Abstract
Levels of recombination vary among species, among chromosomes within species, and among regions within chromosomes in mammals. This heterogeneity may affect levels of diversity, efficiency of selection, and genome composition, as well as have practical consequences for the genetic mapping of traits. We compared the genetic maps to the genome sequence assemblies of rat, mouse, and human to estimate local recombination rates across these genomes. Humans have greater overall levels of recombination, as well as greater variance. In rat and mouse, the size of the chromosome and proximity to telomere have less effect on local recombination rate than in human. At the chromosome level, rat and mouse X chromosomes have the lowest recombination rates, whereas human chromosome X does not show the same pattern. In all species, local recombination rate is significantly correlated with several sequence variables, including GC%, CpG density, repetitive elements, and the neutral mutation rate, with some pronounced differences between species. Recombination rate in one species is not strongly correlated with the rate in another, when comparing homologous syntenic blocks of the genome. This comparative approach provides additional insight into the causes and consequences of genomic heterogeneity in recombination.
Recombination is a fundamentally important process in evolution, and along with mutation it is responsible for producing the patterns of genetic diversity seen in extant populations. Genomic variability in the intensity of recombination interacts with, and can moderate the effects of, other genome features. For example, recombination and natural selection can jointly affect patterns of nucleotide variation, including neutral polymorphism levels (Maynard Smith and Haigh 1974; Begun and Aquadro 1992; Charlesworth et al. 1993), rates of protein evolution (Pál et al. 2001; Betancourt and Presgraves 2002), distribution of transposable elements (Rizzon et al. 2002), and magnitude of codon bias (Comeron et al. 1999; Marais and Piganeau 2002). Understanding the factors responsible for the variation in recombination rate across the genome is essential for explaining the patterns of variation observed in extant species (Reich et al. 2002), for increasing the power of association studies and linkage disequilibrium mapping of complex disease (Arnheim et al. 2003), as well as for evolutionary genomic studies and the identification of genomic regions recently impacted by selection (Aquadro 1997). In addition, the recombination rate directly affects our ability to genetically map and dissect complex traits in animal models of disease.
Recombination is known to occur nonuniformly across the genomes of mammals (Nachman and Churchill 1996; Broman et al. 1998; Yu et al. 2001; Kong et al. 2002), but we are far from understanding how and why. Humans have about twice as much recombination as mouse and rat, and in many mammalian species females recombine more than males. The rate of recombination in humans is reduced near the centromeres and elevated near the telomeres, although regional variation in recombination is only partly explained by this (Yu et al. 2001; Kong et al. 2002). Recombination rate covaries with the neutral mutation rate (Hardison et al. 2003), but whether recombination is mutagenic is unresolved; it may be that mutation and recombination covary with a third variable. Similarly, recombination rate covaries with local GC content, with higher recombining regions being more GC-rich (Birdsell 2002; Marais 2003), although the causative relationship between these variables remains elusive. If recombination increases the local GC content, genomic heterogeneity in recombination could produce the isochore structure of vertebrate genomes (Bernardi 1993; Montoya-Burgos et al. 2003). Alternatively, GC content may be moderating recombination (Petes 2001). It is difficult to infer the direction of causation in the correlation with many of these variables, because multiple genome sequence variables all covary together (Hardison et al. 2003).
The effects of these relationships with recombination will also depend on how conserved the local rate of recombination is over time. Measuring recombination rates in homologous regions of closely related and more distantly related species can provide an answer to this question, but to date such estimates of recombination have only been available for invertebrates (True et al. 1996). The recent availability of the complete genome sequence of the rat (Rattus norvegicus; Rat Genome Sequencing Project Consortium [RGSPC] 2004), mouse (Mus musculus; MGSC 2002), and human (Homo sapiens; IHGSC 2001), combined with publicly available genetic maps of these species (Dietrich et al. 1996; Broman et al. 1998; Steen et al. 1999; Kong et al. 2002) provides powerful new opportunities for examining genomic levels and patterns of recombination in mammals. To date, attempts to do so have focused on single species; here we employ a comparative approach to refine our understanding of variation in recombination rates in the rat, mouse, and human genomes.
RESULTS
Characterization of Recombination Rates
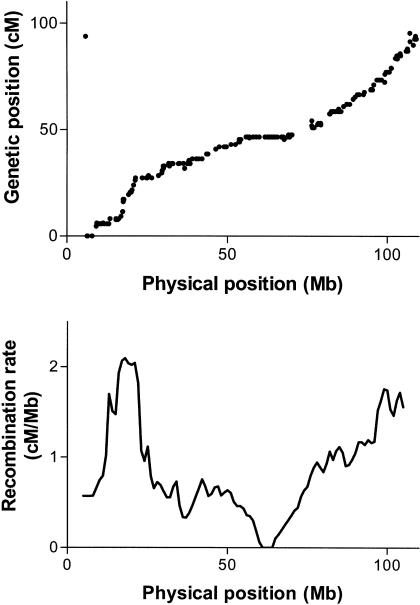
Of the 3824 markers in the rat SHRSPxBN F2 intercross map (Steen et al. 1999), 3323 were placed on the rat genome assembly (v3.1, a.k.a. June 2003 freeze). After filtering for consistent order between the genetic map and the sequence position, 2305 remained. Of the 6336 markers in the mouse OBxCAST F2 intercross map (Dietrich et al. 1996), 5602 were placed on the mouse genome assembly (February 2003 freeze; NCBI build 30), with 4880 remaining after filtering. Of the 5136 markers from the human Iceland pedigree map (Kong et al. 2002), 5114 were placed on the essentially finished human genome sequence (April 2003 freeze; NCBI build 33) of the human genome assembly. Positions of all markers are available from the UCSC genome browser, table browser, or ftp site (http://www.genome.ucsc.edu; Kent et al. 2002). Recombination rates were estimated by comparing the genetic distance (cM) between markers to the physical (Mb) distance in 5 Mb and 10 Mb windows (Fig. 1, Supplemental Figs. A–I available online at www.genome.org). Nonoverlapping windows were used for most of the analyses below. Windows with more than 50% “N”s were discarded, as were windows covering sequence at the beginning or end of chromosomes that contained no markers. For the 5 Mb windows, this resulted in the removal of 26, 13, and 38 windows in rat, mouse, and human, respectively. For 10 Mb windows, we removed 12, 0, and 13 windows in rat, mouse, and human, respectively. In addition, those windows in the rat consisting of the first 20 Mb of chromosome 6, the first 15 Mb of chromosome 13, and the first 40 Mb of chromosome X were discarded due to large discrepancies between the genetic and sequence maps, most likely a result of assembly errors. The rates for the remaining windows are available as Supplemental Tables A–F. Rates of all three species are also displayed on their respective genomes in the UCSC genome browser.
Figure 1.
Plots of genetic position vs. physical position for all markers placed on the telocentric rat chromosome 10 (top), and recombination rate vs. physical position for rat chromosome 10 (bottom) after removing inconsistently placed markers. Similar plots for all chromosomes in all species are available as Supplemental Figures A–I.
The total length of the rat, mouse, and human genetic maps are 1509 cM, 1361 cM, and 3615 cM, respectively (Dietrich et al. 1996; Steen et al. 1999; Kong et al. 2002). Taking the size of the genome of these same species to be 2.72 Gb, 2.58 Gb, and 3.02 Gb (IHGSC 2000; MGSC 2002; RGSPC 2004), the genome-wide average recombination rates are 0.555 cM/Mb, 0.528 cM/Mb, and 1.20 cM/Mb for rat, mouse, and human, respectively. A more accurate measure including only distances measured between placed markers (i.e., not counting portions of chromosomes before and after the first and last markers), gives genome-wide estimates of 0.60 cM/Mb for rat, 0.56 cM/Mb for mouse, and 1.26 cM/Mb for human. Humans have about twice as much recombination per generation as the rodents, with rat and mouse experiencing similar rates of recombination.
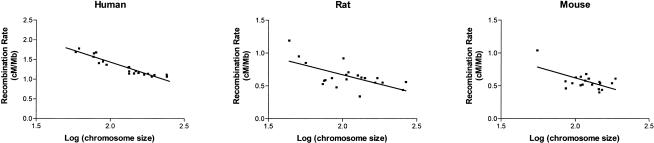
Substantial variation among chromosomes is seen in all three mammals (Table 1 and Suppl. Fig. J). In the two rodents, the X chromosome has the lowest recombination rate (lower than the autosomal average for rat, P < 0.05, one-tailed t-test with single observation; Sokal and Rohlf 1995; for mouse, P < 0.05 only after removal of the outlier chromosome 19), whereas human chromosome X has a rate very near the human genome average. In the rat and mouse, the smallest chromosome in each genome has the highest recombination rate. The same is nearly true in human, except that chromosome 22 has a slightly higher recombination rate than chromosome 21, although the latter is slightly smaller than the former. Note that the chromosome sizes here are based on the size of the genome sequence assemblies, which in the rodents may exclude satellite DNA and other heterochromatic regions. Therefore for example, rat chromosome 12 is the smallest by these criteria but may not be the smallest physically, as the p-arm of this chromosome is satellited (Levan 1974) and contains an NOR (Sasaki et al. 1986). There is a strong negative correlation between chromosome size and chromosome recombination rate in rats and in humans; the relationship is weaker in rat and mouse (Fig. 2).
Table 1.
Physical and Genetic Lengths of Chromosomes, and Chromosome Wide Recombination Rates
|
Rat
|
Mouse
|
Human
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr | Assembly size (Mb)a | Physical size (Mb)b | Genetic length (cM)c | Recombination rate (cM/Mb)d | Assembly size (Mb) | Physical size (Mb) | Genetic length (cM) | Recombination rate (cM/Mb) | Assembly size (Mb) | Physical size (Mb) | Genetic length (cM) | Recombination Rate (cM/Mb) |
| 1 | 268.1 | 267.4 | 149.3 | 0.56 | 195.9 | 187.6 | 113.6 | 0.61 | 245.2 | 241.0 | 270.3 | 1.12 |
| 2 | 258.2 | 255.3 | 112.4 | 0.44 | 181.4 | 177.5 | 96.2 | 0.54 | 243.3 | 241.6 | 257.5 | 1.07 |
| 3 | 171.0 | 166.0 | 91.5 | 0.55 | 160.7 | 151.9 | 66.7 | 0.44 | 199.4 | 199.0 | 220.8 | 1.11 |
| 4 | 187.4 | 186.1 | 102.2 | 0.55 | 152.9 | 147.4 | 79.8 | 0.54 | 191.6 | 190.6 | 204.5 | 1.07 |
| 5 | 173.1 | 171.6 | 105.7 | 0.62 | 149.7 | 145.7 | 82.0 | 0.56 | 181.0 | 180.3 | 205.7 | 1.14 |
| 6 | 147.6 | 134.2 | 85.2 | 0.63 | 150.0 | 144.7 | 65.6 | 0.45 | 170.7 | 169.7 | 189.6 | 1.12 |
| 7 | 143.1 | 141.6 | 88.5 | 0.62 | 134.4 | 130.0 | 67.8 | 0.52 | 158.4 | 155.1 | 179.3 | 1.16 |
| 8 | 129.1 | 126.8 | 84.1 | 0.66 | 128.9 | 123.7 | 75.4 | 0.61 | 145.9 | 144.6 | 166.1 | 1.15 |
| 9 | 113.7 | 109.5 | 78.3 | 0.71 | 124.5 | 116.8 | 67.7 | 0.58 | 134.5 | 133.9 | 160.0 | 1.20 |
| 10 | 110.7 | 101.2 | 92.8 | 0.92 | 130.7 | 122.9 | 75.4 | 0.61 | 135.5 | 134.2 | 176.0 | 1.31 |
| 11 | 87.8 | 73.6 | 39.0 | 0.53 | 122.9 | 118.4 | 80.9 | 0.68 | 135.0 | 134.0 | 152.5 | 1.14 |
| 12 | 46.6 | 43.6 | 51.9 | 1.19 | 114.5 | 108.5 | 55.7 | 0.51 | 133.5 | 132.8 | 172.0 | 1.30 |
| 13 | 111.3 | 75.1 | 43.9 | 0.58 | 116.2 | 112.1 | 57.9 | 0.52 | 114.2 | 94.4 | 130.0 | 1.37 |
| 14 | 112.2 | 105.7 | 70.9 | 0.67 | 115.8 | 108.9 | 69.9 | 0.64 | 105.3 | 83.8 | 118.5 | 1.41 |
| 15 | 109.8 | 106.1 | 63.8 | 0.60 | 104.1 | 100.9 | 63.4 | 0.63 | 100.1 | 77.5 | 128.8 | 1.66 |
| 16 | 90.2 | 76.6 | 45.5 | 0.59 | 99.0 | 95.4 | 51.4 | 0.54 | 90.0 | 89.2 | 128.9 | 1.45 |
| 17 | 97.3 | 91.0 | 43.9 | 0.48 | 93.5 | 85.8 | 49.2 | 0.57 | 81.7 | 80.2 | 135.1 | 1.68 |
| 18 | 87.3 | 84.7 | 52.3 | 0.62 | 91.0 | 86.3 | 39.3 | 0.46 | 77.8 | 77.0 | 120.6 | 1.57 |
| 19 | 59.2 | 56.4 | 48.1 | 0.85 | 61.1 | 55.5 | 57.9 | 1.04 | 63.8 | 61.5 | 109.7 | 1.78 |
| 20 | 55.3 | 50.6 | 48.2 | 0.95 | 63.6 | 58.4 | 98.6 | 1.69 | ||||
| 21 | 47.0 | 30.1 | 61.9 | 2.06 | ||||||||
| 22 | 49.5 | 31.4 | 65.9 | 2.10 | ||||||||
| X | 160.8 | 130.8 | 44.6 | 0.34 | 150.0 | 145.6 | 57.9 | 0.40 | 152.6 | 148.5 | 179.0 | 1.21 |
| Total | 2719.9 | 2553.9 | 1542.1 | 0.60 | 2577.3 | 2465.6 | 1373.7 | 0.56 | 3019.6 | 2888.5 | 3630.5 | 1.26 |
| Autosomes | 2559.1 | 2423.1 | 1497.4 | 0.62 | 2427.3 | 2320.0 | 1315.8 | 0.57 | 2866.9 | 2740.0 | 3451.5 | 1.26 |
Assembly size is the size of the chromosome based on the complete genome assembly
Physical size is the distance in Mb between the first and last mapped markers
Genetic length is the distance in cM between the first and last mapped markers
Recombination rate is the genetic length divided by the physical size
Figure 2.
Chromosomal recombination rate as a function of chromosome size. The regression slope is no longer significantly different than zero in mouse if the smallest chromosome (chr. 19) is removed. Similarly, in rat the regression is not significant if the three smallest chromosomes are removed. Note that the y-axis of the human graph is scaled differently than for the rodents.
There is also smaller-scale variation in the levels of recombination along each chromosome in each genome (e.g., Fig. 1), as seen with most genomic parameters (IHGSC 2001; MGSC 2002; RGSPC 2004). The ranges of recombination rates for 5 Mb and 10 Mb windows in the rat are 0–2.59 cM/Mb and 0–1.80 cM/Mb, respectively. For these same size windows the ranges in mouse are 0–2.52 cM/Mb and 0–1.91 cM/Mb, respectively, and for human the ranges are 0.029–4.26 cM/Mb and 0.108–3.39 cM/Mb. The observation that the rodent genomes have minima of zero whereas the human does not may be partly attributable to the fewer number of meioses used to construct the rodent genetic maps. The human genetic map we used was constructed with 1257 meioses (Kong et al. 2002), whereas the rat and mouse maps were made with only 90 and 92 meioses, respectively (Dietrich et al. 1996; Steen et al. 1999). However, this reduced resolution may not hinder our efforts to detect genomic patterns at these scales as judged by a comparison between recombination rates estimated from the high-resolution human map and those from a lower-resolution map (188 meioses; Broman et al. 1998). The correlation between rates estimated from the two human maps using 10 Mb nonoverlapping windows is 0.90 for the autosomes.
Rat and mouse display significantly less genomic variation in recombination rate than human: the variance in these rodents is about one-third that of human (Table 2). This does not seem to be caused by a reduced resolution of the rodent maps (resulting from the smaller number of meioses and associated increased sampling error), as a similar discrepancy is observed when using recombination rates estimated from the Marshfield map. Levels of variation are very similar between rat and mouse (for example, on the 10 Mb scale, variancerat = 0.117; variancemouse = 0.113). In all species greater variance is seen when using 5 Mb windows than 10 Mb windows, suggesting either that recombination rate varies at a scale finer than 10 Mb, or that estimating recombination rate with fewer markers leads to larger error. The greater heterogeneity in human recombination rate appears to be related to a higher average rate; on a relative scale (as measured by the coefficient of variation), human variation is actually reduced (Table 2). Thus, the data from these three species suggest that the level of variation in recombination rate and the average recombination rate are positively correlated, that is, variance scales with the mean. We further investigated this hypothesis by comparing the variance and mean in recombination rates on a chromosomal scale. Chromosomes with higher average recombination rates show higher variances (human 10 Mb windows, Spearman's correlation coefficient (rs) = 0.55, P = 0.006; human 5 Mb, rs = 0.47, P = 0.024; mouse 10 Mb, rs = 0.70, P = 0.0007; mouse, 5 Mb, rs = 0.74, P = 0.0002, rat 5 Mb, rs = 0.49, P = 0.026). If the outlier chromosome 12 is excluded in rat, this species also shows a significant correlation at 10 Mb (rs = 0.47, P = 0.036).
Table 2.
Variance and Coefficient of Variation in Recombination Rates
| Genomic variance in recombination rate | ||||
|---|---|---|---|---|
| Species | Scale (Mb) | Estimate | Lower CLa | Upper CLa |
| Rat | 10 | 0.117 | 0.096 | 0.139 |
| 5 | 0.191 | 0.160 | 0.223 | |
| Mouse | 10 | 0.113 | 0.090 | 0.138 |
| 5 | 0.207 | 0.173 | 0.243 | |
| Human (DeCode) | 10 | 0.396 | 0.317 | 0.476 |
| 5 | 0.513 | 0.436 | 0.594 | |
| Human (Marshfield) | 10 | 0.421 | 0.344 | 0.504 |
| 5 | 0.602 | 0.518 | 0.690 | |
| Genomic coefficients of variation in recombination rateb | ||||
|---|---|---|---|---|
| Species | Scale (Mb) | Estimate | Lower CLa | Upper CLa |
| Rat | 10 | 58.6 | 53.5 | 63.5 |
| 5 | 74.1 | 68.9 | 79.3 | |
| Mouse | 10 | 63.5 | 57.5 | 69.4 |
| 5 | 82.3 | 76.4 | 88.2 | |
| Human (DeCode) | 10 | 51.2 | 46.8 | 55.2 |
| 5 | 56.1 | 52.6 | 59.4 | |
| Human (Marshfield) | 10 | 56.6 | 51.7 | 61.5 |
| 5 | 66.6 | 62.4 | 70.9 | |
Lower CL and Upper CL provide 95% confidence limits estimated with 10,000 bootstrap replicates
The coefficient of variation is expressed as a percentage
Chromosomal and Sequence Correlates of Recombination Rates
In order to determine what factors may be affecting the patterns of recombination we looked for correlations between recombination rate and both chromosome and sequence features for 5 Mb and 10 Mb windows in the rat, mouse, and human genomes (Table 3). Among the chromosomal variables examined (chromosome size, arm size, proportional distance from the centromere, and proportional distance from the center of the chromosome), for rat and human the strongest correlation was distance from the center of the chromosome, and for mouse it was distance from the centromere, regardless of window size. These correlations were all positive, reflecting a higher rate of recombination near the telomeres in all species, and reduced recombination near the center of the chromosome, as demonstrated by others for the human genome (Nachman and Churchill 1996; Payseur and Nachman 2000; Yu et al. 2001; Kong et al. 2002). This telomere effect is strongest in human, with Spearman correlation coefficients (rs) greater than 0.5, and weakest in mouse.
Table 3.
Correlationsa of Chromosomal Variables and Recombination Rate in Nonoverlapping Windows
| 5 Mb
|
|||
|---|---|---|---|
| Variable | Rat | Mouse | Human |
| ChrSizeb | -0.150 | -0.074 (n.s.) | -0.216 |
| ArmSizeb | -0.143 | -0.074 (n.s.) | -0.316 |
| DistCentro | 0.154 | 0.209 | 0.530 |
| DistCenter | 0.273 | 0.205 | 0.589 |
| 10 Mb
|
|||
|---|---|---|---|
| Variable | Rat | Mouse | Human |
| ChrSizeb | -0.162 | -0.081 (n.s.) | -0.217 |
| ArmSizeb | -0.158 | -0.081 (n.s.) | -0.317 |
| DistCentro | 0.186 | 0.299 | 0.598 |
| DistCenter | 0.353 | 0.149 | 0.631 |
Spearman nonparametric correlation coefficients (rs). Nonsignificant correlations are indicated by “n.s”
Chromosome size and arm size are identical for mouse, because all mouse chromosomes are telocentric
Nearly all of the genome sequence variables included in the analyses (see Methods) were significantly correlated with recombination rates in all species using either 5 Mb or 10 Mb nonoverlapping windows; however, most of the associations were weak. In other words, although most of the correlations have low P-values, they also have low correlation coefficients, and most of these sequence variables are also correlated with each other. The top six sequence correlates with recombination rates for 5 Mb windows are shown in Table 4, with scatterplots of selected variables and recombination rates shown in Supplemental Figures K–M. Several of the parameters with the highest correlations in human, such as CpG fraction, GC%, and polypurine/polypyrimidine tract fraction, had been previously shown by others to be moderately correlated with recombination rate (Yu et al. 2001; Kong et al. 2002). That our analyses reveal slightly higher correlations than those of Kong et al. (2002) probably derives from our use of a newer version of the human genome assembly (with 5114 markers placed on the genome instead of 4690), and larger windows (5 Mb instead of 3 Mb). We observed slightly higher correlation coefficients with 10 Mb windows than with 5 Mb windows for most, but not all, sequence parameters.
Table 4.
Strongest Correlations of Sequence Variables and Recombination Rate in 5-Mb Nonoverlapping Windows
|
Rat
|
Mouse
|
Human
|
|||
|---|---|---|---|---|---|
| Variablea | rsb | Variablea | rsb | Variablea | rsb |
| CpG | 0.386 | LINEs | -0.514 | (CA)n≥20 | 0.511 |
| SimpleRpts | 0.376 | Total IRs | -0.506 | CpG | 0.498 |
| Rn≥30/Yn≥30 | 0.370 | CpG | 0.503 | GC% | 0.449 |
| Total IRs | -0.362 | An≥4/Tn≥4 | -0.473 | Rn≥30/Yn≥30 | 0.446 |
| LINEs | -0.355 | GC% | 0.464 | LINEs | -0.443 |
| (CA)n≥20 | 0.343 | (CA)n≥20 | 0.460 | Sn≥20 | 0.428 |
See Methods section explanation of sequence variable abbreviations
Spearman nonparametric correlation coefficient
Several of the top correlates with recombination are shared across all three species, including GC content and the fraction of the window comprised of CpG dinucleotides, LINEs, total interspersed repeats, tracts of polypurine or polypyrimidine, and CA repeats with greater than 20 units. For all sequence parameters, the rat shows weaker correlations with recombination rate than does mouse or human, probably reflecting greater error in the estimation of recombination rate in rat. Nevertheless, this increased noise does not seriously obscure the relationships between these sequence parameters and recombination rate, as similar variables are identified in rat, mouse, and human. In general, neither the lower variance in recombination rate, nor the reduced resolution of the genetic maps in rodents, compromises our ability to identify associated sequence motifs, as evidenced by the similarity in correlation coefficients between mouse and human (Table 4).
Although many of the sequence parameters are similarly correlated in all three species, there are some notable differences. First, none of the sequence parameters most highly correlated with recombination rate in human (CA repeat fraction, CpG density, and GC%) are as strongly correlated as the chromosomal position variables, whereas the same is certainly not true for rat or mouse. In human, the single best predictor of recombination rate is proximity to the telomere, whereas in mouse and rat this variable explains much less variance than many of the sequence parameters. Second, the negative correlation between total fraction occupied by interspersed repetitive elements is much stronger in rat and mouse, with rs = -0.362 and rs = -0.506 respectively, compared to human (rs = -0.214) using 5 Mb windows. Third, the relationship between the fraction of the window containing stretches of only A or T (i.e., Wn) and recombination is reasonably strong in mouse, but weak or nonsignificant in rat and human, most prominently with longer Wn tracts and larger windows. For example, with 10 Mb windows the correlation coefficient with Wn≥10 is -0.541 in mouse (P < 0.001), but in rat and human rs = -0.287 and rs = -0.297, respectively. For Wn≥30, rs = -0.454 (P < 0.001) in mouse with no significant correlation in either human or rat. Finally, the behavior of the correlation between recombination and tracts of only A (An)oronly T (Tn) is complex. As reported by Kong et al. (2002), there is a relatively strong and significant negative correlation between recombination and fraction of An≥4/Tn≥4 in human, and we show here the same is true for rat (rs = -0.319, P < 0.001 at 5 Mb; rs = -0.319, P < 0.001 at 10 Mb) and mouse (rs = -0.473, P < 0.001 at 5 Mb; rs = -0.556, P < 0.001 at 10 Mb). However, if we restrict the sequence motif to only include tracts greater or equal to 8 bp long (An≥8/Tn≥8), the correlation switches from negative to positive in human (rs = 0.214, P < 0.001 at 5 Mb; rs = 0.340, P < 0.001 at 10 Mb) and rat (rs = 0.212, P < 0.001 at 5 Mb; rs = 0.232, P < 0.001 at 10 Mb), whereas in mouse for a variety of lengths from An≥8/Tn≥8 to An≥30/Tn≥30 the correlations are either very weak or in most cases not significant. Negative correlations for An/Tn tract fraction and recombination are stronger for human and rat when 4 ≤ n ≤ 6 (rs = -0.494, P < 0.001 and rs = -0.346, P < 0.001, respectively for 10 Mb windows), than for n ≥ 4 (rs = -0.305, P < 0.001 and rs = -0.319, P < 0.001, respectively for 10 Mb windows).
To determine what combination of chromosomal and sequence variables best predicts recombination rates in each species we performed multiple linear regression. For 5 Mb windows, the best combination of chromosomal variables in rat was chromosome size and distance from the center of the chromosome (R2 = 0.111). In human these same two variables explained a much larger amount of the variance (R2 = 0.440), whereas in mouse the combination of chromosome size and distance from the centromere explained less of the variation in recombination (R2 = 0.0553). In mouse, chromosome size was not significantly correlated with recombination in either size of the nonoverlapping windows, whereas in the multiple regression model it did add significantly when combined with the distance from the centromere. As was seen at the whole-chromosome level and in the simple correlation analyses, the chromosomal parameters more strongly affect recombination in human than they do in the rodents.
The combination of five sequence variables explaining the largest proportion of the variance in recombination in the rat genome at 5 Mb windows is CpG fraction, GC%, Wn≥10, fraction of window made of all dinucleotide repeats, and density of (CA)n≥10 (R2 = 0.236). As previously shown for human (Kong et al. 2002), when CpG is included in this multiple regression model, the sign of the coefficient of GC% becomes negative, whereas it was positively correlated in a simple bivariate correlation. Including both sequence and chromosomal position variables, the combination of the above five variables with the proportional distance from the center of the chromosome brings the explained variance in rat to R2 = 0.240. In mouse for the same window size, the sequence variables that in combination explain the greatest variance are CpG, total interspersed repetitive elements, and (CA)n≥20, with R2 = 0.274. For the mouse, adding any of the chromosomal variables either alone or in combination did not significantly improve the model, nor did inclusion of GC%. Finally, in human the combination of the sequence variables GC%, fractions of CpG, (CA)n≥10, Wn≥10, Rn≥30, A4≤n≤6/T4≤n≤6, and DNA transposons yields R2 = 0.505 with all seven variables adding significantly to the model. These sequence variables, when combined with the variables chromosome size and distance from the center, explain over half of the variance in recombination rates in humans (R2 = 0.589). Using 10 Mb windows yielded very similar results, with a slight increase in R2 values.
We asked whether there was a correlation between recombination rates in these mammals and the neutral mutation rate determined by the rate of substitution in ancestral repeats along each branch in the human–mouse–rat (HMR) phylogenetic tree. Using 5 Mb windows, there is a significant positive correlation between recombination rate and substitution rate along the branch from the HMR ancestor to present-day humans (rs = 0.114), consistent with previous studies (Hellman et al. 2003 and references therein). Similarly, in rat there is a significant positive correlation between recombination rate and substitution rate along the branch to rat from the MR ancestor (rs = 0.138). Interestingly, in the mouse genome the relationship between recombination rate and mutation is also significant, for substitutions from both the HMR ancestor and the MR ancestor, but in both cases the correlation is negative (rs = -0.128 and rs = -0.127, respectively), not positive. Because previous research uncovered a complex relationship between GC content and different measures of the neutral mutation rate (Hardison et al. 2003), and a well established correlation between GC content and recombination rate (Birdsell 2002), we included GC content and substitution rate in a multiple linear regression model with recombination rate as the dependent variable. In mouse, the substitution rate did not contribute significantly to explaining recombination rate after GC content was included in the model, although the sign of the coefficient of substitution rate became positive.
Conservation of Recombination Across Mammals
Perhaps the most interesting question from an evolutionary standpoint that can be addressed for the first time with our data is whether recombination rates in homologous genomic regions are correlated across mammalian species. Answers to this question, restricting the analyses to homologous blocks with no breakpoints, are shown in Table 5. Except for the rat–human comparison at 10 Mb windows, all species pairs show significant correlations in recombination rates. However, the correlation coefficients are small, suggesting clear evolutionary divergence in recombination rate for genomic regions with generally similar sequences. The species pair with the strongest correlation is human–mouse, which may reflect greater error in recombination rate estimates in rat, or a lineage-specific acceleration in evolutionary divergence of recombination rate in the rat genome. Although the correlations are weak they yield a clear phylogenetic signal: Rat and mouse are more highly correlated with each other than rat is with human.
Table 5.
Correlations Between Homologous Segments
| 5 mb | rs | P |
|---|---|---|
| Rat-human | 0.166 | <0.002 |
| Rat-mouse | 0.221 | <0.001 |
| Rat-mouse telocentrics | 0.200 | <0.002 |
| Human-mouse | 0.300 | <0.001 |
| 10 mb | ||
|---|---|---|
| Rat-human | 0.071 | n.s. |
| Rat-mouse | 0.249 | <0.001 |
| Rat-mouse telocentrics | 0.261 | <0.005 |
| Human-mouse | 0.356 | <0.001 |
Because chromosomal size and position seem to be important variables, it may not be surprising to find only weak correlation between species, as even mouse and rat have undergone many chromosomal rearrangements since their divergence (Nilsson et al. 2001; RGSPC 2004). To determine whether this is a major factor, we examined individual rat–mouse chromosome pairs with strong syntenic conservation. Although rat chromosome 8 and mouse chromosome 9 have nearly complete conservation of gene order across the entire length of these chromosomes and are both telocentric, we found no significant correlation of recombination in homologous blocks. Similarly, rat chromosome 15 and mouse chromosome 14 have strongly conserved gene order, but different morphology (rat chromosome 15 is metacentric). This chromosomal pair also has no significant correlation of recombination rates. Finally, when all rat chromosomes with a similar morphology to mouse chromosomes (i.e., the telocentric chr. 2, 4–10, and X) are used, no clear pattern is observed, with a slightly decreased correlation coefficient at 5 Mb and a slightly increased coefficient at 10 Mb.
DISCUSSION
Estimation of Recombination Rates
We provide the first published genome-wide estimates of recombination rates in the rat and mouse genomes, based on comparisons between genetic maps and genome sequence assemblies. In a comparative perspective, these new data provide insights into the variation within and among species in patterns of recombination and genome evolution in general. The rat and mouse genetic maps used in our analyses were constructed from intercrosses using 90 and 92 meioses, respectively, and the human map was made with over 1200 meioses. The increased number of meioses in human provides a greater resolution by reducing the error in the estimates of genetic distance between markers. Such high resolution is needed in order to map recombination hotspots (true hotspots, i.e., regions of dramatically increased recombination over neighboring regions, on the order of several kb; Jeffreys et al. 2001; Jeffreys and Neumann 2002) at a fine scale (Arnheim et al. 2003); however, our data suggest that when looking at phenomena occurring at a larger scale (i.e., 5–10-Mb intervals), the increased resolution provides minimal increase in accuracy as judged by the comparisons between the high-resolution deCODE map (Kong et al. 2002) and the lower-resolution Marshfield map (Broman et al. 1998; Yu et al. 2001). It remains an open question as to whether regional variation in recombination is the result of variation in the density, or in the intensity, of hotspots, or whether regional variation in recombination is unrelated to the specific behavior of small hotspots (Petes 2001). The confidence in the placement of genetic markers on the human genome, and therefore also in the estimates of recombination, improved incrementally with each release of the human genome assembly (Kong et al. 2002), and by analogy we anticipate a similar increase in the reliability of estimates of recombination in the rat and mouse genomes. However, we still found recombination rate to covary with many of the same sequence parameters in rat as in mouse and human. Finally, the recombination rate estimates provided herein for rat and mouse are based on intercrosses between specific inbred strains, which provide little information about the variation within species, or whether these results will hold for the species generally. This may be especially relevant for the mouse genome, because the genetic map was built using an intersubspecies cross which could theoretically experience a suppression of recombination because of sequence divergence (Dietrich et al. 1996). It was shown previously that there is variation in the amount of recombination within and between individual humans (Broman et al. 1998; Kong et al. 2002) and mice (Reeves et al. 1990; Koehler et al. 2002).
Causes and Consequences of Recombination
The most pronounced difference between rat, mouse, and human with respect to recombination is the twofold greater amount of overall recombination in humans. A mechanistic explanation that has been suggested is that recombination is proportional to the number of chromosome arms (Pardo-Manuel de Villena and Sapienza 2001), as proper disjunction requires at least one chiasma per arm. For example, with a larger number of chromosomes and mostly metacentric chromosomes, humans have about twice as many chromosome arms and about twice as much recombination as mice. Our data show that although rats are intermediate in the number of chromosome arms they have the same, or perhaps only slightly greater, levels of recombination as mice. Additionally, in comparing recombination at the chromosome level, metacentric chromosomes in the rat have less than half as much recombination as similarly sized human metacentrics (rat chr. 16–20, human chr. 16–20; Table 1), demonstrating that there is more to recombination rate evolution than changes in the number of chromosome arms. The data used by Pardo-Manuel de Villena and Sapienza (2001) from rat to support their hypothesis (an ∼2200 cM genome) are unrealistic and incompatible with most recent genetic maps in the rat (Bihoreau et al. 1997; Steen et al. 1999; Dracheva et al. 2000). If chromosomal morphology does play a role, the reduced recombination rate in mouse and rat could be a neutral byproduct of chromosomal evolution, or alternatively chromosomal rearrangements themselves may be selected for in order to modulate recombination rates (Qumsiyeh 1994; Dumas and Britton-Davidian 2002).
In addition to increased overall recombination, humans show more genomic heterogeneity than do the two rodents. Whether this pattern is due to an increase in the number of recombination hotspots, their intensity, or a baseline increase in recombination remains an open question. The existence of hotspots in all three species may be responsible for the observed positive correlation between variance in recombination rate and average recombination rate at the chromosomal level; further theoretical work on the effects of hotspots and the acquisition of data on a finer scale, such as that afforded by sperm genotyping (Cullen et al. 2002; Jeffreys and Neumann 2002; Yauk et al. 2003) is required to evaluate this possibility. The high similarity in levels of variation in recombination rate in rat and mouse indicates conservation of the mechanisms that generate diversity in recombination rate, regardless of the nature of these mechanisms, across millions of years of evolution.
All three species show a positive correlation of local recombination rates with CpG fraction and with GC%, with CpG always yielding a larger correlation coefficient than GC%. Such a correlation between GC% and recombination was seen previously in a wide variety of eukaryotes, and at both fine and gross scales (Fullerton et al. 2001; Birdsell 2002). The question of causation is unresolved. GC-rich regions of the genome, particularly those on a larger scale, may stimulate recombination through increased binding of the recombinational machinery or through structural conformational changes associated with high GC content (Petes 2001; Petes and Merker 2002). Alternatively, recombination may increase local GC content through biased gene conversion (Birdsell 2002 and references therein), whereby base-pair mismatches at heterozygous sites between homologous chromosomes are resolved with a bias toward GC. Genomic heterogeneity in recombination can therefore lead to genomic heterogeneity in GC content, or isochores; substantial empirical data emerged recently supporting this hypothesis (Eyre-Walker and Hurst 2001; Birdsell 2002; Montoya-Burgos et al. 2003). Although the correlation between recombination and GC% is not novel, our observation of an even higher correlation between CpG fraction and recombination in all three mammalian species may be seen as providing evidence to support the biased gene conversion hypothesis, in that CpG dinucleotides may be especially sensitive to biased gene conversion (Marais 2003), although alternative hypotheses involving mechanisms whereby CpG dinucleotides stimulate recombination either structurally or through protein-binding cannot be rejected.
In rat and mouse, one of the variables with the strongest correlation in the bivariate analysis is the fraction of the window made up of interspersed repetitive elements, with fewer such elements in regions of higher recombination. This same relationship was seen in the human genome, but less strongly. This negative correlation is predicted if we assume that transposable elements act as very slightly deleterious mutations (Rizzon et al. 2002). The efficacy of purifying selection in removing these repetitive elements will be proportional to the strength of recombination, under most models of the interaction between selection and recombination (Hill and Robertson 1966). Alternatively, the same pattern is also predicted under a model of selection against chromosomal rearrangements caused by ectopic exchange between interspersed repetitive elements at nonhomologous locations (Bartolomé et al. 2002). Finally, an alternative neutral explanation is that repetitive elements somehow act mechanistically to suppress recombination. Testing these different hypotheses will be difficult, as the relationship between these variables is complex, as are the effects of the different classes of interspersed repeats and the covariation with other genomic sequence parameters. Similarly, why the negative correlation between interspersed repeat density and recombination rate is stronger in the rodents than in human remains an open question, but may be related to the observation that a substantially greater fraction of the rodent genomes (nearly one-quarter of the rat genome) is derived from the L1 LINE element (RGSPC 2004). Furthermore, if we hypothesize that LINEs act to suppress recombination, or there is selection against recombination near LINEs, the proliferation of these elements in rodents may be sufficiently responsible for their genome-wide decrease in recombination— although we are far from a position to test this hypothesis.
At the chromosomal level, the rat and mouse X chromosomes have the lowest levels of recombination, whereas human chromosome X has a rate near its genome average, and about what would be expected based on its size. In all species, chromosome X has reduced GC content and CpG density compared to the autosomes, but in rat and mouse this difference is much more pronounced. The GC% of rat and mouse chromosome X is even less than the human X, despite an ∼1% greater GC% in the rodents at the whole-genome level (MGSC 2002; RGSPC 2004). Again, the direction of causation in the relationship between recombination and GC% or CpG density is unknown, but the observation of a striking effect on the X chromosome suggests that whatever the mechanism is, it is not restricted to male meiosis. Although the X chromosome is generally highly conserved, showing few rearrangements across many mammalian orders, it has been much more active specifically in the rodent lineage (Bourque et al. 2004; RGSPC 2004). Whether and how this increased chromosomal evolution is related to the decreased recombination of the rodent X chromosome are unknown. Data from additional mammals, especially more distantly related rodents, may help to answer this question. Finally, it may be of interest that an X-linked locus has been proposed to explain genome-wide levels of variation in recombination, and in controlling differences between male and female recombination rates, in mouse (de la Casa-Esperón et al. 2002).
We observed that the chromosomal position of a region, and the size of the chromosome that it is on, have a greater effect on its recombination rate in human than in rat or mouse. Although all of our data are based on sex-averaged genetic maps, it has been shown in humans that the increased recombination at the telomeres is largely due to an increase in male recombination (Broman et al. 1998). Therefore, it is likely that the difference between species in this respect may be restricted to male meioses; future studies investigating high-resolution sex-specific recombination in rat and mouse may provide a definitive answer. That chromosome size does not have a significant effect on recombination in mouse windows may be due to the fact that except for chromosome 19, there is not much variation in size among the mouse chromosomes, relative to human and rat.
It is essential to understand the relationship between mutation and recombination, as these potentially interacting variables can confound interpretations in studies of the effects of recombination and in estimating costs involved in the evolution of sex and recombination. For example, the positive correlation in humans between recombination rate and nucleotide diversity has been explained as the result of background selection or hitch-hiking (Nachman 2001; Lercher and Hurst 2002), although subsequent studies with larger data sets show that this correlation may be entirely due to a correlation between the mutation and recombination rates (Hellman et al. 2003), with the possibility that recombination is mutagenic. Our results show that at face value the positive correlation between recombination and mutation is not a universal feature of mammals; in fact, in mouse the correlation is negative. Is it possible that recombination is mutagenic in rat and human, but antimutagenic in mouse? Perhaps, but the complex relationships between these variables and other genomic factors such as GC content suggest that the difference may lie in the nature of interactions between multiple factors and may depend on which estimate of the neutral mutation rate is used (Hardison et al. 2003). For example, when the effects of GC% on substitution and recombination rates are taken into account, the correlation between these variables becomes positive in mouse. Although the nature of these relationships and causative explanations remain elusive for now, the strengths of a multiple-species comparative approach are obvious, especially with the observation that the closely related mouse and rat differ in these respects.
Conservation of Recombination Across Mammals
When comparing syntenic homologous blocks across species, we found only a slight positive correlation between recombination rates of different species. On one hand, this was somewhat surprising because sequence parameters that covary with recombination such as GC% tend to be highly correlated across species (MGSC 2002). On the other hand, multiple genomic rearrangements have occurred among rat, mouse, and human (RGSPC 2004), placing homologous regions into different chromosomal environments in each species. Also, it has been predicted on theoretical grounds—and later with empirical support—that recombination hotspots will tend to drive themselves to extinction, resulting in a rapid turnover of hotspots (Boulton et al. 1997; Jeffreys and Neumann 2002). The genomic scale and the time scale at which such a phenomenon occurs are unknown, as the relationship between true hotspots and large-scale regional variation in recombination is unknown.
Understanding the relationships between recombination rate and various genomic parameters (such as nucleotide composition, mutation rate, efficiency of natural selection, rate of protein evolution, and molecular diversity within species) from an evolutionary perspective requires information about the tempo of evolution in recombination rate. Theories addressing the origin and maintenance of sexual reproduction also identify the rate of divergence in recombination as a key parameter. In the future, combining our data with those from several additional closely related species could provide the first estimates of this parameter, paving the way for studies of coevolution with other genomic variables and providing empirical benchmarks for theories about the evolution of recombination and sexual reproduction.
Consequences for Mapping Traits
If multiple genes each with a small to moderate effect in the same direction on a quantitative trait are clustered together in a region of low recombination, this region may show up with strong and significant linkage to the trait in a typical genome scan in an animal model. This is because even though each gene contributes only a small effect to the phenotype, the lack of recombination will cause such a cluster of genes to act as a single large-effect gene (Noor et al. 2001). Conversely, if these multiple genes are tightly linked but alleles have opposite effects on a trait, it may be difficult to detect the effect of any single gene in a genome scan. Once a QTL is identified for a complex trait in rat, it is common to attempt to positionally clone the responsible gene through the construction of congenic and subcongenic rats where, for example, several substrains are developed that contain small portions of the QTL introgressed from the normal strain onto the background of a strain susceptible to a complex disease (Markel et al. 1997). This process relies entirely on identifying the rare recombinant occurring within the QTL. We identified multiple “cold spots” of recombination in the rat genome, where it may be difficult to dissect a QTL through the breeding of subcongenic rats. With the quantification of the amounts and patterns of recombination in the rat genome herein it should be possible to incorporate this information into the planning of future research projects, and to develop an optimized set of markers to maximize the information content from a genome scan.
Future Directions
We anticipate that the accuracy and resolution of the estimates of recombination in the rat, mouse, and human will continue to improve along with the improvements in each new iteration of the genome assembly, and with the construction of higher-resolution genetic maps with larger numbers of progeny. This will be important for the future of complex trait mapping in animal models if the strategies of using linkage disequilibrium being developed in human are to be eventually applied to the rat and mouse (Arnheim et al. 2003). The high-resolution recombination rate maps will also be of tremendous value for investigating questions of genome evolution. Understanding the causes and consequences of recombination rate variation will also be enhanced with accurate estimates of sex-specific recombination in rat and mouse. This is crucial, because there is obviously more to the regulation of recombination than sequence motifs and chromosome location; human females have 1.65 times as much recombination as males with same genome sequence (Broman et al. 1998; Kong et al. 2002). Finally, as we have shown, data from multiple species provide new insights into the factors that covary with, and therefore may be affecting or affected by, recombination that are seen in only one species. Data from more mammals will likely reveal lineage-specific patterns in the evolution of recombination. Genetic maps are available for a number of additional species (Swinburne et al. 2000; Maddox et al. 2001; Dukes-McEwan and Jackson 2002; Slate et al. 2002), and soon genome sequence will be also.
The effect of recombination on long-term evolutionary patterns has received considerable attention in recent years. For example, covariation with nucleotide diversity may reveal the effects of background selection (Charlesworth et al. 1993) and genetic hitchhiking (Maynard Smith and Haigh 1974; Begun and Aquadro 1992; Nachman 2002), or may reflect an association between recombinational and mutational processes (Lercher et al. 2001; Hellman et al. 2003). Similarly, purifying or directional selection is proposed to be more efficient in regions of higher recombination, with detectable patterns in extant genomes (Pál et al. 2001; Betancourt and Presgraves 2002). Methods that include multiple species of greater or lesser amounts of divergence (i.e., a “bushy tree”), incorporate lineage-specific estimates of substitution, and that utilize node-specific estimates of recombination will have substantially greater power to detect such patterns compared to simple human–mouse comparisons. Therefore, our rat–mouse–human comparative approach provides a beginning toward what will be a much more complete understanding of the evolution of recombination and how it interacts with other features of mammalian genome evolution.
METHODS
Placement of Markers and Estimates of Recombination
The following genetic maps were used: rat SHRSPxBN F2 intercross map (Steen et al. 1999), mouse OBxCAST F2 intercross map (Dietrich et al. 1996), human Icelandic family map (Kong et al. 2002), and human CEPH family map (Broman et al. 1998). All maps were made using the Kosambi map function. The versions of the genome assemblies used were: rat June 2003 freeze (v3.1), mouse February 2003 freeze (NCBI build 30), and human April 2003 freeze (NCBI build 33). Locations of individual markers for each of the rat, mouse, and human genomes were determined based on alignments of the full sequence of the marker (when available) using BLAT (Kent 2002) and also using primer sequence information using e-PCR (Schuler 1997) and BLAT. Placement information is available for download from the UCSC Genome Browser (Kent et al. 2002; http://www.genome.ucsc.edu). Markers placed to different chromosomes in the genetic versus genome sequence were discarded. We filtered the complete set of markers placed on the genomic sequences to include the maximal set for each map such that the order of the markers in both the genetic and sequence maps agreed.
From the maximally consistent set of markers derived for a particular genetic map, the recombination rate between all pairs of adjacent markers was calculated by simply dividing the distance between the markers in the genetic map (in centimorgans, cM) by the distance between the markers in the sequence map (in megabases, Mb). For simplicity, the location of a marker in the sequence map is set to the midpoint of the alignment of that marker. Each base pair in the interval between adjacent markers is then assigned the calculated rate. To approximate the recombination rate for any window of sequence of arbitrary size and location, we summed the rates corresponding to each base in the window and divided by the size of the window. Following this, windows were removed that contained more than 50% “N” in the sequence assembly, as were windows at the beginning or end of chromosomes with zero markers placed in them. Finally, a few windows with large discrepancies between the genetic map and the sequence assembly were removed. Recombination rates for individual chromosomes were calculated by dividing the genetic length (cM) by the sequence length (Mb) between the first and last marker placed on each chromosome.
Measuring Chromosomal and Sequence Features and Substitution Rates
For each window in each species, we calculated the proportional distance from the center of the chromosome to the center of the window (absolute distance divided by half the chromosome length) and proportional distance from the centromere to the center of the window (absolute distance divided by the length of the chromosomal arm). For this, the position of the centromere in rat was estimated by comparing the positions of markers mapped by fluorescent in situ hybridization (data from RatMap: http://www.ratmap.gen.gu.se) to their assigned position on the rat genome, as well as from the locations of centromeric repeats (RGSPC 2004). The centromeres rat chromosomes 3, 11, and 12 were assigned to base pair position zero, because the p-arms of these chromosomes are NOR containing satellited DNA and therefore are presumed to not be in the current genome assembly (RGSPC 2004). The centromeres of rat and mouse telocentrics are also placed at position zero. Positions of human centromeres were estimated based on the cytogenetic band mapping to the genome (Furey and Haussler 2003).
The fraction of each window (after correcting for the number of “N”s) comprised of the following sequences were calculated (in units of bp/Mb): G or C (GC%); CpG dinucleotides; A or Tn where n ≥ 4, 6, 8, 10, 15, 20, 15, 30, and 4 ≤ n ≤ 6; polypurine or polypyrimidine stretches (R or Yn, where R = A or G, Y = C or T); stretches of the weak nucleotides (Wn, where W = A or T); stretches of the strong nucleotides (Sn, where S = C or G); short interspersed repetitive elements (SINEs); long interspersed repetitive elements (LINEs); long terminal repeats (LTRs); DNA-based transposons; total interspersed repeats (IRs); di-, tri-, tetra-, and pentanucleotide repeats; 6–10 mer and 11–100 mer repeats; total simple repeats; and (CA)n where n = 10, 20, and 30. Data for the repetitive elements were taken from the RepeatMasker (A. Smit and P. Green, unpubl.) track of the UCSC Genome Browser (Kent et al. 2002). The remaining sequence motifs were calculated with custom perl scripts.
Ancestral repeat (AR) sites from retro- or DNA-transposons were inserted in the human–rodent ancestral genome before the human–rodent split and appear in syntenic positions in all species (A. Smit and P. Green, unpubl.). Alignments of AR sites with the human, mouse, and rat genomes were found using the BLASTZ programs (Schwartz et al. 2003). Using the general time-reversible model of base substitution (REV; Tavaré 1986; Yang et al. 1994; Whelan et al. 2001), we used the frequencies of observed changes to estimate the number of substitutions per AR site. The human–mouse–rat phylogenetic tree used for this model was constructed using maximum likelihood methods (Siepel and Haussler 2003).
Variance and CV Calculations
Genomic levels of variation in recombination rate were estimated, separately on the 10 Mb and 5 Mb scales, by calculating variance and the coefficient of variation (CV). Ninety-five percent confidence limits were estimated as the 2.5% and 97.5% quantiles of these statistics across 10,000 data sets generated by resampling with replacement from the original data set of recombination rates. This approach measures uncertainty associated with estimation of variance and CV from genomic variation but does not address the underlying error associated with assigning genetic and physical map positions.
Correlation and Multiple Regression
We used nonparametric Spearman correlation coefficients (rs) to assess covariation between recombination rates and the above chromosome and sequence variables. To determine what combination of variables were contributing to the variance in recombination and how they may interact, we performed multiple linear regression with the above variables, excluding those not contributing significantly through the use of the t-statistic and with backward stepwise regression. All coefficients of multiple determination (R2) reported for multiple linear regressions were adjusted for the number of variables (Radj2).
Syntenic Homology Mapping
Chained BLASTZ pairwise alignments were obtained from the UCSC Genome Browser. For each nonoverlapping 5 Mb and 10 Mb window in one species, we determined whether there existed a chained alignment from the target species to the corresponding paired species such that: The chained alignment spanned at least 50% of the window in the target species, the chained alignment spanned a region in the query species that was at least 50% but no more than 150% of the size of the window, and the number of bases aligned were at least 10% of the size of the window. Recombination rates were then calculated for the homologous regions as described above.
Acknowledgments
We thank George Weinstock, Kim Worley, and Richard Gibbs at the Baylor College of Medicine for including us in the analysis team of the Rat Genome Sequencing Project Consortium (RGSPC), and all of the groups in the consortium who produced data and made it publicly available. We also thank three anonymous reviewers who made several helpful suggestions. M.I. Jensen-Seaman is supported by NIH grant 1F32HL70527-01. T.S.F., Y.L., and D.H. are supported by a grant from NHGRI and the Howard Hughes Medical Institute (HHMI). B.A.P. is supported by an NSF Integrative Graduate Education and Research Training (IGERT) Grant in Genomics. K.M.R. is an HHMI Predoctoral Fellow.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1970304.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Aquadro, C.F. 1997. Insights into the evolutionary process from patterns of DNA sequence variability. Curr. Opin. Genet. 7: 835-840. [DOI] [PubMed] [Google Scholar]
- Arnheim, N., Calabrese, P., and Nordborg, M. 2003. Hot and cold spots of recombination in the human genome: The reason we should find them and how this can be achieved. Am. J. Hum. Genet. 73: 5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomé, C., Maside, X., and Charlesworth, B. 2002. On the abundance and distribution of transposable elements in the genome of Drosophila melanogaster. Mol. Biol. Evol. 19: 926-937. [DOI] [PubMed] [Google Scholar]
- Begun, D.J. and Aquadro, C.F. 1992. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature 356: 519-520. [DOI] [PubMed] [Google Scholar]
- Betancourt, A.J. and Presgraves, D.C. 2002. Linkage limits the power of natural selection in Drosophila. Proc. Natl. Acad. Sci. 99: 13616-13620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birdsell, J.A. 2002. Integrating genomics, bioinformatics, and classical genetics to study the effects of recombination on genome evolution. Mol. Biol. Evol. 19: 1181-1197. [DOI] [PubMed] [Google Scholar]
- Bernardi, G. 1993. The vertebrate genome: Isochores and evolution. Mol. Biol. Evol. 10: 186-204. [DOI] [PubMed] [Google Scholar]
- Bihoreau, M.-T., Sebag-Montefiore, L., Godfrey, R.F., Wallis, R.H., Brown, J.H., Danoy, P.A., Collins, S.C., Rouard, M., Kaisaki, P.J., Lathrop, M., et al. 1997. A high-resolution consensus linkage map of the rat, integrating radiation hybrid and genetic maps. Genomics 75: 57-69. [DOI] [PubMed] [Google Scholar]
- Boulton, A., Myers, R.S., and Redfield, R.J. 1997. The Hotspot conversion paradox and the evolution of meiotic recombination. Proc. Natl. Acad. Sci. 94: 8058-8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque, G., Pevzner, P.A., and Tesler, G. 2004. Reconstructing the genomic architecture of ancestral mammals: Lessons from human, mouse, and rat genomes. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Broman, K.W., Murray, J.C., Sheffield, V.C., White, R.L., and Weber, J.L. 1998. Comprehensive human genetic maps: Individual and sex-specific variation in recombination. Am. J. Hum. Genet. 63: 861-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B., Morgan, M., and Charlesworth, D. 1993.The effect of deleterious mutations on neutral molecular variation. Genetics 134: 1289-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron, J.M., Kreitman, M., and Agaudé, M. 1999. Natural selection on synonymous sites is correlated with gene length and recombination in Drosophila. Genetics 151: 239-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen, M., Perfetto, S.P., Klitz, W., Nelson, G., and Carrington, M. 2002. High-resolution patterns of meiotic recombination across the human major histocompatibility complex. Am. J. Hum. Genet. 71: 759-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Casa-Esperón, E., Loredo-Osti, J.C., Pardo-Manuel de Villena, F., Briscoe, T.L., Malette, J.M., Vaughan, J.E., Morgan, K., and Sapienza, C. 2002. X chromosome effect on maternal recombination and meiotic drive in the mouse. Genetics 161: 1651-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich, W.F., Miller, J., Steen, R., Merchant, M.A., Damron-Boles, D., Husain, Z., Dredge, R., Daly, M.J., Ingalls, K.A., O'Connor, T.J., et al. 1996. A comprehensive genetic map of the mouse genome. Nature 380: 149-152. [DOI] [PubMed] [Google Scholar]
- Dracheva, S.V., Remmers, E.F., Chen, S., Chang, L., Gulko, P.S., Kawahito, Y., Longman, R.E., Wang, J., Du, Y., Shepard, J., et al. 2000. An integrated genetic linkage map with 1137 markers constructed from five F2 crosses of autoimmune disease-prone and -resistant inbred rat strains. Genomics 63: 202-226. [DOI] [PubMed] [Google Scholar]
- Dukes-McEwan, J. and Jackson, I.J. 2002. The promises and problems of linkage analysis by using the current canine genome map. Mamm. Genome 13: 667-672. [DOI] [PubMed] [Google Scholar]
- Dumas, D. and Britton-Davidian, J. 2002. Chromosomal rearrangements and evolution of recombination: Comparison of chiasma distribution patterns in standard and Robertsonian populations of the house mouse. Genetics 162: 1355-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker, A. and Hurst, L.D. 2001. The evolution of isochores. Nat. Rev. Genet. 2: 549-555. [DOI] [PubMed] [Google Scholar]
- Fullerton, S.M., Bernardo Carvalho, A., and Clark, A.G. 2001. Local rates of recombination are positively correlated with GC content in the human genome. Mol. Biol. Evol. 18: 1139-1142. [DOI] [PubMed] [Google Scholar]
- Furey, T.S. and Haussler, D. 2003. Integration of the cytogenetic map with the draft human genome sequence. Hum. Mol. Genet. 12: 1037-1044. [DOI] [PubMed] [Google Scholar]
- Hardison, R.C., Roskin, K.M., Yang, S., Diekhans, M., Kent, W.J., Weber, R., Elnitski, L., Li, J., O'Conner, M., Kolbe, D., et al. 2003. Covariation in frequencies of substitution, deletion, transposition, and recombination during Eutherian evolution. Genome Res. 13: 13-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellman, I., Ebersberger, I., Ptak, S.E., Paääbo, S., and Przeworski, M. 2003. A neutral explanation for the correlation of diversity with recombination rates in humans. Am. J. Hum. Genet. 72: 1527-1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, W.G. and Robertson, A. 1966. The effect of linkage on the limits to artificial selection. Genet. Res. 8: 269-294. [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium (IHGSC). 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Jeffreys, A.J. and Neumann, R. 2002. Reciprocal crossover asymmetry and meiotic drive in a human recombination hot spot. Nat. Genet. 31: 267-271. [DOI] [PubMed] [Google Scholar]
- Jeffreys, A.J., Kauppi, L., and Neumann, R. 2001. Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nat. Genet. 29: 217-222. [DOI] [PubMed] [Google Scholar]
- Kent, W.J. 2002. BLAT—The BLAST-like alignment tool. Genome Res. 12: 656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, W.J., Sugnet, C.W., Furey, T.S., Roskin, K.M., Pringle, T.H., Zahler, A.M., and Haussler, D. 2002. The human genome browser at UCSC. Genome Res. 12: 996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler, K.E., Cherry, J.P., Lynn, A., Hunt, P.A., and Hassold, T.J. 2002. Genetic control of mammalian meiotic recombination. I. Variation in exchange frequencies among males from inbred mouse strains. Genetics 162: 297-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, A., Gudbjartsson, D.F., Sainz, J., Jonsdottir, G.M., Gudjonsson, S.A., Richardsson, B., Sigurdardottir, S., Barnard, J., Hallbeck, B., Masson, G., et al. 2002. A high-resolution recombination map of the human genome. Nat. Genet. 31: 241-247. [DOI] [PubMed] [Google Scholar]
- Lercher, M.J. and Hurst, L.D. 2002. Human SNP variability and mutation rate are higher in regions of high recombination. Trends Genet. 18: 337-340. [DOI] [PubMed] [Google Scholar]
- Lercher, M.J., Williams, E.J., and Hurst, L.D. 2001. Local similarity in evolutionary rates extends over whole chromosomes in human–rodent and mouse–rat comparisons: Implications for understanding the mechanistic basis of the male mutation bias. Mol. Biol. Evol. 18: 2032-2039. [DOI] [PubMed] [Google Scholar]
- Levan, G. 1974. Nomenclature for G-bands in rat chromosomes. Hereditas 77: 37-52. [DOI] [PubMed] [Google Scholar]
- Maddox, J.F., Davies, K.P., Crawford, A.M., Hulme, D.J., Vaiman, D., Cribiu, E.P., Freking, B.A., Beh, K.J., Cockett, N.E., Kang, N. et al. 2001. An enhanced linkage map of the sheep genome comprising more than 1000 loci. Genome Res. 11: 1275-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais, G. 2003. Biased gene conversion: Implications for genome and sex evolution. Trends Genet. 19: 330-338. [DOI] [PubMed] [Google Scholar]
- Marais, G. and Piganeau, G. 2002. Hill-Robertson interference is a minor determinant of variations in codon bias across Drosophila melanogaster and Caenorhabditis elegans genomes. Mol. Biol. Evol. 19: 1399-1406. [DOI] [PubMed] [Google Scholar]
- Markel, P., Shu, P., Ebeling, C., Carlson, G.A., Nagle, D.L., Smutko, J.S., and Moore, K.J. 1997. Theoretical and empirical issues for marker-assisted breeding of congenic mouse strains. Nat. Genet. 17: 280-284. [DOI] [PubMed] [Google Scholar]
- Maynard Smith, J. and Haigh, J. 1974. The hitch-hiking effect of a favorable gene. Genet. Res. 23: 23-35. [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium (MGSC). 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- Montoya-Burgos, J.I., Boursot, P., and Galtier, N. 2003. Recombination explains isochores in mammalian genomes. Trends Genet. 19: 128-130. [DOI] [PubMed] [Google Scholar]
- Nachman, M.W. 2001. Single nucleotide polymorphisms and recombination rate in humans. Trends Genet. 17: 481-485. [DOI] [PubMed] [Google Scholar]
- Nachman, M.W. 2002. Variation in recombination rate across the genome: Evidence and implications. Curr. Opin. Genet. Dev. 12: 657-663. [DOI] [PubMed] [Google Scholar]
- Nachman, M.W. and Churchill, G.A. 1996. Heterogeneity in rates of recombination across the mouse genome. Genetics 142: 537-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson, S., Helou, K., Walentinsson, A., Szpirer, C., Nerman, O., and Ståhl, F. 2001. Rat–mouse and rat–human comparative maps based on gene homology and high-resolution zoo-FISH. Genomics 74: 287-298. [DOI] [PubMed] [Google Scholar]
- Noor, M.A.F., Cunningham, A.L., and Larkin, J.C. 2001. Consequences of recombination rate variation on quantitative trait locus mapping studies: Simulations based on the Drosophila melanogaster genome. Genetics 159: 581-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pál, C., Papp, B., and Hurst, L.D. 2001. Does the recombination rate affect the efficiency of purifying selection? The yeast genome provides a partial answer. Mol. Biol. Evol. 18: 2323-2326. [DOI] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena, F. and Sapienza, C. 2001. Recombination is proportional to the number of chromosome arms in mammals. Mamm. Genome 12: 318-322. [DOI] [PubMed] [Google Scholar]
- Payseur, B.A. and Nachman M.W. 2000. Microsatellite variation and recombination rate in the human genome. Genetics 156: 1285-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes, T.D. 2001. Meiotic recombination hot spots and cold spots. Nat. Rev. Genet. 2: 360-368. [DOI] [PubMed] [Google Scholar]
- Petes, T.D. and Merker, J.D. 2002. Context dependence of meiotic recombination hotspots in yeast: The relationship between recombination activity of a reporter construct and base composition. Genetics 162: 2049-2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qumsiyeh, M.B. 1994. Evolution of number and morphology of mammalian chromosomes. J. Hered. 85: 455-465. [DOI] [PubMed] [Google Scholar]
- Rat Genome Sequencing Project Consortium (RGSPC). 2004. Genome sequence of the Brown Norway Rat yields insights into mammalian evolution. Nature (in press). [DOI] [PubMed]
- Reeves, R.H., Crowley, M.R., O'Hara, B.F., and Gearhart, J.D. 1990. Sex, strain, and species differences affect recombination across an evolutionarily conserved segment of mouse chromosome 16. Genomics 8: 141-148. [DOI] [PubMed] [Google Scholar]
- Reich, D.E., Schaffner, S.F., Daly, M.J., McVean, G., Mullikin, J.C., Higgins, J.M., Richter D.J., Lander, E.S., and Altshuler, D. 2002. Human genome sequence variation and the influence of gene history, mutation and recombination. Nat. Genet. 32: 135-142. [DOI] [PubMed] [Google Scholar]
- Rizzon, C., Marais, G., Guoy, M., and Biémont, C. 2002. Recombination rate and the distribution of transposable elements in the Drosophila melanogaster genome. Genome Res. 12: 400-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, M., Nishida, C., and Kodama, Y. 1986. Characterization of silver-stained nucleolus organizer regions (Ag-NORs) in 16 inbred strains of the Norway rat, Rattus norvegicus. Cytogenet. Cell Genet. 41: 83-88. [DOI] [PubMed] [Google Scholar]
- Schuler, G.D. 1997. Sequence mapping by electronic PCR. Genome Res. 7: 541-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, S., Kent, W.J., Smit, A., Zhang, Z., Baertsch, R., Hardison, R.C., Haussler, D., and Miller, W. 2003. Human–mouse alignments with BLASTZ. Genome Res. 13: 103-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel, A. and Haussler, D. 2003. Combining phylogenetic and hidden Markov models in biosequence analysis. Proceedings of the Seventh Annual International Conference on Computational Molecular Biology (RECOMB 2003). p. 277-286. [DOI] [PubMed]
- Slate, J., van Stijn, T.C., Anderson, R.M., McEwan, K.M., Maqbool, N.J., Mathias, H.C., Bixley, M.J., Stevens, D.R., Molenaar, A.J., Beever, J.E., et al. 2002. A deer (subfamily Cervinae) genetic linkage map and the evolution of ruminant genomes. Genetics 160: 1587-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal, R.R. and Rohlf, F.J. 1995. Biometry, p. 228. W.H. Freeman, NY.
- Steen, R.G., Kwitek-Black, A.E., Glenn, C., Gullings-Handley, J., Van Etten, W., Atkinson, O.S., Appel, D., Twigger, S., Muir, M., Mull, T., et al. 1999. A high-density integrated genetic linkage and radiation hybrid map of the laboratory rat. Genome Res. 9: AP1-8. [PubMed] [Google Scholar]
- Swinburne, J., Gerstenberg, C., Breen, M., Aldridge, V., Lockhart, L., Marti, E., Antczak, D., Eggleston-Stott, M., Bailey, E., Mickelson, J., et al. 2000. First comprehensive low-density horse linkage map based on two 3-generation, full-sibling, cross-bred horse reference families. Genomics 66: 123-134. [DOI] [PubMed] [Google Scholar]
- Tavaré, S. 1986. Some probabilistic and statistical problems in the analysis of DNA sequences. Lect. Math. Life Sci. 17: 57-86. [Google Scholar]
- True, J.R., Mercer, J.M., and Laurie, C.C. 1996. Differences in crossover frequency and distribution among three sibling species of Drosophila. Genetics 142: 507-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan, S., Lio, P., and Goldman, N. 2001. Molecular phylogenetics: State-of-the-art methods for looking into the past. Trends Genet. 17: 262-272. [DOI] [PubMed] [Google Scholar]
- Yang, Z., Goldman, N., and Friday, A. 1994. Comparison of models for nucleotide substitution used in maximum-likelihood phylogenetic estimation. Mol. Biol. Evol. 11: 316-324. [DOI] [PubMed] [Google Scholar]
- Yauk, C.L., Bois, P.R., and Jeffreys, A.J. 2003. High-resolution sperm typing of meiotic recombination in the mouse MHC Eβ gene. EMBO J. 22: 1389-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, A., Zhao, C., Fan, Y., Jang, W., Mungal, A.J., Deloukas, P., Olsen, A., Doggett, N.A., Ghebranious, N., Broman, K.W., et al. 2001. Comparison of human genetic and sequence-based physical maps. Nature 409: 951-953. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.genome.ucsc.edu; University of California–Santa Cruz Genome Bioinformatics site.
- http://www.ratmap.gen.gu.se; RatMap.