Abstract
Metagenomic analysis of colonic mucosa-associated microbes has been complicated by technical challenges that disrupt or alter community structure and function. In the present study, we determined the feasibility of laser capture microdissection (LCM) of intact regional human colonic mucosa-associated microbes followed by phi29 multiple displacement amplification (MDA) and massively parallel sequencing for metagenomic analysis. Samples were obtained from the healthy human subject without bowel preparation and frozen sections immediately prepared. Regional mucosa-associated microbes were successfully dissected using LCM with minimal contamination by host cells, their DNA extracted and subjected to phi29 MDA with a high fidelity, prior to shotgun sequencing using the GS-FLX DNA sequencer. Metagenomic analysis of approximately 67 million base pairs of DNA sequences from two samples revealed that the metabolic functional profiles in mucosa-associated microbes were as diverse as those reported in feces, specifically the representation of functional genes associated with carbohydrate, protein, and nucleic acid utilization. In summary, these studies demonstrate the feasibility of the approach to study the structure and metagenomic profiles of human intestinal mucosa-associated microbial communities at small spatial scales.
Keywords: Laser capture microdissection, Metagenomics, Mucosa-associated microbes, Multiple displacement amplification, Pyrosequencing, Host-microbe interactions
Introduction
Gut mucosa-associated microbes are defined as the microbial community that reside in the colonic epithelial cell surface and constitute a biofilm with a polysaccharide-rich mucus gel layer (Bollinger et al. 2007; Sonnenburg et al. 2004). Mucosa-associated microbes are in intimate contact with the host, affecting pivotal functions such as immune activation, epithelial growth and development, and mucus production (Atuma et al. 2001; Sonnenburg et al. 2004). Perturbations in the gut microbiota may be associated with several human diseases, particularly inflammatory bowel diseases which often affect mucosal inflammation in a regional as well as a spatially dependent manner (Swidsinski et al. 2005, 2002).
For many years, the ability to study the human enteric microbiome was restricted to cultivation-based technologies, which was problematic since more than 80% of the estimated species present cannot be readily cultivated (Eckburg et al. 2005). Application of culture-independent molecular methods, based on retrieval of 16S rRNA-encoding gene sequences, allows a more comprehensive phylogenetically derived definition of mucosa-associated microbial populations to be possible (Schmidt and Relman 1994). These methods generally involve amplification of 16S rRNA-encoding genes by PCR, followed by either clone library construction and DNA sequencing, the generation of community “fingerprints” by terminal restriction fragment length polymorphism (T-RFLP) (Sakamoto et al. 2003), or denaturing gradient gel electrophoresis (Green et al. 2006). However, these 16S rRNA gene-based approaches provide little functional information.
Recently, massively parallel high-throughput sequencing that circumvents the need for gene-specific PCR amplification or cloning has been applied to sequence whole microbial-community DNA. This information can be used to assess both the taxonomic and functional diversity of a microbial sample. To date, massively parallel high-throughput sequencing of microbial metagenomes has been used to describe the enormous functional diversity of environmental samples such as soil and marine samples, as well as human stool (Gill et al. 2006; Tringe et al. 2005). However, the application of DNA sequencing technologies to the study of mucosa-associated enteric microorganisms has been hampered by numerous challenges: (1) significant contamination by host DNA that confounds interpretation of the data, especially when short sequence reads are obtained; (2) methods that mechanically separate mucosa-associated microbes from mucosal samples significantly disrupt the structure and composition of microbial communities; (3) acquisition of adequate amounts of microbial DNA for the sequencing of the metagenome through pyrosequencing (which requires microgram quantities).
In this study, we developed a system by laser capture microdissection (LCM) to cleanly separate intact and representative communities of mucosa-associated microbes from freshly frozen human biopsy samples obtained from a healthy individual not subjected to any colon preparation solutions. Genomic DNA was then extracted and amplified by phi29 multiple displacement amplification to yield sufficient quantities for metagenomic analysis by using pyrosequencing with the GS-FLX instrument (454 Life Sciences—Roche Applied Science). The sequencing data was then analyzed based on the annotation produced by the Rapid Annotation using Subsystems Technology for meta-genomes (MG-RAST) system (Meyer et al. 2008). The MG-RAST-based analysis revealed significant metabolic functions pertaining to the microbial metabolisms of carbohydrate, protein, and nucleic acid. Potential differences in the carbohydrate utilization profiles of the mucosa-associated microbes between the left and right colon were also observed that may be relevant to regional difference in host physiology and function.
Materials and methods
Biopsy specimen collection
The biopsy specimens for LCM were taken from the ascending and descending colons of a healthy individual using a standard colonoscopy procedure with no bowel preparation. This individual had not used antibiotics or any other medications for 6 months prior to specimen collection. The fresh tissues were embedded in OCT compound (Sakura Finetechnical, Tokyo, Japan) immediately, snap frozen, and stored at −80°C until use.
Laser capture microdissection and DNA extraction
Frozen tissue sections (8 um thick) were cut on a cryostat, CM 1900 microtome (Leica, Milton Keynes, UK). Sections were thawed and mounted on a MembraneSlide (PEN-Membrane 2.0 um, Leica), and then fixed at room temperature in 95% ethanol for 1 min. A quick double-staining protocol was performed by the following procedure to show the mucus and epithelial cells. Sections were immersed in Alcian blue solution (Sigma–Aldrich, Inc. St. Louis, MO) for 1 min at room temperature, washed with tap water, and then immersed in 0.1% nuclear fast red solution (Sigma–Aldrich, Inc. St. Louis, MO) for 10 s. The sections were then air dried. Laser microdissection (LMD) was performed using a Leica AS LMD system to harvest the mucus layer.
After microdissection, the microdissected samples were carefully collected into a PCR tube. DNA used for the downstream phi29 multiple displacement amplification and metagenomic analysis was extracted by using QIAamp DNA micro kit (Qiagen Inc. Valencia, CA) according to the manufacturer’s instructions. DNA extraction for biopsy tissues which was used for amplification of 16S rRNA gene was performed as described previously (Wang et al. 2009). Briefly, samples were lysed in 1 ml extraction buffer [50 mM Tris (pH 7.4), 100 mM EDTA (pH 8.0), 400 mM NaCl, 0.5% SDS] containing 20 ul proteinase K (20 mg/ml) with overnight water bath incubation at 55°C. After overnight digestion, DNA was then extracted with an equal volume of phenol:chloroform:isoamyl alcohol, and precipitated with ethanol. Isolated DNA was dissolved in TE buffer and stored in −80°C. DNA concentration was measured by using the Quant-iT double-stranded-DNA HS assay kit and the Qubit fluorometer (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions.
phi29 multiple displacement amplification
Laser capture microdissected DNA used for metagenomic analysis was amplified using Genomiphi V2 DNA amplification kit (GE Healthcare, UK) according to the manufacturer’s instructions. Briefly, different amounts (1–10 ng) of template DNA were added to a reaction volume of 50 μl. Multiple displacement amplification (MDA) was carried out during several different durations of time (1, 2, 3, and 4 h). Amplified material from the different time points were verified by agarose gel electrophoresis of aliquots from amplified reactions (1 μl) in 1.0% agarose. After optimized amplification, DNA was further purified by ethanol precipitation.
16S rRNA gene PCR and T-RFLP
Fidelity of phi29 multiple displacement amplification reaction was assessed by 16S rRNA gene PCR and T-RFLP. 16S rDNA was amplified from DNA samples using universal primers 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) labeled with 6′ carboxyfluorescein (6-FAM) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) as described previously (Wang et al. 2009). PCR products were verified for size by agarose gel electrophoresis, precipitated with ethanol, and resuspended in TE buffer. Aliquots of the precipitated 16S PCR reactions were digested using MspI (New England Biolabs). Samples were desalted using cellulose membranes on water, then mixed with size marker to load on the Applied Biosystems DNA sequencer 3130 and analyzed on the genescan mode. For T-RFLP data, fragment length and abundance were determined using GeneMapper software (Applied Biosystems) enhanced with semi-automatic quality control tools for peak calling developed in our Systems Biology Institute. Briefly, terminal restriction fragment (TRF) data generated by GeneMapper were binned and filtered by the method developed by Abdo et al. (2006). Based on the normalized T-RFLP profile, the number and height of peaks were treated as number and abundance of bacterial phylotypes represented in samples. Calculation of the overall TRF richness and evenness indices were calculated as described previously (Wang et al. 2009). Pairwise Bray–Curtis distances were calculated (Michie 1982) and the relationship between communities was examined by the construction of dendrograms using the Molecular Evolutionary Genetics Analysis package (MEGA) (http://www.megasoftware.net) (Kumar et al. 2008).
16S rRNA gene library cloning and sequencing
Unlabeled PCR primers 8F and 1492R were used to amplify 16S rRNA gene sequences from the samples using the same protocol as those for T-RFLP analysis. PCR products were purified by QIAquick gel extraction kit (Qiagen, Valencia, CA) and cloned into pCR-2.1-TOPO® vectors (Invitrogen, Carlsbad, CA) using the TOPO-TA Cloning Kit for sequencing according to the manufacturer’s instructions. From each library, 200 clones were picked randomly and processed for sequencing. Plasmid inserts were sequenced in one direction by using 8F as the primer.
Analysis of 16S rRNA gene sequences was performed as described previously (Wang et al. 2009). Raw DNA sequence chromatogram files were uploaded to the Ribosomal Database Project II (RDP-II) website at http://rdp.cme.msu.edu/ (Cole et al. 2008). Automated base calling, vector removal, quality trimming, and sequence alignment were performed via myRDP (available through the RDP website). The RDP Classifier tool was used to assign 16S rRNA sequences to the ROP taxonomic hierarchy. Distance matrices were downloaded from the RDP and the program DOTUR used to assign sequences to operational taxonomical units (OTUs or phylotypes) and perform diversity analysis. Dendrograms were constructed by the neighbor-joining distance matrix method in the Clustal X program (Thompson et al. 1997) with 100 bootstrap replicates and displayed using MEGA. For Unifrac analysis, all 16S rRNA gene sequences were imported into the ARB software package and aligned into a phylogenetic tree with sequences most similar to each other in closest proximity and connected by branch points indicating which sequences were phylogenetically related. This tree was annotated to indicate the sample origin for each sequence and used to measure the difference between bacterial communities in samples using the online version of UniFrac (http://bmf2.colorado.edu/unifrac/index.psp) (Lozupone et al. 2006). All 16S rRNA gene sequences in this study were deposited in GenBank under the accession numbers HQ236723-HQ237454.
Sequencing and annotation of mucosa-associated microbial metagenomes
DNA samples from phi29 amplification were precipitated using ethanol. Shotgun libraries were then prepared as previously described for sequencing using the GS-FLX instrument (454 Life Sciences—Roche Applied Science) (Pfister et al. 2010). Annotation of the sequence data was performed with MG-RAST as described in Meyer et al. (2008). The MG-RAST system is a high-throughput analysis pipeline based on the SEED framework for comparative genomics. The pipeline produces automated functional assignments of sequences present in the metagenome-derived dataset by comparing both protein and nucleotide databases. Both phylogenetic and functional summaries are generated, and tools for comparing multiple datasets can simultaneously be viewed. Metagenomic sequencing data for this study were deposited in MG-RAST under the links http://mg-rast.mcs.anl.gov/linkin.cgi?metagenome=4441655.3 for right colon and http://mg-rast.mcs.anl.gov/linkin.cgi?metagenome=4441648.3 for left colon.
Results
Laser capture microdissection and DNA extraction of human mucosa-associated microbiota
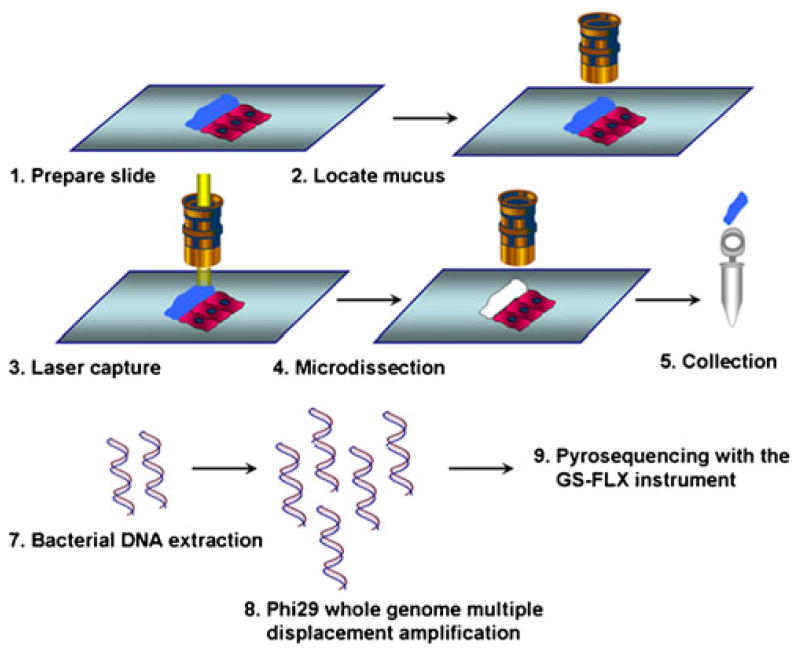
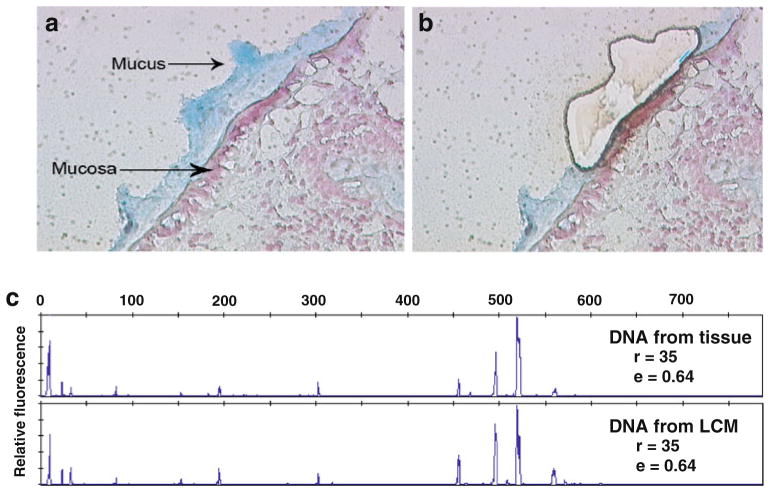
The strategy used in this study to analyze metagenomic information from mucosa-associated microbes was outlined in Fig. 1. Human biopsy samples were collected via standard pinch biopsy forceps from the colons of one healthy human individual who underwent colonoscopy without any lavage preparation. This protocol was approved by the Institutional Review Board. Specimens were then immediately embedded in OCT. Using a rapid double-staining protocol with Alcian blue and nuclear fast red, tissues samples were frozen (no fixation was used), sectioned on a membrane slide, and visualized under a microscope. As shown in Fig. 2a, the mucus layer with mucosa-associated bacteria was well preserved (blue) and distinct from human mucosal cells (pink). Using the Leica AS laser microdissection instrument, the mucus layer and its associated microbes were then harvested by microdissection (Fig. 2b). The advantage of LCM is its ability to separate intact mucosa-associated microbial communities cleanly from host eukaryotic cells. As illustrated in Electronic supplementary Fig. 1, a host epithelial cell sluffed into the mucus layer was removed by LCM before acquisition of the microbial community. LCM therefore provides a viable approach for clean separation of microbial communities with minimal contamination by host eukaryotic DNA, an essential pre-condition for metagenomic analysis.
Fig. 1.
Procedure for the study of mucosa-associated microbial communities using metagenomic methods
Fig. 2.
Laser capture microdissection (LCM) to harvest mucosa-associated microbes (×400). a By Alcian blue and nuclear fast red staining, the mucus layer (blue) is clearly different from mucosal epithelial cells (pink). b Mucosa-associated microbes are separated by LCM. c 16S rRNA gene PCR and T-RFLP analysis shows that representative microbes are collected by LCM from human colon mucosa. Richness (r) and evenness (e) were calculated based on the number and height of peaks in each profile
After LCM, the relative yield of bacterial DNA from LCM sections was evaluated. Approximately 3 ng of microbial DNA (measured by Qubit quantitation platform) can be harvested from a combination of ten 0.8 um thick LCM mucosal sections (all the blue staining areas in each section were dissected and collected; Fig. 1). After DNA extraction, phylogenetic profiles of the microdissected microbial-community DNA were assessed by PCR amplification of the 16S rRNA gene and T-RFLP analysis. The T-RFLP approach is highly reproducible and showed that the same profiles can be obtained from the different reactions of an environmental DNA sample (Electronic supplementary Fig. 2). Using this technique, similar profiles were observed from LCM-derived DNA (which was obtained by dissecting half of the biopsy tissue) and DNA extracted directly from the other half. As shown in Fig. 2c, the richness (the number of distinct peaks in each sample, (r)) and evenness (a measure of the distribution of the peak area through the T-RFLP profile (e)) of the T-RFLP profiles, were used to compare quantitative differences between samples. The T-RFLP profiles representing tissue-derived DNA (r=35, e=0.64) were the same as those calculated from the LCM DNA profiles (r=35, e=0.64). These data support for the application of the LCM approach for harvesting representative microbial populations from colonic biopsies without significant distortion of mucosa-associated microbial community.
Fidelity of phi29 polymerase multiple displacement amplification
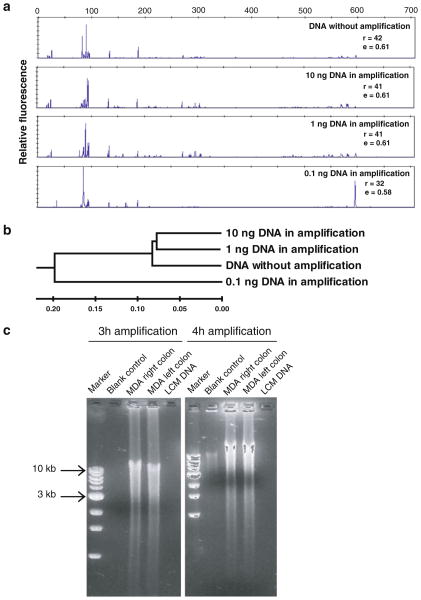
To determine the fidelity and optimal conditions for linear amplification of metagenomic DNA from the LCM samples using phi29 multiple displacement amplification, human stool-derived genomic DNA samples were used as the DNA templates. We examined the DNA concentration required to maintain an even amplification of the templates present in the stool-derived DNA. A dilution series of stool-derived DNA (0.1–10 ng) were added to the reactions and amplified by phi29 MDA for the same length of time. 16S rRNA-encoding genes from different MDA reactions were amplified by PCR and analyzed to test for amplification bias. As shown in Fig. 3a, high fidelity of MDA amplification from increasing amounts of stool-derived DNA was observed with T-RFLP analysis. When more than 1 ng DNA template was used in the reaction, 16S rRNA genes from different species were amplified evenly. Similar patterns of T-RFLP profiles were observed for products collected from phi29 MDA reactions using 10 and 1 ng DNA template as compared with DNA that had not been amplified with phi29. However, skewed T-RFLP profiles were observed when less than 0.1 ng DNA template was used in the phi29 MDA reaction (Fig. 3a). To quantitatively show the similarity between reactions based on peak composition and relative abundance, the T-RFLP profiles were further analyzed by the calculation of pairwise Bray–Curtis distances, then displayed as a dendrogram. As shown in Fig. 3b, the phi29 MDA reactions using 10 and 1 ng DNA template were grouped with DNA without phi29 amplification within a distance of 0.07. However, reaction using 0.1 ng DNA template could only be clustered with the other three reactions at the distance of 0.19. This indicates skewing of amplicons at lower template input amounts.
Fig. 3.
Optimization of phi29 multiple displacement amplification (MDA) with complex DNA templates. a 16S rRNA gene PCR and T-RFLP analysis of fidelity of MDA with the different amount of DNA templates. High fidelity of MDA is observed in the reactions using more than 1 ng DNA as template. Richness (r) and evenness (e) were calculated to show the differences between samples b Dendrogram based on similarity analysis of T-RFLP profiles shows the relationships among samples. The scale bar shows the distance of similarity. c Optimization of Phi29 MDA time on laser capture microdissected DNA samples. Within 3-h amplification, microgram quantity DNA is harvested with minimum endogenous background amplification
Since endogenous background amplification could be induced in phi29 MDA by prolonged incubation of the reaction (Zhang et al. 2006), we further optimized the amplification time without compromising yields. Sufficient template DNA was thus generated for downstream metagenomic-based analysis with the minimal incubation time for the MDA reaction. As shown in Fig. 3c, approximately 50 μg of DNA was readily obtained during a 3 h period with phi29 MDA, starting from approximately 1 ng DNA templates obtained through LCM. Under the same conditions, no amplified DNA could be detected in blank controls by gel electrophoresis (i.e., no endogenous background amplification, Fig. 3c).
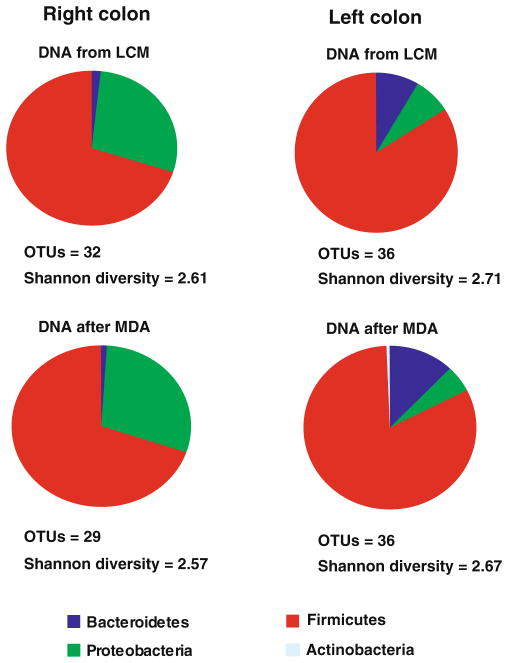
With optimized MDA conditions, the fidelity of the phi29-mediated amplification of DNA from the micro-dissected human mucosa-associated microbial community was further tested by 16S rRNA gene-based library cloning and sequencing. Phi29-based amplification yielded a similar phylogenetic composition and diversity of the sample as judged by comparison of the 16S rRNA gene sequences from the non-amplified LCM DNA and phi29-amplified DNA. As shown in Fig. 4, consistent bacterial compositions were detected in phi29-amplified DNA (3 ng) samples as those detected in two LCM DNA samples (3 ng) from human right and left colon. Based on the diversity analysis (Fig. 4), the number of observed OTUs and Shannon diversity were calculated and found to be similar between non-amplified and phi29-amplified DNA for both samples. To further demonstrate the fidelity of phi29 amplification, phylogenetic trees of all representative clones in the libraries from the non-amplified and phi29-amplified DNA were constructed. The phylogenetic tree indicates that for all the OTUs identified in the non-amplified DNA, the same phylotypes or their closely related sequences could also be found in the DNA after phi29 amplification for both left and right colon (Electronic supplementary Fig. 3). Moreover, no significant differences were found between two 16S rRNA gene libraries constructed before and after phi29 MDA for both left and right colon samples (right colon P=0.26; left colon P=0.06) by Unifrac analysis. These data demonstrate the feasibility of phi29-based DNA amplification with high fidelity for providing sufficient material for 454 pyrosequencing and metagenomic analysis. Unifrac analysis also showed that there was a significant difference of mucosa-associated microbes between human left and right colon (P=0.001). However, since only one pair of right and left colon samples were compared in this study, more samples need to be analyzed to more completely establish the differences.
Fig. 4.
Overview of fidelity of phi29 MDA on laser capture micro-dissected DNA samples using 16S rRNA gene library cloning and sequencing. Bacterial composition is compared before and after phi29 MDA, based on the alignment with RDP. Operational taxonomic units and Shannon diversity were calculated based on the 16S rRNA gene sequences at the 97% cutoff value
Sequencing and analysis of mucosa-associated microbial metagenome
Representative biopsy samples from the right and left colon of a healthy human volunteer were subjected to LCM and MDA using the optimized conditions described in the previous sections. Fifty micrograms of DNA from the right colon and 50 μg from the left colon were amplified from the respective mucosal sections, and adaptor libraries for pyrosequencing were prepared according to the manufacturer’s protocol (input of 5 μg). A total of 160,790 sequences (with an average read length of 219 bp) were recovered from the right colon sample and 141,721 from the left (230 bp average read length). These sequences were then annotated using the MG-RAST system, which assigned them to functional categories, including identification of 16S rRNA-encoding sequences.
In each sample, few sequences were identified to be encoding 16S rRNA (using an E value cutoff of 10−5, and a minimal 50 bp of alignment). The distribution in the right colon sample (total of 33 sequences; 0.02% of the total sequences) was dominated by Proteobacteria (75.76%), whereas for the left colon sample (total of eight sequences; 0.01% of the total) there was an even distribution by the members of the Proteobacteria and Actinobacteria (37.5% each) and the Firmicutes (25%). (The compositions described here were based on comparison with the Ribosomal Database Project; similar results were found in comparisons with both the Greengenes and SILVA databases, data not shown). These results appear more stochastic than the clone libraries, owing to the small sample size associated with the random nature of the shotgun sequencing (Pham et al. 2008).
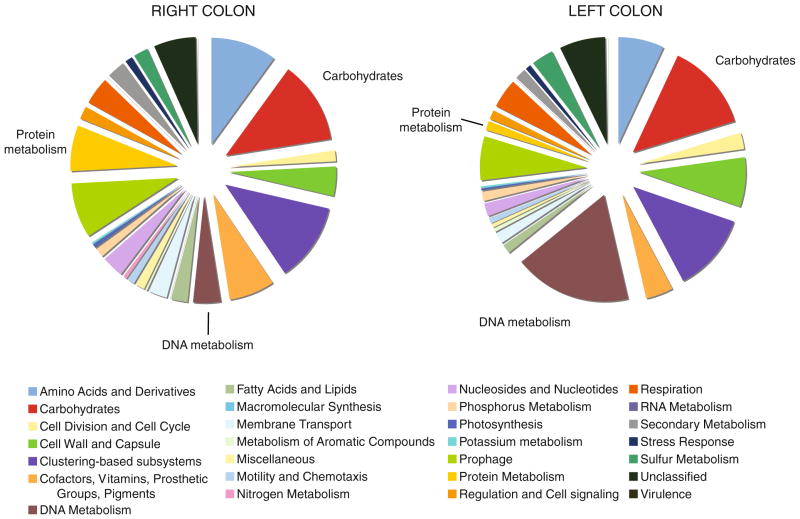
An overview of the metabolic subsystems was also performed on the mucosa-associated microbes (Fig. 5). At the broadest level of categorization, the largest proportion of sequences in the right colon sample was composed of sequences related to DNA metabolism (17.9% of all classified sequences, compared with only 4.2% in the left colon sample). All 102 of the sequences classified in this category for the right colon were assigned the functional role of DNA-binding protein HU, a histone-like DNA-binding protein found in the Eubacteria (Drlica and Rouviere-Yaniv 1987; Swinger and Rice 2004). Carbohydrate metabolism encoding sequences were detected in relatively equal amounts between both samples (13.5% of classified sequences in the right colon sample, and 12.4% in the left). This is not surprising, considering the elevated density of microorganisms residing in the colon relative to other portions of the gastrointestinal tract, and the opportunity for fermentation of a variety of substrates. However, once the stringency of the parameters for filtering of the data was relaxed to a maximum E value of 0.01, the distribution of the types of carbohydrate metabolism encoding sequences was largely different between the two samples. The distributions appear inverted for most of the classes (Electronic supplementary Fig. 4). The shift to sequences representing metabolism of di- and oligosaccharides in the left colon may be indicative of the release of simple sugars from complex carbohydrates in that region of the colon.
Fig. 5.
Distribution of classified metagenomic-derived sequences obtained from human colon samples. Genomic DNA was isolated from the mucosa of biopsy samples and subjected to phi29 polymerase multiple displacement amplification (MDA). Note the difference in representation of genes associated with DNA and protein metabolism between the samples obtained from the right colon relative to the left colon sample
Discussion
Numerous studies have shown that the mucosa-associated microbiota are significantly different from those in the luminal content of the human digestive tract and the former are thought to be pivotal in the maintenance of regional normal functions of colonic epithelial cells. Despite many technological advances in exploring the diversity and function of the microbiota within the human colon, there are still significant limitations in our ability to study the mucosa-associated enteric microbiome. Among such challenges are the practical issues of obtaining representative microbial samples and determining the function of microbes that are tightly attached to the colonic mucosal surface. The methods described here allow for the harvest of region-specific samples of mucosal associated microbes in their natural state from the human colon without lavage preparation, and demonstrates the potential for a more precise and comprehensive picture of the mucosa-associated microbiota.
LCM was first developed as a laser-based, microscope-aided method of tissue microdissection (Simone et al. 1998); LCM applications have significantly broadened the opportunities to study gene and protein expression of selected cell populations in situ. This method allows cells to be removed undistorted from their natural environments, with fewer artifacts from cell culture and bulk isolation protocols (Blatt and Srinivasan 2008). Although LCM has been used to harvest and analyze the mucosa-associated bacteria in human biopsy samples (Klitgaard et al. 2005; Molbak et al. 2006), two major limitations were observed in these studies. Firstly, biopsy samples collected to perform LCM were fixed by formalin, which could crosslink genomic DNA and bias the downstream PCR amplification. Secondly, whether LCM could harvest the representative microbial population adherent to one biopsy sample was not fully investigated. In the present study, there are two major technical achievements that are critical to the success of using LCM to separate microbes from host epithelial cells in situ: (1) freezing of sections to effectively preserve the mucus layer, the natural habitat of colonic microbes and microbial DNA. After comparing the thickness of the mucus layers between frozen section and Carnoy-fixed section in this study, we did not see a significant difference in the thickness of mucus layers between these two methods (data not shown); (2) a quick double-immunostaining protocol with alcian blue and nuclear fast red sensitive enough to distinguish the mucus layer and embedded microbes from underlying host mucosal epithelial cells. The advantages of LCM performed in this study include: (1) sampling is very specific and preserves the community diversity and dynamics; (2) samples can be easily retrieved from any part of the unprepared (i.e., no mechanical lavage solution) human colon; (3) LCM can cleanly separate microbial from host DNA, a confounding factor when performing metagenomic analysis.
Owing to its ability to amplify the whole genome of a single cell, MDA is a valuable technique to provide sufficient quantity of DNA for subsequent 454 sequencing reaction. We validated MDA from complex microbial DNA templates by using the 16S rRNA gene as a genome marker to demonstrate even amplification of genome sequences from mixed microbial DNA. Our data showed that the relative abundance of the 16S rRNA genes identified by PCR-based clone library sequencing from LCM-derived DNA was consistent with their patterns in the DNA sample of phi29 MDA. The 16S rRNA gene taxonomic classification of the clone library sequences were nearly the same from DNA samples before and after phi29 amplification. Although the degree of chromosomal amplification from different species is difficult to characterize using a single genome marker, our experimental protocol was optimized to minimize this bias at the minimum. For example, we determined that at least 1 ng DNA template is necessary to maintain consistency of phi29 amplification. Amplification contamination, a known confounding variable in studying a single cell genome, was also addressed in our study. We also demonstrated that endogenous background amplification could be avoided by limiting the amplification duration to less than 3 h when at least 1 ng of DNA template was used in the reaction.
The ability to harvest pure microbial populations and extract DNA from a LCM sample provides an opportunity to perform metagenomic analysis of mucosa-associated microbes at small spatial scales. Since the quantity of DNA available from these samples is only at the nanogram level, the phi29 MDA shows its utility in evenly amplifying the metagenome of a complex microbial DNA template to the amount required for pyrosequencing with the GS-FLX platform. Although there are still limitations in this system, such as the lack of an efficient tool to check the overall fidelity of MDA of microbial communities, LCM in combination with whole genome amplification open up new avenues for acquiring metagenomic-derived information from mucosa-associated microbes. It also advances DNA sequencing and metagenomic analysis of substantial numbers of previously uncharacterized members of the human mucosa-associated enteric microbiome. Moreover, this system could be further applied in future studies to explore the differences and functions of mucosa-associated microbes from both healthy and different disease states.
Supplementary Material
Acknowledgments
We thank Kevin White and the Institute for Genomics and Systems Biology at the University of Chicago for their support and technical assistance. We would like to acknowledge the following grant support: Digestive Disease Research Core Center (P30 DK42086; EBC), R21HG004858 (EBC), R01 HG004906 (VY), UH3 UH3DK083993 (VY), The Goldgraber fellowship foundation (LL), R01 5R01GM062344-11 (JA), and Research Fellowship Award (YW) from the Crohn’s and Colitis Foundation of America. We would also like to thank the Gastrointestinal Research Foundation of Chicago and Peter and Carol Goldman for supporting the microbiome research.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00253-010-2921-8) contains supplementary material, which is available to authorized users.
Contributor Information
Yunwei Wang, Department of Medicine, Knapp Center for Biomedical Discovery, University of Chicago, Rm 9031, 900 E. 57th Street, Chicago, IL 60637, USA.
Dionysios A. Antonopoulos, Department of Medicine, Knapp Center for Biomedical Discovery, University of Chicago, Rm 9031, 900 E. 57th Street, Chicago, IL 60637, USA. Institute for Genomics and Systems Biology, Argonne National Laboratory, 9700 S. Cass Avenue, Argonne, IL 60439, USA
Xiaorong Zhu, Department of Medicine, Knapp Center for Biomedical Discovery, University of Chicago, Rm 9031, 900 E. 57th Street, Chicago, IL 60637, USA.
Laura Harrell, Department of Medicine, Knapp Center for Biomedical Discovery, University of Chicago, Rm 9031, 900 E. 57th Street, Chicago, IL 60637, USA.
Ira Hanan, Department of Medicine, Knapp Center for Biomedical Discovery, University of Chicago, Rm 9031, 900 E. 57th Street, Chicago, IL 60637, USA.
John C. Alverdy, Department of Surgery, University of Chicago, 5841 S. Maryland Ave, Chicago, IL 60637, USA
Folker Meyer, Institute for Genomics and Systems Biology, Argonne National Laboratory, 9700 S. Cass Avenue, Argonne, IL 60439, USA.
Mark W. Musch, Department of Medicine, Knapp Center for Biomedical Discovery, University of Chicago, Rm 9031, 900 E. 57th Street, Chicago, IL 60637, USA
Vincent B. Young, Division of Infectious Diseases, University of Michigan, 4618D Med Sci II SPC 5623, 1150 W. Medical Center Dr, Ann Arbor, MI 48109, USA
Eugene B. Chang, Email: echang@medicine.bsd.uchicago.edu, Department of Medicine, Knapp Center for Biomedical Discovery, University of Chicago, Rm 9031, 900 E. 57th Street, Chicago, IL 60637, USA
References
- Abdo Z, Schuette UM, Bent SJ, Williams CJ, Forney LJ, Joyce P. Statistical methods for characterizing diversity of microbial communities by analysis of terminal restriction fragment length polymorphisms of 16S rRNA genes. Environ Microbiol. 2006;8:929–938. doi: 10.1111/j.1462-2920.2005.00959.x. [DOI] [PubMed] [Google Scholar]
- Atuma C, Strugala V, Allen A, Holm L. The adherent gastrointestinal mucus gel layer: thickness and physical state in vivo. Am J Physiol Gastrointest Liver Physiol. 2001;280:G922–G929. doi: 10.1152/ajpgi.2001.280.5.G922. [DOI] [PubMed] [Google Scholar]
- Blatt R, Srinivasan S. Defining disease with laser precision: laser capture microdissection in gastroenterology. Gastroenterology. 2008;135:364–369. doi: 10.1053/j.gastro.2008.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollinger RR, Barbas AS, Bush EL, Lin SS, Parker W. Biofilms in the normal human large bowel: fact rather than fiction. Gut. 2007;56:1481–1482. [PMC free article] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2008;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drlica K, Rouviere-Yaniv J. Histonelike proteins of bacteria. Microbiol Rev. 1987;51:301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green GL, Brostoff J, Hudspith B, Michael M, Mylonaki M, Rayment N, Staines N, Sanderson J, Rampton DS, Bruce KD. Molecular characterization of the bacteria adherent to human colorectal mucosa. J Appl Microbiol. 2006;100:460–469. doi: 10.1111/j.1365-2672.2005.02783.x. [DOI] [PubMed] [Google Scholar]
- Klitgaard K, Molbak L, Jensen TK, Lindboe CF, Boye M. Laser capture microdissection of bacterial cells targeted by fluorescence in situ hybridization. Biotechniques. 2005;39:864–868. doi: 10.2144/000112024. [DOI] [PubMed] [Google Scholar]
- Kumar S, Nei M, Dudley J, Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Hamady M, Knight R. UniFrac–an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinform. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F, Paarmann D, D’Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, Wilkening J, Edwards RA. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinform. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michie MG. Use of the Bray-Curtis similarity measure in cluster analysis of foraminiferal data. Math Geol. 1982;14:661–667. [Google Scholar]
- Molbak L, Klitgaard K, Jensen TK, Fossi M, Boye M. Identification of a novel, invasive, not-yet-cultivated Treponema sp. in the large intestine of pigs by PCR amplification of the 16S rRNA gene. J Clin Microbiol. 2006;44:4537–4540. doi: 10.1128/JCM.01537-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister CA, Meyer F, Antonopoulos DA. Metagenomic profiling of a microbial assemblage associated with the california mussel: a node in networks of carbon and nitrogen cycling. PLoS ONE. 2010;5:e10518. doi: 10.1371/journal.pone.0010518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham VD, Konstantinidis KT, Palden T, DeLong EF. Phylogenetic analyses of ribosomal DNA-containing bacterioplankton genome fragments from a 4000 m vertical profile in the North Pacific Subtropical Gyre. Environ Microbiol. 2008;10:2313–2330. doi: 10.1111/j.1462-2920.2008.01657.x. [DOI] [PubMed] [Google Scholar]
- Sakamoto M, Hayashi H, Benno Y. Terminal restriction fragment length polymorphism analysis for human fecal micro-biota and its application for analysis of complex bifidobacterial communities. Microbiol Immunol. 2003;47:133–142. doi: 10.1111/j.1348-0421.2003.tb02796.x. [DOI] [PubMed] [Google Scholar]
- Schmidt TM, Relman DA. Phylogenetic identification of uncultured pathogens using ribosomal RNA sequences. Meth Enzymol. 1994;235:205–222. doi: 10.1016/0076-6879(94)35142-2. [DOI] [PubMed] [Google Scholar]
- Simone NL, Bonner RF, Gillespie JW, Emmert-Buck MR, Liotta LA. Laser-capture microdissection: opening the microscopic frontier to molecular analysis. Trends Genet. 1998;14:272–276. doi: 10.1016/s0168-9525(98)01489-9. [DOI] [PubMed] [Google Scholar]
- Sonnenburg JL, Angenent LT, Gordon JI. Getting a grip on things: how do communities of bacterial symbionts become established in our intestine? Nat Immunol. 2004;5:569–573. doi: 10.1038/ni1079. [DOI] [PubMed] [Google Scholar]
- Swidsinski A, Ladhoff A, Pernthaler A, Swidsinski S, Loening-Baucke V, Ortner M, Weber J, Hoffmann U, Schreiber S, Dietel M, Lochs H. Mucosal flora in inflammatory bowel disease. Gastroenterology. 2002;122:44–54. doi: 10.1053/gast.2002.30294. [DOI] [PubMed] [Google Scholar]
- Swidsinski A, Weber J, Loening-Baucke V, Hale LP, Lochs H. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J Clin Microbiol. 2005;43:3380–3389. doi: 10.1128/JCM.43.7.3380-3389.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinger KK, Rice PA. IHF and HU: flexible architects of bent DNA. Curr Opin Struct Biol. 2004;14:28–35. doi: 10.1016/j.sbi.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW, Podar M, Short JM, Mathur EJ, Detter JC, Bork P, Hugenholtz P, Rubin EM. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- Wang Y, Hoenig JD, Malin KJ, Qamar S, Petrof EO, Sun J, Antonopoulos DA, Chang EB, Claud EC. 16S rRNA gene-based analysis of fecal microbiota from preterm infants with and without necrotizing enterocolitis. ISME J. 2009;3:944–954. doi: 10.1038/ismej.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Martiny AC, Reppas NB, Barry KW, Malek J, Chisholm SW, Church GM. Sequencing genomes from single cells by polymerase cloning. Nat Biotechnol. 2006;24:680–686. doi: 10.1038/nbt1214. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.