Abstract
Completion of the Rattus norvegicus genome sequence enabled a global inventory and analysis of the nuclear receptors (NRs) in three mammalian species. Forty-nine NR members were found in mouse, 48 in human. Forty-seven were found in the rat, with gaps at the locations expected for the other two. Pairwise comparisons of their distribution in rat, mouse, and human identified 11 syntenic NR gene blocks, including three small clusters of two or three closely related genes, each spanning 40 kb to 1700 kb. The exon structure of the ligand-binding domain suggests that exon shuffling has played a role in the evolution of this family. An invariant splice junction in all members of the NR family except LXRβ suggests a functional role for the intron. The ligand-binding domains of PXR and CAR are among the most divergent in the family. Their higher nucleotide substitution rates may be related to the central role played by these two NRs in the metabolism of the foreign compounds and may have resulted from limited positive selection.
Nuclear receptors (NRs) are transcription factors capable of exerting regulation of gene expression in the nucleus in response to various extracellular and intracellular signals (Tsai and O'Malley 1994; Mangelsdorf et al. 1995). They are activated by binding of small hydrophobic compounds, such as steroids, retinoids, and thyroid hormones. Ligand binding triggers a conformational change in the receptor proteins, which enables an interaction with cofactors and specific cis-regulatory DNA sequences called hormone response elements (HREs) to subsequently modify gene expression. Cognate ligands are not identified for all nuclear receptors. Those that currently lack identified ligand molecules are termed “orphan” NRs (Giguere 1999). Because NRs bind small molecules which can be easily modified by drug design, and regulate a group of diverse and crucial biological functions such as metabolism, homeostasis, development, and disease, they have become promising pharmacological targets.
NRs share a similar modular domain structure, which includes, from N-terminus to C-terminus, the variable modulatory A/B domain, the DNA-binding domain (DBD), the hinge D-region, the ligand-binding domain (LBD), and an F-domain that is not found in all NRs. The DBD contains two zinc fingers in tandem that encompass ∼80 amino acid residues in total and are directly involved in recognition of the cognate HRE. The LBD harbors a hydrophobic ligand-binding pocket, deep within its core, that is specific to and thus variable among different receptors. The DBD and LBD are the two most conserved domains of NRs and, as a result, are regarded as dual signatures of this protein family.
NRs constitute one of the largest groups of transcription factors in animals. Twenty-one NR genes are identified in the complete sequence of the Drosophila melanogaster genome (Adams et al. 2000), and over 270 are found in Caenorhabditis elegans (Sluder and Maina 2001). The latest estimate of the number in the human genome sequence, based on sequence alignment and phylogenetic analysis, is 49 NR genes and three NR pseudogenes (Robinson-Rechavi et al. 2001). In a detailed study of the evolutionary relationship among NRs (Laudet 1997), the majority of them were assigned to six well defined subfamilies whose inter-relationships remain unresolved. As a result of the work of Laudet, a systematic naming convention was proposed (Nuclear Receptors Committee 1999) including the creation of a new subfamily 0, which consists of the nuclear receptors lacking either the DBD or the LBD.
With the draft rat genome sequence available (Rat Genome Sequencing Project Consortium 2004), it is now possible to conduct a three-way study of the NR genes comparing the human, mouse, and rat. To gain new insights into the structure, regulation, and evolution of this fascinating family we sought to determine their genomic location and their gene structure, and re-evaluate their phylogenetic relationships in Homo sapiens and the two most medically important model systems.
RESULTS AND DISCUSSION
Nuclear Receptor Inventory in Rat, Mouse, and Human Genomes
The presence of six NR domains was examined in the rat, mouse, and human genomic sequences using GENEWISEDB (see Methods). The numbers of NR domains identified in the three genomes are summarized in Table 1 (see Supplemental Table 1 for a detailed inventory and genomic coordinates). Grouping and subsequent assignment of these domains to different NRs by BLAST revealed that most of the known mammalian NR genes are present in the current three genome sequences (Suppl. Table 1, Fig. 1); however, the sequences encoding several receptors are partially or completely missing in the rat and mouse genomes. The absence of the sequences encoding Rev-erbβ (NR1D2) and PNR (NR2E3) and the LBD of TLX (NR2E1) in the rat genome and the DBD of LXRβ (NR1H2) in the mouse genome can be explained by gaps in these two assemblies at the expected syntenic locations. The final tally of complete or partially identified NR genes was 48 for human, 49 for mouse, and 47 for rat.
Table 1.
Numbers of Sequences Encoding Nuclear Receptors Found in Rat, Mouse, and Human Genomesa
| Sequence | Rat | Mouse | Human |
|---|---|---|---|
| NR genes | 47b | 49 | 48 |
| NR genes, aberrantc | 0 | 1 | 1 |
| NR pseudogenes | 3 | 4 | 3 |
| Domain singletonsd | |||
| DBD | 1 | 1 | 0 |
| LBD | 0 | 1 | 0 |
| SMDe | 0 | 0 | 1 |
Genome versions: human, April 2003; mouse, February 2003; rat, April 2003
All complete and partial genes are included in the tally (see text). Not tallied are NR1D2 and NR2E3, whose complete absence correlates with sequence gaps in the rat genome assembly at the location predicted by synteny with the mouse and human orthologs
One aberrant NR gene structure was identified in each of the human and the mouse genomes
Domain singletons not including DAX-1 and SHP, which are subfamily 0 NR genes
Steroid modulatory A/B domain. One GCR domain singleton was found in the human genome
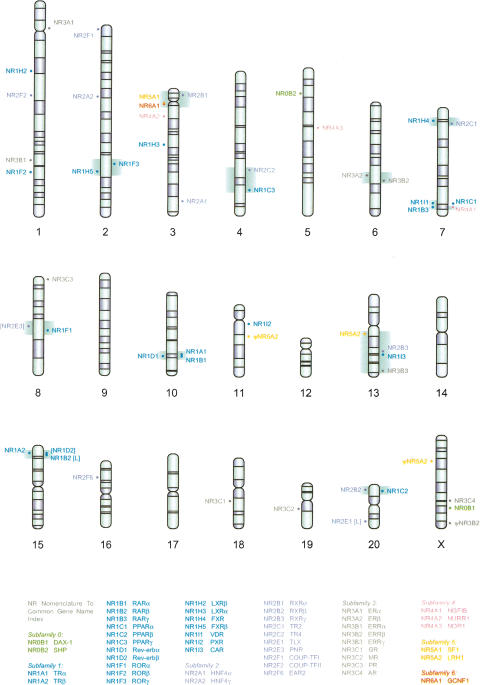
Figure 1.
The chromosomal landscape of rat nuclear receptor genes. NR genes on the forward strand were placed on the right of the chromosomes, and NR genes on the reverse strand were placed on the left. NR1D2, NR2E3, and the sequences encoding the LBDs of NR1B2 and NR2E1 are missing due to sequence gaps in the current rat genome assembly. Their genomic locations are indicated in the square brackets (L for LBD). The syntenic blocks containing NR genes are highlighted in green (see also Table 3).
Among the NR genes are also “domain singletons,” the genomic sequences encoding NR domains without nearby sequences, or gaps, to make complete NR genes (Suppl. Table 1). They do not share sequence similarity with the single-domain DAX-1 (NR0B1) and SHP (NR0B2), two NRs known to lack a DBD.
Although some domain singletons might be a result of false positive identifications, others defy so quick a dismissal and remain puzzling. For example, a 522-bp sequence identified on human chromosome 16 encodes a partial A/B domain of the glucocorticoid receptor (GR, NR3C1), and is 95% identical to a portion of the first coding exon of GR. The observation that the intron downstream of the first coding exon of GR harbors a potentially active family-Y Alu element (Batzer et al. 1990) and that the 522-bp partial copy is immediately surrounded by two Alu elements from the families Y and C, suggested that the creation of this GR domain singleton may be related to the retrotransposition activity of the nearby Alu-Y element.
NR pseudogenes (Ψ) were identified in each species (Table 2). Our results confirmed the existence of the three known pseudogenes in the human genome including ΨFXRβ, the only unprocessed pseudogene (Maglich et al. 2001; Robinson-Rechavi et al. 2001). Because the mouse and rat orthologs of ΨFXRβ are expressed (identified and experimentally proven to be active by one of the authors, J.W., and Otte et al. 2003) and because FXRβ may share some functions with FXR in cholesterol metabolism, it remains unclear under what circumstances FXRβ was silenced and how its loss was tolerated and fixed in the ancestral primate population.
Table 2.
Human and Rodent Nuclear Receptor Pseudogenes
|
Truncationa
|
|||||
|---|---|---|---|---|---|
| Genome | Pseudogene | Location | Type | 5′ | 3′ |
| Human | ψFXRβ | chr1+:114480335 | unprocessed | no | no |
| ψHNF4γ | chr13—:55510764 | processed | yes | yes | |
| ψERRα | chr13—:19064728 | processed | no | no | |
| Mouse | ψRev-erbβ | chr19+:39472488 | semiprocessed | no | no |
| ψPNR | chr15+:35760537 | processed | yes | no | |
| ψLRH1 | chr3+:145441245 | semiprocessed | yes | no | |
| ψLRH1 | chr6—:119298331 | processed | yes | no | |
| Rat | ψERRβ | chrX+:146791239 | processed | no | no |
| ψLRH1 | chr11+:48636215 | processed | yes | no | |
| ψLRH1 | chrX—:31030327 | processed | yes | no | |
Truncation is relative to the coding sequences
Four pseudogenes were detected in the mouse genome and three in the rat genome. Although there are two LRH1 pseudogenes in both the mouse and rat genomes, it is likely that the two sets were created independently because there are no syntenic pairings, and they have marked differences in their sequence features (data not shown).
Genomic Distribution of Nuclear Receptors
The genomic locations of NRs were mapped onto the rat karyogram (Fig. 1). NR genes were distributed throughout the rat genome except for chromosomes 9, 12, 14, and 17. Although rat chromosome Y was unavailable, no NR genes are found on the human Y chromosome, and none were expected there for rat or mouse. The Poisson test rejected the random distribution (P < 0.001) of NRs in the rat genome. We identified 11 syntenic blocks common to all three genomes; that is, in each block, the same set of NR genes locate on a single chromosome in all three genomes (Table 3). The sizes of these 11 blocks vary from 0.21 Mb to 54.33 Mb. Except for the blocks I, II, and IV, all syntenic blocks have similar sizes in all three genomes. RORγ (NR1F3) and FXRβ (NR1H5) in block IV are less than 9 Mb apart in the rodent genomes; however, they are separated by a 34-Mb interval that includes the centromere in human.
Table 3.
Syntenic Blocks Containing NR Genes in Rat, Mouse, and Human Genomes
|
Genomic location and size (Mb)
|
|||||||
|---|---|---|---|---|---|---|---|
| Block | Rat | Mouse | Human | ||||
| I | NR1C2, NR2B2 | chr20 | 1.6 | chr17 | 5.6 | chr6 | 2.2 |
| II | NR1C3, NR2C2 | chr4 | 25.2 | chr6 | 23.5 | chr3 | 2.6 |
| III | NR1F1, NR2E3 | chr8 | 10.0a | chr9 | 9.4 | chr15 | 11.3 |
| IV | NR1F3, NR1H5 | chr2 | 8.8 | chr3 | 8.6 | chr1 | 34.1 |
| V | NR1H4, NR2C1 | chr7 | 5.1 | chr10 | 4.7 | chr12 | 5.5 |
| VI | NR3A2, NR3B2 | chr6 | 11.8 | chr12 | 10.3 | chr14 | 12.2 |
| VII | NR1A1, NR1B1, NR1D1 | chr10 | 0.2 | chr11 | 0.2 | chr17 | 0.3 |
| VIII | NR1A2, NR1B2, NR1D2 | chr15 | 1.34b | chr14 | 1.8 | chr3 | 1.5 |
| IX | NR1B3, NR1I1, NR4A1 | chr7 | 4.4 | chr15 | 4.5 | chr12 | 5.4 |
| X | NR2B1, NR5A1, NR6A1 | chr3 | 11.8 | chr2 | 11.4 | chr9 | 10.1 |
| XI | NR1I3, NR2B3, NR3B3, NR5A2 | chr13 | 54.3 | chr1 | 51.1 | chr1 | 55.4 |
The size of the rat span was estimated from the location of the gap situated at the expected location of NR2E3 based on syntenic flanking mouse gene Pkm2 and NR1F1
The size of the rat span was estimated from the location of the gap situated at the expected location of NR1D2 based on syntenic flanking mouse gene Rpl15 and NR1A2 (see Fig. 2 legend)
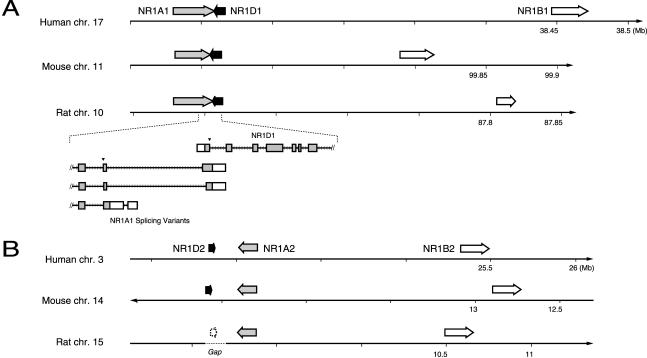
Three tightly linked NR gene clusters stand out within the syntenic blocks: cluster i composed of TRα (NR1A1), RARα (NR1B1), and Rev-erbα (NR1D1) from block VII; cluster ii of TRβ (NR1A2), Rev-erbβ (NR1D2), and RARβ (NR1B2) from block VIII; and cluster iii of SF1 (NR5A1) and GCNF1 (NR6A1), a subset of block X. They span 270 kb, 1700 kb, and 40 kb, respectively, in the rat genome. Salient features of clusters i and ii in the human and rat genomes were described previously (Laudet et al. 1992; Koh and Moore 1999). They are composed of closely related paralogous triplets that must have arisen by duplication of an ancestral TR, Rev-erb, and RAR gene cluster. The most remarkable feature of cluster i, the overlap of the 3′-most exons of one variant of TRα with Rev-erbα (Lazar et al. 1989), has not been observed in the chicken (Forrest et al. 1990; Bonnelye et al. 1994). In cluster ii TRβ and Rev-erbβ do not share terminal exons (Koh and Moore 1999).
The genome sequences bring details of this organization into focus. The gene order, spacing, and orientations are different in the extant clusters i and ii (Fig. 2). Although TR and Rev-erb maintain the same tail-to-tail orientation, the pair is inverted relative to RAR in the two clusters. Among these six genes, only TRα has two splice variants with downstream extended 3′ coding exons, that is, the ones overlapping Rev-erbα (see inset, Fig. 2A). This would suggest that the TRβ gene structure reflects the ancestral state of TR and therefore recruitment of the terminal exon occurred as a result of the juxtaposition of the two NR genes. It will be interesting to determine whether this is a mammalian invention, as suggested by the negative findings in chicken, or is a general feature of vertebrates.
Figure 2.
Chromosomal location of related NR gene clusters i and ii. Genes are labeled on each figure, and closely related paralogs are similarly shaded. Coordinate positions for each chromosome are indicated below the number lines. Gene arrow lengths are proportional to the size of each gene. (A) Cluster i spans ∼270 kb. The inset gives a scale drawing of the relationship of the NR1A1 and 1D1 genes. The three known variants of NR1A1 are shown. Coding exons are shaded boxes, 3′ UTRs are open. A filled inverted triangle marks the splice acceptor of the invariant LBD splice junction (see also Fig. 3B). (B) Cluster ii, ∼1.4 Mb. The rat gene for NR1D2 is only presumed to exist at the indicated position. Sequences for this gene are absent from the assembly, and a gap exists at this position (indicated by the broken line). The rat NR1B2 is a partial gene, containing a DBD but not an LBD, most likely as a result of incomplete assembly of this draft genome. Note that the 1A and 1D genes are on opposite strands in each cluster, in the same orientation relative to each other; their order changes relative to the 1B gene.
Given the propensity for processes of chromosomal rearrangement to scatter the majority of the NR genes, it is interesting that both clusters remained closely linked, suggesting that natural selection favors the clusters. All other syntenic groups of NR genes found here belong to a set of large syntenic blocks shared by the rat, mouse, and human genomes and may simply reflect the current state of the chromosomal organization on the whole-genome scale. Studies of the segmental duplication suggest that the recent segmental duplication events have contributed little to the evolution of the NRs in human, mouse, and rat, as no NR genes or their functional domains are found in the large duplicated regions in the human and rat genomes (Bailey et al. 2002; Tuzun et al. 2004).
Phylogenetic Analysis
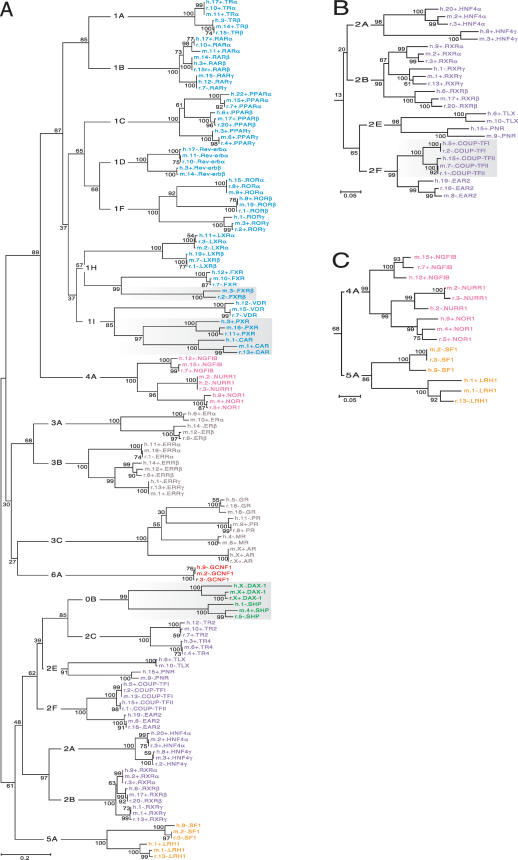
The NR DBDs and LBDs were tested separately to reinvestigate the possibility that recombination between the two domains accounted for some of the diversity in the NR family (see Laudet et al. 1992), and to enable investigation of the relationships with the NR0B group. The overall topologies of the two trees gave the expected subfamily and group clades, upon which their systematic nomenclature is based (Laudet 1997). They differed in small details in a way that could possibly be consistent with one or more exchange events between these two domains early in NR history. For example, subfamily NR4 was closer to NR1 in the LBD tree (Fig. 3A) but was closer to NR5 in the DBD tree (Fig. 3C). However, 68% bootstrap support was marginal for the DBD configuration. This issue warrants further investigation.
Figure 3.
Unrooted phylogenetic trees of the NR family. The same color scheme for NR subfamilies is used as in Fig. 1. Group-level designations (e.g., 0B, 1A, 1B,..., 6A) label the interior branches, but common gene names label the terminal branches. Bootstrap values expressed in percentage are indicated at the nodes (branch bifurcations). (A) A complete tree constructed from the multiple sequence alignment of the LBDs of all NRs found in rat, mouse, and human. Shading highlights groups exhibiting rapid evolution. (B) NR2 subfamily clade (orphan receptors) taken from the DBD tree. Shading highlights a group exhibiting increased conservation. (C) Portion of DBD tree showing the relationship between subfamilies NR4 and NR5.
SHP (small heterodimer partner) and DAX-1 (dosage-sensitive sex and AHC critical region on the X, gene 1) of the NR0B group were thought to possibly represent an ancient gene structure (cf. Guo et al. 1996) because of their high degree of divergence from other NRs. Our results place the NR0B group most closely to NR2C (TR2 and TR4) with strong bootstrap support for this configuration suggesting that they arose, by the loss of the DBD, during or after the duplications that expanded the NR2 subfamily. They subsequently evolved much more rapidly than the other NR2 members, as indicated by long branch lengths after divergence from NR2C, freed from functional constraints presumably imposed by the DNA binding requirement. They now act as modulators of other NRs through a variety of protein–protein interactions (e.g., Johansson et al. 2000; Zhang and Chiang 2001; Gurates et al. 2003).
The LBDs of most NR members have changed little since the divergence of humans and rodents. This is manifested in the tree as extremely short terminal branch lengths, that is, those branches representing the last common ancestor of the three species. However, three groups, (NR1I2-3, NR1H5, and NR0B1-2, see Fig. 3A, shaded groups) were significantly more divergent among the three species. Nucleotide substitution analysis revealed that the synonymous rates in the LBDs of CAR (NR1I3) and PXR (NR1I2) are average for the family, whereas the nonsynonymous rates were 6.4 and 3.7 times higher than the average (Suppl. Fig. 1).
Evaluation of the terminal branch lengths of all NR members revealed cases where the rat sequence was closer to human than the mouse was: RARα (NR1B1), GR (NR3C1), and LXRα (NR1H3). For many others members, the human, rat, and mouse were virtually indistinguishable. These observations may be of practical value in choosing model systems for pharmacological studies.
There was too little variation in the ∼80-aa DBD to form a well resolved tree, so DNA sequences were used for this domain. The terminal branches were again of most interest to the inter-species comparison. Long terminal branches were observed for all but two NR members: COUP-TFI (NR2F1) and COUP-TFII (NR2F2; Fig. 3B, shaded portion). The relative absence of inter-species variation in the DNA encoding these two NR domains suggested the possibility of selection operating on the DNA sequence itself. Conserved regulatory sequences could be one explanation for this observation. It may therefore be significant that the DBD is uninterrupted by introns in these two NRs (see below).
The KA/KS ratios of the LBD domains indicate that the NRs are subject to strong purifying selection. No positive selection was detected by Student's t-test. However, because the KA/KS ratios of the LBDs of PXR and CAR were 4.0 and 5.6 times greater than the averages, respectively, these two domains may have experienced limited positive selection in the context of the NR evolution. For PXR and CAR, the increased KA/KS ratios in the LBDs could be more readily explained by their biological functions. PXR, an orphan NR preferentially expressed in the liver and intestine, responds to potentially harmful chemicals by activating the expression of cytochrome P-450 genes crucial for the detoxification of a wide variety of structurally diverse xenobiotics and endobiotics (Kliewer et al. 1998; Lehmann et al. 1998). The KA/KS ratios of the remaining orphans were much more conserved and thus their ligands, if any, are not likely to be species-specific.
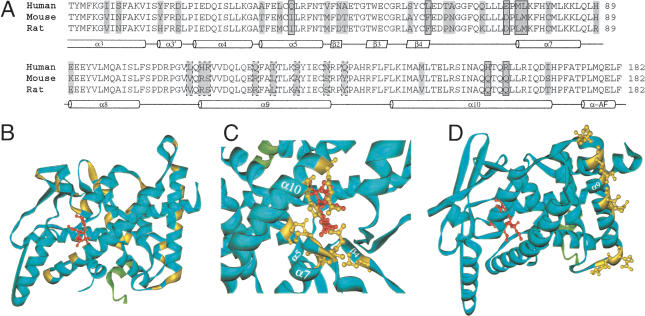
We investigated the structural implications of the LBD sequence variation in the PXR group. Thirty-three variable sites in the multiple sequence alignment of the LBDs of PXR from human, mouse, rat, rhesus, pig, rabbit, dog, chicken, and zebrafish were mapped on the tertiary structure of the LBD of the human PXR (Watkins et al. 2003). Seven sites line the inner surface of the ligand-binding pocket (Watkins et al. 2001); eight variable sites were distributed along α-helix 9 (α9), which is involved in protein–protein interactions; the remaining sites were distributed uniformly throughout the LBD (Fig. 4). The set lining the ligand-binding pocket was in a position that could possibly form direct contacts with the bound ligand and may therefore contribute to the difference between the ligand-binding properties of the human and rodent PXRs. The set distributed along α9 was outwardly oriented.
Figure 4.
Variable sites in the LBD of PXR. (A) The variable sites, highlighted in gray, in the protein sequence alignment of the LBDs of the human, mouse, and rat PXRs. The corresponding secondary structure is indicated below the sequence alignment: the α-helix is represented by the cylinder, and the β-sheet by the parallelogram. The seven variable sites at which the amino acid residues line the ligand-binding pocket and the ones found in the α-helix 9 and its vicinity are boxed with the solid line and broken line, respectively. Other available LBD sequences of the rhesus, pig, rabbit, dog, chicken, and zebrafish PXRs are omitted from the sequence alignment presented here, because the inclusion of them does not introduce changes to the general variation pattern. (B) The same sites, highlighted in yellow, in the tertiary structure of the LBD of the human PXR (the blue solid ribbon). The agonist is shown as the small red molecular structure bound in the receptor's ligand-binding pocket, and the coactivator in fragment is shown as the green solid ribbon. Details of the variable sites, with the side chains of the amino acid residues at these sites shown in yellow, in the ligand-binding pocket (C) and in the α-helix 9 and its vicinity (D) are shown.
By contrast, the longest α-helix, α10, has only four variable sites, all extending toward the interior protein. The tertiary structure of the PPAR-RXR heterodimer (Gampe Jr. et al. 2000) reveals that the outer surface of α10 is involved in the interaction with RXR. α10 probably functions similarly in other heterodimeric partners of the RXR, including PXR. Thus variation of the outward face of PXR α10 may be constrained by this important function.
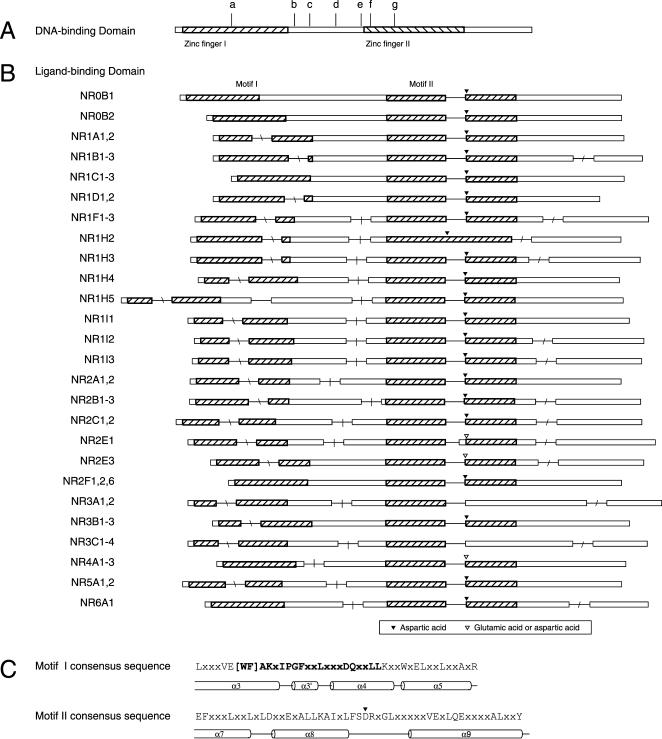
Exon Structures of DBD and LBD
Information on the splice junctions, derived from BLAT alignments of amino acid sequences of NR mRNAs to the genome, was used to further characterize the family. All splice junctions were conserved among orthologs of the various NR family members. When comparing paralogous members, informative patterns of conservation emerge within these two domains (Fig. 5).
Figure 5.
The gene structure encoding the DBD and LBD domains of the NR genes. Open bars are exons, drawn to scale; line segments, drawn at fixed length, give intron locations. (A) DBD splice junctions. Sequences are 75–78 aa in length. The shaded boxes indicate the location of the two C4 zinc finger motifs within this highly conserved domain. Introns may be found at seven different locations in the DBD across the entire family, or may be absent. Vertical hash marks indicate the location of junctions that were shared in the following groupings: a, NR2B, NR2C1,2; b, NR1A,1B,1C,1D,1F,1H4-5, NR5A; c, NR1I, NR4A; d, NR3A,3B,3C; e, NR2E3; f, NR2A, NR2F6; g, NR2E1; and not shown are group h, NR1H, NR2F, and NR6A, which have no intron in the DBD. (B) LBD splice junctions. Sequences are 170 (NR1D2) to 208 (NR0B1) aa in length. Each row is a schematic drawing giving the relative location of the splice junction and the group of NRs sharing the splice junction pattern. The position of splice junctions in orthologs was always the same, and thus species designations are omitted. Two conserved motifs (I and II, see text) in the LBDs are shown as the hatched areas. The location of a highly conserved negatively charged amino acid residue (aspartic acid or glutamic acid) in motif II is marked by an inverted triangle. The four regions within which introns were found are indicated by slash marks: “\” in motif 1, “|” intermotif region, no slash in motif II, and “/” after motif II (see text). (C) The consensus sequences of motifs I and II. The secondary structure of the corresponding part of the LBD, derived from crystallographic studies, is indicated below the sequence. Letters in bold correspond to the residues of the NR signature, involved in stabilizing the canonical fold of the NR LBDs (see Wurtz et al. 1996).
Eight patterns are evident in the DBD splice junctions (Fig. 5A). The junction is located at various positions in between the two zinc finger motifs in four of the eight groups. It is located in the first zinc finger motif in the NR2B1-3, NR2C1&2 group, and it is located at different positions within the second zinc finger in NR2A1&2, NR2F6, and NR2E1 groups.
The splice junction was lost from NR1H2&3, NR2F1&2, and NR6A1. Because these do not form a monophyletic group in the tree (Fig. 3A), the intron was probably lost in three separate events. Members of subfamilies NR1 and NR3 show little variation in junction location, whereas subfamily NR2 has several variants. In two cases members of different subfamilies shared the same splice junction: NR1 and NR5, and NR1I and NR4. This result, taken together with the phylogenetic results described above, may suggest a complex evolutionary relationship between the subfamilies NR1, 4, and 5 (see Phylogenetic Analysis above). Alternatively, there could be preferred sites for acquiring introns. Elucidation of the principles governing the dynamics of intron acquisition and change over long evolutionary timescales is needed to understand these relationships.
The LBD was less conserved, overall, than the DBD compared across the whole family (alignments in Suppl. Fig. 2). Within it, three sequence motifs were identified (see Methods), although none of those were as conserved as the zinc finger motifs in the DBD. Motif I, spanning α-helices 3 and 4, was previously described (see Wang et al. 1989 and Wurtz et al. 1996 for definition of helices in LBD). Motif II, α-helices 7–9, was described in part (Wang et al. 1989 identified conservation in helices 7 and 8). Steroid receptor groups NR3A and NR3C differed from all others in that motif II was not detected. However, sequence similarity to the left half of the consensus pattern (Fig. 5C) was easily observable (note crosshatching of NR3A1-3 and NR3C1-4 in Fig. 5B, and see Suppl. Fig. 2; see also Wang et al. 1989). Motif III spans most of α-helices 10 and 11. Motifs I and II are indicated schematically in Figure 5B (motif III is altered or deleted in some NR isoforms, so we have omitted it from the figure pending further analysis of all of the individual splice variants).
Up to four splice junctions were found in the peptide sequences of the region of the LBDs to which our analysis was confined (see the Pfam profiles used to identify LBD, and Methods). The locations of the four introns were confined to distinct regions of the LBD as defined by the aforementioned structural motifs. The first is within motif I; second, between motifs I and II; third, within motif II; and fourth, downstream from motif II. Introns were lost multiple times at regions 1, 2, and 4. Moreover, the precise location of introns 1, 2, and 4 was variable. In distinct contrast, intron 3 was invariant in that it was present in all of the NRs except LXRβ (NR1H2), and it was always a phase-1 intron at the same amino acid position in motif II. Although conserved in position and phase, this intron varied in size from 123 bp in mouse NR2A2 to 53,000 bp in human NR5A2. Except in TLX and the steroid hormone receptor genes, on the splice acceptor side of the intron, there was a highly conserved aspartic acid (occasionally replaced by glutamic acid) which contributes to the polar interactions involved in the NR dimerization of those members in which it is present.
The conserved LBD splice junction was likely to have originated early in the family and was subsequently conserved in evolution: it was also observed in the LBD of the Danio rerio SVP46 (NR2F5, data not shown). The selective pressure maintaining the splice junction could arise from conservation of amino acid sequence. The aforementioned aspartic acid codon is split by this phase-1 splice junction. However, motif II is much less conserved than the zinc finger motifs or motif I, and some NR sub-families have neither the aspartic acid nor a glutamic acid at the splice junction. Thus, some sequence or structural motif in the NR mRNA involved in its regulation, processing, or stability may be the determinant of the conservation of this splice junction. Because LXRβ (NR1H2)—as the single exception—lost this splice junction, the comparison of its expression to other NR genes may shed light on this phenomenon.
Comparison of the 26 different splicing patterns of the sequence encoding the LBD (Fig. 5B) conveys the sense that large-scale sequence changes, intron loss or gain, and exon addition or substitution played an important role in shaping the evolution of this family. The loss or gain of the first, second, or fourth introns in the LBD occurred within many NRs. Large-scale innovative changes in the coding sequence of the LBD region may have contributed significantly to the rise of some new NR genes. FXRβ (NR1H5) appears to have added one exon between the conserved motifs I and II. In the steroid receptors (ER of the group NR3A; GR, MR, PR, and AR of the group NR3C), the helix 9 homology of motif II abruptly disappears beginning with the loss of the conserved aspartic acid on the splice acceptor side of the exon (see above). Although the downstream C-terminal boundary of the loss of homology is difficult to determine, this observation could be neatly explained by substitution of the exon downstream from the conserved third splice junction, an event leading to specificity for steroid ligands in groups NR3A and 3C.
Further variation in LBD splice junction patterns may exist in other isoforms, and thus a full accounting of all isoforms, in these and other species, will be important.
Conclusion
The genomic comparison of the NR families from three related mammals affords new insight and raises new questions about the structure, function, and evolution of this important family of transcription factors. Despite the high degree of conservation among the NR sequences, there was clearly distinguishable species-specific variation in three groups. Among them, PXR and CAR, in group NR1I, share some ligands (Moore et al. 2000) and regulate overlapping but distinct sets of genes involved in xenobiotic detoxification (Maglich et al. 2002). Given the central role of CAR and PXR in the xenobiotic metabolism and ingestion, these two NRs may have evolved faster in response to different sets of environmental challenges encountered by humans, mice, and rats. The nature of the species-specific adaptations represented by the other two rapidly evolving groups, NR1H5 and NR0B1-2, awaits improved understanding of their functional roles.
Paralogous NR family members exhibit a variety of different exon structures in both their DBDs and LBDs. Among the variation, the conserved location of the splice junction in the second motif of the LBD stands out as a peculiar phenomenon. It may prove to be a more reliable signature for the NR genes than the C4 zinc finger. Very similar findings are reported in other gene families, for example, chemoreceptor superfamily (Robertson et al. 2003) and DEAD helicase genes (Boudet et al. 2001). An understanding of the selective constraints that preserve such ancient introns may lead to new understanding of protein or mRNA structure and processing.
METHODS
Identification of Nuclear Receptor Genes in Human, Mouse, and Rat Genomes
Six structural and functional domains specific for members of the NR family were obtained from Pfam (Bateman et al. 2002). They are the ligand-binding domain (Pfam database entry name: hormone_rec), found in all members of the family, the C4-type zinc finger DNA-binding domain (zf-C4), found in all but two members, and the four modulator A/B domains, each specific for a given steroid receptor: androgen receptor (Androgen_recep), glucocorticoid receptor (GCR), estrogen receptor (Oest_recep), and progesterone receptor (Prog_receptor). The DBD sequence corresponded to a 75–78-residue segment, starting at the location two amino acid residues before the first conserved cysteine, and encompassing both C4 zinc fingers. The LBD began at the twelfth residue of α-helix 3 and extended through α-helix 10 (Wurtz et al. 1996; Greschik et al. 1999).
The mRNA and protein sequences of 62 representative NRs (Robinson-Rechavi et al. 2001) were downloaded from GenBank. If the human gene sequence of an NR was available, the mouse and rat gene sequences of the same NR were also retrieved, if available. The Pfam domains present in these 62 NRs were identified using HMMPFAM (HMMER 2.3.1; Eddy 1998). Because the E-values of the identification of the NR domains are 1020 ∼ 1050 times less than those of other domains identified, their identification and presence in NRs were unambiguous.
The human, mouse, and rat genomic sequences used in this study were human genome build 34 of June 2003, mouse genome build of February 2003, and rat genome build of April 2003. To take advantage of parallel computing, each of these three genomes was partitioned into 750-kb segments with 2-kb overlaps. Only domains of the NRs identified at the previous step with stringently high E-values were searched in the genomic sequences using GENEWISEDB (Wise 2.2.0; Birney and Durbin 2000). Usually GENEWISEDB can predict the presence of a domain in a genome based on the domain profile in Pfam without any modifications to the genomic sequence, but occasionally it introduced one or more frameshifts to make sensible prediction alignments. Although GENEWISEDB labeled such predictions as pseudogenes, we treated them with extra care because the necessity of introducing frameshifts may well result from sequencing errors in the genome.
Domains identified in each genome were grouped together based on their orientation and the coordinates of their genomic locations, and were compared to the 62 NR protein sequences using the best BLASTP hit as the identity. The GENEWISEDB search results were also parsed to create custom annotation tracks in the UCSC genome browser (http://genome.ucsc.edu/) to depict the exon-intron structure of the predicted domains and to enable cross-examination with mRNA/EST evidence, synteny, and genomic sequence conservation across species.
Pseudogenes were identified among NR genes which had more than one copy in a genome and when the sequence of the mRNA transcript of this gene or its orthologs was available. The mRNA sequence was aligned using TBLASTN and BLAT (Kent 2002) to the genomic sequences at the locations where the different copies of multiple NR genes were found. A copy of an NR gene in the genome was considered to be a pseudogene if frameshifts or nonsense mutations were found in its sequence which could not be credibly attributed to the sequencing errors.
Statistical Test for Clustering of Nuclear Receptor Genes in the Rat Genome
The spatial distribution of the NRs in the rat genome was tested for clustering by χ2 (Zar 1984). The rat genome was divided into nonoverlapping 2.25-Mb segments, and the number (X) of NRs was tallied in each segment. The observed frequency (fo) of X was tallied, and the corresponding expected frequency (fe) was calculated from the Poisson probability P(X). The χ2 value was χ2 = 17.702, degrees of freedom 1. Because χ20.001, 1 = 10.828, the random distribution is rejected (P < 0.001).
Sequence Analyses
The peptide sequences of the DBD and the LBD were identified and extracted from genomic sequence using GENWISEDB. Sequences in each set were aligned using CLUSTALW, and the multiple sequence alignments were then inspected and manually refined in BioEdit. The C-terminal 5–10 residues were incorrect in about half of all sequences extracted from the genomes by GENEWISEDB. They were corrected to match the corresponding GenBank sequence. Nucleotide sequences of each domain were aligned in accordance with their corresponding amino acid sequence alignment.
Corrected but unaligned LBD peptide sequences were searched for conserved sequence motifs (http://blocks.fhcrc.org/). BLOCKMAKER returns two sets of motifs generated by complimentary methods of detecting ungapped regions of similarity (Henikoff et al. 1995). Positive identifications required that both methods agreed on the presence and location of the given motif within the sequence. Three motifs were obtained for all sequences except the steroid receptors and TLX (data not shown). Motif I included the “canonical LBD signature” spanning α-helices 3–5 (see Fig.4B,C; Wang et al. 1989; Wurtz et al. 1996) of 12 helices in the LBD. Motif II corresponded to a central portion of the LBD spanning helices 7–9, involved in dimerization (see Fig. 4B,C). Conservation in the first half of this domain, up to the aspartic acid (Fig. 4C) was observed by Wang et al. (1989), but others have noted extended conservation (Laudet et al. 1992). Motif III spanned helices 10–11. This region is subject to alternate splicing in some NR genes, so it was set aside pending complete description of the family isoforms.
CLUSTALW correctly aligned the residues corresponding to motifs I and III but not motif II. In particular, the subfamily NR0B alignment was greatly improved using motif II as a guide; minor adjustments were required in some other subfamilies. Phylogenetic tree reconstruction of both the protein and DNA alignments was performed using an implementation of the neighbor-joining method in the PAUP*4.0 software package (Swofford 2003) together with a bootstrap of 1000 replicates.
KA/KS of every orthologous gene pair was calculated as the measure of sequence evolution (Li et al. 1985). Student's t-test was used to detect positive Darwinian selection.
Splice Junction Analysis
Splice junctions in the coding sequences were located using BLAT to match all protein sequences (62 representative members from GenBank described above) to the corresponding genome. The BLAT exon segments were manually aligned in a manner that brought into register the DBD and LBD of each protein from the three genomes using EXCEL. This enabled rapid curation of exons found by BLAT, which included elimination of false positive exons due to such things as single-residue indels, missing small N-terminal exons, and other splice site ambiguities that may have tricked BLAT.
Acknowledgments
We thank Hugh M. Robertson for helpful discussions. A.J.C., R.B.L., and F.A.P. were supported in part by the NIH NURSA orphan receptor program, grant #U19DK62434-01. This work was supported by grants 5-U54 HG02051 (Human/MB) from the NHGRI and 5-U54 HG02345 (Rat) from the NHGRI/NHLBI to R.A.G.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2160004.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Adams, M.D., Celniker, S.E., Holt, R.A., Evans, C.A., Gocayne, J.D., Amanatides, P.G., Scherer, S.E., Li, P.W., Hoskins, R.A., Galle, R.F., et al. 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185-2195. [DOI] [PubMed] [Google Scholar]
- Bailey, J.A., Gu, Z., Clark, R.A., Reinert, K., Samonte, R.V., Schwartz, S., Adams, M.D., Myers, E.W., Li, P.W., and Eichler, E.E. 2002. Recent segmental duplications in the human genome. Science 297: 1003-1007. [DOI] [PubMed] [Google Scholar]
- Bateman, A., Birney, E., Cerruti, L., Durbin, R., Etwiller, L., Eddy, S.R, Griffiths-Jones, S., Howe, K.L., Marshall, M., and Sonnhammer, E.L. 2002. The Pfam protein families database. Nucleic Acids Res. 30: 276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer, M.A., Kilroy, G.E., Richard, P.E., Shaikh, T.H., Desselle, T.D., Hoppens, C.L., and Deininger, P.L. 1990. Structure and variability of recently inserted Alu family members. Nucleic Acids Res. 18: 6793-6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney, E. and Durbin, R. 2000. Using GeneWise in the Drosophila annotation experiment. Genome Res. 10: 547-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnelye, E., Vanacker, J.M., Desbiens, X., Begue, A., Stehelin, D., and Laudet, V. 1994. Rev-erb β, a new member of the nuclear receptor superfamily, is expressed in the nervous system during chicken development. Cell Growth Differ. 5: 1357-1365. [PubMed] [Google Scholar]
- Boudet, N., Aubourg, S., Toffano-Nioche, C., Kreis, M., and Lecharny, A. 2001. Evolution of intron/exon structure of DEAD helicase family genes in Arabidopsis, Caenorhabditis, and Drosophila. Genome Res. 11: 2101-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy, S. 1998. Profile hidden Markov models. Bioinformatics 14: 755-763. [DOI] [PubMed] [Google Scholar]
- Forrest, D., Sjoberg, M., and Vennstrom, B. 1990. Contrasting developmental and tissue-specific expression of α and β thyroid hormone receptor genes. EMBO J. 9: 1519-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gampe Jr., R.T., Montana, V.G., Lambert, M.H., Miller, A.B., Bledsoe, R.K., Milburn, M.V., Kliewer, S.A., Willson, T.M., and Xu, H.E. 2000. Asymmetry in the PPARγ/RXRα crystal structure reveals the molecular basis of heterodimerization among nuclear receptors. Mol. Cell 5: 545-555. [DOI] [PubMed] [Google Scholar]
- Giguere, V. 1999. Orphan nuclear receptors: From gene to function. Endocr. Rev. 20: 689-725. [DOI] [PubMed] [Google Scholar]
- Greschik, H., Wurtz, J.-M., Hublitz, P., Kohler, F., Moras, D., and Schule, R. 1999. Characterization of the DNA-binding and dimerization properties of the nuclear orphan receptor germ cell nuclear factor. Mol. Cell. Biol. 19: 690-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, W., Burris, T.P., Zhang, Y.H., Huang, B.L., Mason, J., Copeland, K.C., Kupfer, S.R., Pagon, R.A., and McCabe, E.R. 1996. Genomic sequence of the DAX1 gene: An orphan receptor responsible for X-linked adrenal hypoplasia congenital and hypergonadotropic hypogonadism. J. Clin. Endocrinol. Metab. 81: 2481-2486. [DOI] [PubMed] [Google Scholar]
- Gurates, B., Amsterdam, A., Tamura, M., Yang, S., Zhou, J., Fang, Z., Amin, S., Sebastian, S., and Bulun, S.E. 2003. WT1 and DAX-1 regulate SF-1-mediated human P450arom gene expression in gonadal cells. Mol. Cell Endocrinol. 208: 61-75. [DOI] [PubMed] [Google Scholar]
- Henikoff, S., Henikoff, J.G., Alford, W.J., and Pietrokovski, S. 1995. Automated construction and graphical presentation of protein blocks from unaligned sequences. Gene 163: GC17-26. [DOI] [PubMed] [Google Scholar]
- Johansson, L., Thomsen, J.S., Damdimopoulos, A.E., Spyrou, G., Gustafsson, J.A., and Treuter, E. 2000. The orphan nuclear receptor SHP utilizes conserved LXXLL-related motifs for interactions with ligand-activated estrogen receptors. Mol. Cell. Biol. 20: 1124-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, W.J. 2002. BLAT: The BLAST-like Alignment Tool. Genome Res. 4: 656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliewer, S.A., Moore, J.T., Wade, L., Staudinger, J.L., Watson, M.A., Jones, S.A., McKee, D.D., Oliver, B.B., Willson, T.M., Zetterstrom, R.H., et al. 1998. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell 92: 73-82. [DOI] [PubMed] [Google Scholar]
- Koh, Y.S. and Moore, D.S. 1999. Linkage of the nuclear hormone receptor genes NR1D2, THRB, and RARB: Evidence for an ancient, large-scale duplication. Genomics 57: 289-292. [DOI] [PubMed] [Google Scholar]
- Laudet, V. 1997. Evolution of the nuclear receptor superfamily early diversification from an ancestral orphan receptor. J. Mol. Endocrinol. 19: 207-226. [DOI] [PubMed] [Google Scholar]
- Laudet, V., Hanni, C., Coll, J., Catzeflis, F., and Stehelin, D. 1992. Evolution of the nuclear receptor gene superfamily. EMBO J. 11: 1003-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar, M.A., Hodin, R.A., Darling, D.S., and Chin, W.W. 1989. A novel member of the thyroid/steroid hormone receptor family is encoded by the opposite strand of the rat c-erbA α transcriptional unit. Mol. Cell. Biol. 9: 1128-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann, J.M., McKee, D.D., Watson, M.A., Willson, T.M., Moore, J.T., and Kliewer, S.A. 1998. The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J. Clin. Invest. 102: 1016-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W.-H., Wu, C.I., and Luo, C.C. 1985. A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol. Biol. Evol. 2: 150-174. [DOI] [PubMed] [Google Scholar]
- Maglich, J.M., Sluder, A., Guan, X., Shi, Y., McKee, D.D., Carrick, K., Kamdar, K., Willson, T.M., and Moore, J.T. 2001. Comparison of complete nuclear receptor sets from the human, Caenorhabditis elegans and Drosophila genomes. Genome Biol. 2: RESEARCH0029. [DOI] [PMC free article] [PubMed]
- Maglich, J.M., Stoltz, C.M., Goodwin, B., Hawkins-Brown, D., Moore, J.T., and Kliewer, S.A. 2002. Nuclear pregnane X receptor and constitutive androstane receptor regulate overlapping but distinct sets of genes involved in xenobiotic detoxification. Mol. Pharmacol. 62: 638-646. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf, D.J., Thummel, C., Beato, M., Herrlich, P., Schutz, G., Umesono, K., Blumberg, B., Kastner, P., Mark, M., Chambon, P., et al. 1995. The nuclear receptor superfamily: The second decade. Cell 83: 835-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, L.B., Parks, D.J., Jones, S.A., Bledsoe, R.K., Consler, T.G., Stimmel, J.B., Goodwin, B., Liddle, C., Blanchard, S.G., Willson, T.M., et al. 2000. Orphan nuclear receptors constitutive androstane receptor and pregnane X receptor share xenobiotic and steroid ligands. J. Biol. Chem. 275: 15122-15127. [DOI] [PubMed] [Google Scholar]
- Nuclear Receptors Committee. 1999. A unified nomenclature system for the nuclear receptor subfamily. Cell 97: 1-20. [DOI] [PubMed] [Google Scholar]
- Otte, K., Kranz, H., Kober, I., Thompson, P., Hoefer, M., Haubold, B., Remmel, B., Voss, H., Kaiser, C., Albers, M., et al. 2003. Identification of farnesoid X receptor β as a novel mammalian nuclear receptor sensing lanosterol. Mol. Cell Biol. 23: 864-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rat Genome Sequencing Project Consortium. 2004. Genome sequence of the Brown Norway Rat yields insights into mammalian evolution. Nature (in press). [DOI] [PubMed]
- Robertson, H.M., Warr, C.G., and Carlson, J.R. 2003. Molecular evolution of the insect chemoreceptor superfamily in Drosophila melanogaster. Proc. Natl. Acad. Sci. 100: 14537-14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson-Rechavi, M., Carpentier, A.-S., Duffraisse, M., and Laudet, V. 2001. How many nuclear hormone receptors in the human genome? Trends Genet. 17: 554-556. [DOI] [PubMed] [Google Scholar]
- Sluder, A.E. and Maina, C.V. 2001. Nuclear receptors in nematodes: Themes and variations. Trends Genet. 17: 206-213. [DOI] [PubMed] [Google Scholar]
- Swofford, D.L. 2003. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sinauer Associates, Sunderland, MA.
- Tsai, M.J. and O'Malley, B.W. 1994. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu. Rev. Biochem. 63: 451-486. [DOI] [PubMed] [Google Scholar]
- Tuzun, E., Bailey, J.A., and Eichler, E.E. 2004. Recent segmental duplications in the working draft assembly of the brown Norway rat. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Wang, L.H., Tsai, S.Y., Cook, R.G., Beattie, W.G., Tsai, M.J., and O'Malley, B.W. 1989. COUP transcription factor is a member of the steroid receptor superfamily. Nature 340: 163-166. [DOI] [PubMed] [Google Scholar]
- Watkins, R.E., Wisely, G.B., Moore, L.B., Collins, J.L., Lambert, M.H., Williams, S.P., Willson, T.M., Kliewer, S.A., and Redinbo, M.R. 2001. The human nuclear xenobiotic receptor PXR: Structural determinants of directed promiscuity. Science 292: 2329-2333. [DOI] [PubMed] [Google Scholar]
- Watkins, R.E., Davis-Searles, P.R., Lambert, M.H., and Redinbo, M.R. 2003. Coactivator binding promotes the specific interaction between ligand and the pregnane X receptor. J. Mol. Biol. 331: 815-828. [DOI] [PubMed] [Google Scholar]
- Wurtz, J.M., Bourguet, W., Renaud, J.P., Vivat, V., Chambon, P., Moras, D., and Gronemeyer, H. 1996. A canonical structure for the ligand-binding domain of nuclear receptors. Nat. Struct. Biol. 3: 87-94. [DOI] [PubMed] [Google Scholar]
- Zar, J.H. 1984. The Poisson distribution and randomness. Biostatistical analysis, 2nd ed. Prentice-Hall, Inc., Englewood Cliffs, NJ.
- Zhang, M. and Chiang, J.Y. 2001. Transcriptional regulation of the human sterol 12α-hydroxylase gene (CYP8B1): Roles of heaptocyte nuclear factor 4α in mediating bile acid repression. J. Biol. Chem. 276: 41690-41699. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://genome.ucsc.edu/; UCSC genome bioinformatics.
- http://blocks.fhcrc.org/; blocks WWW server.