus (GEO) database under accession number GSE44936.
Supplemental Information including Supplemental Experimental Procedures, eight Supplemental Figures, and 2 Supplemental Tables can be found with this article online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco MA, LeRoy G, Khan Z, Aleckovic M, Zee BM, Garcia BA, Kang Y. Global secretome analysis identifies novel mediators of bone metastasis. Cell research. 2012;22:1339–1355. doi: 10.1038/cr.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation. Nature. 2003;423:337–342. doi: 10.1038/nature01658. [DOI] [PubMed] [Google Scholar]
- Carlos TM, Harlan JM. Leukocyte-endothelial adhesion molecules. Blood. 1994;84:2068–2101. [PubMed] [Google Scholar]
- Chaffer CL, Dopheide B, McCulloch DR, Lee AB, Moseley JM, Thompson EW, Williams ED. Upregulated MT1-MMP/TIMP-2 axis in the TSU-Pr1-B1/B2 model of metastatic progression in transitional cell carcinoma of the bladder. Clin Exp Metastasis. 2005;22:115–125. doi: 10.1007/s10585-005-5141-3. [DOI] [PubMed] [Google Scholar]
- Clezardin P. Therapeutic targets for bone metastases in breast cancer. Breast Cancer Res. 2011;13:207. doi: 10.1186/bcr2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey RA, Turner AG, McManus JF, Chiu WS, Tjahyono F, Moore AJ, Atkins GJ, Anderson PH, Ma C, Glatt V, et al. Calcitonin receptor plays a physiological role to protect against hypercalcemia in mice. J Bone Miner Res. 2008;23:1182–1193. doi: 10.1359/JBMR.080310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ell B, Kang Y. SnapShot: Bone Metastasis. Cell. 2012;151:690–690. e691. doi: 10.1016/j.cell.2012.10.005. [DOI] [PubMed] [Google Scholar]
- Flamini E, Mercatali L, Nanni O, Calistri D, Nunziatini R, Zoli W, Rosetti P, Gardini N, Lattuneddu A, Verdecchia GM, Amadori D. Free DNA and carcinoembryonic antigen serum levels: an important combination for diagnosis of colorectal cancer. Clin Cancer Res. 2006;12:6985–6988. doi: 10.1158/1078-0432.CCR-06-1931. [DOI] [PubMed] [Google Scholar]
- Gonzalo P, Guadamillas MC, Hernandez-Riquer MV, Pollan A, Grande-Garcia A, Bartolome RA, Vasanji A, Ambrogio C, Chiarle R, Teixido J, et al. MT1-MMP is required for myeloid cell fusion via regulation of Rac1 signaling. Dev Cell. 2010;18:77–89. doi: 10.1016/j.devcel.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H, Kukita T, Kukita A, Iwamoto Y, Iijima T. Involvement of lymphocyte function-associated antigen-1 and intercellular adhesion molecule-1 in osteoclastogenesis: a possible role in direct interaction between osteoclast precursors. Endocrinology. 1998;139:3967–3975. doi: 10.1210/endo.139.9.6171. [DOI] [PubMed] [Google Scholar]
- Hemingway F, Taylor R, Knowles HJ, Athanasou NA. RANKL-independent human osteoclast formation with APRIL, BAFF, NGF, IGF I and IGF II. Bone. 2011;48:938–944. doi: 10.1016/j.bone.2010.12.023. [DOI] [PubMed] [Google Scholar]
- Hershey CL, Fisher DE. Mitf and Tfe3: members of a b-HLH-ZIP transcription factor family essential for osteoclast development and function. Bone. 2004;34:689–696. doi: 10.1016/j.bone.2003.08.014. [DOI] [PubMed] [Google Scholar]
- Hodgkinson CA, Moore KJ, Nakayama A, Steingrimsson E, Copeland NG, Jenkins NA, Arnheiter H. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell. 1993;74:395–404. doi: 10.1016/0092-8674(93)90429-t. [DOI] [PubMed] [Google Scholar]
- Holmbeck K, Bianco P, Caterina J, Yamada S, Kromer M, Kuznetsov SA, Mankani M, Robey PG, Poole AR, Pidoux I, et al. MT1-MMP-deficient mice develop dwarfism, osteopenia, arthritis, and connective tissue disease due to inadequate collagen turnover. Cell. 1999;99:81–92. doi: 10.1016/s0092-8674(00)80064-1. [DOI] [PubMed] [Google Scholar]
- Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, Guise TA, Massague J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- Kettritz R, Choi M, Rolle S, Wellner M, Luft FC. Integrins and cytokines activate nuclear transcription factor-kappaB in human neutrophils. J Biol Chem. 2004;279:2657–2665. doi: 10.1074/jbc.M309778200. [DOI] [PubMed] [Google Scholar]
- Kim CH, Lee KH, Lee CT, Kim YW, Han SK, Shim YS, Yoo CG. Aggregation of beta2 integrins activates human neutrophils through the IkappaB/NF-kappaB pathway. J Leukoc Biol. 2004;75:286–292. doi: 10.1189/jlb.0103038. [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Korpal M, Yan J, Lu X, Xu S, Lerit DA, Kang Y. Imaging transforming growth factor-beta signaling dynamics and therapeutic response in breast cancer bone metastasis. Nat Med. 2009;15:960–966. doi: 10.1038/nm.1943. [DOI] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Kurachi T, Morita I, Murota S. Involvement of adhesion molecules LFA-1 and ICAM-1 in osteoclast development. Biochim Biophys Acta. 1993;1178:259–266. doi: 10.1016/0167-4889(93)90202-z. [DOI] [PubMed] [Google Scholar]
- Le XF, Merchant O, Bast RC, Calin GA. The Roles of MicroRNAs in the Cancer Invasion-Metastasis Cascade. Cancer microenvironment : official journal of the International Cancer Microenvironment Society. 2010;3:137–147. doi: 10.1007/s12307-010-0037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelekakis M, Moseley JM, Martin TJ, Hards D, Williams E, Ho P, Lowen D, Javni J, Miller FR, Slavin J, Anderson RL. A novel orthotopic model of breast cancer metastasis to bone. Clin Exp Metastasis. 1999;17:163–170. doi: 10.1023/a:1006689719505. [DOI] [PubMed] [Google Scholar]
- Lomaga MA, Yeh WC, Sarosi I, Duncan GS, Furlonger C, Ho A, Morony S, Capparelli C, Van G, Kaufman S, et al. TRAF6 deficiency results in osteopetrosis and defective interleukin-1, CD40, and LPS signaling. Genes Dev. 1999;13:1015–1024. doi: 10.1101/gad.13.8.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Mu E, Wei Y, Riethdorf S, Yang Q, Yuan M, Yan J, Hua Y, Tiede BJ, Haffty BG, et al. VCAM-1 promotes osteolytic expansion of indolent bone micrometastasis of breast cancer by engaging alpha4beta1-positive osteoclast progenitors. Cancer Cell. 2011;20:701–714. doi: 10.1016/j.ccr.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M, Barad O, Agami R, Geiger B, Hornstein E. miRNA-based mechanism for the commitment of multipotent progenitors to a single cellular fate. Proc Natl Acad Sci USA. 2010 doi: 10.1073/pnas.0915022107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi F, Izu Y, Hayata T, Hemmi H, Nakashima K, Nakamura T, Kato S, Miyasaka N, Ezura Y, Noda M. Osteoclast-specific Dicer gene deficiency suppresses osteoclastic bone resorption. J Cell Biochem. 2010;109:866–875. doi: 10.1002/jcb.22228. [DOI] [PubMed] [Google Scholar]
- Nakano K, Okada Y, Saito K, Tanaka Y. Induction of RANKL expression and osteoclast maturation by the binding of fibroblast growth factor 2 to heparan sulfate proteoglycan on rheumatoid synovial fibroblasts. Arthritis Rheum. 2004;50:2450–2458. doi: 10.1002/art.20367. [DOI] [PubMed] [Google Scholar]
- Quattrocchi CC, Dell’Aia P, Errante Y, Occhicone F, Longo D, Virzì V, Tonini G, Napoli N, Santini D, Zobel BB. Differential effect of zoledronic acid on normal trabecular and cortical bone density in oncologic patients with bone metastases. J Bone Oncology. 2012;1:24–29. doi: 10.1016/j.jbo.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi N, Dai X, Winter CG, Kang Y. Tumor-derived JAGGED1 promotes osteolytic bone metastasis of breast cancer by engaging notch signaling in bone cells. Cancer Cell. 2011;19:192–205. doi: 10.1016/j.ccr.2010.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SM, Bronisz A, Hu R, Patel K, Mansky KC, Sif S, Ostrowski MC. MITF and PU.1 recruit p38 MAPK and NFATc1 to target genes during osteoclast differentiation. J Biol Chem. 2007;282:15921–15929. doi: 10.1074/jbc.M609723200. [DOI] [PubMed] [Google Scholar]
- Sugatani T, Hruska KA. MicroRNA-223 is a key factor in osteoclast differentiation. J Cell Biochem. 2007;101:996–999. doi: 10.1002/jcb.21335. [DOI] [PubMed] [Google Scholar]
- Sugatani T, Hruska KA. Impaired micro-RNA pathways diminish osteoclast differentiation and function. J Biol Chem. 2009;284:4667–4678. doi: 10.1074/jbc.M805777200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugatani T, Vacher J, Hruska KA. A microRNA expression signature of osteoclastogenesis. Blood. 2011 doi: 10.1182/blood-2010-10-311415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teitelbaum SL, Ross FP. Genetic regulation of osteoclast development and function. Nat Rev Genet. 2003;4:638–649. doi: 10.1038/nrg1122. [DOI] [PubMed] [Google Scholar]
- Weilbaecher KN, Guise TA, McCauley LK. Cancer to bone: a fatal attraction. Nat Rev Cancer. 2011;11:411–425. doi: 10.1038/nrc3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weilbaecher KN, Motyckova G, Huber WE, Takemoto CM, Hemesath TJ, Xu Y, Hershey CL, Dowland NR, Wells AG, Fisher DE. Linkage of M-CSF signaling to Mitf, TFE3, and the osteoclast defect in Mitf(mi/mi) mice. Mol Cell. 2001;8:749–758. doi: 10.1016/s1097-2765(01)00360-4. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhao H, Chen J, Xia B, Jin Y, Wei W, Shen J, Huang Y. Interferon-beta-induced miR-155 inhibits osteoclast differentiation by targeting SOCS1 and MITF. FEBS Lett. 2012;586:3255–3262. doi: 10.1016/j.febslet.2012.06.047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.