Abstract
Bacteria can acquire genetic diversity, including antibiotic resistance and virulence traits, by horizontal gene transfer. In particular, many bacteria are naturally competent for uptake of naked DNA from the environment in a process called transformation. Here, we used optical tweezers to demonstrate that the DNA transport machinery in Bacillus subtilis is a force-generating motor. Single DNA molecules were processively transported in a linear fashion without observable pausing events. Uncouplers inhibited DNA uptake immediately, suggesting that the transmembrane proton motive force is needed for DNA translocation. We found an uptake rate of 80 ± 10 bp s−1 that was force-independent at external forces <40 pN, indicating that a powerful molecular machine supports DNA transport.
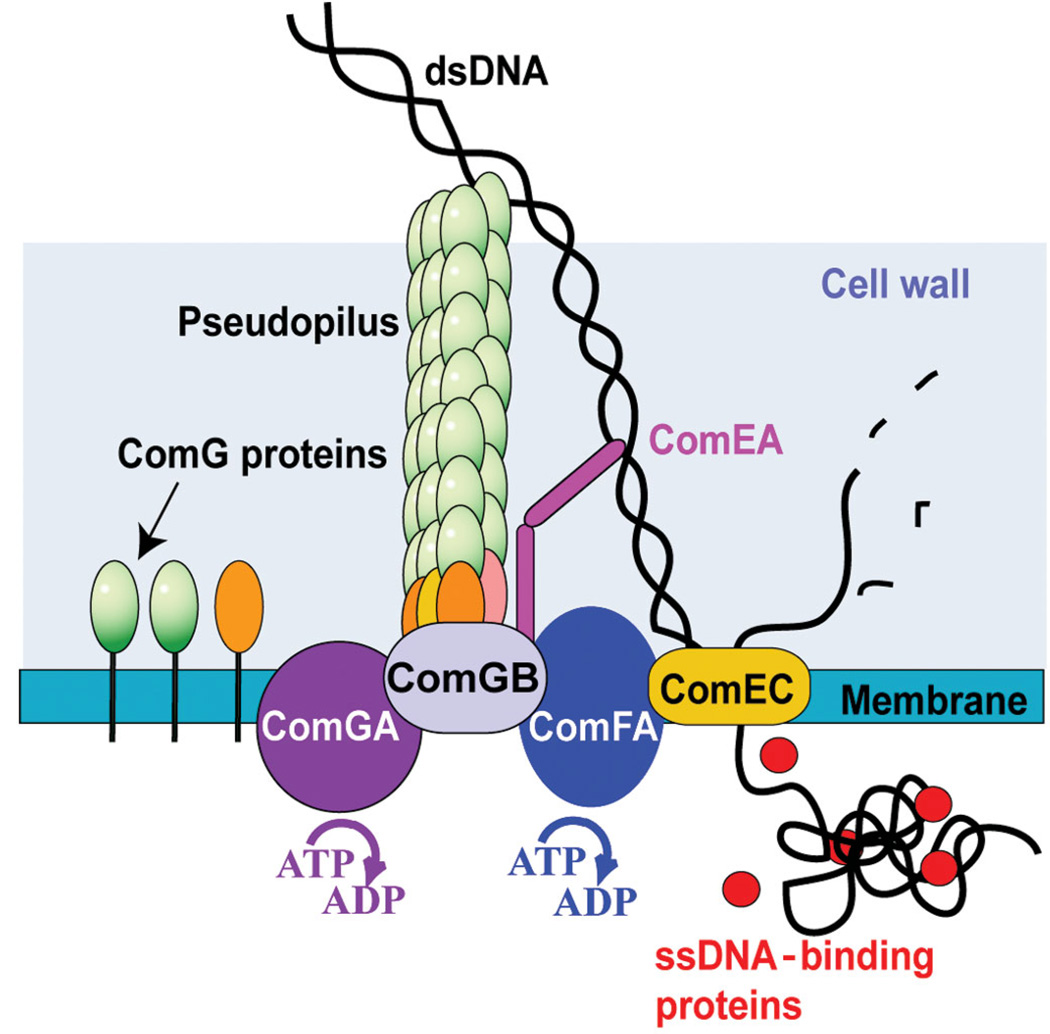
Natural competence for DNA transformation is a genetically programmed, physiological state. Competent cells express specialized proteins that assemble into a DNA transport complex that moves macromolecular DNA through the bacterial cell wall and membrane1,2. These competence proteins are most likely surface-exposed DNA receptors, which facilitate DNA translocation through the cell wall, membrane pores and motor molecules that power DNA transport (reviewed in ref. 3; Fig. 1). The first characterized step in the process of transformation of B. subtilis is the irreversible binding of DNA to the cell surface receptor, ComEA. For DNA to gain access to this receptor, the products of the comG operon must be present, although these proteins do not themselves seem to bind DNA. The ComG proteins resemble those involved in the assembly of type IV pili and in type II secretion, and may form a ‘pseudopilus’ that traverses the thick cell wall to permit access to ComEA. However, no structure resembling a true pilus (in the sense of a fiber that extends beyond the cell surface) has been described in B. subtilis. After binding, DNA is cleaved by the membrane-localized nuclease NucA. The newly introduced end of one strand then initiates transport across the membrane. In addition to ComEA, which seems to present the DNA end to the internalization machinery, transport requires the putative channel proteins ComEC and the ATPase ComFA. Genetic tests have demonstrated that DNA crosses the membrane in linear fashion, as the uptake of a linked pair of markers is delayed compared to that of either single marker.
Figure 1.
Molecular model for DNA transport through the cell envelope in Bacillus subtilis. The traffic NTPase ComGA is a member of a widespread family of proteins implicated in the formation and retraction of the type IV pilus and in protein secretion. These proteins contain Walker A and B nucleotide-binding motifs. It is reasonable to assume that ComGA participates in remodeling a pilus-like structure formed by the major pseudopilin ComGC and possibly also including the minor pseudopilins that may allow DNA to access the DNA receptor ComEA. One DNA strand is degraded and only one single strand enters the cytosol through the membrane channel ComEC. The ATP-binding protein ComFA is required for the transport of DNA through the membrane and has been proposed to contribute energetically to this process2,3.
Although many proteins that constitute the DNA transport machine have been identified, little is known about the molecular mechanism, rates, processivity, energetics and force generation of DNA transfer through the cell envelope. Manipulation of single molecules has been used to characterize these physical properties of translocation through nanometer-sized pores in bacteriophages4, Xenopus laevis eggs5 or artificial membranes6,7. Using laser tweezer technology, we found that a powerful molecular machine drives the transport of naked DNA by B. subtilis. Proton uncouplers rapidly block DNA transport, which implicates proton motive force as a major energy source. These features as well as the fact that the transport shows no pauses or slippage indicate that the bacterial DNA transport system differs substantially from other macromolecular transport systems.
RESULTS
DNA transport observed at the single-molecule level
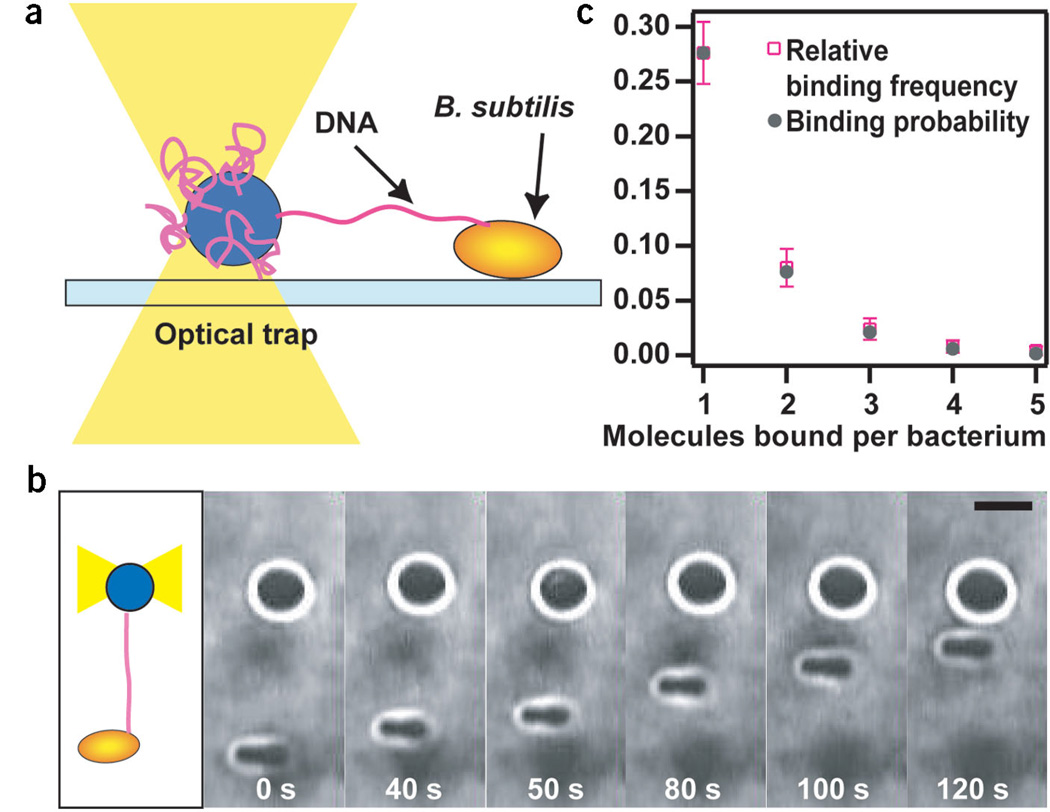
We designed an assay to investigate the transport of single DNA molecules by B. subtilis in vivo. To measure the rate at which single DNA molecules are transported, we immobilized nonflagellated bacilli on a hydrophobic glass surface (Fig. 2a). To increase the number of competent cells, we used a strain lacking Rok, which downregulates the competence regulator ComK8. DNA was bound to a bacillus by bringing a 2-µm bead coated with DNA to the bacterium at a trap stiffness of 0.003–0.005 pN nm−1. After ~15 s, we pulled the bead away from the bacterium until the photodetector detected a deflection of the bead from the center of the optical trap. The deflection indicated that a DNA tether between the bacterium and the bead had formed and that the DNA was under tension. The bead was then relocated to a position where the tension had just relaxed and was held stationary until the bacterium started to pull on the bead. In most cases, the tether broke within seconds. When the bacterium started to pull actively, however, we never observed breaking of the tether at forces <20 pN. We propose that there are at least two different mechanisms of DNA binding. First, DNA may bind transiently to the bacterial surface, explaining our observation that most tethers broke within several seconds. After this step, DNA binds stably to a functional DNA uptake complex, leading to DNA uptake. Moreover, we observed that only one in ~30 attempts led to DNA uptake. This observation is also in agreement with the assumption that initial binding is reversible. A typical time series of a DNA uptake event is shown in Figure 2b. As the bacterium pulled DNA into the intracellular space, the DNA tether between the bacterium and the bead shortened continuously. The position of the bacterium was readjusted by moving the microscopy stage to keep the bead in the linear force range of the laser tweezers.
Figure 2.
Experimental setup. (a) One extremity of a DNA molecule was attached to a microscopic bead immobilized by an optical trap. DNA bound to the surface of competent B. subtilis. Deflection of the bead from the center of the laser trap was measured during DNA transport. (b) Time series of a DNA uptake event. As the DNA tether shortened, the distance between the bacterium and the bead decreased. Scale bar, 2 µm. (c) Relative binding frequency and binding probability of DNA binding to one bacterium. ktrap = 0.03–0.05 pN nm−1. n = 250.
We tested whether the beads were bound by single or multiple DNA tethers. It is usually assumed that DNA uptake proceeds from newly formed ends and not from pre-existing ends2. A nuclease, NucA, cleaves the DNA and the newly formed ends are delivered to the membrane pore ComEC9. Thus, it is highly unlikely that we observed two DNA tethers of exactly the same length that were transported by the bacterium simultaneously. To verify this hypothesis, we investigated whether we could detect multiple binding events by holding a DNA tether until the attachment broke and then checking for further attachments. We assumed that the relative frequency of one binding occurrence was equal to its probability, P1 = relative frequency. The latter assumption should hold for a probability of 70% within an error of 0.07, n = 250 (Bernoulli law of large numbers). Then Pn = P1n. The number of experimentally detected bonds agreed very well with the number of expected bonds (Fig. 2c), indicating that single DNA uptake events are monitored by our technique.
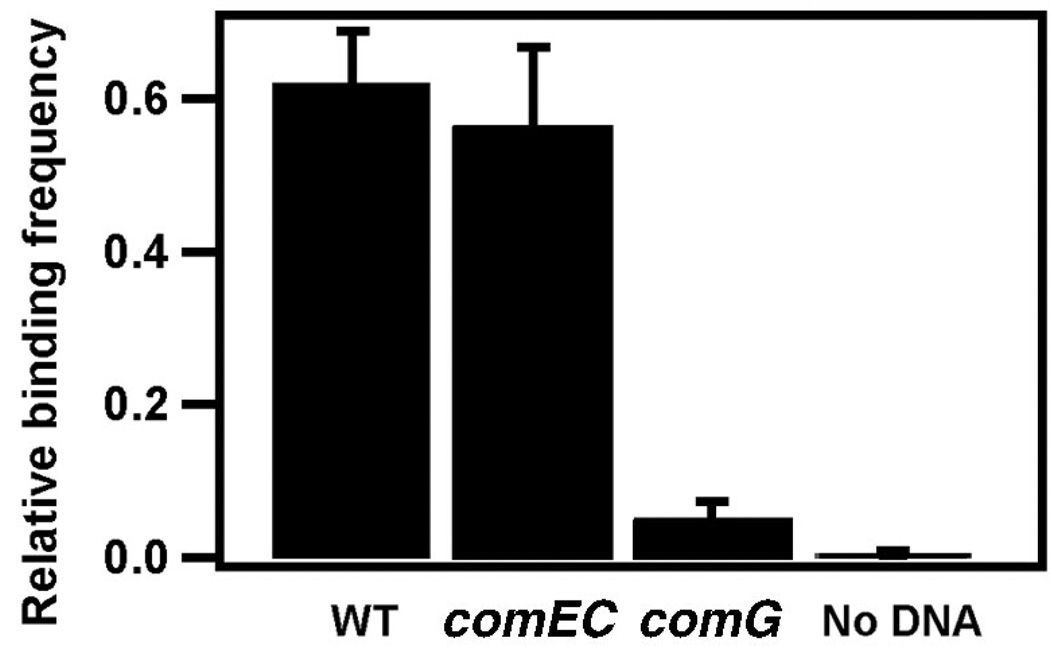
To distinguish between nonspecific and specific binding of DNA to B. subtilis or the silanized surface, we compared the frequency of DNA-coated bead binding to a bacterium expressing a complete DNA transport complex (BD3458) and a mutant lacking ComG proteins (BD3484). Binding requires the presence of proteins encoded by the comG operon, including ComGC, GD, GE and GG, which resemble type IV pilins. We assume that they form a structure that facilitates DNA transport through the proteoglycan layer and provides access to the DNA receptor ComEA10. The binding efficiency of the strain expressing an intact DNA transport complex was 12× higher than that of the comG mutant, indicating that most binding events are dependent on the ComG proteins (Fig. 3).
Figure 3.
DNA binding. Relative frequency of DNA binding to wild type (WT; BD3458) (n = 100), bacteria lacking the presumed membrane channel ComEC (BD3476) (n = 50), bacteria lacking the ComG proteins (BD3484) (n = 100) and beads without biotinylated DNA (BD3458) (n = 225). In most cases a DNA tether formed between the bead and the bacteria, but sometimes the bead was firmly stuck to the bacterium. Because the direct contact of the bead with the bacterium is irrelevant for our experiments we counted only the tethers.
ComEC, a membrane protein, supposedly forms a channel that allows the DNA to traverse the bacterial membrane. Deletion of ComEC inhibits DNA transport2. As a further control, we tested a comEC mutant (BD3476) for transport. The relative binding frequency was normal in BD3476 cells (Fig. 3); however, no uptake event was observed in 250 attempts. As we observed one DNA transport event in 30 attempts for BD3458, we conclude that ComEC is required for shortening of the DNA tether.
As a further control to show that binding was caused by DNA as opposed to another cell organelle, we measured the relative binding frequency with naked streptavidin beads. The relative binding frequency was strongly reduced as compared with the experiments with saturating DNA concentration (Fig. 3). The residual binding is probably due to DNA that was released by bacterial lysis and bound nonspecifically to the bead.
Kinetics of DNA translocation
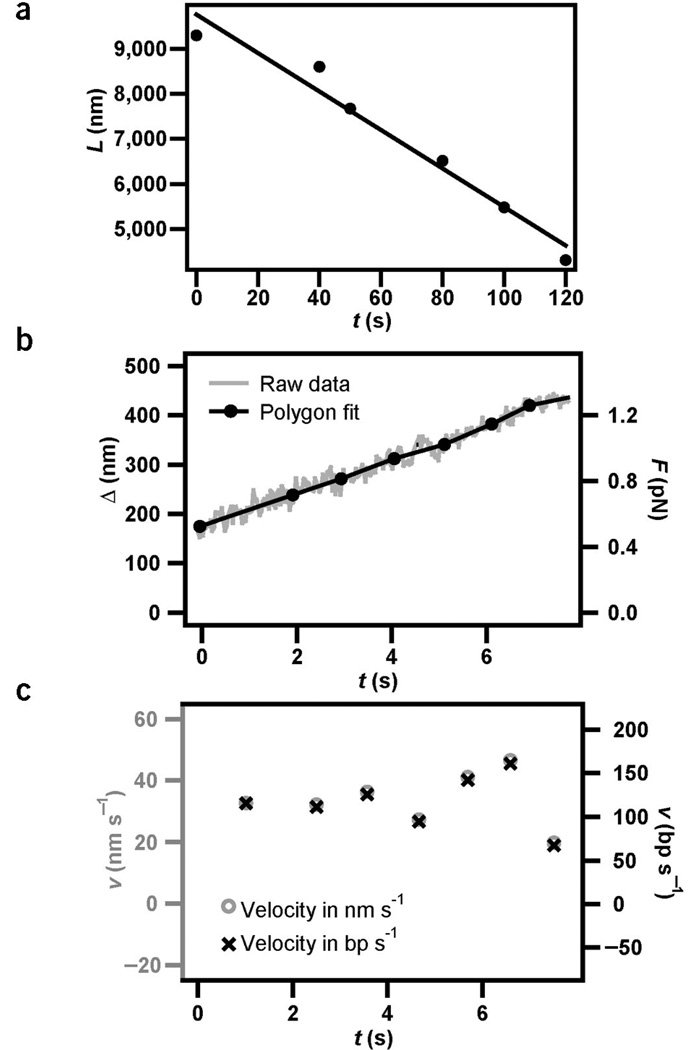
To determine the DNA transport rate, we recorded the length of the DNA tether as a function of time (Figs. 4a and 2b). On a long timescale (several minutes), the length of the DNA tether between the bacterium and the bead shortened continuously, indicating that DNA transport was highly processive. The bacterium shown in Figure 2b internalized DNA with a contour length of 7 µm within 2 min. The transport velocity did not change substantially over time, suggesting that the DNA uptake kinetics did not depend on the length of intracellular DNA as observed during DNA uptake into bacteriophage4. The average velocity was vlong = 43 ± 5 nm s−1.
Figure 4.
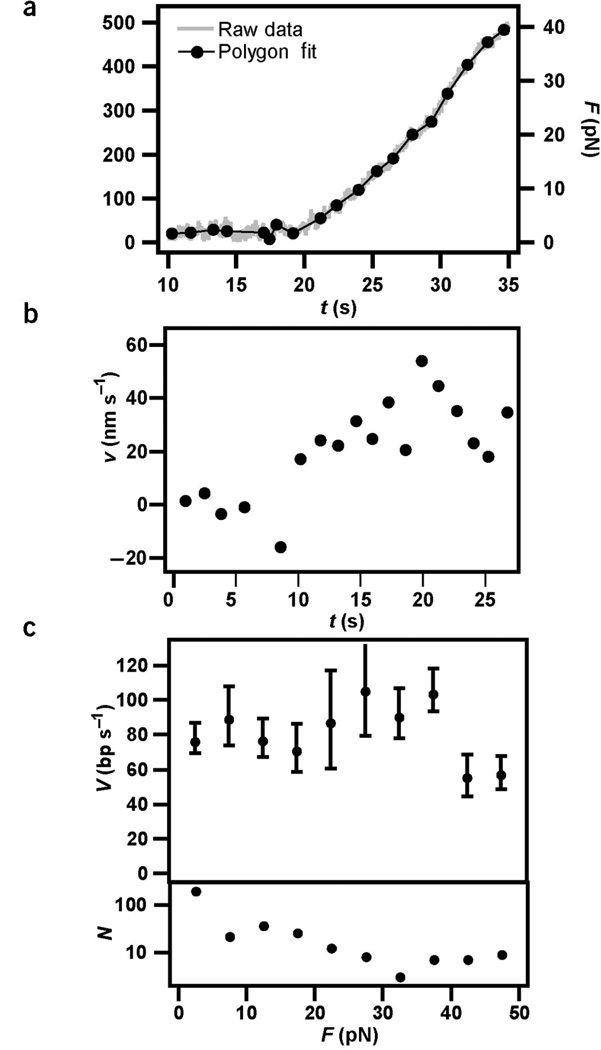
Kinetics of DNA uptake. The stiffness of the optical trap was ktrap = 0.004 pN nm−1. (a) The distance between the bead and the bacterium, L, decreased as a function of time during DNA uptake. The full line is the best linear fit, yielding an average uptake rate of vshort = 43 ± 5 nm s−1. (b) Example for tether shortening, Δ, as a function of time during DNA uptake and best polygon fit with an average time interval of 1 s. (c) Uptake velocity derived from b. The average uptake rate of the bacterium in a was vshort = 34 ± 4 nm s−1 (n = 6). Taking into account the elastic properties of DNA, positional information was converted into uptake velocity measured in base pairs per second (Supplementary Fig. 2 online).
To investigate pausing and reversibility we recorded shortening of the DNA tether Δ (Supplementary Fig. 1 online), the decrease of distance between the bead and the bacterium, with a temporal resolution of 1 s. On a timescale of t ≥ 1 s (a length scale of l ≥ 80 bp) we did not observe pauses, reversal or slips during DNA transport (Fig. 4b). To calculate the velocity of DNA uptake, the data was averaged over 1 s by fitting a polygon to the experimental data. The average value was vshort = 34 ± 4 nm s−1 (Fig. 4c) for six individual retraction curves from the bacterium in Figure 4a. The slight difference between the velocity measured on a long and short timescale is most likely due to the relaxed state of the DNA between short timescale measurements—that is, DNA transport could be affected by DNA bending.
Owing to its short persistence length, unperturbed λ-DNA with a contour length of 16 µm forms a tight coil in which the average endto- end distance is near zero11. By pulling a DNA tether between the bacterium and the bead, force was exerted between the extremities of the DNA molecule. With increasing force the end-to-end distance increased and, as a consequence, the positional information obtained in our experiments (Fig. 4c) accounts for a different number of base pairs at varying force. We independently measured the end-to-end distance of DNA under varying force under competence conditions. Although proteins are likely to bind to the DNA molecule we did not observe a substantial change in the elastic properties of DNA as compared with the elastic properties of DNA protein-free buffer11 (Supplementary Fig. 2 online). The end-to-end distance versus force curve in Supplementary Fig. 2 online was used to convert positional data into transport rate (Fig. 4c). The average rate was v = 80 ± 10 bp s−1 at F<4 pN.
DNA uptake proceeds against large external force
To investigate the effect of external force on the velocity of DNA transport, we measured the velocity of DNA transport at an optical trap stiffness of ktraP =0.16 pN nm−1. At forces >4 pN the bacterium sometimes moved continuously toward the bead. Therefore the bacterial movement was subtracted from the bead movement to yield the change in tether length Δ (Supplementary Fig. 1 online). Even when an external force in the range of 40 pN was applied, we did not observe pauses, backward steps or slipping on a timescale >1 s (Fig. 5a). Application of F < 40 pN on the DNA did not substantially reduce the DNA transport rate (Fig. 5b,c), although the slight decrease in velocity around 40 pN may mark the onset of the force-induced decrease of translocation velocity.
Figure 5.
Force generation during DNA transport. ktrap = 0.16 pN nm−1. (a) Example of tether shortening as a function of time. The full line is the best polygon fit with an average time interval of 1.3 s. DNA uptake started at t = 10 s. The force was calculated from the deflection of the bead from the center of the laser trap. Owing to continuous bacterial movement in this force range, the latter was <Δ (Supplementary Fig. 1 online). (b) Uptake velocity derived from a. (c) Top graph, velocity of DNA transport averaged over 31 retraction curves and a force interval of 5 pN. Bottom graph, number of samples per force interval.
Effect of proton motive force
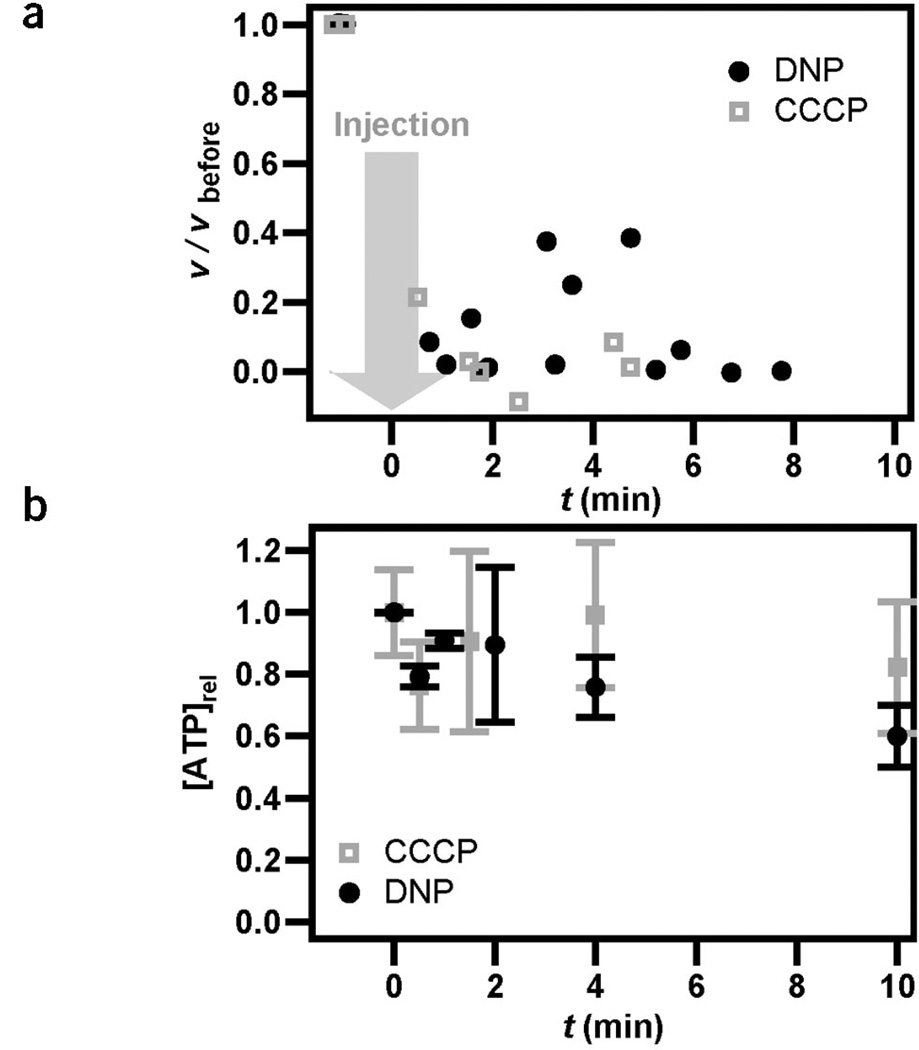
It has been suggested that proton motive force powers DNA transport. Experiments with bulk-competent cultures have shown that DNA transport is dependent on the proton gradient, but it has not been resolved whether the inhibition was caused by proton motive force or by the secondary effect of inhibition of F0F1 ATPase12. To address this question, we applied protonophores during the process of DNA transport. The effectiveness of the protonophores was measured by observing flagellar rotation (Methods). When carbonyl cyanide m-chlorophenyl hydrazone (CCCP) or 2,4-dinitrophenol (DNP) was added during retraction, the rate of DNA transport substantially decreased within 60 s (Fig. 6a). We next investigated whether this inhibition was caused directly by an effect on proton motive force or by a secondary effect on ATP synthesis due to inhibition of F0F1 ATPase. For this, we measured the ATP pool after addition of protonophores. The concentration of ATP did not substantially decrease within 4 min (Fig. 6b), indicating that proton motive force was a driving force for DNA uptake. These experiments do not exclude an additional requirement for other energy sources like ATP; the proton motive force is necessary but perhaps not sufficient to drive DNA uptake.
Figure 6.
DNA uptake is dependent on proton motive force. (a) During DNA retraction, 200 µl of 1 mM CCCP (concentration at bacterium, 200 µM) or 200 µl of 20 mM DNP (concentration at bacterium, 4 mM) was injected into the flow chamber (at t = 0). After injection of CCCP or DNP, the bead was still attached to the bacterium via the DNA tether but the speed of DNA uptake was substantially reduced. n = 2. (b) Time course of average ATP concentration per ~106 bacteria after injection of CCCP (200 µM) or DNP (4 mM) determined using a luciferase assay. n = 3.
In control experiments with bulk cultures, we investigated whether the inhibition of transformation by DNP and CCCP was reversible. First we confirmed that the exposure of competent cells to either inhibitor during incubation with DNA severely inhibited transformation, as expected. The inhibition with CCCP was ~400-fold and with DNP was ~60-fold. To test reversibility, competent cells were treated with CCCP or DNP for 5 min and were then washed to remove the inhibitors. The washed cells were incubated with transforming DNA. In the case of DNP, full transformability and viability were recovered. In the case of CCCP, the transformation frequency was recovered to a level only seven-fold below that of the untreated control. It is likely that washing did not remove all of the CCCP, as viability was also compromised (about ten-fold). We conclude that the inhibition of transformation due to the addition of DNP was completely reversible and that due to CCCP was at least partially reversible, consistent with our conclusion that they inhibit DNA uptake by collapsing the proton gradient.
DISCUSSION
We designed a single-molecule assay to study the kinetics and force generation of the molecular motor complex that powers DNA transport. Several control experiments indicated that the normal competence- specific DNA transport process caused the shortening of a tether between a bacillus and a small bead. First, formation of the tether was dependent on the presence of DNA on the beads. Second, DNA binding was dependent on ComG proteins. The latter are known to be necessary for DNA binding before DNA transport. Third, transport but not binding was inhibited in mutants lacking the presumed pore protein ComEC. Fourth, DNA shortening was shown to be an active cellular process that was inhibited by depleting the proton gradient across the bacterial membrane. Fifth, the transport rate observed in our experiments (80 ± 10 bp s−1) agrees reasonably well with previous estimates of the DNA transport rate using genetic markers. Multiple binding sites on one DNA molecule may explain the higher rate of v = 180 bp s−1 in bulk experiments in B. subtilis2. For instance, a DNA molecule carrying two genetic markers may bind at two sites, permitting the uptake of the markers by two transporters. Other molecules may bind at a single site from which both markers are taken up. The time between entry of the two markers is an average of all the molecules that bind at one or at more than one site and this average will be less than expected for single-site binding. This consideration is not a factor in the single-molecule experiment. In fact, it has been observed in bulk culture experiments with Streptococcus pneumoniae that the entry time does not increase linearly with DNA length in bulk experiments, presumably owing to simultaneous entry from more than one site on some of the DNA molecules13. Taking this into account, a transport rate of v = 90–100 bp s−1 has been estimated13, in further agreement with our single-molecule measurements.
Multiple energy sources may power the translocation of macromolecules through nanometer-sized pores2,14–16. Although our experiments strongly suggest that the transmembrane proton gradient is involved in DNA transport, we cannot exclude the possibility that other molecular motors may support the translocation process. For instance, ComFA, which shares similarities with DEAD helicases17 is required for DNA transport and ATP hydrolysis may directly participate in transport.
Comparison with other DNA-translocating molecular motors
Pausing times >4 s are very common in processive molecular motors that move along DNA, such as a viral portal motor4, exonucleases18, RNA polymerases19 and DNA polymerases20. All of the DNA translocases mentioned above have shown a substantial decrease in translocation rates at external forces ≥20 pN. The strongest DNA translocation motor that has been characterized so far, the portal motor of the B. subtilis phage Ω29, can work against F = 57 pN. However, even at low external forces, the uptake rate decreases substantially. Moreover, the portal motor regularly loses its grip on the DNA, leading to slipping. Even though we observed stretches of DNA uptake >7 µm, we never encountered pauses, reversals or slips during DNA uptake for t > 1 s (on a length scale of 80 bp). Furthermore, we found no substantial decrease of the DNA uptake rate at an external force of 40 pN. Thus the kinetics and force generation characteristics of the motor that drives DNA translocation through the bacterial envelope do not resemble those of other characterized DNA translocation motors.
DNA uptake and type IV pilus retraction
The mechanical characteristics of DNA transport observed here are reminiscent of type IV pilus retraction in Neisseria gonorrhoeae21. Type IV pili are polymeric filaments of 6-nm diameter that protrude several micrometers from the bacterial surface. These dynamic polymers elongate and retract, most likely by polymerization and depolymerization, into the inner membrane22. Pilus retraction does not exhibit marked pausing events and generates exceptionally high molecular forces. There is also no substantial decrease in retraction rate for forces <40 pN (ref. 21).
These kinetic similarities may suggest a role for pseudopilus retraction in DNA uptake. This suggestion is supported by the fact that many of the proteins involved in transformation are highly conserved in both Gram-positive and Gram-negative transformation systems and resemble those required for the assembly of type IV pili2. However, there is currently little genetic evidence for such a role. Type IV pilus systems generally have two related traffic ATPases, one for retraction (such as PilT)21 and another for pilus assembly. In contrast, the B. subtilis transformation system has only one, ComGA, which is more closely related to the assembly ATPase (PilF). Conceivably, retraction of the hypothetical competence pseudopilus could be driven directly by proton motive force, in the absence of a PilT ortholog. We conclude that DNA uptake and type IV pilus retraction share notable similarities in kinetics and the generation of large forces, although details of the molecular mechanisms need to be unveiled.
Concluding remarks
One explanation for the high efficiency of the DNA uptake motor, as compared with other DNA-translocating motors, may be that our experiments characterize fully functional complexes of molecular motors in vivo in contrast to in vitro experiments on isolated motor molecules. We anticipate that this whole-cell single-molecule approach will be useful not only for further unraveling the molecular mechanism of bacterial DNA uptake but also for other translocation processes of micron-sized molecules through nanopores.
METHODS
Bacterial strains and media
The nonflagellated (hag) B. subtilis strains BD3458 (rok8), BD3773 (rok comGA-gfp), BD3484 (rok comGA1223), BD3476 (rok comEC) and the flagellated strain BD2528 (multicopy comS24) were grown to T2, shock-frozen and stored at −80 °C. T2 is the time point 2 h after entry into stationary growth phase. The transformation frequency of BD3458 and BD3773 was 0.2 ± 0.1%. Immediately before an experiment was carried out, bacteria were thawed quickly, washed with Spizizen salts25 and adsorbed to a silanized coverslip for 10 min. Subsequently, unbound bacteria were washed away and competence medium26 with DNA-coated beads was added. Experiments were done at 37 ± 2 °C. A green fluorescent protein fusion to ComGA in BD3773 was used to monitor the development of competence microscopically.
DNA transformation in bulk
Transformation, selective and growth media, and the growth of strains to competence were as described or referenced23. For the measurement of transformation, DNA from strain BD170 was used with selection for leucine prototrophy.
Bacterial viability and energy supply
To ensure that our experimental conditions did not affect energy supply or viability, flagellated bacteria (BD2528) were adsorbed to a silanized coverslip. Some surface-bound bacteria rotated. The nonmotile bacteria were assessed for flagella rotation by approaching them with a bead at low laser trap stiffness; rotating flagella generated bead movement. At 20–80 min after plating, <20% of the surface-bound bacteria were nonmotile. At 180 min after plating, the percentage of nonmotile bacteria increased to 60%. Experiments were carried out <120 min after plating.
Time course of proton gradient depletion
Proton motive force powers flagella rotation27. We found that addition of 200 µM CCCP to flagellated B. subtilis stopped the rotation of the bacterium immediately. The addition of 4 mM DNP caused a gradual decrease in the velocity of flagella rotation between 1 min and 4 min after injection, and rotation stopped completely after 4 min.
Determination of ATP concentration
We used a luciferase ATP determination kit (Molecular Probes) to determine the relative ATP concentration of ~106 cells. Bacteria were thawed, washed and resuspended in competence medium. CCCP or DNP was added at t = 0. Cells were mixed with the luciferase kit components and subsequently incubated with 2% (v/v) Triton X-100 for 2 min. Luminescence was quantified using a scintillation counter.
DNA binding
λ-DNA was labeled with biotin using an end-filling procedure with biotin dUTP (Roche). Streptavidin-coated latex (Polysciences) beads (2.2 µm) were washed in PBS and incubated with biotinylated DNA overnight. Unbound DNA was removed by centrifugation.
A bead with multiple DNA molecules was held on top of a bacterium for 15 s at a trap stiffness of ktraP = 0.03–0.05 pN nm−1. Afterwards, the bead was pulled away from the bacterium. In some cases, the bead adhered firmly to the bacterium. In other cases, a DNA tether of several microns was observed between the bead and the bacterium. The attachment was considered positive for binding if the bead was either pulled out of the laser trap and flipped back on top of the bacterium or if the bacterium detached from the glass surface and followed the movement of the bead. Slipping events were ignored.
The standard error for the experimental value was calculated according to s.e.m. = (p(1 − p)(n − 1)−1)0.5, where p is the relative frequency of occurrence and n the sample size.
Measuring the DNA uptake rate
The optical tweezers system consisted of a Nd:YAG laser (2 W), an inverted Zeiss microscope and a movable mirror system that allowed computer-controlled deflection of the laser beam. We used an image acquisition system that contained a video camera (Nuvicon), a S-VHS video recorder and an SGI workstation running Isee particle tracking software (Inovision). For real-time monitoring of the bead deflection, acquisition of positional data was done as described28 at 1 kHz time resolution using a quadrant photodiode (Hamamatsu) and LabView software (National Instruments). We calibrated the trap stiffness by a spectrum analysis of the bead’s brownian motion and verified the force by viscous drag.
At forces >4 pN, the bacterium sometimes moved toward the bead. Thus, we determined the uptake rate using a differential signal (by subtracting the trace of the bacterium from the trace of the bead; Supplementary Fig. 1 online). Eventually, the bacterium detached and slipped over the surface. When this event occurred we terminated the acquisition. Because the rate of DNA uptake turned out to be low compared with video frequency, we analyzed the DNA uptake rate obtained from video recordings.
To determine the local rate of DNA transport v(t), one has to average over fluctuations in the bead position. Because filtering Δ(t) would smear out pauses, we chose to fit the distance signal Δ(t) (0 < t < tmax) to a polygon of M vertices20. The coordinates {ti,li} of the polygon vertices were adjusted to minimize the fitting error (χ2 test) while keeping ti − ti − 1 ≈ ΔT (solid line in Fig. 4b). ΔT was set as follows: the standard deviation σn of a bead that is subject to brownian motion decreases with the inverse of the square root of the number of data points n, as σn = α n−0.5. For a free bead in the optical trap at a stiffness k = 0.16 pN we found α = 5 (data not shown). Allowing an error on the velocity of 5% or ~1 nm s−1, we can determine the number of data points per vertex n from Δx ΔT−1 = (α n−0.5)(1 / nΔt) ≈ 1 nm s−1, where Δt = 0.0333 s is the time per data point. We found n = 30 or ΔT = 1 s. At lower trap stiffness k = 0.004 pN nm−1 the error was 20% for ΔT = 1 s. This estimation provides a maximum error, as fluctuations will be reduced when the DNA tether is straightened. As shown previously, in our procedure the averaging interval ΔT does not influence the average values of velocity21.
The external force increased continuously during each retraction event because the deflection of the bead from the center of the laser trap increased. The external force increased linearly at deflections between 0 and 400 nm and data beyond this range were disregarded. The average velocity in a force interval was obtained by averaging the velocity values from various retraction curves.
We assume that the major error in the measurement of DNA uptake rates arises from the finite size of the bacterium and thus from a velocity and force component in the vertical direction, defined as perpendicular to the coverslip. The bead was located ~1 µm above the coverslip and the length of the DNA tether varied between 2.5 µm and 10 µm. We corrected the uptake rate for the vertical movement of the bead at the respective length of the DNA tether. With an uncertainty of δh ≈ 0.5 µm in the height of the bead we estimate a maximum error of 5% in the uptake velocity. Although the DNA most likely attaches on the top of the bacterium, there is a possibility that attachment occurs near the glass surface. We account for this uncertainty by adding an additional error of 5% toward higher velocity
DNA is a highly flexible molecule. To convert the deflection of the bead from the center of the optical trap into a rate of base pairs per second (bp s−1) we needed to take into account the elastic properties of DNA in competence medium. Eventually the end of λ-DNA bound nonspecifically to the silanized coverslip. Using a piezo stage we measured the force versus extension curve of double-stranded DNA under competence conditions. A fit to the WLC model29 yielded lP = 54 ± 5 nm (Supplementary Fig. 2 online). Therefore the latter was used for the conversion of nanometers into base pairs for F < 10 pN and the experimental value was used for conversion at higher forces. From repeated elasticity measurements we estimate an error of 0.8%.
The error in the retraction velocity Δv was calculated as a sum of the statistical error Δvs = σs n−0.5, the geometrical error Δvg = 0.1v and the elastic error Δve = 0.008v; that is, Δv = Δvs + Δvg + Δve, where σs is the s.d. and n the number of measurements.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Hahn for strain BD3773 and invaluable discussions. The project was supported by grant GM3677 and D.D. by grant GM43756. B.M. acknowledges support by the Deutsche Forschungsgemeinschaft (Emmy Noether Programm).
Footnotes
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
References
- 1.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 2.Dubnau D. DNA uptake in bacteria. Annu. Rev. Microbiol. 1999;53:217–244. doi: 10.1146/annurev.micro.53.1.217. [DOI] [PubMed] [Google Scholar]
- 3.Chen I, Dubnau D. DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2004;3:241–249. doi: 10.1038/nrmicro844. [DOI] [PubMed] [Google Scholar]
- 4.Smith DE, et al. The bacteriophage straight phi29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 5.Salman H, Zbaida D, Rabin Y, Chatenay D, Elbaum M. Kinetics and mechanism of DNA uptake into the cell nucleus. Proc. Natl. Acad. Sci. USA. 2001;98:7247–7252. doi: 10.1073/pnas.121067698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sauer-Budge AF, Nyamwanda JA, Lubensky DK, Branton D. Unzipping kinetics of double-stranded DNA in a nanopore. Phys. Rev. Lett. 2003;90:238101. doi: 10.1103/PhysRevLett.90.238101. [DOI] [PubMed] [Google Scholar]
- 7.Li J, et al. Ion-beam sculpting at nanometre length scales. Nature. 2001;412:166–169. doi: 10.1038/35084037. [DOI] [PubMed] [Google Scholar]
- 8.Hoa TT, Tortosa P, Albano M, Dubnau D. Rok (YkuW) regulates genetic competence in Bacillus subtilis by directly repressing comK. Mol. Microbiol. 2002;43:15–26. doi: 10.1046/j.1365-2958.2002.02727.x. [DOI] [PubMed] [Google Scholar]
- 9.Provvedi R, Chen I, Dubnau D. NucA is required for DNA cleavage during transformation of Bacillus subtilis. Mol. Microbiol. 2001;40:634–644. doi: 10.1046/j.1365-2958.2001.02406.x. [DOI] [PubMed] [Google Scholar]
- 10.Provvedi R, Dubnau D. ComEA is a DNA receptor for transformation of competent Bacillus subtilis. Mol. Microbiol. 1999;31:271–280. doi: 10.1046/j.1365-2958.1999.01170.x. [DOI] [PubMed] [Google Scholar]
- 11.Bouchiat C, et al. Estimating the persistence length of a worm-like chain molecule from force-extension measurements. Biophys. J. 1999;76:409–413. doi: 10.1016/s0006-3495(99)77207-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nieuwenhoven MH, Hellingwerf KJ, Venema G, Konings WN. Role of proton motive force in genetic transformation of Bacillus subtilis. J. Bacteriol. 1982;151:771–776. doi: 10.1128/jb.151.2.771-776.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mejean V, Claverys JP. DNA processing during entry in transformation of Streptococcus pneumoniae. J. Biol. Chem. 1993;268:5594–5599. [PubMed] [Google Scholar]
- 14.Neupert W, Brunner M. The protein import motor of mitochondria. Nat Rev Mol. Cell. Biol. 2002;3:555–565. doi: 10.1038/nrm878. [DOI] [PubMed] [Google Scholar]
- 15.Chauwin JF, Oster G, Glick BS. Strong precursor-pore interactions constrain models for mitochondrial protein import. Biophys. J. 1998;74:1732–1743. doi: 10.1016/S0006-3495(98)77884-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oster G, Wang H. Rotary protein motors. Trends Cell Biol. 2003;13:114–121. doi: 10.1016/s0962-8924(03)00004-7. [DOI] [PubMed] [Google Scholar]
- 17.Londono-Vallejo JA, Dubnau D. Mutation of the putative nucleotide binding site of the Bacillus subtilis membrane protein ComFA abolishes the uptake of DNA during transformation. J. Bacteriol. 1994;176:4642–4645. doi: 10.1128/jb.176.15.4642-4645.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perkins TT, Dalal RV, Mitsis PG, Block SM. Sequence-dependent pausing of single lambda exonuclease molecules. Science. 2003;301:1914–1918. doi: 10.1126/science.1088047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davenport RJ, Wuite GJ, Landick R, Bustamante C. Single-molecule study of transcriptional pausing and arrest by E. coli RNA polymerase. Science. 2000;287:2497–2500. doi: 10.1126/science.287.5462.2497. [DOI] [PubMed] [Google Scholar]
- 20.Maier B, Bensimon D, Croquette V. Replication by a single DNA polymerase of a stretched single-stranded DNA. Proc. Natl. Acad. Sci. USA. 2000;97:12002–12007. doi: 10.1073/pnas.97.22.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maier B, et al. Single pilus motor forces exceed 100 pN. Proc. Natl. Acad. Sci. USA. 2002;99:16012–16017. doi: 10.1073/pnas.242523299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaiser D. Bacterial motility: how do pili pull? Curr. Biol. 2000;10:R777–R780. doi: 10.1016/s0960-9822(00)00764-8. [DOI] [PubMed] [Google Scholar]
- 23.Albano M, Breitling R, Dubnau DA. Nucleotide sequence and genetic organization of the Bacillus subtilis comG operon. J. Bacteriol. 1989;171:5386–5404. doi: 10.1128/jb.171.10.5386-5404.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu L, Nakano MM, Lee OH, Zuber P. Plasmid-amplified comS enhances genetic competence and suppresses sinR in Bacillus subtilis. J. Bacteriol. 1996;178:5144–5152. doi: 10.1128/jb.178.17.5144-5152.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anagnostopoulos C, Spizizen J. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 1961;81:3389–4302. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Albano M, Hahn J, Dubnau D. Expression of competence genes in Bacillus subtilis. J. Bacteriol. 1987;169:3110–3117. doi: 10.1128/jb.169.7.3110-3117.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fung DC, Berg HC. Powering the flagellar motor of Escherichia coli with an external voltage source. Nature. 1995;375:809–812. doi: 10.1038/375809a0. [DOI] [PubMed] [Google Scholar]
- 28.Simmons RM, Finer JT, Chu S, Spudich JA. Quantitative measurements of force and displacement using an optical trap. Biophys. J. 1996;70:1813–1822. doi: 10.1016/S0006-3495(96)79746-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marko JF, Siggia ED. Stretching DNA. Macromolecules. 1995;28:8759–8770. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.