Abstract
We aligned Incyte ESTs and publicly available sequences to the rat genome and analyzed rat chromosome 1q43-54, a region in which several quantitative trait loci (QTLs) have been identified, including renal disease, diabetes, hypertension, body weight, and encephalomyelitis. Within this region, which contains 255 Ensembl gene predictions, the aligned sequences clustered into 568 Incyte genes and gene fragments. Of the Incyte genes, 261 (46%) overlapped 184 (72%) of the Ensembl gene predictions, whereas 307 were unique to Incyte. The rat-to-human syntenic map displays rearrangement of this region on rat chr. 1 onto human chromosomes 9 and 10. The mapping of corresponding human disease phenotypes to either one of these chromosomes has allowed us to focus in on genes associated with disease phenotypes. As an example, we have used the syntenic information for the rat Rf-1 disease region and the orthologous human ESRD disease region to reduce the size of the original rat QTL to only 11.5 Mb. Using the syntenic information in combination with expression data from ESTs and microarrays, we have selected a set of 66 candidate disease genes for Rf-1. The combination of the results from these different analyses represents a powerful approach for narrowing the number of genes that could play a role in the development of complex diseases.
The recent publication of the rat genome sequence makes it possible to study genes in the context of their genomic location as well as their syntenic association to genes characterized on the human and mouse genomes. This is of particular value for the identification of genes responsible for disease phenotypes (Leo et al. 2002). More than 200 disease-related quantitative trait loci (QTL) have been genetically mapped in rat (http://ratmap.gen.gu.se/). Further analyses of such regions via positional cloning strategies are typically used to identify the genes responsible for a particular disease. This often involves tedious timeconsuming approaches, such as construction of a physical map and the production of congenic and subcongenic rat strains. However, the availability of the rat genomic sequence allows for the identification and functional study of genes across chromosomal locations and species, which can accelerate the assignment of a disease phenotype to a particular gene or set of genes. Furthermore, syntenic mapping between two species with a similar disease phenotype, such as renal failure, which is examined in this study, can help to identify genes responsible for a disease.
The Ensembl database provides information on known rat genes as well as rat gene predictions that are derived by an automated pipeline using a combination of ab-initio Genscan predictions, GeneWise protein homology alignments, or a set of aligned rat and mouse cDNAs. However, computationally based gene predictions have inherent limitations (Burge and Karlin 1997; Solovyev and Salamov 1997), and a majority of these predictions do not contain rat cDNA evidence (Rat Genome Sequencing Project Consortium 2004). The addition of cDNA evidence for these gene predictions is critical for their evaluation and identification of potential disease genes. Furthermore, alignment of cDNA sequences to the rat genome can be used to identify novel gene loci that cannot be found by computational methods and can provide a powerful means to infer organspecific gene expression through the associated cDNA library information.
In addition to the characterization of genes on the rat genome, the identification of human orthologs is critical to link identified disease genes across species. Reciprocal BLAST sequence comparison between genes from two species is a commonly used approach to identify orthologs. However, this method is most reliable when used in species with complete transcriptomes, which are not yet available in either human or rat (Tatusov et al. 1997). This method also has limitations when comparing genes with high-sequence similarity between family members. For such genes, unique orthologous gene pairs are difficult to identify via reciprocal BLAST, as multiple candidate genes are typically obtained among family members. However, these candidates can be further evaluated by integrating information on syntenic genes across two species, thus allowing for the retrieval of more putative orthologous gene pairs.
Rat chromosome 1 (chr. 1) at 1q43-54 encompasses several different QTLs, including renal failure 1 (Rf-1; Brown et al. 1996; Shiozawa et al. 2000), non-insulin-dependent diabetes mellitus (Niddm; Jacob et al. 1995; Galli et al. 1996, Wei et al. 1999), hypertension (Bp42; Kovacs et al. 1997), body weight (Weight3; Kovacs et al. 1998), and experimental allergic encephalomyelitis (Eae7; Bergsteinsdottir et al. 2000, Dracheva et al. 2000). When a particular disease phenotype affects a specific organ, this information can be utilized to help to select or exclude potential candidate genes for participation in a disease phenotype on the basis of their organ expression. Thus, we have provided rat cDNA microarray data and expression information based on EST clone frequency for known and novel genes in this region.
We use the segmentation of the syntenic map between rat and human to reduce the size of the rat Rf-1 QTL region. Furthermore, by using rat EST alignments to rat chr. 1q43-54, we have provided transcriptional evidence for rat genes, as well as information on organ expression, which was previously unavailable for a large set of these genes. We demonstrate a distinctive approach in the study of candidate disease genes that combines syntenic and disease linkage information across two or more species, as well as expression information, to reduce the number of candidate disease genes.
RESULTS
Genomic Alignments
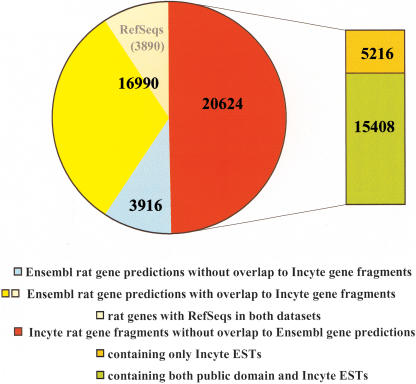
To determine EST-based genes, we aligned and clustered 1.2 M Incyte and 0.3 M public rat ESTs, 4248 rat Refseq, and 5605 protein-containing GenPept mRNA sequences from GenBank to the rat genomic sequence. A total of 4248 RefSeq sequences generated 3890 rat genes. In addition, 35,860 EST-inferred gene fragments that contained three or more clones were formed. Some of these rat gene fragments represent underclustered segments of the same genes and need to be studied to derive a final number of genes. Of the Incyte EST-inferred genes, 20,452 (57%) were composed of mixed public and Incyte EST content, whereas 15,408 (43%) contained only Incyte ESTs, which could potentially represent novel genes. To further validate Incyte EST-based genes, we examined protein predictions and homology to human genes. A total of 16,017 (45%) of the Incyte rat gene fragments contained protein predictions and 26,061 (72%) had human syntenic homologs. To assess how many of these genes are not represented in the Ensembl database, we compared overlaps between our set of genes and the 20,906 rat gene predictions found in the Ensembl database. The number of genes overlapping in both the Ensembl and the Incyte data set is shown in Figure 1. The Incyte data set contains EST-based gene evidence for 81% of the Ensembl gene predictions, whereas more than half of the Incyte gene fragments do not overlap any of the Ensembl gene predictions.
Figure 1.
Overlap between the Ensembl and Incyte gene data set.
Aligned Sequences on Rat chr. 1q43-54
Within the rat chr. 1 coordinates, 232 Mb (1q43) to 264 Mb (1q54), a total of 11,827 Incyte and 2891 public domain ESTs from 435 libraries were aligned (from 11,357 and 2704 clones, respectively). In addition to EST sequences, 42 RefSeq and 61 mRNA sequences with corresponding protein sequences in GenPept aligned to rat chr. 1q43-54. The RefSeq sequences and GenPept-derived mRNA sequences clustered into 42 genes containing 4129 clones. A total of 9932 additional clones formed 526 gene fragments, containing three or more clones or at least one mRNA splice site.
As RefSeq sequences are mapped in both the Incyte and Ensembl data set, we compared the agreement between the RefSeq-containing genes in both data sets. The Ensembl database contained 50 known rat genes and 205 novel gene predictions in the same region of chr. 1q43-54. Of the known genes, 46 referenced a RefSeq sequence, but two genes at two separate locations shared the same RefSeq sequence (NM_012963 and NM_017272), whereas RefSeq locations in the Incyte database are unique. A total of 40 of the RefSeq sequences in both data sets mapped to corresponding same gene locations. The ensembl data set contained three additional RefSeq sequences that Incyte did not map to this region, because the sequence alignments had low identity over the coding region (NM_020094—91%, NM_020095—92%, NM_053646—60%). In addition, Ensembl localized RefSeq NM_033539 to chr. 1. However, in Incyte's database, this RefSeq was mapped with higher identity (100%) and coverage (100%) to rat chromosome 9. Moreover, NM_033539 maps to 17 different locations on the genome, and thus, the locus on chr. 1 is questionable. Another difference between the data sets was Incyte's mapping of NM_138512 to this region on chromosome 1q, which is a unique alignment of this RefSeq sequence to the genome. Furthermore, NM_017042 was also mapped to chr. 1q, whereas this RefSeq is mapped to chr. 15 in the Ensembl data set. We chose the mapping of this RefSeq to chr. 1 over the mapping to chr. 15 due to higher coverage (97%) and identity (95%).
Clustering of Aligned Sequences Into Genes and Gene Fragments
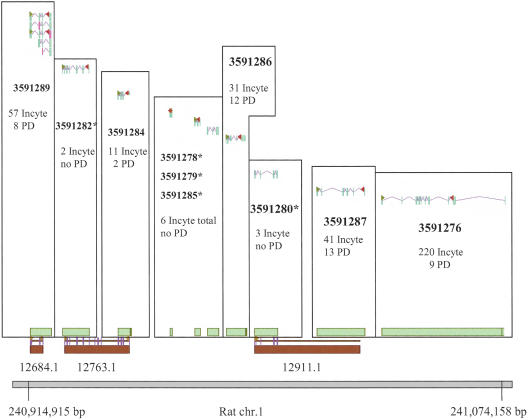
The total of 14 K sequences that aligned to chr. 1q43-54 clustered into 568 Incyte genes and gene fragments. A total of 261 of these genes overlapped 184 Ensembl gene predictions, which includes the overlap of 40 RefSeq containing genes in both data sets. In 90 cases, there was a 1:1 match between an Incyte and Ensembl gene. For the remaining comparisons, we investigated the clustering of genes in both data sets. A total of 171 Incyte genes overlapped 94 Ensembl genes. Most of the Ensembl genes overlapped two Incyte genes, however, one gene overlapped eight Incyte genes. In contrast, 16 Incyte genes were found to overlap two or more Ensembl gene predictions, indicating some potential for underclustering in the Ensembl data set or overclustering in the Incyte data set. The complete set of Incyte genes and gene fragments, their coordinates, and overlap to Ensembl genes are presented in Supplemental file 1 available online at www.genome.org. An example distribution of Incyte and Ensembl genes and their overlap is shown in Figure 2.
Figure 2.
Browser shot of Incyte and Ensembl genes in a subregion of chr. 1q43-54. Incyte genes IDs are given in bold and are presented as green boxes, whereas the Ensembl genes are represented as brown boxes with the distinct nuremical part of the Ensembl gene ID (all have prefix of ENSRNOG000000*) below the boxes. Incyte transcripts are presented in blue. The number of Incyte ESTs (Incyte) and ESTs from the public domain (PD) are given below each gene ID. Protein sequence start and stop locations are indicated by arrows in the transcripts. (*) Genes composed of Incyte ESTs only.
We identified 307 rat genes and gene fragments that did not overlap with Ensembl gene predictions. To derive further information about these gene fragments, we examined their BLAST hit comparisons to other species, their EST and splice site content, and potential protein content. Overall, 210 gene fragments contained transcripts with one or more significant BLAST sequence comparisons to other species. A total of 204 and 125 genes and gene fragments had significant BLAST sequence homology to mouse and to human genes, respectively. Most notably, 17 of the 307 genes were composed of Incyte ESTs only, had an ORF of >60 amino acids, had sequence homology to human or mouse cDNA, and showed splicing when aligned to the genome (Table 1). In contrast, two other gene fragments were identified as potential pseudogenes, as they contained single exon copies of multiexon RefSeq sequences, thus indicating a potential retrotransposition.
Table 1.
Incyte EST-Inferred Gene Fragments Without Overlap to Ensembl Genes but With Syntenic Human Homologs and Protein Sequence
| Gene ID | Chromosomal start coordinate | Number of splice sites | Number of clones | ORF length [aa] | BLAST hits to other speciesa |
|---|---|---|---|---|---|
| INCY:3591278 | 240,961,913 | 2 | 1 | 93 | Hs, Mm, Cf |
| INCY:3591279 | 240,970,160 | 2 | 2 | 115 | Hs, Mm, Cf |
| INCY:3591474 | 241,478,869 | 3 | 7 | 97 | Hs, Mm, Cf |
| D1Mit34 | 242,893,444 | - | - | - | - |
| INCY:3591684 | 245,269,246 | 5 | 2 | 253 | Hs, Mm, Cf |
| INCY:3591769 | 246,839,356 | 3 | 2 | 83 | Hs, Mm, Cf |
| INCY:3592034 | 249,499,454 | 1 | 7 | 283 | Hs, Mm, Cf |
| INCY:3592151 | 251,569,065 | 4 | 4 | 266 | Hs, Cf |
| INCY:3592138 | 251,577,289 | 4 | 1 | 165 | Hs, Mm |
| INCY:3592142 | 251,966,650 | 5 | 6 | 404 | Hs, Mm, Cf, Mf |
| D1RAT156 | 253,994,157 | - | - | - | - |
| INCY:3592461 | 254,525,849 | 8 | 3 | 342 | Hs, Mm, Cf |
| INCY:3592603 | 255,411,038 | 2 | 2 | 127 | Hs |
| INCY:3592604 | 255,428,657 | 5 | 1 | 240 | Hs |
| INCY:3592608 | 255,822,094 | 2 | 2 | 177 | Hs, Mm, Cf |
| INCY:3592707 | 257,051,672 | 1 | 2 | 92 | Hs, Mm |
| INCY:3592766 | 258,817,247 | 6 | 8 | 142 | Hs, Mm, Cf |
| INCY:3592947 | 259,518,570 | 8 | 6 | 284 | Hs, Mm, Cf |
| INCY:3592944 | 260,048,191 | 2 | 4 | 133 | Mm |
(Hs) Human; (Mm) mouse; (Cf) dog
Verification of Ensembl Novel Gene Predictions
Of the 255 Ensembl rat gene predictions in chr. 1q43-54, 184 (72%) had overlaps to Incyte gene and gene fragments. Of these genes with overlap to EST-containing Incyte genes, 134 were novel Ensembl predictions. To validate the Ensembl gene predictions, we examined the EST content provided by the overlapping Incyte ESTs as well as the splice sites and BLAST hits to other species. A total of 133 (65%) of the Ensembl novel genes overlapped gene fragments that had sequence homology to human, mouse, or dog sequences, as identified by BLAST sequence comparisons; 44 of the novel Ensembl gene predictions overlapped Incyte gene fragments that were composed of Incyte ESTs only and could not be confirmed with any public domain ESTs; 94 (64%) Ensembl novel genes had splice sites that could be verified by Incyte gene fragments (for example, see Fig. 2); and 40 had exonic overlaps only. Two of the Ensembl genes that overlapped Incyte EST sequences were identified as potential pseudogenes, as they overlapped Incyte genes that contained a single-exon copy of a multiexon RefSeq sequence, thus indicating a possible retrotransposon event. In addition to Ensembl genes with overlap to Incyte gene fragments, the chr. 1q43-54 region contained 71 Ensembl gene predictions that did not overlap an exon of an Incyte gene.
Annotation Comparison of Ensembl Gene Predictions Overlapping Incyte Genes and Gene Fragments
Of the 50 Ensembl known genes, 46 (92%) had annotations that agreed with the overlapping Incyte genes or gene fragments. Of the remaining Ensembl known genes, three lacked annotation and one gene, ENSRNOG00000018277, shared exons with an Ensembl-known gene on the opposite strand, suggesting that it might be an artifact. Of the 134 Ensembl novel gene predictions overlapping an Incyte gene, only 69 had annotation agreement with the Incyte gene. A total of 33 Ensembl genes lacked annotation, but overlapped an Incyte gene with annotation (see Table 2 for examples with shared overlapping coding regions). In addition, five Ensembl novel predictions had inconsistent annotation with their Incyte counter parts. These differences in annotation were a result of differing gene-coding regions rather than true differences in the way the annotation was derived.
Table 2.
Addition of Title Lines for Ensembl Gene Predictions Using Overlap to EST-Inferred Genes
| Ensemble gene | Description/protein family | Incyte gene | Description |
|---|---|---|---|
| G00000011806.1 | No description/UNKNOWN | INCY:3591102 | Calcineurin A β isoform, catalytic subunit of Ca2+/calmodulin-regulated protein phosphatase, plays role in regulating activity of transcription factor NFAT |
| G00000017298.1 | No description/UNKNOWN | INCY:3591616 | Strong similarity to (Homo sapiens) CGI-32: Member of the CutC family, which are involved in copper transport, has moderate similarity to uncharacterized C. elegans ZK353.7 |
| G00000020248.1 | No description/AMBIGUOUS | INCY:3592445 | α-internexin, a neurofilament protein that may function in neurogenesis and brain development; human INA may play a role in HTLV-associated neurological disorders and may be a candidate for infantile onset spinocerebellar ataxia |
| G00000019509.1 | No description/AMBIGUOUS | INCY:3592299 | Very strong similarity to (Mus musculus) 0710008C12Rik: Protein containing an F-box domain, which serve as a link between a target protein and a ubiquitin-conjugating enzyme, has low similarity to F-box leucine-rich repeat 3 (rat Fbxl2), which may target proteins to the SCF complex |
Differential Expression and Electronic Northern for Genes on Chromosome 1q43-54
As organ expression information can indicate the potential association of a gene to one of the disease phenotypes localized on rat chr. 1q43-54, we examined the anatomical expression of the genes and gene fragments in this region using both cDNA microarrays and electronic Northern analysis. Microarray analysis was performed using RNA isolated from 27 different organs and run in competitive hybridizations against a pool of all of the RNAs. The microarrays used contained 275 clones that mapped to 177 of the gene and gene fragments on rat chr. 1q43-54. A significant differential expression of twofold or greater for an individual organ relative to the pool of all organs was seen for 41 genes and gene fragments (Table 3).
Table 3.
Genes with Significant Increase in Differential Expression as Well as High Specificity to One Organ in the Electronic Northern Evaluation
| Gene ID | Microarray | Electronic northern | Number of clones |
|---|---|---|---|
| INCY:3590992 | LIVER | Liver | 7 |
| INCY:3591185 | OVARY | Ovary | 15 |
| INCY:3591223 | BRAIN | Brain | 23 |
| INCY:3591251 | - | Brain | 9 |
| INCY:3591252 | - | Brain | 3 |
| INCY:3591272 | LUNG | Lung | 8 |
| INCY:3591277 | - | Kidney | 11 |
| INCY:3591280 | - | Liver | 3 |
| INCY:3591289 | - | Kidney | 10 |
| INCY:3591292 | - | Liver | 63 |
| INCY:3591460 | - | Kidney | 17 |
| INCY:3591461 | OVARY | - | 0 |
| INCY:3591465 | - | Bones | 5 |
| INCY:3591472 | BLADDER/Lung | -/- | 0/2 |
| INCY:3591479 | MUSCLE | - | 1 |
| INCY:3591480 | KIDNEY | Kidney | 7 |
| INCY:3591484 | - | Liver | 3 |
| INCY:3591486 | TESTIS/- | Testis/Brain | 3/13 |
| INCY:3591488 | KIDNEY | Kidney | 35 |
| INCY:3591555 | MUSCLE/TONGUE/- | Muscle/-/Kidney | 4/1/25 |
| INCY:3591558 | - | Brain | 4 |
| INCY:3591587 | - | Intestine | 3 |
| INCY:3591616 | LIVER/KIDNEY | Liver/- | 5/1 |
| INCY:3591631 | STOMACH | Stomach | 5 |
| INCY:3591768 | LUNG/BLADDER/- | -/-/Esophagus | 0/0/3 |
| INCY:3591769 | LUNG | - | 1 |
| INCY:3591989 | HEART | - | 2 |
| INCY:3591991 | - | Ear | 3 |
| INCY:3592018 | STOMACH/BLADDER | -/- | 1/1 |
| INCY:3592074 | - | Liver | 118 |
| INCY:3592077 | - | Liver | 313 |
| INCY:3592078 | - | Liver | 19 |
| INCY:3592088 | HEART/LIVER | -/- | 0/1 |
| INCY:3592109 | - | Liver | 163 |
| INCY:3592110 | - | Liver | 357 |
| INCY:3592111 | - | Liver | 3 |
| INCY:3592113 | - | Liver | 16 |
| INCY:3592114 | LIVER | Liver | 3 |
| INCY:3592125 | - | Liver | 3 |
| INCY:3592127 | PROSTATE/OVARY/BRAIN/ADRENAL | Prostate/Ovary/Brain/- | 13/5/197/1 |
| INCY:3592129 | TRACHEA/LIVER | Trachea/Liver | 11/43 |
| INCY:3592144 | - | Liver | 5 |
| INCY:3592150 | - | Salivary Glands | 3 |
| INCY:3592218 | - | Kidney | 13 |
| INCY:3592225 | HEART | Heart | 8 |
| INCY:3592281 | LIVER/- | Liver/Kidney | 41/11 |
| INCY:3592284 | LIVER/- | Liver/Kidney | 8/10 |
| INCY:3592285 | STOMACH | Stomach | 4 |
| INCY:3592296 | TESTIS/- | -/Kidney | 2/11 |
| INCY:3592298 | SALIVARY GLANDS/- | -/Lung | 0/6 |
| INCY:3592299 | - | Spinal Cord | 5 |
| INCY:3592340 | - | Kidney | 16 |
| INCY:3592388 | PANCREAS/- | -/Liver | 0/9 |
| INCY:3592445 | BRAIN/- | Brain/Spinal Cord | 4/21 |
| INCY:3592487 | - | Brain | 5 |
| INCY:3592613 | STOMACH | - | 2 |
| INCY:3592766 | BRAIN | Brain | 4 |
| INCY:3592789 | BRAIN | Brain | 9 |
| INCY:3592927 | LIVER | Liver | 100 |
| INCY:3592950 | HEART/BLADDER/- | Heart/Bladder/Kidney | 6/4/7 |
| INCY:3592983 | LIVER/- | Liver/Kidney | 325/65 |
| INCY:3592984 | LIVER/- | Liver/Kidney | 245/13 |
| INCY:3593074 | PITUITARY/OVARY/SALIVARY GLAND/ADRENAL | Pituitary/-/-/- | 4/0/1/1 |
| INCY:3593080 | MUSCLE | - | 0 |
| INCY:3593083 | LIVER/- | Liver/Kidney | 226/7 |
| INCY:3593093 | LIVER | - | 1 |
| INCY:3593141 | LIVER | Liver | 328 |
Because not all of the rat genes in this region contained clones that were represented on the microarrays, the source organs for clones that map to each gene were examined. Mapped clones were derived from 322 Incyte libraries and 95 public domain libraries. Across all aligned clones, 40 organs were represented, of which brain (2346 clones) and liver (3405 clones) were the most abundant. A total of 57 rat genes and gene fragments showed a high specificity for one or more organs. Overall, 22 organs with gene expression information were found on chr. 1q43-54, including brain, pancreas, and heart, which represent some of the organs that might be related to the QTLs found in this region.
To assess how redundant or additive the information of these two approaches is, we compared the results from the electronic Northern analyses and microarray experiments (Table 3). Of the 55 occurrences of increased organ expressions in the hybridization experiment, 31 (56%) were confirmed by electronic Northern. In 15 cases, the number of clones for that particular organ in the gene was below three, our threshold for deriving electronic Northern data. In nine instances, none of the clones in the gene were derived from the organ that was shown to have increased expression in the microarray experiments. The electronic Northern approach revealed an additional 39 gene-to-organ associations that were not found by the microarray data analyses.
Synteny and Homologs
The human-to-rat syntenic map covers 2.60 Gb of the human genome and 2.45 Gb of the rat genome. The map is constructed from chained syntenic anchors that represent high-confidence mutually unique alignments between regions of the rat and human genomes. The average distance between the 567,779 syntenic anchors comprising our map is 4 kb. A genomic expansion factor of 1.06 in human compared with rat can be observed, which is similar to the one observed in mouse (Waterston et al. 2002) and is also described in rat (Rat Genome Sequencing Project Consortium 2004). Rat chr. 1q43-54 is syntenic to segments on human chr. 9, 10, and X and on mouse chr. 19 and X.
The syntenic map was used to verify sequence homology pairs between rat and human. The presence of a syntenically confirmed homolog in human increases the confidence level for a particular rat gene as it reveals not only BLAST hit similarity, but consistent location of the genes on both genomes, which indicates that these might be orthologous genes. A total of 274 of the rat Incyte genes and gene fragments had overlaps to a syntenic homolog in human. Most of these rat Incyte genes with syntenic homologs also had overlaps to rat Ensembl genes, with only 89 of them being Incyte unique genes. In total, 160 human genes were classified as syntenic homologous to rat genes. Of these human genes, 142 contained protein-coding sequences found in GenBank, and 18 genes contained EST sequences from GenBank and Incyte. Thus, rat genes on chr. 1q43-54 did not reveal syntenic homologs in human that represented potential novel human genes. The mouse region syntenic to rat chr. 1q43-54 contained a total of 263 gene predictions, of which 143 were known genes (54%). In contrast, the rat Ensembl database contains only 20% known genes within the corresponding syntenic region. Of these mouse genes, 195 were homologous to the rat genes on chr. 1q43-54, and can thus be used for additional validation of the rat genes.
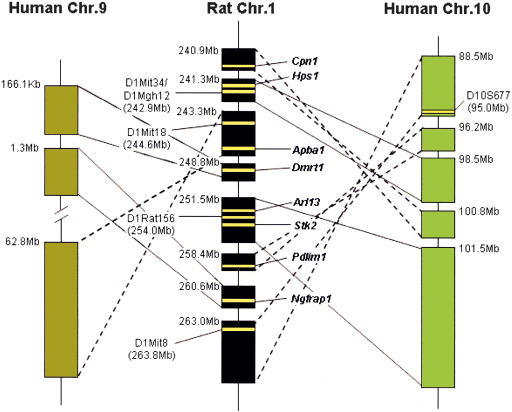
Human chr. 10 was shown to be linked to end-stage renal disease (ESRD) which is similar to the Rf-1 disease found on rat chr. 1 (Freedman 2002; Freedman et al. 2002). In contrast, mouse does not have a similar phenotype reported to be associated within the mouse syntenic regions on chr. 19 or chr. X. Hence, only the rat-to-human syntenic map was considered for the search of candidate disease genes for Rf-1. The rat syntenic region is rearranged into eight different segments on the human genome, five of which are on human chr. 10, whereas three are on human chr. 9 (Fig. 3). Only human chr. 10 carries the linkage to ESRD, which is similar to the Rf-1 phenotype, thus, only five of the eight segments are likely to carry genes that are involved in Rf-1. The size of the rat Rf-1 QTL can thus be reduced to a total of 11.5 Mb that span those five rat segments, whereas originally, the complete region considered in rat was roughly 20 Mb. Thus, the segmentation and rearrangement of the genomic sequence in the syntenic map of two species can be an asset for the study of QTLs.
Figure 3.
Syntenic map for rat chr. 1q43-54. Markers associated with Rf-,Niddm24, Bp42, Weight3, and Eae7 (D1Mit8, D1Mit18, D1mit34, D1Rat156, and D1Mgh12) and for human ESRD (D10S677), are presented as bold yellow lines. Some of the known gene symbols were added for orientation. Syntenic segments that changed orientation on human chromosomes are linked by broken lines to rat chr1.
Candidate Rf-1 Genes
The Rf-1 phenotype is likely to be caused by the misfunction of a gene expressed in kidney (Churchill et al. 1997). In addition, the rat Rf-1 phenotype is believed to be comparable with the human ESRD disease phenotype on human chr. 10. This led us to investigate rat genes with expression in kidney that have a syntenic human homolog on chr. 10.
To investigate genes that meet this criteria, we identified all genes expressed in kidney via either microarray analyses or clone origin of the genes. In microarray studies, three genes were found to have significant differential increase of expression in the kidney (see Table 3). The human homologs for all three of these genes contained kidney EST sequences as well. In addition, we also studied microarray analyses available in the gene expression omnibus database (GEO; http://www.ncbi.nlm.nih.gov/geo/). In this database, no rat study was performed that included the use of normal kidney tissues. However, the human microarray experiments included the hybridization of kidney tissues to one of the human platforms (GDS181). This platform contained 7860 human clone ids that were mapped to the human genomic backbone. A total of 22 of these clones mapped to 21 genes within the human region on chr. 10 that is syntenic to rat chr. 1q43-54, and two of these human genes contained clones that showed increased expression in kidney on the GEO platform. These two human genes had two rat syntenic homologs each, which were rat INCY:3592143 and INCY:3592176 for one of the human genes and INCY:3592453, INCY:3592473 for the other human gene. None of these were found to have a significant increase in kidney expression, but the latter two Incyte genes contained four clones derived from kidney tissues.
As comparison between microarray and electronic Northern results showed that additive expression information could be derived by electronic Northern, we also included the analysis of clone content of the rat genes for the identification of genes potentially expressed in kidney tissues. The total number of clones from 23 rat kidney libraries was 74,787. Of these, 1029 mapped to rat chr. 1q43-54 (941 Incyte and 88 public, respectively). We selected gene or gene fragments that contained clones from at least two different kidney libraries for verification of organ expression. A total of 90 of the Incyte novel gene fragments had clones from two or more kidney libraries. As the Rf-1 orthologous human gene is believed to map to human chr. 10q, we considered only genes and gene fragments that were confirmed to have a human syntenic homolog. Thus, we derived 66 rat genes that (1) are located close to the rat markers for Rf-1 as well as human markers for ESRD, (2) have a verified human syntenic homolog, and (3) contain clones from more than two kidney libraries, or (4) had significant differential expression in the microarray studies (Supplemental file 2).
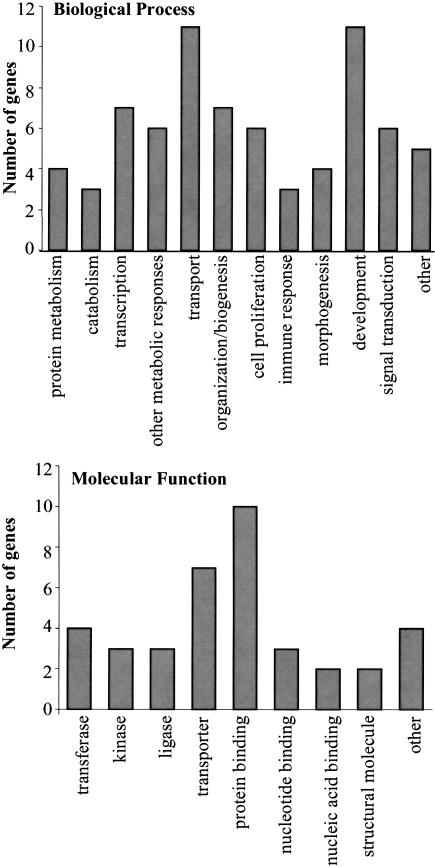
For further validation of these genes and their potential involvement in the Rf-1 phenotype, we studied functional gene ontology association of these genes. The gene ontology associations found for these genes are summarized in Figure 4. The candidate disease genes contained a cluster of genes that are predicted to be involved in plasma membrane transport, which might be associated with kidney function, as well as gene products predicted to be involved in protein binding and signal transduction. Furthermore, two predicted kinases and two predicted structural proteins are represented in this set of genes. This information, as well as title lines and annotation, is provided for each gene in Supplemental file 2.
Figure 4.
GO terms predicted for candidate genes for Rf-1. (Top) GO terms for biological processess. (Bottom) GO terms for molecular function.
DISCUSSION
We have studied a region on rat chr. 1 that contains QTLs for several disease phenotypes, including renal failure, diabetes, hypertension, encephalomyelitis, and body weight. We validated the rat genes by identification of EST-inferred genes that are found both in rat and human. In addition, we studied microarray experiments and electronic Northern data to generate organexpression information. Using the rat-to-human syntenic map, we significantly reduced the size of the rat Rf-1 QTL locus, and thus, the number of candidate disease genes. Through the combination of expression information as well as syntenic mapping and disease linkage data for the renal failure phenotype, we present a unique approach for the selection of candidate disease genes. Within the large pool of genes mapping to a disease region, we thus highlight 66 candidate disease genes for Rf-1.
The rat chr. 1q43-54 region contained a total of 307 Incyte rat gene or gene fragments that did not overlap any of the Ensembl gene predictions, and approximately half of these were composed of Incyte ESTs only. Manual curation and analysis of 31 of these fragments to evaluate the amount of underclustering suggests a gene fragmentation ratio of 3:1. In addition, multiple rat genes were verified syntenically as homologs of the same human gene, which also indicates underclustering in the data set. Thus, the 307 gene fragments could represent a set of 100 additional rat genes on rat chr. 1q43-54.
The use of chromosomal synteny has allowed us to avoid the problems inherent with a reciprocal best-hit approach. Rat-to-human syntenic comparisons enabled the identification of 26 k rat-human syntenic homolog pairs genome wide that have EST evidence in both rat and human. This is more than the ortholog pairs derived via reciprocal BLAST hits (∼12 k; Rat Genome Sequencing Project Consortium 2004; Incyte analyses). All of the human genes that were identified as syntenic homologs to rat genes on chr. 1q43-54 contained either proteins available at GenBank or at least several public domain EST sequences, and thus, did not represent novel human gene content. The rat gene fragments for which we could not identify a syntenic homolog in human could potentially represent rat-specific and novel genes. However, the abundance of rat-specific exons over the genome is low. It is possible that these gene fragments represent untranslated gene regions with low conservation across two species, or that some of these resulted from genomic contamination of cDNA libraries. Further sequencing is required to obtain longer mRNA sequences to identify the validity of these fragments.
The complete sequencing of the rat genome enabled a number of rat gene-prediction studies to be performed, including those predictions found in the Ensembl database (Rat Genome Sequencing Project Consortium 2004). Computationally based gene prediction algorithms, however, have known limitations (Burge and Karlin 1997; Solovyev and Salamov 1997), thus, confirmation of gene predictions using cDNA-derived data is essential. Of the 205 Ensembl novel gene predictions in the 1q43-54 region, 35% could not be verified using cDNA-derived data. Reasons for this could include the lack of sequences for those genes in Incyte's database, strict uniquing of sequences from close family members, or invalid Ensembl gene predictions. Only a few of these (<1%) had annotation, and 25% were single exon genes. Further study of these rat Ensembl predictions is needed to verify their validity.
We have shown how the rearrangement of syntenic segments in human compared with rat is a valuable characteristic that can be exploited for the selection of candidate disease genes. The Rf-1 locus was described in the fawn-hooded hypertensive rat, which uniformly develops end-stage renal disease (Brown et al. 1996; Shiozawa et al. 2000) and is syntenic to human chr. 10q, which contains the linkage region to human ESRD (Yu et al. 1999; Freedman 2002; Freedman et al. 2002). Using the syntenic map for rat chr. 1q43-54 and the linkage information between rat Rf-1 and human ESRD, we reduced the size of the QTL for Rf-1, and thus, the number of candidate disease genes that might be responsible for this phenotype. We have further validated these genes by searching for syntenic homologs in our human data set, which is also based on EST alignments, thus doubling the EST evidence for these genes. We have also determined which genes have kidney expression, as transplant studies in rat have indicated that the responsible gene is probably expressed in the kidney (Churchill et al. 1997).
The mapping of rat clones to the genomic sequence establishes a rat transcriptome (Panda et al. 2003) that can be used for the examination of organ expression of the gene loci using cDNA microarrays and by analyzing the source organ of the constituent clones for the genes, termed electronic Northern analysis. A comparison between microarray and electronic Northern expression information showed significant overlaps between the two sets of results, but also exclusive expression information for each method. Microarray studies compare relative expression with a set of other genes, whereas electronic Northern is an approximate measure of organ abundance in a gene, therefore, the discrepancies between the two sets of expression information is expected and do not invalidate either measure. However, equal expression information for a gene in both sets of results can be used as an additional confirmation for the expression pattern of that gene. Furthermore, both microarray experiments and electronic Northern analyses utilize only a subset of the clones that aligned to the genes and are subject to false negative results. Expression information can thus be used to select genes of higher interest, but not to completely exclude genes from the set of candidate disease genes.
The impact of expression information is increased if more expression experiments can be included that address a larger number of the sequences studied. Thus, we have also investigated the experiments presented in the GEO database. This approach added two potential genes that might be of interest. Unfortunately, this expression information was limited to human, and no appropriate rat hybridization data could be found in the GEO data set. It would be interesting to extend this comparison to additional expression experiments, especially if the number of expression experiments in rat increases.
The selected rat candidate disease genes were further evaluated for their potential function and association to biological processes. To date, the information on Rf-1 phenotype is not conclusive enough to be able to associate it with a particular type of gene. The human marker associated with ESRD (D10S677; Freedman et al. 2002) mapped to the PLCE1 gene, which encodes a bifunctional phospholipase C that hydrolyzes PIP(2) to generate inositol 1,4,5 trisphophate and diacylglycerol (Song et al. 2001). PLCE1, however, has not been identified as the gene responsible for the renal failure phenotype (Freedman et al. 2002). In addition, rat Plce1 is located outside of the region designated Rf-1B that was used to generate congenic rats, which display the renal failure phenotype (Provoost et al. 2002). Thus, either PLCE1 is not directly responsible for this phenotype, or several genes in this region contribute to the phenotype.
The histological evaluation of affected rat Rf-1 kidney tissues indicates that this phenotype represents a lack of resistance to long-term high blood pressure in the renal glomerula. This might well be accounted for by differences in the extracellular matrix components, which resist the pressure of the blood that is being filtered at the basal lamina of the glomerula. Thus, the candidate disease genes characterized as structural proteins might be of higher interest. It is also perceivable that the four candidate disease genes that were classified to have a potential role as plasmamembrane transporters might play a crucial role in kidney function. Furthermore, two rat genes, which are the syntenic homologs of human Hps1 and Hps6, have clones from kidney and are located within this region. These genes in human have been thought to be associated with the Hermansky-Pudlak syndrome, a heterogeneous syndrome complex that involves different severe symptoms, including renal failure. It is conceivable that a separate mutation of one of these genes could lead to renal failure only, without the other associated symptoms of the syndrome. Further studies evaluating these genes are necessary to determine which gene(s)' misfunction is responsible for the Rf-1 phenotype.
To date, no mouse phenotype involved in renal failure has been shown to be localized to the mouse region syntenic to rat Rf-1 and human ESRD. However, several mouse phenotypes of gene knockouts localized on chr. 19, such as cyclooxygenase-2 and apolipoprotein-E, have been described to be associated with kidney impairment (Norwood et al. 2000; Chen et al. 2001). Even though it is possible that misfunctions in the same gene might lead to quite distinct phenotypes in different species, it is difficult to link the kidney impairments described in these knockout mice to the Rf-1 or ESRD phenotype. Further study of disease linkage to mouse chr. 19 is needed to verify whether an Rf-1-like phenotype in this region is conserved in the mouse.
We have shown how the rearrangement of genomic segments between rat and human can be used to reduce the size of a QTL region, and thus, also the number of candidate disease genes. Furthermore, by using alignments of rat ESTs to rat chr. 1q43-54, which contains several QTLs, we have provided transcriptional evidence for rat genes as well as information on organ expression, which was to date unavailable for a large set of these genes. We have shown how this expression information in combination with synteny and disease-linkage information can be used to select candidate disease genes for the rat Rf-1 QTL.
METHODS
cDNA Library Construction and Sequencing
The Incyte Database contains 876,507 rat cDNA clones from 417 libraries that represent 40 different organs (Supplemental file 3) from Sprague-Dawley rats. From the Incyte clones, 1.2 M EST sequences were derived. In addition, 300 k public rat sequences were downloaded from GenBank (release 134). Low-quality regions, sequencing artifacts, vector sequences, and 3′ polyadenylated termini were clipped from cDNA sequences; low-complexity regions and repetitive elements were masked. A total of 4248 RefSeq (3/2/2003) and 5605 nucleotide sequences derived from GenPept entries were downloaded from GenBank (release 134).
Alignment of Sequences to the Rat Genome and Clustering Into Potential Gene Loci
The rat cDNA sequences were aligned to the rat genomic contigs from the draft rat genome sequence at NCBI (January, 2003; Baylor v.2.1) using GENSCRIPT (an internal unpublished sequence algorithm) that was modified to optimize cDNA to genomic DNA alignments. To minimize alignment errors, low-confidence terminal exons were clipped and clones with excessively large introns as well as EST alignments with <95% identity over 95% of the sequence length were removed. RefSeq sequences that did not exhibit at least 95% identity over 80% of the protein-coding sequence were removed. As sequences align with high identity and coverage to locations of recent genome duplications (Sankoff 2001), we resolved that multiple alignments to the genome by removing any alignments at a lower percent identity or lower percent coverage if the percent identity was equal. Detection of expressed pseudogenes was performed by assessment of single exon alignments of RefSeq sequences that had at least one multiexon alignment to the rat genome. Alignment strand was assigned by splice-site orientation or by using cDNA library directional information. The Incyte EST sequences that mapped to rat 1q43-54 were released to GenBank.
Analysis of Additional EST-Inferred Gene Fragments That Do Not Overlap Ensembl Genes
Incyte gene fragments that did not overlap an Ensembl gene, but had evidence of three or more clones or splicing RNA, were studied further for their potential gene content. Transcripts generated in these gene fragments were analyzed for ORF containment and homology to other species. The homologous hits were also studied for their syntenic evidence to potentially assign them as human homologs for these gene fragments. To evaluate the potential underclustering of these gene fragments, distance to neighboring genes as well as homologous hits was assessed, and manual curation was performed to determine which gene fragments could be merged.
Verification of Ensembl Genes
Ensembl known and predicted gene and transcript exon coordinates on rat chr. 1q43-54, and the mouse syntenic segments on chr. 19 and X were downloaded from Ensembl (http://www.ensembl.org). Incyte rat genes, gene fragments, and transcripts were generated by exonic clustering of EST sequences, RefSeq sequences, and mRNAs from GenBank that have a corresponding protein entry in GenPept, which were aligned to the rat genomic sequence. These genes were designated EST-inferred genes due to their EST-alignment content. Genes and gene fragments containing sequences from three or more clones, or that had evidence of RNA splicing, were characterized as EST-inferred high-evidence genes and considered for further analysis. Ensembl known genes were verified by RefSeq or other annotation content. Ensembl gene predictions were assessed for their overlap with Incyte genes and segregated into groups of evidence on the basis of mapping overlap with Incyte sequences. Exonic overlap and confirmation of RNA splice sites between Incyte EST-inferred transcripts and Ensembl predictions were done by manual study of the alignments (see Fig. 2). If more than one Incyte EST-inferred gene or gene fragment overlapped one Ensembl gene, or if more than one Ensembl gene overlapped one Incyte gene, the genes were investigated manually for potential of overclustering or underclustering. If several Incyte genes spanned one Ensembl gene and had similar BLAST hits to other species and a lack of complete clone sequence, they were considered underclustered. If one or more clones in an Incyte gene spanned several Ensembl genes and verified splice sites in both genes, then the Ensembl genes were considered to be underclustered. If a gene contained nonoverlapping coding sequence, it was considered overclustered. Incyte gene fragments that contained RefSeq sequences or that had exons overlapping Ensembl gene exons were considered as genes. Additional EST clusters that did not overlap Ensembl genes or contain RefSeq sequences were considered gene fragments until further study of potential clustering.
Gene Annotation
The annotation of Ensembl genes in 1q43-54 was compared manually with that of Incyte genes and assessed for consistency and completeness. Ensembl gene homology to Homo sapiens, Mus musculus, Danio rerio, and Takifugu rubripes was derived from Ensembl and compared with Incyte-available cross-species homologies. The presence of pfam annotation was compared with Incyte Pfam presence or absence, and the Ensembl gene descriptions and family assignments were compared with Incyte gene title lines.
Title lines and predictive Gene Ontology (GO; Ashburner et al. 2000) properties were curated manually for the rat Rf-1 candidate genes by Incyte's proprietary BioKnowledge Transfer (BKT) process. Annotation is based on family membership and domain structure (Pfam analysis), and on similarity to characterized proteins in the Proteome BioKnowledge Library (BKL) database (BLAST analysis). GO properties are predicted with three methods: Direct Transfer, Consensus Transfer, and Pfam Transfer. The validity of the BKT process has been tested by blind curation of characterized proteins.
BKT Pfam analysis uses HMMer (version 2.1.1; Krogh et al. 1994), to compare protein sequences from the BKL with all Pfam domain/family seed sequences (Pfam version 9.0; Bateman et al. 2000). A query protein is considered to contain a particular domain or belong to a particular protein family only if the HMMer alignment score is less than or equal to the trusted cut-off score defined by Pfam for each Pfam seed sequence. For each Pfam protein family or protein domain, Incyte generates a descriptive phrase, and where possible, assigns GO properties, based upon manual curation. These descriptive phrases and associated GO properties (Pfam-to-GO mappings) are applied to the uncharacterized protein.
The Direct Transfer method uses BLAST analysis to identify the best BLAST hit among all BKL proteins. Proteins are ranked according to their Smith-Waterman scores, and preference is given to the highest-ranking characterized protein. If there are no characterized proteins among the BLAST targets, an uncharacterized protein is selected as the best BLAST target and is used in the title line, but no GO properties are captured by Direct Transfer. If the best BLAST hit is a characterized protein, GO properties from the BLAST hit are filtered to obtain more general parent GO properties, and these are applied to the uncharacterized protein. The Consensus Transfer method considers the entire set of BLAST target proteins with an E-value of 1e-10 or less, in order to identify the most predominant GO properties shared among the list of targets.
Title lines for uncharacterized proteins are written to contain both the standard phrases to describe Pfam membership, and information about similarity to the best BLAST target, with a phrase describing the function of the BLAST target. The level of similarity to the best BLAST target (very strong, strong, high, moderate, low, weak; Sander and Schneider 1991) is indicated by controlled vocabulary that accounts for both identity and overlap. The title line phrase “Protein of unknown function” is applied to proteins having no Pfam hits and having only BLAST hits that are either uncharacterized and/or have <70% overlap with the uncharacterized protein.
Incyte transcripts generated by clustering of ESTs aligned to the genome were analyzed for ORFs using a 6-frame translation. Transcripts were allowed to have more than one ORF and were scored as ORF containing if they had either a terminal ORF with length ≥60 amino acids, consistent with the cDNA containing the tail end of a complete protein or internal ORF with length ≥100 amino acids. The data were examined in a strand-dependent manner for spliced gene fragments, and a strand-independent manner for unspliced gene structures.
Differential Expression Analysis
Differential expression was generated using Incyte's Human and Rat LifeArray microarrays. RNA was isolated from 27 organs from six young adult Sprague Dawley (Cr1:CD(SE)BR) rats that were 33–57 d old. Gender-specific organs were derived from three donors each. A pooled reference was created using RNA from all of the organs (Supplemental file 4). The individual organs and the RNA pool were labeled with Cy3 and Cy5. The labeled samples were then run in hybridizations across cDNA microarrays. The data from a minimum of three donors was averaged together to generate the differential expression ratios. Significant differential expression was defined as greater than a twofold change.
Furthermore, the GEO database was searched for rat or human microarray experiments that used kidney tissues. Experiments with treated or tumorigenic tissues were excluded. Data from hybridization experiments with kidney samples GSM2830, GSM2843, and GSM2871 to human platform GDS181 were examined for clones that showed a significant increase in kidney tissue. Genes derived from clones found on this platform were identified in the human EST-based gene set and analyzed for their synteny to rat genes on chr. 1q43-54.
Electronic Northern
Electronic Northern analysis of Incyte genes was performed by counting the number of clones from each cDNA library that contributed to a specific gene or gene fragment. Combining this information with the organ information associated with each library enables construction of an expression profile. Because normalized, amplified, and subtracted libraries are skewed toward low-abundance genes, clones from these libraries were not included in this analysis. To examine organ specificity of the individual genes, the clone counts were normalized on the basis of the total number of clones from the organ. For an organ to have significant specificity, the percent specificity within a gene needed to be 40% or greater and be represented by three or more clones from that organ. In addition, the genes were analyzed for their total content of clones from a specific organ without exclusion of normalized libraries. Clones from two or more kidney libraries present in the same gene or gene fragment were considered as evidence for kidney expression.
Syntenic Map and Syntenic Confirmation of Rat-to-Human Homologs
A genome-wide syntenic map between rat (January 2003, Baylor build 2.1) and human (NCBI, build 31) was generated. BLASTZ (Schwartz et al. 2003) alignments of the two genomes were used to generate a large number of high-scoring local alignments (HSPs). These HSPs were filtered (Schwartz et al. 2003) and then uniqued across both genomes. As a result, each of the filtered nonoverlapping HSPs represents a 1:1 syntenic anchor between the two genomes. An iterative algorithm was then used to build colinear syntenic segments using these syntenic anchors and to coalesce adjacent segments if possible. Each iteration involved the chaining of adjacent anchors into segments, the deletion of noise anchors, and the coalescence of adjacent segments if possible. Segments with less than three syntenic anchors and <300 kb were filtered out. The remaining anchors are considered high in the context of coancestry. The rat-to-mouse syntenic map was derived from the Ensembl database for the respective genomic versions used in the gene analysis.
To leverage the syntenic map to determine a set of putative orthologous gene pairs, the rat EST-derived transcript sequences were compared with a corresponding set of human known and Incyte genes (Incyte LifeSeq Foundation Release 11). BLASTN (Altschul et al. 1990; Myers and Durbin 2003) was used for this comparison with a cutoff of 10e-8 normalized to a database size of 2.1 Gb. The top five BLAST hits for each rat transcript were then considered for syntenic confirmation. A pair of genes was considered syntenically confirmed if both sequences overlapped the corresponding syntenic anchors or were localized between the same corresponding pairs of syntenic anchors.
Verification of Genes in the Rf-1 Disease Region
STS marker mappings to the genome were derived from UniSTS (http://www.ncbi.nlm.nih.gov/genome/sts/). The region studied was rat chr. 1 between 232 and 264 Mb. This encompassed markers associated with Rf-1, D1Mit18 (uniSTS:120257), D1Mit8 (uniSTS:118508) (Brown et al. 1996), D1Mit34 (uniSTS:227028), and D1Rat156 (uniSTS:120298; Provoost et al. 2002). Furthermore, central to this region is the mapping of D1Mgh12 (242.9 Mb), a marker that is associated with hypertension, diabetes, body weight, and encephalomyelitis.
Acknowledgments
We thank George Weinstock and Kim Worley, Baylor Collegue of Medicine, for their support. We acknowledge the support of our past and present Incyte colleagues during the course of this work. We thank Richard Goold, Robert Lagace, and Brent Louie for assistance in gene analysis. We thank Scott Anderson, Qixin Bei, John Blanchard, Anissa Jones, Sarah Mullahy, Iqbal Panesar, Srikanth Patury, Pierre Rioux, Wayne Wonchoba, and Mingham Wu for providing algorithms for the analyses. We thank Cindy Dole for her editing assistance.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1932304.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403-410. [DOI] [PubMed] [Google Scholar]
- Ashburner, M., Ball, C.A., Blake, J.A., Botstein, D., Butler, H., Cherry, J.M., Davis, A.P., Dolinski, K., Dwight, S.S., Eppig, J.T., et al. 2000. Gene Ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25: 25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman, A., Birney, E., Durbin, R., Eddy, S.R., Howe, K.L., and Sonnhammer, E.L.L. 2000. The Pfam Protein Families Database. Nucleic Acids Res. 28: 263-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsteinsdottir, K., Yang, H.T., Pettersson, U., and Holmdahl, R. 2000. Evidence for common autoimmune disease genes controlling onset, severity, and chronicity based on experimental models for multiple sclerosis and rheumatoid arthritis. J. Immunol. 164: 1564-1568. [DOI] [PubMed] [Google Scholar]
- Brown, D.M., Provoost, A.P., Daly, M.J., Lander, E.S., and Jacob, H.J. 1996. Renal disease susceptibility and hypertension are under independent genetic control in the fawn-hooded rat. Nat. Genet. 12: 44-51. [DOI] [PubMed] [Google Scholar]
- Burge, C. and Karlin, S. 1997. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268: 78-94. [DOI] [PubMed] [Google Scholar]
- Chen, G., Paka, L., Kako, Y., Singhal, P., Duan, W., and Pillarisetti, S. 2001. A protective role for kidney apolipoprotein E. Regulation of mesangial cell proliferation and matrix expansion. J. Biol. Chem. 276: 49142-49147. [DOI] [PubMed] [Google Scholar]
- Churchill, P.C., Churchill, M.C., Bidani, A.K., Griffin, K.A., Picken, M., Pravenec, M., Kren, V., Lezin, E., Wang, J.M., Wang, N., et al. 1997. Genetic susceptibility to hypertension-induced renal damage in the rat. Evidence based on kidney-specific genome transfer. J. Clin. Invest. 100: 1373-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dracheva, S.V., Remmers, E.F., Chen, S., Chang, L., Gulko, P.S., Kawahito, Y., Longman, R.E., Wang, J., Du, Y., Shepard, J., et al. 2000. An integrated genetic linkage map with 1,137 markers constructed from five F2 crosses of autoimmune disease-prone and -resistant inbred rat strains. Genomics 63: 202-226. [DOI] [PubMed] [Google Scholar]
- Freedman, B.I. 2002. End-stage renal failure in African Americans: Insights in kidney disease susceptibility. Nephrol. Dial. Transplant. 17: 198-200. [DOI] [PubMed] [Google Scholar]
- Freedman, B.I., Rich, S.S, Yu, H., Roh, B.H., and Bowden, D.W. 2002. Linkage heterogeneity of end-stage renal disease on human chromosome 10. Kidney Int. 62: 770-774. [DOI] [PubMed] [Google Scholar]
- Galli, J., Li, L.S., Glaser, A., Ostenson, C.G., Jiao, H., Fakhrai-Rad, H., Jacob, H.J., Lander, E.S., and Luthman, H. 1996. Genetic analysis of non-insulin dependent diabetes mellitus in the GK rat. Nat. Genet. 12: 31-37. [DOI] [PubMed] [Google Scholar]
- Jacob, H.J., Brown, D.M., Bunker, R.K., Daly, M.J., Dzau, V.J., Goodman, A., Koike, G., Kren, V., Kurtz, T., Lernmark, A., et al. 1995. A genetic linkage map of the laboratory rat, Rattus norvegicus. Nat. Genet. 9: 63-69. [DOI] [PubMed] [Google Scholar]
- Kovacs, P., Voigt, B., and Kloting, I. 1997. Novel quantitative trait loci for blood pressure and related traits on rat chromosomes 1, 10, and 18. Biochem. Biophys. Res. Commun. 18: 343-348. [DOI] [PubMed] [Google Scholar]
- —. 1998. Congenic strain confirms putative quantitative trait locus for body weight in the rat. Mamm. Genome 9: 294-296. [DOI] [PubMed] [Google Scholar]
- Krogh, A., Brown, M., Mian, I.S., Sjolander, K., and Haussler, D. 1994. Hidden Markov models in computational biology: Applications to protein modeling. J. Mol. Biol. 235: 1501-1531. [DOI] [PubMed] [Google Scholar]
- Leo, C.P., Hsu, S.Y., and Hsueh, A.J. 2002. Hormonal genomics. 2002. Endocr. Rev. 23: 369-381. [DOI] [PubMed] [Google Scholar]
- Myers, G. and Durbin, R. 2003. A table-driven, full-sensitivity similarity search algorithm. J. Comput. Biol. 10: 103-117. [DOI] [PubMed] [Google Scholar]
- Norwood, V.F., Morham, S.G., and Smithies, O. 2000. Postnatal development and progression of renal dysplasia in cyclooxygenase-2 null mice. Kidney. Int. 58: 2291-2300. [DOI] [PubMed] [Google Scholar]
- Panda, S., Sato, T.K., Hampton, G.M., and Hogenesch, J.B. 2003. An array of insights: Application of DNA chip technology in the study of cell biology. Trends. Cell. Biol. 13: 151-156. [DOI] [PubMed] [Google Scholar]
- Provoost, A.P., Shiozawa, M., Van Dokkum, R.P., and Jacob, H.J. 2002. Transfer of the Rf-1 region from FHH onto the ACI background increases susceptibility to renal impairment. Physiol. Genomics 8: 123-129. [DOI] [PubMed] [Google Scholar]
- Rat Genome Sequencing Project Consortium. 2004. Genome sequence of the Brown Norway Rat yields insights into mammalian evolution. Nature (in press). [DOI] [PubMed]
- Sander, D. and Schneider, D. 1991. Database of homology-derived protein structures and the structural meaning of sequence alignment. Proteins 9: 56-68. [DOI] [PubMed] [Google Scholar]
- Sankoff, D. 2001. Gene and genome duplication. Curr. Opin. Genet. Dev. 11: 681-684. [DOI] [PubMed] [Google Scholar]
- Schwartz, S., Kent, W.J., Smit, A., Zhang, Z., Baertsch, R., Hardison, Haussler, D., and Miller, W. 2003. Human–mouse alignments with BLASTZ. Genome Res. 13: 103-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozawa, M., Provoost, A.P., van Dokkum, R.P., Majewski, R.R., and Jacob, H.J. 2000. Evidence of gene–gene interactions in the genetic susceptibility to renal impairment after unilateral nephrectomy. J. Am. Soc. Nephrol. 11: 2068-2078. [DOI] [PubMed] [Google Scholar]
- Solovyev, V. and Salamov, A. 1997. The Gene-Finder computer tools analysis of human and model organisms genome sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 5: 294-302. [PubMed] [Google Scholar]
- Song, C., Hu, C.D., Masago, M., Kariyai, K., Yamawaki-Kataoka, Y., Shibatohge, M., Wu, D., Satoh, T., and Kataoka, T. 2001. Regulation of a novel human phospholipase C, PLCepsilon, through membrane targeting by Ras. J. Biol. Chem. 276: 2752-2757. [DOI] [PubMed] [Google Scholar]
- Tatusov, R.L., Koonin, E.V., and Lipman, D.J. 1997. A genomic perspective on protein families. Science 278: 631-637. [DOI] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- Wei, S., Wei, K., Moralejo, D.H., Ogino, T., Koike, G., Jacob, H.J., Sugiura, K., Sasaki, Y., Yamada, T., and Matsumoto, K. 1999. Mapping and characterization of quantitative trait loci for non-insulin-dependent diabetes mellitus with an improved genetic map in the Otsuka Long-Evans Tokushima fatty rat. Mamm. Genome 10: 249-258. [DOI] [PubMed] [Google Scholar]
- Yu, H., Sale, M., Rich, S.S., Spray, B.J., Roh, B.H., Bowden, D.W., and Freedman, B.I. 1999. Evaluation of markers on human chromosome 10, including the homologue of the rodent Rf-1 gene, for linkage to ESRD in black patients. Am. J. Kidney. Dis. 33: 294-300. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://ratmap.gen.gu.se/; The rat genome database.
- http://www.ncbi.nlm.nih.gov/genome/sts/; UniSTS:
- http://www.ncbi.nlm.nih.gov/geo/; GEO
- http://www.ensembl.org/; Ensembl