Abstract
The mouse V1R putative pheromone receptor gene family consists of at least 137 intact genes clustered at multiple chromosomal locations in the genome. Species-specific pheromone receptor repertoires may partly explain species-specific social behavior. We conducted a genomic analysis of an orthologous pair of mouse and rat V1R gene clusters to test for species specificity in rodent pheromone systems. Mouse and rat have lineage-specific V1R repertoires in each of three major subfamilies at these loci as a result of postspeciation duplications, gene loss, and gene conversions. The onset of this diversification roughly coincides with a wave of Line1 (L1) retrotranspositions into the two loci. We propose that L1 activity has facilitated postspeciation V1R duplications and gene conversions. In addition, we find extensive homology among putative V1R promoter regions in both species. We propose a regulatory model in which promoter homogenization could ensure that V1R genes are equally competitive for a limiting transcriptional structure to account for mutually exclusive V1R expression in vomeronasal neurons.
The mammalian vomeronasal organ (VNO) recognizes pheromones that provide information about the social and reproductive status of other individuals (Aujard 1997; Keverne 1999; Krieger et al. 1999; Curtis et al. 2001; Del Punta et al. 2002). The putative pheromone receptors expressed in the VNO are encoded by at least two G-protein-coupled receptor gene families: V1R receptors are expressed in the apical compartment of the VNO, and V2R receptors are expressed in the basal domain (Dulac and Axel 1995; Herrada and Dulac 1997; Matsunami and Buck 1997; Ryba and Tirindelli 1997; Pantages and Dulac 2000). The mouse V1R gene family consists of at least 137 intact genes from at least 12 distinct subfamilies (Rodriguez et al. 2002). V1R genes are clustered at multiple loci in the genome; large V1R clusters have been described on mouse Chromosomes 6, 7, 13, and 17 (Lane et al. 2002; Rodriguez et al. 2002).
Distinct social and sexual behaviors exhibited by different species are likely to be reflected in differences in the repertoire of pheromones and their receptors (Lane et al. 2002; Rodriguez and Mombaerts 2002). Species-specific pheromone recognition can contribute to prezygotic barriers that reduce the frequency of unproductive mating. Experimental results affirm that sensory cues used in assortive mating behaviors (“won't mate”) arise shortly after the establishment of zygotic incompatibility (“can't mate”; Dobzhansky and Pavlovsky 1971; Noor 1995). The implication of these observations is that pheromone systems might be subject to abrupt postspeciation diversification.
Individual neurons in the VNO express one allele of a single V1R gene such that the function of the sensory cell is defined by the receptor gene that is transcribed (Rodriguez et al. 2002). Unlike olfactory receptors expressed in the main olfactory epithelium of the nose, V1Rs are thought to interact with a narrow range of odorants, if not single pheromone substances (for review, see Zufall et al. 2002). Therefore, duplication and/or deletion of individual V1R genes during evolution could result in gain and/or loss of the ability to detect specific pheromones, resulting in distinct differences in social behavior.
We have conducted a comparative genomic analysis of orthologous mouse and rat V1R clusters to test for species specificity in V1R repertoires. We find that the clusters in these two species have diverged along species lines shortly after the mouse–rat split by a combination of duplications, deletions, and gene conversions. We propose that this postspeciation V1R divergence was driven by a wave of retrotransposition into the loci that began during or shortly after the mouse–rat split. We also find that the V1R genes at these loci have a putative promoter region that is highly conserved in both species. We suggest a model in which promoter homogenization could be beneficial in the coregulation of these genes.
RESULTS AND DISCUSSION
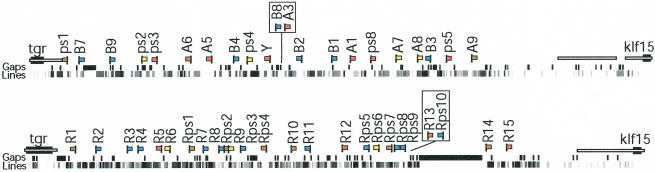
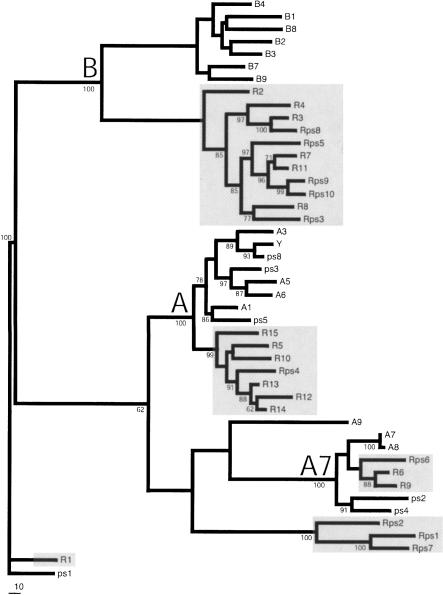
We analyzed orthologous V1R gene clusters on mouse Chromosome 6 and rat Chromosome 4 (Fig. 1). The mouse locus contains ≥21 V1R genes, including 15 intact open reading frames (ORF), and the rat locus contains ≥25 V1R genes, also including 15 intact ORFs. Both loci have gaps in the sequence assembly, and therefore the complete gene content is not known. A maximum likelihood tree partitions the majority of these mouse and rat V1R genes into three major subfamily clades that diverged from each other before the mouse–rat split (Fig. 2, A, A7, and B). Both loci also contain divergent genes that do not belong to the three major subfamilies. Notably, each of the major subfamilies delineates into species-specific clades, suggesting that subfamily expansions occurred independently in the two species after the mouse–rat split. The mouse and rat loci have also diverged by postspeciation V1R gene deletion and/or pseudogenization. For example, the rat counterpart of the mouse A9 gene has either been deleted or is missing within gaps, the mouse ortholog of the rat R1 gene is a pseudogene (ps1), and the clade of rat Rps1, Rps2, and Rps7 pseudogenes has no mouse counterpart.
Figure 1.
Synteny maps depicting rodent V1R clusters on mouse Chromosome 6 (90.2–90.9 Mb, February 2003 release, Mouse Genome Sequencing Consortium 2002) and rat Chromosome 4 (123.9–125.0 Mb, June 2003, Rat Genome Sequencing Project Consortium 2004). The V1R clusters in both species are immediately flanked by single-copy orthologs: the thioredoxin and glutathione reductase (tgr) gene and the Kruppel-like factor 15 gene (klf15). V1R genes are indicated by flags; the subfamily is indicated by color (red, A; blue, B; yellow, A7; gray, others). Pseudogenes are named with a “ps” and “Rps” prefix in mouse and rat, respectively. Mouse V1R nomenclature is derived from previous reports (Lane et al. 2002; Rodriguez et al. 2002), and the rat V1R genes are numbered according to position in the cluster. The mouse B8 and A3 genes and rat Rps10 and R13 genes (boxed) are not present in the current assembly and are positioned according to previous assemblies (Lane et al. 2002 for mouse, November 2002 and January 2003 sequence releases for rat). Because these maps are derived from draft sequences, the relative position and orientation of some of the genes should be considered tentative. L1 repeat content and gaps in the assembly are as reported in the UCSC genome browser (http://genome.ucsc.edu) and are shown beneath the maps. The relative order and orientation are known for all contigs. Contiguous synteny between the mouse and rat loci as determined by dot plots (data not shown) is indicated by unfilled bars on both maps; synteny extends beyond the tgr and klf15 genes.
Figure 2.
Maximum likelihood phylogram of V1R genes and pseudogenes in mouse Chromosome 6 and rat Chromosome 4 clusters. Rat V1R gene clades are shaded gray. To construct this tree, 675 bp of nucleotide sequence was manually aligned such that gaps were minimized. The tree is rooted by the ps1/R1 orthologs based on an outgroup analysis using more distant V1Rs (data not shown). The root branches for the three major subfamily clades (B, A7, and A) are indicated. Bootstrap values (1000 parsimony replicas) are shown on branches if >50%.
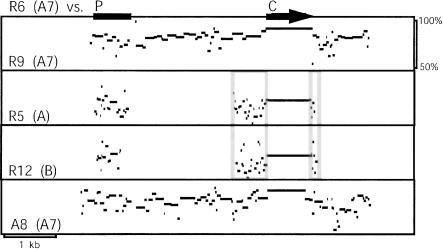
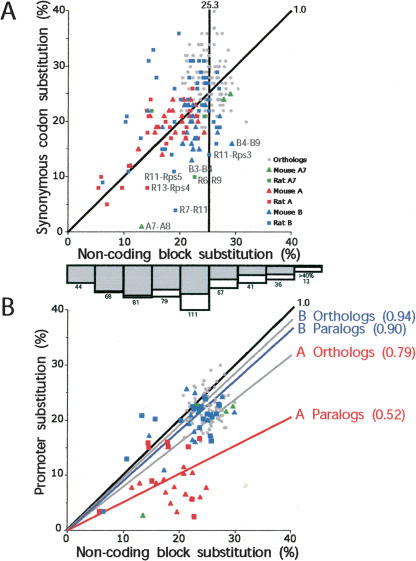
Sequence similarity between subfamily members extends over 2–25 kb blocks of duplicated material (e.g., Fig. 3, R6 vs. R9). We compared noncoding portions of these blocks to further investigate the timing of subfamily expansions. Orthologous mouse and rat sequences not under selection are expected to differ by ∼25% (Li et al. 1987; Bulmer et al. 1991). The maximum pairwise divergence observed between paralogs within the three major V1R subfamilies is 30%, and 77% of the paralogous pairs within subfamilies exhibit 15%–25% divergence (Fig. 4A). These data suggest that paralogs within each subfamily coalesce around the time of the mouse–rat split, and that many of these paralogous duplications occurred shortly after the two species diverged.
Figure 3.
PipMaker plots of the rat R6 gene (A7 subfamily) versus the rat R9 (A7 subfamily), rat R5 (A subfamily), rat R12 (B subfamily), and mouse A8 (A7 subfamily) genes. Homology is plotted according to position and percent identity. Homology does not extend beyond the regions shown. The coding region (C) is indicated by a black horizontal arrow; note that sequences immediately surrounding the coding regions are also apparently under selection (outlined gray in cross-subfamily comparisons). The conserved promoter regions (P) are indicated by a black bar.
Figure 4.
Scatterplots of substitution levels in pairwise comparisons within subfamilies of noncoding and coding regions (A) and noncoding and promoter regions (B). Noncoding block substitutions were calculated using RepeatMasked sequences not apparently under selection; thus, we excluded the conserved promoter regions, conserved coding regions, as well as regions immediately upstream of the coding regions (see Fig. 3). The substitution levels are Jukes-Cantor adjusted (Jukes and Cantor 1969) for multiple substitutions. For coding regions, we considered only substitutions in synonymous sites. Comparisons among A, B, and A7 subfamily paralogs are plotted in red, blue, and green, respectively; comparisons among mouse paralogs are plotted with triangles, and rat paralogs are plotted with squares. Cross-species intrafamily comparisons are plotted with gray circles. The diagonal black lines (slope = 1.0) indicate equal noncoding block and synonymous codon position (or promoter) substitution levels. (A) The average orthologous block divergence within subfamilies is indicated by the vertical line (25% ± 3%). The average paralogous block divergence within subfamilies is 20% (SD ± 5%; 25-th percentile = 17%, 50-th percentile = 22%, 75-th percentile = 24%). On the whole, B subfamily paralogous pairs (average = 22% ± 5%; 25-th percentile = 20%, 50-th percentile = 23%, 75-th percentile = 25%) exhibit greater divergence than A subfamily paralogous pairs (average = 18% ± 5%; 25-th percentile = 15%, 50-th percentile = 18%, 75-th percentile = 21%). Gene pairs that exhibit at least twofold difference in their synonymous and noncoding block substitution levels are labeled. The bar graph at the bottom of the A panel is the age distribution of L1 repeats that integrated in the mouse and rat loci as determined by RepeatMasker (% divergence from consensus, Jukes-Cantor-adjusted). The numbers of L1 repeats identified in each bin is indicated. The gray shading indicates the fraction of these L1s located between duplicated gene blocks. (B) The average orthologous and paralogous promoter:noncoding block substitution ratios are indicated for the A (red labels) and B (blue labels) subfamilies, respectively. These average ratios range between 0.52 (A paralogs) and 0.94 (B orthologs), indicating a range of apparent selective pressures. Note: most pseudogenes are missing the putative promoter sequence, and therefore, fewer pairwise comparisons are shown in B than in A.
The species-specific clades of major V1R subfamilies at this locus could be the result of postspeciation duplications and/or gene conversion events among subfamily members. To investigate the possibility of gene conversions, we examined portions of paralogous blocks for marked differences in substitution levels. Several pairs of coding regions exhibit substantially lower synonymous substitution (dS) levels than the average substitution levels of the surrounding noncoding portions of these blocks (Fig. 4A). Such low relative dS levels suggest either that purifying selection acts at the nucleotide level in coding sequences or that the ancestry of the coding and noncoding portions of some blocks differ. We found statistically significant evidence (p ≤ 0.05) of nine gene conversion events by applying Sawyer's GeneConv algorithm (Sawyer 1989) to sets of genes from within each subfamily (Table 1). These events occurred in the promoter, coding, and intervening noncoding regions of the gene blocks. Some of these gene conversion events could explain the low relative dS levels observed for two of the gene pairs marked in Figure 4A; an event involving a third pair, R7–R11, reaches statistical significance when a different portion of their noncoding sequence is used as GeneConv input (data not shown). Therefore, coalescence times within subfamilies appear more recent than speciation because of a combination of gene conversion events and species-specific duplications. Because gene conversion operates at these loci, more than one gene could have been present in each subfamily in the common ancestor.
Table 1.
Pairs of Genes with GeneConv Statistical Support for Gene Conversion Events
| Gene pair | Regions involved | Length (bp) | P-value |
|---|---|---|---|
| Mouse | |||
| A5—A6 | Noncoding to coding | 3042 | 0.00097 |
| A5—A1 | Noncoding to coding | 487 | 0.0092 |
| A6—ps3 | Noncoding | 430 | 0.0064 |
| A7—A8 | Promoter to coding | 7221 | 0.0098 |
| B1—B2 | Promoter | 633 | 0.0024 |
| B3—B1 | Coding | 442 | 0.031 |
| B4—B1 | Coding | 397 | 0.034 |
| Rat | |||
| R10—R5 | Coding | 234 | 0.0041 |
| R6—R9 | Coding | 418 | 0.0021 |
Cross-subfamily comparisons reveal a large region (∼1 kb) of highly conserved noncoding upstream sequence likely under strong selection (Fig. 4B; Supplemental Fig. 1, and examples in Fig. 3 [R6 vs. R5 and R12]). We previously mapped mouse transcription start sites by RACE-PCR to these upstream homologous regions (Lane et al. 2002), indicating that they might be promoters (Supplemental Fig. 1 available online at www.genome.org). All intact rat V1Rs at this locus also exhibit this putative promoter homology, suggesting that these V1Rs are regulated by common mechanisms in both species.
The selection acting on A subfamily promoter sequences appears greater than on B subfamily promoters (Fig. 4B). Several A subfamily promoters are even more conserved than corresponding coding regions (data not shown). The substitution rates in the promoters of A subfamily members average 79% and 52% of those of substitution rates in noncoding blocks when orthologs or paralogs are compared, respectively. The corresponding values for the B subfamily are 94% and 90%, indicating significantly higher conservation of promoters than surrounding blocks for the B subfamily orthologs and paralogs (p-values of 0.003 and 0.0005, respectively, in two-tailed paired T-test). Thus, both A and B subfamily promoters appear to be under selection.
Each vomeronasal neuron expresses only a single V1R (or V2R) gene (Rodriguez et al. 1999). A competition-based model, in which promoter regions must compete for limiting regulatory factors to achieve this singular gene choice, could account for the high conservation of these regions (Lane et al. 2001). The fact that only genes within this particular V1R gene cluster exhibit this promoter homology (data not shown) suggests that the regulatory significance of this homology is locus-specific. It will be interesting to test whether V1R genes, unlike several tested OR transgenes (e.g., Qasba and Reed 1998; Vassalli et al. 2002), are regulated by locus-dependent mechanisms.
However, such a broad region of high and contiguous similarity remains a conundrum, seeming to be too large to be caused by conservation of individual transcription-factor-binding sites. The extensive homology within promoter regions could be due partly to gene conversions. This hypothesis is consistent with paralogous promoters being more conserved relative to sequences in the surrounding block than orthologous promoters (Fig. 4B), because orthologous sequences cannot gene-convert. Gene conversion events involving B1 (Table 1) could explain the proximity of the B1 and B2 genes in the promoter tree (Supplemental Fig. 1) as compared with the coding sequence tree (Fig. 2). However, the topologies in the two trees are generally in good agreement. Therefore, if gene conversions are largely responsible for the extensive promoter similarity, then these gene conversions are mostly occurring between pairs of genes that are already nearest neighbors in the coding tree.
Therefore, the similarity of these V1R promoters can be due to both selective pressure and gene conversions. We propose that gene conversions have been more prevalent among the A subfamily promoters to account for the significantly higher degree of similarity in paralogous versus orthologous promoters of this group relative to their surrounding sequence (promoter: block = 0.52 and 0.79 for paralogs and orthologs, respectively). Yet, even with gene conversions at play, these promoter sequences are likely under selective pressure because orthologous promoter sequences, which cannot gene-convert, are significantly more conserved than surrounding block sequences, both within and between subfamilies. The unexpected large size of these conserved regions may provide insight into the nature of V1R transcriptional machinery, or more generally, into the findings of large conserved nongenic blocks (>100 bp) elsewhere in the genome (Dermitzakis et al. 2002; Thomas et al. 2003).
Both the mouse and rat V1R clusters are densely packed with L1 repeats (∼44% and ∼52% of the mouse and rat sequence, respectively). This L1 density is far greater than that of the surrounding non-V1R territory (∼6% and ∼5%, respectively) and at least double the L1 density expected from whole-genome surveys (∼22% in areas with A–T base-pair compositions comparable to this region [59%]; Mouse Genome Sequencing Consortium 2002). We estimated the age of each L1 repeat by examining divergence from consensus L1 sequences. Using this measure, the peak time for L1 integration began approximately when members of each major V1R subfamily coalesce (Fig. 4A).
The temporal correlation between the onset of retrotransposition and local gene duplications/conversions suggests a possible cause–effect relationship. The reiterating patterns of V1Rs embedded among numerous L1 repeats are ideal templates for unequal meiotic crossover events. However, meiotic crossovers mediate tandem duplication (Tharapel et al. 1999), whereas the closest V1R paralogs are typically not adjacent. V1Rs in tandem sets with similar orientations, subfamily representation, and divergence are also not observed. If all duplications happened in tandem, we would need to invoke extensive postduplication rearrangement of the clusters and/or gene conversion between nonadjacent paralogs to explain the current gene arrangement. Another mechanism that could generate the structure of the cluster is nonallelic strand invasion during DNA repair, which is known to be able to cause duplication (Smithies and Kim 1994; Ferguson and Holloman 1996; van den Bosch et al. 2002). Such strand invasion could result from gap repair following L1 integration at the V1R locus. This model predicts the observed temporal overlap between L1 activity and V1R duplications.
Eldredge and Gould's theory of punctuated equilibrium is based on interpretation of the fossil record in which morphological divergence associated with speciation occurs abruptly and is followed by long periods of morphological constancy (Eldredge and Gould 1972). This model of speciation predicts that genomes undergo burst-like periods of reorganization. Genomic repeats could facilitate such adaptive bursts, because repeat activity is associated with various forms of genome shuffling (Dehal et al. 2001; Hughes and Coffin 2001; Gilbert and Lutz-Prigge 2002; Symer et al. 2002), retrotransposition is induced during periods of cellular stress (Ratner et al. 1992; Li et al. 1999; Rocha et al. 2002), and periodic repeat activities can lead to adaptive founder effects in populations (Naas et al. 1995).
Our observations fit well with this model of speciation: each of two rodent species has undergone a burst of independent duplications and/or gene conversions, possibly caused by a flurry of L1 repeat activity, that has resulted in divergence of pheromone receptor subfamilies shortly after the mouse–rat split. The postspeciation changes in V1R repertoires might have contributed to distinguishable lines of pheromone communication within, but not between, these two species, requiring that rat and mouse evolve different sensory capabilities. Consistent with this idea, we find that several amino acid residues in these V1R genes show evidence for diversifying (positive) selection. We performed maximum likelihood analysis using PAML (Yang 1997) on a subset of intact V1R genes that are unlikely to have been involved in gene conversion, as such sequence shuffling may cause false positives when testing for diversifying selection (Anisimova et al. 2003). Models of evolution that include a subset of residues under positive selection fit the data much better than corresponding simpler models allowing only purifying and neutral evolution (model 2 vs. 1, p < 10-5; model 3 vs. 1, p < 10-9; model 8 vs. 7, p < 10-4; see Methods). The three models that permit positive selection (models 2, 3, and 8) predict 7% (ω = 3.4), 4% (ω = 3.3), and 11% (ω = 2.3), respectively, of residues to be under positive selection. Four residues in particular (amino acid position 77 in the intracellular loop between the first and second transmembrane [TM] domains, position 152 in the third TM domain, and positions 182 and 188 in the second extracellular loop between the fourth and fifth TM domains) are especially likely (>90% under all three models) to have been subject to diversifying selection. TM3 and especially the extracellular loop between TM4 and TM5 are regions of the predicted protein structure that are thought to be important in G-protein-coupled receptor interactions with ligands (Palczewski et al. 2000; Zhang et al. 2002; Man et al. 2004).
Our results raise several important questions about genome and gene family evolution that could be addressed by extending the study to other loci/species. For example, have other V1R subfamilies undergone similar species-specific expansion since the mouse and rat diverged? Does gene-family expansion at other loci temporally correlate with repeat-integration events? Do subsequent murine speciation events correlate with expansions of different V1R subfamilies? A more global analysis involving additional murine loci and species should provide further insights into the connections between V1R evolution, repeat activity, and speciation.
METHODS
Sequence Assembly
The sequence of the mouse Chromosome 6 (February 2003 release, Mouse Genome Sequencing Consortium 2002) and rat V1R chromosome 4 (June 2003 release) loci were taken from the public draft assemblies available at the UCSC genome browser server (http://genome.ucsc.edu/).
Bioinformatics Tools
Sequences were analyzed for gene content using the UCSC genome browser and associated links (http://genome.ucsc.edu). Repeat content and base-pair composition were analyzed using RepeatMasker (http://ftp.genome.washington.edu/cgi-bin/RepeatMasker). L1 divergence from consensus sequences was provided by RepeatMasker outputs. Alignment of coding and noncoding sequences was aided by PIPMAKER (http://bio.cse.psu.edu/pipmaker/), BLAST (http://www.ncbi.nlm.nih.gov/BLAST/), and CLUSTALW (http://searchlauncher.bcm.tmc.edu/multi-align/multi-align.html). Phylogenetic reconstructions were produced using PAUP (Sinauer Associates). Synonymous substitution levels were determined using SNAP (http://hiv-web.lanl.gov/content/hiv-db/SNAP/WEBSNAP/SNAP.html). We parsed PIPMAKER outputs to calculate average percent divergence for portions of the noncoding block that do not exhibit cross-subfamily homology (i.e., excluding any promoter or coding region homology that is demonstrably under selection). PIPMAKER was used with default settings (no chaining). Boxshade alignments of promoter regions were obtained from http://www.ch.embnet.org/software/BOX_form.html.
GeneConv Analysis
We analyzed all V1R genes and upstream sequence using GeneConv (http://www.math.wustl.edu/~sawyer/geneconv) to identify apparent gene conversion events (Sawyer 1989). GeneConv implements an algorithm that compares an alignment of at least three nucleotide sequences, identifies sites of paralogous variation among them, and for each pair, measures the length of stretches of shared variants. The length of these stretches is compared to a simulated distribution (10,000 iterations) of the same number of sequences with a similar average density of variation and used to calculate a Bonferroni-corrected P-value. A low P-value indicates a high probability that gene conversion occurred.
Error Analysis
All substitution rates used are Jukes-Cantor-adjusted to correct for multiple hits (Jukes and Cantor 1969). We also used the Jin-Nei adjustment method (Jin and Nei 1990) on a randomly selected 10% of the comparisons, and the results were within 0.5% of our Jukes-Cantor results. An estimate of standard error is based on variance of the estimate of each parameter, as described by the following equation for binomial probability: s2 = p(1 – p)/(n – 1), where s is the standard error, p is the fraction of nucleotides that are substituted, and n is the number of nucleotides compared. Binomial sampling errors range from ∼3.9 to ∼2.4% for synonymous sites (152-bp sample size), ∼2.3 to ∼1.5% for promoter regions (483 bp), and ∼5.4 to ∼0.5% for noncoding blocks (78–4305 bp) for 35% to 10% substitution levels, respectively.
PAML Analysis
A codon-based alignment of the B8, B2, B7, B9, R2, R7, A3, Y, R15, R13, R12, R14, A9, and R1 V1R sequences was used to derive a maximum likelihood tree using PAUP's heuristic search algorithm. The tree and alignment were used to test the likelihood of obtaining the observed sequences under various models of evolution using PAML version 3.13 (Yang 1997). Each model allows various classes of residue with different ω values (Yang et al. 2000). ω values represent the likelihood of amino acid change being fixed during evolution, with neutrally evolving residues having ω values of 1, residues under purifying selection having values of less than 1, and residues under diversifying (or positive) selection having values >1. Briefly, model 1 allows two classes of residue—one evolving neutrally (ω = 1) and the other under strict purifying selection (ω = 0). This model was compared with two other models that both allow at least one of the three classes of ω to vary, thereby allowing for the possibility of positive selection at some residues (model 2 has ωs of 0, 1, and a third class with ω estimated for this data set to be 3.4; model 3 has three classes with estimated ωs of 0.3, 1.1, and 3.3). In addition, model 7 (10 classes of residue, in which ωs follow a β distribution with all values between 0 and 1) was compared with model 8 (β distribution plus an eleventh class of positively selected residues with ω estimated at 2.3). Significance of the difference in log-likelihoods was determined as described in Yang et al. (2000). The algorithm converged on the same likelihood values and parameter estimates when run using three different starting ω values (0.2, 1, and 3.14159), indicating that the estimated parameters are likely to represent global rather than local maxima.
Acknowledgments
We thank Elena Linardopoulou for her thoughtful comments, Willie Swanson for advice on the use of PAML, and Joseph Ross for technical support. This work was supported by the National Institutes of Health Grants R01-DC004209 and R01-DC006267.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2117004.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Anisimova, M., Nielsen, R., and Yang, Z. 2003. Effect of recombination on the accuracy of the likelihood method for detecting positive selection at amino acid sites. Genetics 164: 1229-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aujard, F. 1997. Effect of vomeronasal organ removal on male socio-sexual responses to female in a prosimian primate (Microcebus murinus). Physiol. Behav. 62: 1003-1008. [DOI] [PubMed] [Google Scholar]
- Bulmer, M., Wolfe, K.H., and Sharp, P.M. 1991. Synonymous nucleotide substitution rates in mammalian genes: Implications for the molecular clock and the relationship of mammalian orders. Proc. Natl. Acad. Sci. 88: 5974-5978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis, J.T., Liu, Y., and Wang, Z. 2001. Lesions of the vomeronasal organ disrupt mating-induced pair bonding in female prairie voles (Microtus ochrogaster). Brain Res. 901: 167-174. [DOI] [PubMed] [Google Scholar]
- Dehal, P., Predki, P., Olsen, A.S., Kobayashi, A., Folta, P., Lucas, S., Land, M., Terry, A., Ecale Zhou, C.L., Rash, S., et al. 2001. Human chromosome 19 and related regions in mouse: Conservative and lineage-specific evolution. Science 293: 104-111. [DOI] [PubMed] [Google Scholar]
- Del Punta, K., Leinders-Zufall, T., Rodriguez, I., Jukam, D., Wysocki, C.J., Ogawa, S., Zufall, F., and Mombaerts, P. 2002. Deficient pheromone responses in mice lacking a cluster of vomeronasal receptor genes. Nature 419: 70-74. [DOI] [PubMed] [Google Scholar]
- Dermitzakis, E.T., Reymond, A., Lyle, R., Scamuffa, N., Ucla, C., Deutsch, S., Stevenson, B.J., Flegel, V., Bucher, P., Jongeneel, C.V., et al. 2002. Numerous potentially functional bun non-genic conserved sequences on human chromosome 21. Nature 420: 578-582. [DOI] [PubMed] [Google Scholar]
- Dobzhansky, T. and Pavlovsky, O. 1971. Experimentally created incipient species of Drosophila. Nature 230: 289-292. [DOI] [PubMed] [Google Scholar]
- Dulac, C. and Axel, R. 1995. A novel family of genes encoding putative pheromone receptors in mammals. Cell 83: 195-206. [DOI] [PubMed] [Google Scholar]
- Eldredge, N. and Gould, S.J. 1972. Punctuated equilibria: An alternative to phyletic gradualism. In Models in paleobiology (ed. T.J.M. Schopf), pp. 82-115. Freeman, Cooper and Co., San Francisco.
- Ferguson, D.O. and Holloman, W.K. 1996. Recombinatorial repair of gaps in DNA is asymmetric in Ustilago maydis and can be explained by a migrating D-loop model. Proc. Natl. Acad. Sci. 93: 5419-5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, N. and Lutz-Prigge, S. 2002. Genomic deletions created upon LINE-1 retrotransposition. Cell 110: 315-325. [DOI] [PubMed] [Google Scholar]
- Herrada, G. and Dulac, C. 1997. A novel family of putative pheromone receptors in mammals with a topographically organized and sexually dimorphic distribution. Cell 90: 763-773. [DOI] [PubMed] [Google Scholar]
- Hughes, J.F. and Coffin, J.M. 2001. Evidence for genomic rearrangements mediated by human endogenous retroviruses during primate evolution. Nat. Genet. 29: 487-489. [DOI] [PubMed] [Google Scholar]
- Jin, L. and Nei, M. 1990. Limitations of the evolutionary parsimony method of phylogenetic analysis. Mol. Biol. Evol. 7: 82-102. [DOI] [PubMed] [Google Scholar]
- Jukes, T.H. and Cantor, C.R. 1969. Evolution of protein molecules. In Mammalian protein metabolism (ed. H.N. Munro), pp. 21-123. Academic Press, New York.
- Keverne, E.B. 1999. The vomeronasal organ. Science 286: 716-720. [DOI] [PubMed] [Google Scholar]
- Krieger, J., Schmitt, A., Lobel, D., Gudermann, T., Schultz, G., Breer, H., and Boekhoff, I. 1999. Selective activation of G protein subtypes in the vomeronasal organ upon stimulation with urine-derived compounds. J. Biol. Chem. 274: 4655-4662. [DOI] [PubMed] [Google Scholar]
- Lane, R.P., Cutforth, T., Young, J., Athanasiou, M., Friedman, C., Rowen, L., Evans, G., Axel, R., Hood, L., and Trask, B.J. 2001. Genomic analysis of orthologous mouse and human olfactory receptor loci. Proc. Natl. Acad. Sci. 98: 7390-7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane, R.P., Cutforth, T., Axel, R., Hood, L., and Trask, B.J. 2002. Sequence analysis of mouse vomeronasal receptor gene clusters reveals common promoter motifs and a history of recent expansion. Proc. Nat. Acad. Sci. 99: 291-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W.H., Tanimura, M., and Sharp, P.M. 1987. An evaluation of the molecular clock hypothesis using mammalian DNA sequences. J. Mol. Evol. 25: 330-342. [DOI] [PubMed] [Google Scholar]
- Li, T.-H., Spearow, J., Rubin, C.M., and Schmid, C.W. 1999. Physiological stresses increase mouse short interspersed element (SINE) RNA expression in vivo. Gene 239: 367-372. [DOI] [PubMed] [Google Scholar]
- Man, O., Gilad, Y., and Lancet, D. 2004. Prediction of the odorant binding site of olfactory receptor proteins by human–mouse comparisons. Protein Sci. 13: 240-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunami, H. and Buck, L.B. 1997. A multigene family encoding a diverse array of putative pheromone receptors in mammals. Cell 90: 775-784. [DOI] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- Naas, T., Blot, M., Fitch, W.M., and Arber, W. 1995. Dynamics of IS-related genetic rearrangements in resting Escherichia coli K-12. Mol. Biol. Evol. 12: 198-207. [DOI] [PubMed] [Google Scholar]
- Noor, M.A. 1995. Speciation driven by natural selection in Drosophila. Nature 375: 674-675. [DOI] [PubMed] [Google Scholar]
- Palczewski, K., Kumasaka, T., Hori, T., Behnke, C.A., Motoshima, H., Fox, B.A., Le Trong, I., Teller, D.C., Okada, T., Stenkamp, R.E., et al. 2000. Crystal structure of rhodopsin: A G protein-coupled receptor. Science 289: 739-745. [DOI] [PubMed] [Google Scholar]
- Pantages, E. and Dulac, C. 2000. A novel family of candidate pheromone receptors in mammals. Neuron 28: 835-845. [DOI] [PubMed] [Google Scholar]
- Qasba, P. and Reed, R.R. 1998. Tissue and zonal-specific expression of an olfactory receptor transgene. J. Neurosci. 18: 227-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner, V.A., Zabanov, S.A., Kolesnikova, O.V., and Vasilyeva, L.A. 1992. Induction of the mobile genetic element Dm-412 transposition in the Drosophila genome by heat shock treatment. Proc. Natl. Acad. Sci. 89: 5650-5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha, E.P.C., Matic, I., and Taddei, F. 2002. Overexpression of repeats in stress response genes: A strategy to increase versatility under stressful conditions? Nucleic Acids Res. 9: 1886-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, I. and Mombaerts, P. 2002. Novel human vomeronasal receptor-like genes reveal species-specific families. Curr. Biol. 12: R409-R411. [DOI] [PubMed] [Google Scholar]
- Rodriguez, I., Feinstein, P., and Mombaerts, P. 1999. Variable patterns of axonal projections of sensory neurons in the mouse vomeronasal system. Cell 97: 199-208. [DOI] [PubMed] [Google Scholar]
- Rodriguez, I., Del Punta, K., Rothman, A., Ishii, T., and Mombaerts, P. 2002. Multiple new and isolated families within the mouse superfamily of V1R vomeronasal receptors. Nat. Neurosci. 5: 134-140. [DOI] [PubMed] [Google Scholar]
- Ryba, N.J. and Tirindelli, R. 1997. A new multigene family of putative pheromone receptors. Neuron 19: 371-379. [DOI] [PubMed] [Google Scholar]
- Sawyer, S. 1989. Statistical tests for detecting gene conversion. Mol. Biol. Evol. 6: 526-538. [DOI] [PubMed] [Google Scholar]
- Smithies, O. and Kim, H.S. 1994. Targeted gene duplication and disruption for analyzing quantitative genetic traits in mice. Proc. Natl. Acad. Sci. 91: 3612-3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symer, D.E., Connelly, C., Szak, S.T., Caputo, E.M., Cost, G.J., Parmigiani, G., and Boeke, J.D. 2002. Human L1 retrotransposition is associated with genetic instability in vivo. Cell 110: 327-338. [DOI] [PubMed] [Google Scholar]
- Tharapel, A.T., Michaelis, R.C., Velageleti, G.V., Laundon, C.H., Martens, P.R., Buchanan, P.D., Teague, K.E., Tharapel, S.A., and Wilroy, R.S. 1999. Chromosome duplications and deletions and their mechanisms of origin. Cytogenet. Cell Genet. 85: 285-290. [DOI] [PubMed] [Google Scholar]
- Thomas, J.W., Touchman, J.W., Blakesley, R.W., Bouffard, G.G., Beckstrom-Sternberg, S.M., Margulies, E.H., Blanchette, M., Siepel, A.C., Thomas, P.J., McDowell, J.C., et al. 2003. Comparative analyses of multi-species sequences from targeted genomic regions. Nature 424: 788-793. [DOI] [PubMed] [Google Scholar]
- van den Bosch, M., Lohman, P.H., and Pastink, A. 2002. DNA double-strand break repair by homologous recombination. Biol. Chem. 383: 783-892. [DOI] [PubMed] [Google Scholar]
- Vassalli, A., Rothman, A., Feinstein, P., Zapatocky, M., and Mombaerts, P. 2002. Minigenes impart odorant receptor-specific axon guidance in the olfactory bulb. Neuron 35: 681-696. [DOI] [PubMed] [Google Scholar]
- Yang, Z. 1997. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13: 555-556. [DOI] [PubMed] [Google Scholar]
- Yang, Z., Nielsen, R., Goldman, N., and Pedersen, A.M. 2000. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155: 431-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., DeHaven, R.N., and Goodman, M. 2002. NMR and modeling studies of a synthetic extracellular loop II of the κ opioid receptor in a DPC micelle. Biochemistry 41: 61-68. [DOI] [PubMed] [Google Scholar]
- Zufall, F., Kelliher, K.R., and Leinders-Zufall, T. 2002. Pheromone detection by mammalian vomeronasal neurons. Microsc. Res. Tech. 58: 251-260. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://bio.cse.psu.edu/pipmaker/; PipMaker and MultiPipMaker.
- http://ftp.genome.washington.edu/cgi-bin/RepeatMasker; RepeatMasker Server.
- http://genome.ucsc.edu/; UCSC Genome Bioinformatics Home.
- http://hiv-web.lanl.gov/content/hiv-db/SNAP/WEBSNAP/SNAP.html; HIV Sequence Database: SNAP Submission Form.
- http://searchlauncher.bcm.tmc.edu/multi-align/multi-align.html; BCM Search Launcher.
- http://www.ch.embnet.org/software/BOX_form.html; BoxShade Server.
- http://www.math.wustl.edu/~sawyer/geneconv; GENECONV Computer Program Index.
- http://www.ncbi.nlm.nih.gov/BLAST/; NCBI BLAST.