Abstract
Nucleotide insertion and deletion (indel) events, together with substitutions, represent the major mutational processes of gene evolution. Through the alignment of 8148 orthologous genes from human, mouse, and rat, we have identified 1743 indel events within rodent protein-coding sequences. Using human as an out-group, we reconstructed the mutational event underlying each of these indels. Overall, we found an excess of deletions over insertions, particularly for the rat lineage (70% excess). Sequence slippage accounts for at least 52% of insertions and 38% of deletions. We have also evaluated the selective tolerance of identifiable protein structures to indels. Transmembrane domains are the least, and low complexity regions, the most tolerant. Mapping of indels onto known protein structures demonstrated that structural cores are markedly less tolerant to indels than are loop regions. There is a specific enrichment of CpG dinucleotides in close proximity to insertion events, and both insertions and deletions are more common in higher G+C content sequences.
Together with point mutations, insertions and deletions (indels) provide the raw material for evolutionary change in gene sequences (Soding and Lupas 2003). Analyzing the mutational processes that result in insertions and deletions between distantly related proteins is problematic, as indels frequently occur in regions where amino acid sequences are not well conserved, making exact placement of the event difficult. Moreover, the longer the time interval separating the divergence of two orthologous genes, the greater the likelihood that multiple mutational events have occurred, potentially masking the factors that caused the indel events. To reconstruct the evolutionary history of indel events, it is desirable to study closely related sequence pairs. The ideal situation would be to study spontaneous indel events occurring within the genome of a single species. Such a general strategy would require massive sequencing of individuals to identify indel mutations. Cases in which indels lead to disease phenotypes, however, are more readily identifiable, and the mechanisms and sequence features associated with them have been the subject of a recent review (Chuzhanova et al. 2003).
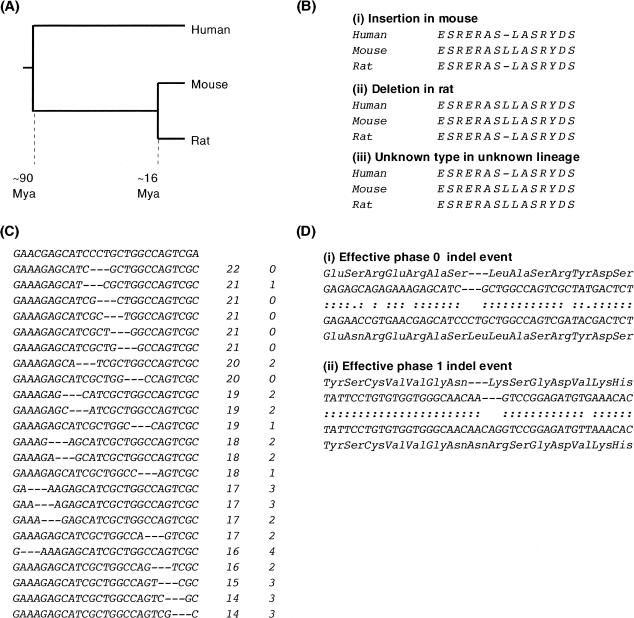
A different approach is to study indel events in gene pairs from closely related genomes of different species. The recently sequenced mouse and rat genomes (Waterston et al. 2002; Rat Genome Sequencing Project Consortium 2004) provide a wealth of such pairs. By aligning the sequences of mouse and rat orthologs, together with their human ortholog as an out-group, we can identify indel events that occurred in the rodent lineage, and infer whether they represent lineage-specific insertions or deletions (Fig. 1).
Figure 1.
Identification of coding indel events. (A) The relative phylogenetic relationship of human, mouse, and rat. Mouse and rat last shared a common ancestor ∼16 million years ago (Mya; Springer et al. 2003). Rodents and humans last shared a common ancestor ∼90 Mya (Springer et al. 2003). (B) An alignment gap between mouse and rat could be caused by an insertion in one rodent or a deletion in the other. By alignment to a homologous sequence from an out-group species (here, human) that shared a common ancestor with rodents prior to the divergence of mouse and rat lineages, the mutational event can, through parsimony, be resolved into either an insertion (i) or a deletion (ii) in a specific lineage. (iii) Alignment gaps in the out-group (human) relative to the rodent sequences cannot be resolved through parsimony. (C) Optimal nucleotide alignment. Amino acid alignment indicates the approximate nucleotide position of an insertion or deletion event, in this case a single amino acid deletion in the mouse sequence. The rat sequence (bold) is aligned with the deletion-containing mouse sequence, where a deletion sized gap (1 codon) is allowed to “slide” between –12 and +12 nt of the position indicated by amino acid alignment. These alignments are scored by nucleotide identity between mouse and rat (column 2) and then the number of transition substitutions (column 3). If there are multiple optimal gap positions, the 5′-most position is arbitrarily used for further analysis. (D) Effective phase of indel events. Alignments show mouse (upper rows) and rat (lower rows) amino acid and nucleotide sequences. Amino acids are gapped as indicated by initial amino acid alignment, nucleotides after optimization as shown in panel C. Colons indicate nucleotide identity and periods show transition substitutions. (i) Optimal nucleotide alignment shows that this deletion event is likely to have deleted the third nucleotide of a serine codon and the first two nucleotides of a leucine codon. It is therefore a phase 2 event. However, because the event did not cause an amino acid substitution as well as a deletion, it can be considered an effective phase 0 event (the same consequence on the encoded protein as a phase 0 event). (ii) Optimal nucleotide alignment indicates that this insertion event is also a phase 2 event. In this case, the indel results both in the insertion of an amino acid and the substitution of an amino acid. This can be considered an effective phase 1 event.
Indel events occur throughout the genome, but those that occur within protein-coding regions are particularly subject to selective constraints: reading frames must be maintained and the structural consequences of an event must preserve protein function if it is to be fixed within a population. Previous studies of indel events within proteins have either focused on compositionally biased regions such as trinucleotide repeats (Hancock et al. 2001; Lai and Sun 2003), or distantly related protein pairs (Pascarella and Argos 1992). We wished to understand the complex interplay of mutation and selection in coding sequence, and so have analyzed orthologous mouse, rat, and human coding sequences that provide an opportunity to investigate indel events that have persisted despite selection. Our results demonstrate that duplicative insertion and deletion of directly repeated sequence (collectively referred to as slippage), even outside the context of extended repeat regions (n > 2), is a major initiator of indel events, and that indels occur nonrandomly in proteins, their genes, and the mammalian genome.
RESULTS
Identification of Insertion and Deletion Events
We aligned the coding sequences of 8148 gene triplets that had previously been shown to have orthologous 1:1:1 relationships between mouse, rat, and human (Rat Genome Sequencing Project Consortium 2004). We identified high-quality regions of alignments that included an insertion or deletion in either of the rodent sequences (see Methods). Only indel events that did not shift the downstream reading frame or introduce an in-frame stop codon would have been selected in this screen. After purging events that could be explained by differences in genomic annotation rather than insertion or deletion of genomic sequence (see Methods), a nonredundant set of 1743 coding indel events in mouse and rat remained. For comparison, there were 3,026,519 nonredundant codons in the aligned data set that passed all filtering criteria applied to indel events (see Methods), which equates to the accumulation of 1 indel event per 1736 codons during the divergence of mouse and rat from a common ancestor. Exactly 1000 examples of rat- or mouse-specific deletions were inferred by the presence of amino acids at these positions in the human out-group sequence. Similarly, the remaining 743 cases were inferred to be rat- or mouse-specific insertions by the presence of gaps at orthologous positions in the human sequence (Fig. 1).
The 1743 indel events were identified in 1282 different genes. Within this set of genes, >98% contained three or fewer indel events, and the mean number of events was 1.4 per gene. These indel events, therefore, are well distributed through coding sequence and do not represent a small number of atypical genes, a conclusion also supported by the data presented in Figure 3 below and Supplemental Figure SF1 available online at www.genome.org.
Figure 3.
The G+C and CpG context of indel events. G+C nucleotide and CpG dinucleotide content was measured in nucleotide windows of 10, 50, 100, 500, and 5000 nt centered around indel events (measurements taken from the event sequence). Ten simulated data sets were used to calculate background levels of nucleotide and dinucleotide frequency. In both charts, error bars indicate standard error of the mean (SEM). Although SEM is plotted for all data points, the bars are too small to resolve on some data points. (A) Average G+C percent (Y-axis) plotted against window sizes used to measure local sequence composition (X-axis, log scale). (B) Average CpG dinucleotide count as a percentage of dinucleotides within window sizes used to measure local sequence composition (X-axis, log scale).
Event Frequency and Size Distribution
We found no significant difference in the total number of indels (counting insertions and deletions in one set) between the two rodent lineages (Table 1). Coding sequence deletion events are more common than insertions in each of the rodent lineages (Table 1; Fig. 2). However, there is a species-specific difference in the extent of this bias: It is substantially greater in rat (deletion to insertion ratio of 1.7:1) than it is in mouse (ratio of 1.1:1). There is also a significant excess of deleted versus inserted codons in the rat but not in mouse (Table 1), despite the mean insertion length being slightly longer (2.0 codons) than the mean deletion length (1.8 codons).
Table 1.
Frequency of Insertion (Ins) and Deletion (Del) Events
| Ins + Del | Ins | Del | Sum Ins | Sum Del | Mean Ins length | Mean Del length | |
|---|---|---|---|---|---|---|---|
| Rat | 855 | 320a | 535a | 664b | 951b | 2.1 | 1.7 |
| Mouse | 888 | 423 | 465 | 828 | 860 | 2.0 | 1.8 |
| Total | 1743 | 743c | 1000c | 1492d | 1811d | 2.0 | 1.8 |
Sum Ins and Sum Del denote the total number of codons inserted and deleted, respectively. Superscript characters indicate significant (P < 0.0001, two-tailed Fisher's exact test) departures from the null hypothesis that insertions and deletions occur with equal frequency (columns Ins and Del) or encompass an equal total length of sequence (columns Sum Ins and Sum Del).
Figure 2.
Frequency distribution of insertion and deletion event lengths. The X-axis shows categories of event length and the Y-axis frequency counts (raw counts) for observed indel events from each event length, type, and species category. The X-axis is truncated at 14 codons for clarity; indels were observed up to 28 codons in length. See Table 1 for the significance estimates of differences evident in the graph.
Indels varied in length from 1 to 28 codons (Fig. 2), with 66% involving a single codon. The frequency of indels decreased approximately exponentially with increasing indel size. Insertions and deletions in both mouse and rat all showed the same excess of small events (Fig. 2). No deletions in excess of 12 codons were found, whereas 1% of insertions extended the open reading frame by at least 13 codons. The 1% of insertions >13 codons in length accounted for all of the difference in mean event length between insertions and deletions (0.2 codons).
Reading Frame Bias of Events
A single DNA insertion or deletion event, as well as leading to the addition or deletion of amino acids, can also cause an amino acid substitution in the protein sequence encoded by the gene. The likelihood of this occurring depends on the phase of the indel event relative to the reading frame of the gene. A phase 0 event (between codons) will have no effect on the translation of adjacent codons. Similarly, a phase 2 event (between second and third codon positions) will usually be synonymous, owing to the degeneracy of the genetic code, creating a codon from the fusion of the first two bases, and the last base of the ancestral codons affected by the indel event. However, a phase 1 event will typically result in the substitution of an adjacent amino acid as well as the gain or loss of amino acids from the event itself. The more drastic effect of phase 1 events indicates that they might be less likely to be observed (Fig. 1).
We directly measured the phase of indel events in the 319 cases in which there was a single optimal nucleotide alignment between mouse and rat (see Methods and Fig. 1), such that the phase could be determined unambiguously. In these data, phase 1 events were found at a lower frequency than expected by chance (p < 0.05, two-tailed χ2 test, based on a null hypothesis of a 1:1:1 distribution of phase 0, 1, and 2 events). However, the prevalence of slippage-like indels (discussed subsequently) indicates that those with a single optimal alignment probably only represent a minor subset of indels. To overcome this issue, we evaluated the consequences of indel events at the amino acid rather than nucleotide level. In 216 of the 1743 events (12%), an amino acid substitution accompanies the indel event; these can be considered to be effectively phase 1 and are subsequently referred to as substitution-accompanied events. Within the simulated data sets (see Methods), 29% of events were substitution-accompanied, demonstrating a significant (p < 0.001, two-tailed χ2 test) underrepresentation of these events in the observed versus the null model (random) represented by the simulated data sets.
Event Sequence Complexity
The relationship of the event sequence to its flanking sequence was studied to gain insight into the mechanism of indel mutation. We were particularly interested in the origins of inserted DNA and wondered if the sequence adjacent to an event could provide a template for its synthesis in a manner similar to that of polymerase slippage in trinucleotide repeat expansion (Strand et al. 1993; for review, see Li et al. 2002). We will subsequently use the term “slippage-like” (Zhu et al. 2000) to collectively refer to duplicative insertion (e.g., TCAGA → TCAGCAGA) and its reciprocal: deletion of a direct repeat (TCAGCAGA → TCAGA). Inserted (from the event sequence) or deleted (from the non-event sequence) nucleotides were compared with the nucleotides immediately adjacent to the site of the indel event. Cases in which the inserted or deleted nucleotides were identical to an adjacent sequence were categorized as slippage-like events.
We considered a null model of indels occurring at random in coding sequence. The prevalence of slippage-like indels was estimated using simulated data sets (see Methods). Considering just single trinucleotide indels, 7.5% of simulated events were categorized as slippage-like. Because a longer nucleotide word is by chance less likely to have an identical word adjacent to it, this was reduced to 4.9% when all event sizes were considered. In total, 770 of the 1743 indel events (44%) have an identical adjacent sequence to that inserted or deleted. This represents a nine-fold enrichment over the null model (p < 0.0001, Table 2). However, the 770 include events occurring within a context of low-complexity nucleotide tracts, sequences that are known to be prone to polymerase slippage (Lai and Sun 2003). If events within low-complexity nucleotide tracts (two or more trinucleotide repeats excluding the inserted or deleted sequence) are excluded, there remains a highly significant (p < 0.0001, Table 2) fivefold enrichment of slippage-like indels over the null model (Table 2).
Table 2.
Detection of Sequence Slippage
| Observed data (% slippage-like) | Simulated data (% slippage-like) | Significance (p) | ||
|---|---|---|---|---|
| All events | Deletions | 38.00% | 4.66% | <0.0001 |
| Insertions | 52.49% | 5.28% | <0.0001 | |
| Single-codon events | Deletions | 37.37% | 7.50% | <0.0001 |
| Insertions | 57.03% | 7.53% | <0.0001 | |
| Nonrepeat single codona | Deletions | 29.55% | 6.94% | <0.0001 |
| Insertions | 38.92% | 6.70% | <0.0001 |
Slippage-like is defined to be when a sequence adjacent to the indel is identical to the inserted or deleted sequence. For the simulated data, slippage status was averaged over all 1000 simulated data sets. P-values were calculated from two-tailed χ2 tests on raw counts.
The nonrepeating single-codon-event data sets have been purged of all events that were observed in the context of trinucleotide repeats of two or more adjacent trinucleotides (excluding the inserted or deleted sequence itself)
The categorization of indel events into insertions or deletions revealed a striking disparity in the proportion of inferred slippage-like events (Table 2). More than 52% of insertions were categorized as slippage-like, whereas a lower proportion (38%) of deletions fell into the same category. We note that the extant sequence in which we observe an indel may not be identical to the ancestral sequence in which it occurred. As it is more likely that a pair of identical sequences will diverge, rather than a pair of nonidentical sequences converge, by nucleotide substitution, the values of 52% and 38% can be considered to be conservative estimates. In total, 380 deletion events and 390 insertion events were annotated as slippage-like by the criteria described (see Methods). This is not significantly different (p > 0.8, two-tailed Fisher's exact test) from the null hypothesis that slippage contributes equally to both insertion and deletion events. Therefore, the excess of deletions over insertions is due to a bias specifically in non-slippage-like rather than slippage-like events.
Sequence Biases in Event Proximity
We investigated the sequences proximal to indel events to determine whether certain motifs were statistically over- or underrepresented in these contexts. Such overrepresentation has been reported previously in the context of deletion events and combined insertion–deletion events in human genes associated with disease (Chuzhanova et al. 2003). Overrepresented motifs may correspond to regions that are particularly prone to copying errors, such as DNA polymerase pause sites (Krawczak and Cooper 1991), or those that are most susceptible to replication slippage (Schlotterer and Tautz 1992; Hartenstine et al. 2000).
The frequencies of three- and four-letter nucleotide words were measured in windows located symmetrically around real and simulated indel events (see Methods). Words were identified that deviated significantly in frequency between real and simulated events (Table 3; Supplemental Tables S1 and S2). The only sequence motif that was found to be significantly underrepresented in the proximity of indels was TTT (p = 2.57 × 10-5), and similar T-rich sequences TAT and ATT were also underrepresented to lesser degrees (p < 0.01; Supplemental Table S1). However, several nucleotide words were found to be significantly overrepresented (Table 3; Supplemental Tables S1 and S2). Several relationships between these words could be identified, including circular permutations and reverse complements. The most plausible groupings that were consistent with the results shown in Table 3 were circular permutations of the trinucleotide CAG; those containing only purines or only pyrimidines, for example, GAGG and CCCC; and high G+C content words containing a CpG dinucleotide. These same groups of overrepresented sequences were identified using multiple window sizes and distributions around the indel and simulated events (ranges 1–12, 7–12, 1–6, 1–4, and 1–3 nt [nucleotides] from the boundaries of the event). The magnitude of differences in frequency between real and simulated events diminished with increasing distance from the site of the event. For example the four-letter word CAGC was 2.5-, 2.2-, 2.0-, and 1.8-fold overrepresented in the ranges of 1–4, 1–6, 1–12, and 7–12 nt around an event, respectively. This signal decay is not simply a measure of the contribution of repetitive sequence. If all indels located within 12 nt of a CAGn trinucleotide repeat (n > 2) are removed from both observed and simulated data sets, the same nucleotide ranges show 1.9-, 1.7-, 1.5-, and 1.4-fold overrepresentation of the CAGC word.
Table 3.
Overrepresented Four-Letter Words in the Proximity of Indels (Nucleotides in the Range 1—4 From the Indel)
|
All indels
|
Insertions
|
Deletions
|
Slippage-like
|
Non-slippage-like
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Worda | Obs | Sim | Obs | Sim | Obs | Sim | Obs | Sim | Obs | Sim |
| GCGG3 | 25 | 8.3 | 15 | 3.5 | 10 | 4.8 | 11 | 2.9 | 6 | 2.9 |
| GGCG3 | 19 | 7.1 | 11 | 3.1 | 8 | 4.0 | 9 | 2.4 | 6 | 2.4 |
| GCCG3 | 23 | 8.7 | 16 | 3.6 | 7 | 5.1 | 11 | 3.0 | 4 | 3.0 |
| GAGG2 | 62 | 24.1 | 33 | 10.4 | 29 | 13.7 | 31 | 8.5 | 9 | 8.0 |
| CAGC1 | 80 | 31.6 | 52 | 13.6 | 28 | 17.9 | 42 | 11.2 | 24 | 10.5 |
| CGGC3 | 23 | 9.3 | 13 | 3.9 | 10 | 5.4 | 12 | 3.1 | 6 | 3.2 |
| CGCC3 | 20 | 9.0 | 14 | 3.8 | 6 | 5.2 | 12 | 3.2 | 5 | 3.0 |
| ACAA | 34 | 15.4 | 11 | 6.5 | 23 | 8.9 | 14 | 5.3 | 13 | 5.3 |
| AGAA2 | 64 | 30.0 | 25 | 13.0 | 39 | 17.0 | 34 | 10.6 | 10 | 10.1 |
| CCTC2 | 44 | 20.9 | 21 | 9.1 | 23 | 11.8 | 22 | 7.4 | 10 | 7.1 |
| CCGC3 | 18 | 8.5 | 13 | 3.6 | 5 | 4.9 | 7 | 2.9 | 4 | 2.9 |
| AGCA1 | 47 | 22.6 | 25 | 9.9 | 22 | 12.7 | 26 | 8.1 | 11 | 7.5 |
| CCCC2 | 35 | 17.0 | 18 | 7.2 | 17 | 9.8 | 14 | 5.8 | 12 | 5.8 |
| GACG3 | 14 | 6.9 | 11 | 2.9 | 3 | 4.0 | 3 | 2.4 | 2 | 2.4 |
| GCAG1 | 56 | 28.7 | 37 | 12.3 | 19 | 16.4 | 28 | 10.1 | 11 | 9.6 |
| CCAC | 35 | 19.1 | 21 | 8.3 | 14 | 10.8 | 14 | 6.7 | 5 | 6.4 |
| AAGA2 | 47 | 27.6 | 17 | 11.8 | 30 | 15.8 | 25 | 9.6 | 8 | 9.4 |
Obs indicates observed frequency counts and Sim the mean frequency counts from 1000 simulated data sets. The complete version of this table (256 rows), showing all four-letter words and additional columns detailing standard deviations and p-values, is available as Supplemental Table S2 (http://www.genome.org). This table shows all rows of the complete table containing significant (p < 0.01) differences between observed and simulated data, indicated by values in bold. Rows are sorted by descending fold overrepresentation for all indels.
Several of the nucleotide words are related. These are indicated by superscript values:
possible permutation of the trinucleotide CAG;
oligo-purine or oligo-pyrimidine tract;
high G+C content words containing a CpG dinucleotide
Categorization of indel events into insertions or deletions, and slippage-like or not (see previous section), allowed the contribution of proximal sequence biases to be studied in greater detail (Table 3; Supplemental Tables S1 and S2). The overrepresentation of high G+C content, CpG-containing words was found to be most prevalent in insertion rather than deletion events (Table 2). Similarly, words based on circular permutations of the CAG trinucleotide were found to be significantly overrepresented in the proximity of insertions but not deletions (Table 3). Considering the sequence contexts of deletion events, two four-letter nucleotide words showed significant (p < 0.001) overrepresentation. One of these was ACAA, a nucleotide word that falls outside the three identified groups of indel-associated sequence. The other deletion-associated word was AGAA, which falls into the poly-purine/pyrimidine category. Several polypurine/pyrimidine words were found to be overrepresented in the context of both insertion and deletion events, but were more specifically associated with slippage-like events (Table 3). It was also noted that poly-purine or poly-pyrimidine words tended to be overrepresented in the proximity of both insertions and deletions, but these were not robust to correction for multiple testing (Supplemental Tables S1 and S2).
The specific association of high G+C content CpG-containing nucleotide words with insertion rather than deletion events indicated a possible general correlation between G+C content and insertion frequency. Moreover, given the general underrepresentation of CpG dinucleotides in mammalian genomes (Lander et al. 2001; Waterston et al. 2002), the specific association of CpGs with small coding insertions was intriguing. These observations were investigated by measuring both C+G content and CpG frequency in nucleotide windows centered on indel events. To consider both local sequence and wider genomic influences on indel frequency, a range of window sizes was used (10 bp, 50 bp, 100 bp, 500 bp, and 5000 bp). The null hypothesis that indel events occur independently of the G+C content or CpG dinucleotide frequency was represented by simulated data sets (see Methods).
We found that all categories of events (insertion, deletion, slippage, and non-slippage) occurred in the context of higher G+C content than the background level found in our simulated data sets (Fig. 3A; data not shown). This G+C content enrichment was detected for all window sizes used, although the effect was greatest in small window sizes (Fig. 3A). Sequences in close proximity (window sizes of 10, 50, and 100 nt) of insertion events, irrespective of their slippage-like status, were significantly enriched for CpG dinucleotides (Table 4; Fig. 3B). The magnitude of this enrichment diminishes with larger window sizes (Table 4; Fig. 3B), indicating that, as with G+C content, it is the sequence in immediate proximity of the event, rather than the more general genomic context, that has most influence on insertion occurrence or persistence. In contrast, sequences in the local proximity of deletion events did not significantly differ in their CpG content from the background level (Table 4; Fig. 3B). In larger windows sizes (500 bp and 5000 bp), both insertions and deletions were enriched in CpG dinucleotides (Fig. 3B). However, this effect in larger window sizes could be a consequence of the more dramatic enrichment of G+C content of all events at these window sizes (Fig. 3A).
Table 4.
Frequency of CpG Dinucleotides in the Proximity of Indel Events
|
10-bp window
|
50-bp window
|
|||||
|---|---|---|---|---|---|---|
| Observed (CpG %) | Simulated (CpG %) | pa | Observed CpG %) | Simulated (CpG %) | pa | |
| Slippage-like | 2.54 | 2.90 | ns | 3.25 | 2.83 | <0.001 |
| Non-slippage-like | 3.53 | 2.90 | <0.01 | 3.28 | 2.83 | <0.001 |
| Insertion | 5.30 | 2.87 | <0.0001 | 3.90 | 2.83 | <0.0001 |
| Deletion | 2.70 | 2.93 | ns | 2.71 | 2.83 | ns |
| Slippage-like insertion | 5.48 | 2.90 | <0.0001 | 3.62 | 2.83 | <0.0001 |
| Slippage-like deletion | 2.54 | 2.90 | ns | 2.75 | 2.83 | ns |
| Non-slippage-like insertion | 5.17 | 2.90 | <0.0001 | 4.44 | 2.83 | <0.0001 |
| Non-slippage-like deletion | 3.02 | 2.90 | ns | 2.91 | 2.83 | ns |
Significance calculated as a non-paired, two-tailed t-test between the distribution of CpG dinucleotide frequencies in observed and simulated data sets. Nonsignificant values (p > 0.01) are denoted by ns
Gene Function and Indel Frequency
The tolerance of a gene to indels might be correlated with protein cellular localization or function. To test this hypothesis, we calculated indel rates for groups of genes from the 1:1:1 ortholog set categorized by a selection of Gene Ontology (GO) terms (Gene Ontology Consortium 2001). We find that genes whose products are annotated as localized in the cytoplasm have accumulated indels at a slower rate than those annotated as extracellular or nuclear (Fig. 4). Similarly, enzymes acquire indels at a slower rate than ligand-binding proteins. As these indel rates exactly match the trends observed for nucleotide substitution evolutionary rates (Waterston et al. 2002), it is clear that the selective constraints on nucleotide substitutions and indels are tightly coupled. In addition, we also observed that mitochondrially localized proteins show the lowest rate of indels, whereas transcriptional and cell cycle regulators are relatively enriched for indels (Fig. 4).
Figure 4.
Comparison of indel frequency between cellular compartments, biological processes, and molecular functions. Y-axis values show indel rates (indels per codon) averaged between mouse and rat for genes categorized by a selection of Gene Ontology (GO) terms. Terms from the three principal GO domains are considered: cellular compartment, biological process, and molecular function. Error bars represent estimates of standard error. Only the (Ensembl; http://www.ensembl.org/) annotation of human genes was considered and applied to the mouse and rat orthologs of those genes. Total indicates the rate based on all genes.
Insertion/Deletion-Tolerant Regions of Proteins
Globular protein domains are likely to be subject to greater purifying selection than sequences, such as low-complexity regions, with fewer structural and functional constraints. This view is consistent with an analysis of nonsynonymous versus synonymous substitution rates within known protein domains and non-domain sequences (Waterston et al. 2002). In addition, different types of nonglobular structure, such as coiled-coil and transmembrane domains are expected to differ in their tolerance of amino acid substitution. Similarly, we expected there would be marked differences in the tolerance of insertion and deletion events between structurally and functionally distinct regions of proteins. Although defined by sequence, low-complexity regions (see Methods) were also considered, as these are often of an unstructured nature and are generally considered to be subject to relatively limited selective constraints. Table 4 summarizes both the background level of low-complexity, transmembrane, coiled-coil, protein domain, and signal peptide sequences in our aligned data set, and the frequency with which these structural features participate in indel events.
As with amino acid substitutions (Waterston et al. 2002), insertion and deletion events are significantly underrepresented in known protein domains. However, the most dramatic underrepresentation of both insertions and deletions is in transmembrane domains, with fivefold fewer indel events occurring in these regions than would be expected by chance. Whereas coiled-coil and signal peptide regions appear to accumulate indels at the background rate, low-complexity regions are enriched for both insertions and deletions (Table 4).
We found that 52 indel-containing sequences could be aligned to known protein structures (see Methods). Regions that aligned to indel events were found to be depleted in regular secondary structure, relative to the structures as a whole: 31.5% of sequence aligning to indels was assigned to secondary structure as opposed to 52.5% of all residues. This result is consistent with indels being more tolerated in loop regions that are not part of the stabilizing core of the protein structure.
DISCUSSION
The availability of rat, mouse, and human genomes (Lander et al. 2001; Waterston et al. 2002; Rat Genome Sequencing Project Consortium 2004) has enabled us to identify indels in rodent protein-coding sequences, discriminate between insertions and deletions, and to begin to understand their underlying causes. We would expect that indel events in protein-coding sequence arise from the same mutational events that cause indels in non-coding DNA. However, selective pressures in protein-coding genes typically are greater than in noncoding sequences, so only a subset of the indel mutational spectrum may be observable through rodent to human protein-coding sequence comparisons.
As a starting point to this work, we used a set of 1:1:1 orthologous genes (Rat Genome Sequencing Project Consortium 2004) so that the relationship between aligned sequences was known and consistent. However, in doing so, very rapidly evolving genes and members of gene families with multiple close paralogs may be underrepresented in the starting data set. Supplemental Figure SF1 summarizes an analysis of the functional bias in this data set compared with all genes. The dominant effect is that unannotated genes are underrepresented in the 1:1:1 data set, and those with annotation are generally enriched (Supplemental Fig. SF1). This probably reflects the amount of information in the public domain to facilitate annotation; if there is little evidence for annotation, there is also likely to be little evidence for the identification of orthologous sequences in other genomes.
Relative to all genes, receptors and immune genes showed evidence for underrepresentation in the 1:1:1 ortholog set. The receptors group contains the odorant receptors, a classic example of a large multigene family (Zhang and Firestein 2002). Immune genes are often subject to diversifying selection and accumulate changes at a higher rate than other sequences (for review, see Hughes and Yeager 1997), making the identification of orthologous sequences problematic. Overall, the 1:1:1 ortholog set appears to be generally representative of most genes, but because subsets that may be accumulating indels at the highest rates are underrepresented, our measures of coding indel rates should be considered conservative in the context of all protein-coding genes.
Several recent lines of evidence are converging on a consensus that human, mouse, and rat genomes have accumulated small deletions at faster rates than small insertions. A small-scale comparison of genes and pseudogenes in humans, mice, and rats identified a 2.5-fold excess of deletions over insertions almost exclusively in the pseudogenes (Ophir and Graur 1997). Waterston et al. (2002) reported an apparent excess of small deletion over insertion events in interspersed repetitive elements of the mouse genome. A 2.9-fold excess of deletions has been reported in a large set of ribosomal protein pseudogenes (Zhang and Gerstein 2003). Recent analysis of indels in interspersed repetitive elements in both mouse and rat shows that both rodents have accumulated deletions at more than twice the rate of insertions (Rat Genome Sequencing Project Consortium 2004). Because insertions and deletions within interspersed repetitive elements and processed pseudogenes are likely to be selectively neutral, these measures provide a good proxy for background rates of insertion and deletion events.
We find that this excess of deletion over insertion is also readily detectable in the rat coding sequence. However, the effect is diminished by selection. A deletion-to-insertion ratio of 3.1:1 (Rat Genome Sequencing Project Consortium 2004) in non-protein-coding (neutral site) sequence is reduced to 1.7:1 in coding sequence. In mouse, the neutral site deletion-to-insertion-ratio of 2.5:1 (Rat Genome Sequencing Project Consortium 2004), is reduced to 1.1:1 in coding sequence. Although we observe more coding sequence deletions than insertions, the reduced excess of deletion events in coding versus noncoding sites indicates that as a proportion of those events that arise by mutation, insertions are more tolerated in protein-coding sequence than deletions.
Selection
We observed mouse and rat indel rates of 5.46 × 10-12 and 5.27 × 10-12 per coding nucleotide per million years, assuming a divergence time of 16 million years (Springer et al. 2003). Based on indel sizes of 3, 6, and 9 nt, Cooper et al. (2004; G. Cooper, pers. comm.) have measured whole-genome indel rates of 4.78 × 10-11 for mouse and 5.20 × 10-11 for rat, again assuming 16 million years as a divergence time. For comparison, if we consider just indels of 1, 2, and 3 codons, the mouse and rat indel rates are 6.11 × 10-12 and 5.89 × 10-12, an order of magnitude lower than the whole-genome rates, indicating that selection has been active in the purification of many coding sequence indels. It is probable that indels with non-3n size multiples are subjected to even greater purifying selection as they would disrupt the reading frame and in many cases abrogate function. Such indels would not have been detected in our screen.
The consequences of purifying selection are most apparent in the distribution of indel events in the encoded protein rather than at the DNA level. In those cases in which indel events could be reliably mapped onto known protein structures, there was enrichment within relatively unstructured regions of the proteins and an underrepresentation of indels within the secondary structure elements, relative to that expected by chance. A previous study of indels found among more divergent sequences than those considered here demonstrated that indels within helices and strands were underrepresented compared with those within loops (Pascarella and Argos 1992). Indels within secondary-structure elements are likely to disrupt multiple interactions within the core of a globular protein fold and destabilize its structure. Consequently, these are more likely to have been removed by purifying selection.
We found that indels are underrepresented in known protein domains (Table 5). This parallels the nucleotide substitution rate differences between domains and nondomains (Waterston et al. 2002). A notable negative result is that there is no substantial difference between the percentage of insertions versus the percentage of deletions found in the context of different protein structural features (Table 5). This indicates that the greater tolerance of insertions relative to deletions, when averaged across all protein sequences, is not more prevalent in any particular protein structural feature (Table 5). Of the five categories of protein structural and sequence features investigated, transmembrane helices showed the least tolerance of indels, a fivefold reduction over that expected by chance. This probably reflects the combined constraints of transmembrane length and the helical register of amino acid side chains within the membrane. Low-complexity regions harbored more indels than expected by chance, reflecting both the higher intrinsic mutability arising from trinucleotide repetition (Lai and Sun 2003; homopolymeric tracts are included in low-complexity regions), and limited selective constraints on these regions (Hancock et al. 2001).
Table 5.
Protein Context of Insertion and Deletion Events
| Background (% neighboring residues) | Ins (%) | Del (%) | Total events (%) | |
|---|---|---|---|---|
| Low complexity (Seg) | 8.80 | 28.94 | 26.40 | 27.48 |
| Transmembrane | 5.13 | 0.94 | 0.80 | 0.86 |
| Coiled coil | 2.69 | 3.10 | 2.10 | 2.52 |
| Pfam domain | 39.30 | 12.65 | 10.10 | 11.18 |
| Signal peptide | 1.00 | 1.88 | 1.30 | 1.54 |
The context of events was measured from annotation of the nonevent sequence (ancestral with respect to the indel). The background measure is the percentage of residues meeting the defined criteria, in the rodent nonredundant protein data set (see Methods).
Analysis of indel rates broken down by GO annotation indicated that in general, cytoplasmic and mitochondrial proteins accumulate indels at a slower rate than either extracellular or nuclear localized proteins (Fig. 4). We also found an excess of indels associated with transcriptional regulators that may relate to the overrepresentation of homopolymeric tracts in transcription factors and DNA-binding proteins (Albà and Guigó 2004). Such tracts would be considered low-complexity regions in our analysis of protein context, and they may be encoded by trinucleotide repeats, sequences that are known to be prone to polymerase slippage (Pearson and Sinden 1998).
Purifying selection was also evident in the reading frame bias of indel events. There was a significant 2.4-fold underrepresentation of indels that resulted in the substitution of amino acids as well as their gain or loss. This indicates that the regions of proteins in which we have identified indel events are not simply minimally constrained regions of proteins that will effectively accommodate any sequence change that maintains the open reading frame. Rather, they are sites in proteins that are subject to a range of constraints but are able to tolerate a specific insertion or deletion and maintain function.
Slippage-Like Events
The majority of insertions (52%) and a substantial fraction of deletions (38%) were found to have a sequence identical to the inserted or deleted sequence, directly adjacent to the indel event (Table 2). This was significantly more than expected by chance and was not simply a consequence of expansion and contraction of repetitive sequences (Table 2) such as microsatellites, which are known to be subject to dynamic changes in tandem repeat copy number (Pearson and Sinden 1998). Similar findings have been reported by Nishizawa and Nishizawa (2002), who used a statistical approach to categorize alignment gaps between gene/pseudogene pairs as slippage-like or non-slippage-like. They reported that ∼80% of insertions in human pseudogenes and ∼50% of insertions in rodent pseudogenes could be categorized as slip-page-like. Zhu et al. (2000) investigated disease-causing human insertion events and found that 70% of insertions could be categorized as duplicative insertions (or slippage-like using our nomenclature). Concordance in estimates of slippage-like indel frequency between studies of neutral (Nishizawa and Nishizawa 2002), selectively tolerated coding (this study) and detrimental coding sequence insertions (∼50% for rodents, 70%–80% for human; Zhu et al. 2000), indicates that the prevalence of slippage-like indels in coding sequence is a consequence of mutational mechanisms rather than selection.
The predominant mutational mechanism of repeat expansion (insertion) and contraction (deletion) is thought to be that of polymerase slippage, also known as slipped strand mispairing (for review, see Li et al. 2002). It is proposed (Strand et al. 1993) that mispairing of DNA strands during replication or recombination can result in a single-stranded loop that, depending on how it is resolved by the DNA repair machinery, may result in an indel mutation. Tandemly repeated sequences are readily able to mispair through the duplexing of nonorthologous repeat units. Although the propensity for polymerase slippage is positively correlated with repeat length (Rose and Falush 1998), even very short (n = 2) repeats exhibit a higher-than-background frequency of insertion and deletion (Bell and Jurka 1997; Nishizawa and Nishizawa 2002). Elevated indel frequency in repeated sequence could account for the higher-than-expected frequency of deletions categorized as slipped, because the ancestral sequence would have had at least two repeat units in direct repeat. Likewise, the high frequency of slippage-like insertions in the context of repetitive sequence can be accounted for. However, this model is difficult to reconcile with the 5.8-fold higher than expected frequency of slippage-like insertions that are not in the context of tandem repeats (i.e., the post event sequence contains a tandem repeat, although none is present in the ancestral sequence). The high prevalence of nonrepeat slippage-like insertions indicates a polymerase-slippage-like mechanism that is partially independent of perfect direct repeats. This is reminiscent of the concept of cryptic simplicity (Tautz et al. 1986), in which scrambled arrangements of repetitive motifs, in addition to simple direct repeats, result in elevated rates of insertion and deletion (Tautz et al. 1986; Hancock and Volger 2000).
Although a smaller fraction of deletion events than insertion events could be considered as slippage-like, there was no significant difference between the total number of slipped insertions and deletions. Therefore, the excess of deletions in the complete data set is not caused by a general bias toward deletion of polymerase slippage. Rather it is caused by an excess of non-slippage-like deletions over non-slippage-like insertions.
Indel-Neighboring Motifs
We identified three categories of motifs that are significantly overrepresented in close proximity to indels: permutations of CAG (for insertions only), oligo-purine and oligo-pyrimidine tracts (insertions and deletions), and high G+C CpG-containing words (insertions only). Both the CAG permutation words and oligo-purine/oligo-pyrimidine tracts were associated specifically with slippage events, and both are known to be particularly prone to polymerase slippage in the context of direct repeats (Sinden et al. 2002). Our results indicate that these sequences are also prone to polymerase slippage even when not in the context of multiple, tandemly arranged direct repeats. In contrast, non-slippage-like indels were not strongly associated with any specific nucleotide words and we did not observe any patterns or correlations that could explain a mechanism for their generation, nor could we subcategorize them further. Non-slippage-like indel events may represent a mechanistically heterogeneous group of mutational events, although it is interesting to note that non-slippage-like deletions are 1.8-fold more frequent than non-slippage-like insertions.
We noted a strong strand asymmetry in the word count results (Table 3). Of the 18 significantly overrepresented four-letter words, 10 contained an A nucleotide whereas only 1 contained a T nucleotide. This is surprising, because for every overrepresented word containing an A, its reverse complement contains a T (all word counts were carried out on the sense strand). With the three-letter words, a similar pattern is observed, but it is interesting to note that several T-rich words were also underrepresented in indel flanking sequence (Supplemental Table S1). Based on our present data, it is not possible to distinguish mutational bias from the consequences of selection in explaining this strand asymmetry. Strand asymmetries in mutational bias have previously been reported for transcribed sequences (Green et al. 2003; Majewski 2003), and there is a plausible mechanism for them to arise, that of transcription-coupled DNA repair (for review, see van den Boom et al. 2002). However, the strand bias could also be explained by biases in codon usage (Sharp et al. 1993) or the amino acid context of indels.
G+C Content
On average, insertions are observed in higher G+C content sequences than deletions, but both are more frequent in higher G+C contexts than simulated events. There is a significant enrichment of CpG dinucleotides in the proximity of insertion, but not deletion, events. Enrichment of CpG dinucleotides in the proximity of insertions is independent of slippage status, and the magnitude of the effect diminishes rapidly with increasing window size around the event: for example, 1.8-fold enrichment at a window size of 10 bp, but a 1.4-fold enrichment at a window size of 50 bp. To our knowledge, a specific correlation of CpG dinucleotide frequency with insertion events has not previously been reported. However, we note that Brock et al. (1999) found that coding trinucleotide repeats located within CpG islands were generally more prone to repeat expansion than those not within CpG islands, a finding that is consistent with our observations. Because this correlation has not previously been investigated in selectively neutral sequence, we are unable to categorically attribute this observation to either mutational bias or the consequence of selection.
Guanine plus cytosine content varies considerably across mammalian genomes (for review, see Bernardi 2000) and has previously been shown to correlate with gene content (Saccone et al. 1996), gene function, expression levels (D'Onofrio 2002), interspersed repeat content, transposition, recombination, nucleotide substitution (Lander et al. 2001; Waterston et al. 2002; Hardison et al. 2003), and deletion rates (Hardison et al. 2003). We find that coding sequence insertions and deletions can also be counted among this number.
Conclusions
The same mutational processes alter coding and noncoding sequences, but selection acts on these discriminately. Our identification and analysis of coding sequence indels has allowed both the mutational origin and selective consequences of indel events to be considered. The consequences of purifying selection on indels are most readily detected at the protein rather than DNA level, with transmembrane domains being particularly intolerant of indels. We find that sequence slippage, probably through a slipped strand mispairing-like mechanism, is a major contributor to the generation of both insertions and deletions, even outside of the context of directly repeated sequences. A subset of nucleotide sequences is particularly prone to slippage, both in repeat and nonrepeat contexts. We have also identified an enrichment of CpG dinucleotides in the proximity of insertion but not deletion events. Further work comparing our results to those based on selectively neutral sites will clarify whether this enrichment can be attributed to mutation or the subsequent action of selection.
METHODS
Amino Acid Alignment
All protein, transcript, and gene structure data sets were obtained from Ensembl (http://www.ensembl.org/; Hubbard et al. 2002; Clamp et al. 2003) version 11, based on whole genome assembly versions 31 (human), 30 (mouse), and 2 (rat). Analyses were based on a set of 8148 genes in which a single orthologous gene could be identified in each of rat, mouse, and human (1:1:1 orthologs). This data set is described elsewhere (Rat Genome Sequencing Project Consortium 2004; provided by L. Goodstadt and A. Ureta-Vidal). The translation of each human transcript of each 1:1:1 ortholog triplet was aligned using BLAST (version 2.2.5, with options -pblastp -m6 -FF -P1; Altschul et al. 1997) to the translations of its orthologous mouse and rat transcripts. Regions were identified where the alignment was gapped between human and one rodent but not between human and the other rodent. To exclude alignment errors, candidate insertion and deletion events were filtered based on local alignment quality. Two windows were positioned symmetrically around each indel event, encompassing residues located from two to seven amino acids from the alignment gap. A minimum of four identical amino acids between mouse and rat and no alignment gaps were required in each window.
Nucleotide Alignment and Filtering
Amino acid alignment coordinates were transformed into genomic sequence coordinates using Ensembl transcript tables. Candidate indel events whose genomic coordinates were within 12 nt of an Ensembl “frame fixing” intron (<6 nt in length) were excluded from further analysis. Amino acid alignment, although an efficient and robust method of identifying indel events, is not able to pinpoint the exact site of an event owing to the triplet and degenerate nature of the genetic code. An optimal nucleotide alignment allowing a single gap identifies the most likely (parsimonious) site of the indel event. Rat and mouse genomic sequences incorporating the candidate indel event were aligned by allowing an indel event sized gap to slide from –12 to +12 nt centered around the coordinate indicated by the amino acid alignment. The optimal alignment was that with the highest identity and, if there were ties, then that with the most transitions (purine to purine, pyrimidine to pyrimidine substitutions) in the alignment. In the event of a further tie, the 5′-most optimal position was arbitrarily selected as the reference coordinate. Redundant events, a legacy of aligning all transcripts of each gene, were filtered out using the reference genomic coordinate. If any of the optimal alignments for an indel placed the alignment gap at a splice site or in intronic sequence, the candidate event was considered to be an artifact because of differences in genomic annotation and was excluded from further analysis.
Simulated Data Sets
The null model of coding sequence indel events is that they occur randomly and are not influenced by reading frame or local sequence composition. This null model was represented by simulations of the observed indel events, matching event organism, type (insertion or deletion) and event length, but scattering the location of events at random over the reference amino acid alignments (see below). Therefore, each simulated event could be considered paired with an observed event, and a simulated data set would contain one paired simulation for every observed indel event. Accordingly, 1000 such simulated data sets were produced. Insertions were simulated by deleting from the nonevent sequence. The phase of simulated events was randomized, giving equal probability of phase 0, 1, or 2 indel events. Each simulated event was filtered in the same manner as real observations for local alignment quality and splice-site proximity. Any simulations failing the filtering criteria were repeated with a new random site. The Perl “rand” function (http://cpan.org) was used to generate random numbers and was reseeded between the generation of data sets.
Word Counts
Nucleotide words of length n were counted using a sliding-window approach. For example, each of the four-letter words CAGC, AGCC, and GCCA would be counted once only in the sequence CAGCCA. These word counts were performed in windows positioned symmetrically around indel events, over the ranges 1–3, 1–4, 1–6, 7–12, and 1–12 nt from the indel event. The frequency (raw count) of a word from observed events was compared with its frequencies (raw counts) in 1000 simulation data sets. p-values were calculated directly from the normal distribution (two-tailed) of word frequency (raw counts) in the simulated data sets (pnorm function of the R statistical package; http://www.r-project.org/). These p-values were Dunn-Sidak-corrected for multiple testing (1 – (1 – p)n, where p is the p-value and n the number of possible nucleotide words, n = 256 in the case of four-letter words).
Detecting Sequence Slippage
The inserted (from the event sequence) or deleted (from the non-event sequence) nucleotide sequence was compared with the immediately adjacent 5′ and 3′ sequence in both event and non-event sequences. Indels were categorized as slippage-like if any of the adjacent words were identical to the inserted or deleted sequence. In cases in which the adjacent word was not identical but one substitution away from the indel sequence, the indel was categorized as “other” for the purposes of slippage analysis, as they are likely to represent a mixture of non-slippage-like indels and slippage-like indels that subsequently underwent substitution. Arrays of tandemly repeated sequences were detected by an extension of the principle where both the event and nonevent sequences were evaluated and a match to either extends the count of adjacent tandem words. The inserted or deleted sequence itself was excluded from the count. The 1000 simulated data sets were used as the null model.
Analysis of Protein Context
Annotations of transmembrane domains, signal peptides, protein domains (Pfam; Bateman et al. 2002), coiled-coil regions, and Gene Ontology (GO) terms were based on Ensembl (http://www.ensembl.org/) annotation. Low-complexity regions were annotated with Seg (Wootton 1994) using default parameters. Annotation of sequence type was not mutually exclusive. For each mouse and rat gene in the aligned data sets, the protein isoforms involved in the highest scoring (BLASTP bit score) was used as a reference (referred to as the rodent nonredundant protein data set) for calculating background percentages of amino acids annotated in each of the five categories of protein sequence type. An indel was considered to be in the context of a sequence type if either of the amino acids flanking the indel coordinate in the nonevent sequence were annotated as that sequence type.
Each indel-event-containing sequence was searched against sequences derived from the Protein Data Bank (PDB, http://www.rcsb.org/pdb/) using BLASTP (Altschul et al. 1997). Cases in which the residues flanking the indel event were aligned within a high-scoring segment pair with a statistical significance of E < 10-10 were mapped onto secondary structures derived from DSSP files. The secondary-structure states of all the residues in the DSSP file that fell between the aligned residues flanking the indel event were counted and compared with the secondary-structure states of the PDB protein as a whole. Regular secondary structure was taken to be (H) α-helix, (G) 310 helix, (E) β-strand, and (B) β-bridges.
Acknowledgments
We thank Leo Goodstadt (MRC-FGU, Oxford), and Abel Ureta-Vidal (EBI, Hinxton) for providing us with sets of orthologous genes, and Gregory Cooper (Stanford) for unpublished results. We are also grateful to Richard Mott (WTCHG, Oxford) and Goncalo Abecasis (University of Michigan) for informative discussions. M.S.T. and R.R.C. are supported by the Wellcome Trust and C.P.P. by the UK Medical Research Council.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1977804.
Footnotes
[Supplemental material is available online at www.genome.org. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: L. Goodstadt, A. Ureta-Vidal, G. Cooper, and the Rat Genome Sequencing Project.]
References
- Albà, M.M. and Guigó, R. 2004. Analysis of amino acid repeats in rodents and humans and relationship to GC content. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman, A., Birney, E., Cerruti, L., Durbin, R., Etwiller, L., Eddy, S.R., Griffiths-Jones, S., Howe, K.L., Marshall, M., and Sonnhammer, E.L. 2002. The Pfam protein families database. Nucleic Acids Res. 30: 276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, G.I. and Jurka, J. 1997. The length distribution of perfect dimer repetitive DNA is consistent with its evolution by an unbiased single-step mutation process. J. Mol. Evol. 44: 414-421. [DOI] [PubMed] [Google Scholar]
- Bernardi, G. 2000. Isochores and the evolutionary genomics of vertebrates. Gene 241: 3-17. [DOI] [PubMed] [Google Scholar]
- Brock, G.J., Anderson, N.H., and Monckton, D.G. 1999. Cis-acting modifiers of expanded CAG/CTG triplet repeat expandability: Associations with flanking GC content and proximity to CpG islands. Hum. Mol. Genet. 8: 1061-1067. [DOI] [PubMed] [Google Scholar]
- Chuzhanova, N.A., Anassis, E.J., Ball, E.V., Krawczak, M., and Cooper, D.N. 2003. Meta-analysis of indels causing human genetic disease: Mechanisms of mutagenesis and the role of local DNA sequence complexity. Hum. Mutat. 21: 28-44. [DOI] [PubMed] [Google Scholar]
- Clamp, M., Andrews, D., Barker, D., Bevan, P., Cameron, G., Chen, Y., Clark, L., Cox, T., Cuff, J., Curwen, V., et al. 2003. Ensembl 2002: Accommodating comparative genomics. Nucleic Acids Res. 31: 38-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, G.M., Brudno, M., Stone, E.A., Dubchak, I., Batzoglou, S., and Sidow, A. 2004. Characterization of evolutionary rates and constraints in three mammalian genomes. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- D'Onofrio, G. 2002. Expression patterns and gene distribution in the human genome. Gene 300: 155-160. [DOI] [PubMed] [Google Scholar]
- Gene Ontology Consortium. 2001. Creating the gene ontology resource: Design and implementation. Genome Res. 11: 1425-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, P., Ewing, B., Miller, W., Thomas, P.J., and Green, E.D. 2003. Transcription-associated mutational asymmetry in mammalian evolution. Nat. Genet. 33: 514-517. [DOI] [PubMed] [Google Scholar]
- Hancock, J.M. and Vogler, A.P. 2000. How slippage-derived sequences are incorporated into rRNA variable-region secondary structure: Implications for phylogeny reconstruction. Mol. Phylogenet. Evol. 14: 366-374. [DOI] [PubMed] [Google Scholar]
- Hancock, J.M., Worthey, E.A., and Santibanez-Koref, M.F. 2001. A role for selection in regulating the evolutionary emergence of disease-causing and other coding CAG repeats in humans and mice. Mol. Biol. Evol. 18: 1014-1023. [DOI] [PubMed] [Google Scholar]
- Hardison, R.C., Roskin, K.M., Yang, S., Diekhans, M., Kent, W.J., Weber, R., Elnitski, L., Li, J., O'Connor, M., Kolbe, D., et al. 2003. Covariation in frequencies of substitution, deletion, transposition, and recombination during eutherian evolution. Genome Res. 13: 13-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartenstine, M.J., Goodman, M.F., and Petruska, J. 2000. Base stacking and even/odd behavior of hairpin loops in DNA triplet repeat slippage and expansion with DNA polymerase. J. Biol. Chem. 275: 18382-18390. [DOI] [PubMed] [Google Scholar]
- Hubbard, T., Barker, D., Birney, E., Cameron, G., Chen, Y., Clark, L., Cox, T., Cuff, J., Curwen, V., Down, T., et al. 2002. The Ensembl genome database project. Nucleic Acids Res. 30: 38-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, A.L. and Yeager, M. 1997. Molecular evolution of the vertebrate immune system. Bioessays 19: 777-786. [DOI] [PubMed] [Google Scholar]
- Krawczak, M. and Cooper, D.N. 1991. Gene deletions causing human genetic disease: Mechanisms of mutagenesis and the role of the local DNA sequence environment. Hum. Genet. 86: 425-441. [DOI] [PubMed] [Google Scholar]
- Lai, Y. and Sun, F. 2003. The relationship between microsatellite slippage mutation rate and the number of repeat units. Mol. Biol. Evol. 20: 2123-2131. [DOI] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Li, Y.C., Korol, A.B., Fahima, T., Beiles, A., and Nevo, E. 2002. Microsatellites: Genomic distribution, putative functions and mutational mechanisms: A review. Mol. Ecol. 11: 2453-2465. [DOI] [PubMed] [Google Scholar]
- Majewski, J. 2003. Dependence of mutational asymmetry on gene-expression levels in the human genome. Am. J. Hum. Genet. 73: 688-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa, M. and Nishizawa, K. 2002. A DNA sequence evolution analysis generalized by simulation and the Markov chain Monte Carlo method implicates strand slippage in a majority of insertions and deletions. J. Mol. Evol. 55: 706-717. [DOI] [PubMed] [Google Scholar]
- Ophir, R. and Graur, D. 1997. Patterns and rates of indel evolution in processed pseudogenes from humans and murids. Gene 205: 191-202. [DOI] [PubMed] [Google Scholar]
- Pascarella, S. and Argos, P. 1992. Analysis of insertions/deletions in protein structures. J. Mol. Biol. 224: 461-471. [DOI] [PubMed] [Google Scholar]
- Pearson, C.E. and Sinden, R.R. 1998. Trinucleotide repeat DNA structures: Dynamic mutations from dynamic DNA. Curr. Opin. Struct. Biol. 8: 321-330. [DOI] [PubMed] [Google Scholar]
- Rat Genome Sequencing Project Consortium. 2004. Genome sequence of the Brown Norway Rat yields insights into mammalian evolution. Nature (in press). [DOI] [PubMed]
- Rose, O. and Falush, D. 1998. A threshold size for microsatellite expansion. Mol. Biol. Evol. 15: 613-615. [DOI] [PubMed] [Google Scholar]
- Saccone, S., Caccio, S., Kusuda, J., Andreozzi, L., and Bernardi, G. 1996. Identification of the gene-richest bands in human chromosomes. Gene 174: 85-94. [DOI] [PubMed] [Google Scholar]
- Schlotterer, C. and Tautz, D. 1992. Slippage synthesis of simple sequence DNA. Nucleic Acids Res. 20: 211-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, P.M., Stenico, M., Peden, J.F., and Lloyd, A.T. 1993. Codon usage: Mutational bias, translational selection, or both? Biochem. Soc. Trans. 21: 835-841. [DOI] [PubMed] [Google Scholar]
- Sinden, R.R., Potaman, V.N., Oussatcheva, E.A., Pearson, C.E., Lyubchenko, Y.L., and Shlyakhtenko, L.S. 2002. Triplet repeat DNA structures and human genetic disease: Dynamic mutations from dynamic DNA. J. Biosci. 1: 53-65. [DOI] [PubMed] [Google Scholar]
- Soding, J. and Lupas, A.N. 2003. More than the sum of their parts: On the evolution of proteins from peptides. Bioessays 25: 837-846. [DOI] [PubMed] [Google Scholar]
- Springer, M.S., Murphy, W.J., Eizirik, E., and O'Brien, S.J. 2003. Placental mammal diversification and the Cretaceous–Tertiary boundary. Proc. Natl. Acad. Sci. 100: 1056-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand, M., Prolla, T.A., Liskay, R.M., and Petes, T.D. 1993. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature 365: 274-276. [DOI] [PubMed] [Google Scholar]
- Tautz, D., Trick, M., and Dover, G.A. 1986. Cryptic simplicity in DNA is a major source of genetic variation. Nature 322: 652-656. [DOI] [PubMed] [Google Scholar]
- van den Boom, V., Jaspers, N.G., and Vermeulen, W. 2002. When machines get stuck—Obstructed RNA polymerase II: Displacement, degradation or suicide. Bioessays 24: 780-784. [DOI] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- Wootton, J.C. 1994. Non-globular domains in protein sequences: Automated segmentation using complexity measures. Comput. Chem. 18: 269-285. [DOI] [PubMed] [Google Scholar]
- Zhang, X. and Firestein, S. 2002. The olfactory receptor gene superfamily of the mouse. Nat. Neurosci. 5: 124-133. [DOI] [PubMed] [Google Scholar]
- Zhang, Z. and Gerstein, M. 2003. Patterns of nucleotide substitution, insertion and deletion in the human genome inferred from pseudogenes. Nucleic Acids Res. 31: 5338-5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y., Strassmann, J.E., and Queller, D.C. 2000. Insertions, substitutions, and the origin of microsatellites. Genet. Res. 76: 227-236. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://cpan.org/; The Comprehensive Perl Archive Network.
- http://r-project.org/; The R-project for statistical computing.
- http://www.ensembl.org/; Ensembl.
- http://www.rcsb.org/pdb/; The Protein Databank.