Abstract
Affinity grids (AG) are specialized EM grids that bind macromolecular complexes containing tagged proteins to obtain maximum occupancy for structural analysis through single-particle EM. In this study, utilizing AG, we show that His-tagged activated PKC βII binds to the small ribosomal subunit (40S). We reconstructed a cryo-EM map which shows that PKC βII interacts with RACK1, a seven-bladed β-propeller protein present on the 40S and binds in two different regions close to blades 3 and 4 of RACK1. This study is a first step in understanding the molecular framework of PKC βII/RACK1 interaction and its role in translation.
Keywords: Affinity grid, Cryo-EM, RACK1, PKC βII, Ribosome
1. Introduction
Structure determination of macromolecular complexes is the goal of single-particle cryo-electron microscopy (cryo-EM). Among the limiting factors toward obtaining this goal is the challenging biochemical preparation and purification of intact macromolecular complexes containing low-affinity binding factors. In order to improve speed and efficiency of structure determination in cryo-EM, monolayer purification methods were introduced. Such methods combine protein purification with cryo-sample preparation, as a fast and easy way to prepare specimens suitable for cryo-EM. A simpler, more effective technique, employing the “Affinity Grid”, was introduced in the same study to further simplify the monolayer purification method (Kelly et al., 2008a). Affinity grids are conventional EM grids coated with lipid layers that contain functionalized head groups, for instance Ni–NTA (Nickel-nitrilotriacetic acid). The Ni–NTA lipid layers bind specifically and with high affinity to His-tagged proteins. In engaging such grid-functionalization, affinity grids provide a rapid and convenient way to prepare cryo-EM specimens of tagged protein complexes (Kelly et al., 2010a,b). In addition, ligand-receptor complexes in which the ligand carries a His-tag can be easily isolated on the affinity grid to maximize occupancy of the ligand in the purified complex. This technique has been successfully used to reconstruct ribosomes containing His-tagged subunits, as well as protein complexes bound to adaptor molecules (Kelly et al., 2010b).
RACK1 (Receptor for Activated C Kinase 1) protein is highly conserved and has homology to the β-subunit of the heterotrimeric G proteins. It contains seven internal WD40 (Trp-Asp 40) repeats devoted to protein–protein interactions (Sondek and Siderovski, 2001). These WD40 repeats fold into a seven-bladed β-propeller structure, with each blade made up of a four-stranded antiparallel β-sheet (Adams et al., 2011). RACK1 is found in free cytoplasmic form (Yatime et al., 2011) as well as residing in the head region of the 40S subunit of the eukaryotic ribosome (Ben-Shem et al., 2010; Li and Roberts, 2001; Nilsson et al., 2004; Sengupta et al., 2004), serving as a docking platform for a range of regulatory proteins. Of particular interest in the current context is Protein Kinase C βII (PKC βII) (Ron et al., 1994), shown to interact with free RACK1 and to play a significant role in regulating translation (Grosso et al., 2008b). Activated PKC βII is composed of four domains connected by unstructured linkers, resulting in an extended ‘beads-on-a-string’ morphology (Leonard et al., 2011), and has been shown to associate with ribosomes (Grosso et al., 2008a,b; Miluzio et al., 2009). Activated PKC βII interacts with RACK1 through two regions: the C2 domain and the unstructured V5 region present in the end of the C-terminus of the kinase domain (Ron et al., 1995; Stebbins and Mochly-Rosen, 2001). RACK1 has been linked to PKC signaling pathways (McCahill et al., 2002; Ron et al., 1999; Schechtman and Mochly-Rosen, 2001) through binding and stabilization of activated PKC βII (Ron et al., 1999). While some biochemical detail of the nature of specific PKC βII/RACK1 interaction is known, there have been no structural studies shedding light on the interaction between PKC βII and RACK1 bound to the ribosome.
2. Affinity grid technique applied for activated PKC βII and 40S subunit
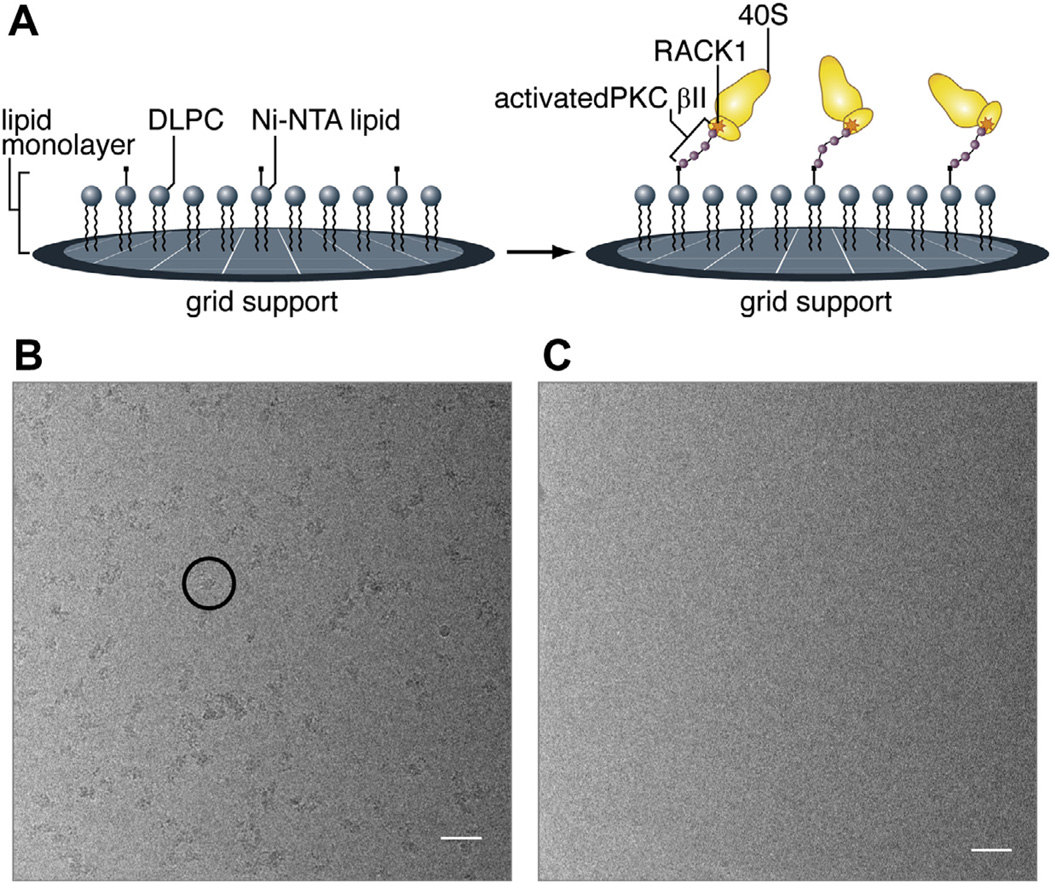
We set out to investigate the binding of activated PKC βII to RACK1 on the 40S subunit by cryo-EM, ultimately with the goal to study its effects on the structure of the 40S subunit. As a first step towards this goal, we assembled complexes formed by activated PKC βII with 40S subunits (PKC-40S) from rabbit reticulocytes (RRL) (See Supporting Information for experimental procedure). PKC-40S was immobilized on Ni–NTA affinity grids (Fig. 1A) (Kelly et al., 2008a,b, 2010a) through an N-terminal His-tag in PKC βII, and the grids were vitrified for cryo-EM. As a negative control, apo-40S subunits were deposited on an identical affinity grid prepared in parallel. Examining micrographs collected from the PKC-40S affinity grids, we observe an even distribution of complexes throughout the micrograph (Fig. 1B), whereas micrographs collected from the negative control apo-40S subunit affinity grid contained no ribosomal subunits (Fig. 1C).
Fig. 1.
Affinity grid preparation of PKC-40S. (A) Schematic illustration of the affinity grid method and how PKC-40S associates on the grid surface. Micrographs of purified 40S subunits (rabbit reticulocytes) on Nickel-affinity grids (B) with and (C) without His-tagged PKC βII placed on affinity grids. The scale on the images is 500 Å. The black circle shows a representative of PKC-40S subunit.
3. Interactions between PKC βII and RACK1 on the 40S subunit
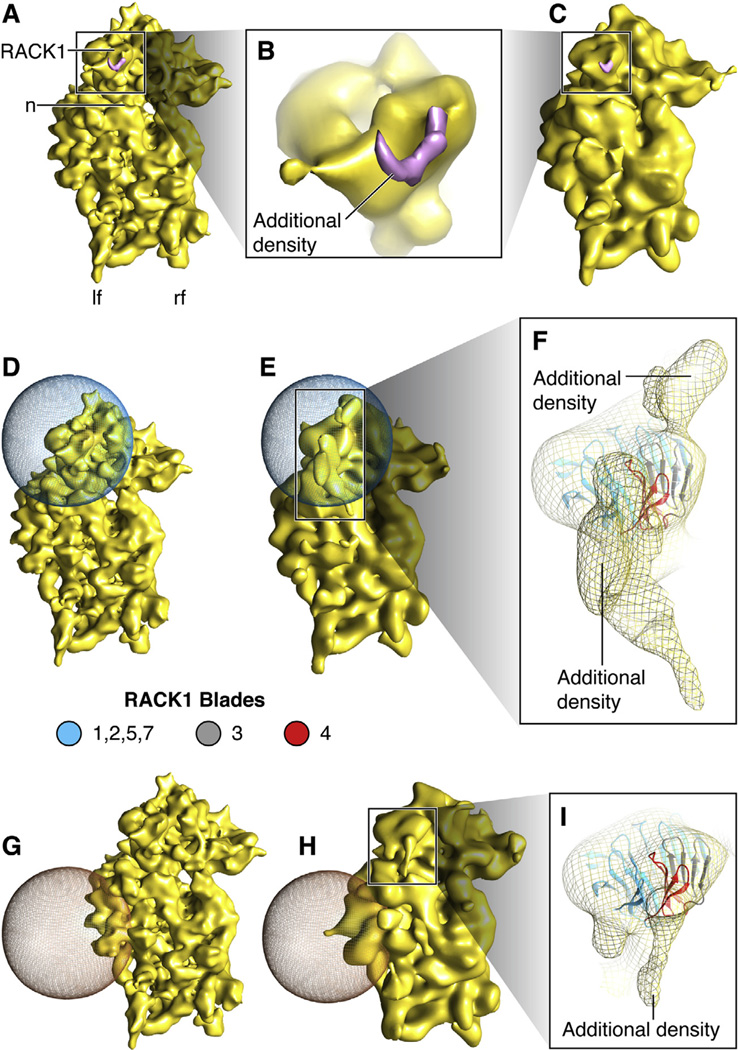
Our PKC-40S data were subjected to single-particle-based reconstruction using SPIDER (Frank et al., 1996), yielding a density map of the 40S subunit at a resolution of 11.4 Å (Fig. 2A–C, S1) (see Supporting Information). We attribute the extra density attached to RACK1, when compared to apo-40S ribosomal subunit, to PKC βII (Fig. 2B, S2G and H) and hypothesized that the rest of PKC βII is invisible as a result of conformational averaging due to its fleximer nature. In an attempt to visualize additional density in the affected region, we performed focused alignment targeting the region of PKC βII-RACK1 interaction. In this procedure, the alignment of projections to the 3D reference is restricted to an area of interest defined by a 3D mask (Fig. 2D–F, S2A–C). Size and placement of the mask ensures that the alignment is governed by the features of interest, not by the position of the ribosomal subunit as a whole. We were able to obtain a total of two additional densities to account for parts of activated PKC βII in its extended beads-on-a-string conformation (Fig. 2E and F, S2B–C), tentatively confirming our hypothesis. As a negative control, we placed an identical 3D mask in a randomly chosen region of the 40S subunit (Fig. 2G–I, S2D–F). Focused alignment was performed in this region by applying the same alignment protocol. Little additional density was observed in the region of the control mask (Fig. 2G and H, S2D–E), whereas more substantial additional density once again was found in the vicinity of the binding site on RACK1 for PKC βII (Fig. 2H and I, S2E–F). As an explanation of the latter result, the angular allowance in the control experiment, with the mask placed far away from RACK1, again resulted in partial alignment of the portion of RACK1-bound PKC affected by interdomain flexibility.
Fig. 2.
Density map of PKC-40S and Focused alignment. (A) Density map of PKC-40S at 11.4 Å. PKC-40S, is viewed from the solvent side. Small ribosomal subunit landmarks: n: neck; lf: left foot; rf: right foot, and RACK1. (B) Close-up showing rigid body docking of RACK1 in PKC-40S density map with additional density. (C) Density map from (A) displayed at same resolution as (E) and (H). (D and E) Focused alignment procedure performed to obtain partial densities of PKC βII near blades 3 and 4 of RACK1; the 3D mask is represented in blue mesh. (F) Close-up showing additional densities in contact with RACK1; the region highlighted in pink is suggested to interact with PKC βII (Grosso et al., 2008b; Ron et al., 1994). (G and H) Focused alignment procedure performed on another part of the 40S subunit, as control, showing very little additional density. (I) Close-up showing additional density generated close to RACK1. RACK1 model is taken from Homo sapiens (PDB ID: 3AOW).
The first additional density of activated PKC βII is seen in close proximity to blade 3 of RACK1, (Fig. 2F, S2C) a position that matches a previous peptide-mapping study, which predicts that PKC βII binds RACK1 at the outer β-strand of blade 3 (Grosso et al., 2008b; Ron et al., 1994). The second additional density of activated PKC βII in our cryo-EM map appears at a novel site, i.e. in very close proximity of blade 4 of RACK1 (Fig. 2F, S2C). Blade 4 has been implicated in homo-dimerization of both yeast and human RACK1 (Liu et al., 2007; Yatime et al., 2011). In yeast, the dimerization process involves flipping of the His-147 residue, which stabilizes the blade-to-blade packing in the monomeric form. When flipped, the two β-strands rearrange to interact with the other monomer, thereby facilitating the formation of the dimeric form, exposing an entirely different surface of the protein to the solvent. Whether flipping of His-147 occurs spontaneously or is dictated by a specific interaction or process is unknown (Yatime et al., 2011). Interestingly, His-147 is replaced by Ser-146 in human RACK1 (and most mammalian RACK1 proteins). It has been shown that phosphorylation of Ser-146 leads to human RACK1 dimerization and therefore can be the target for specific kinases found in higher eukaryotes (Liu et al., 2007). It has been postulated that recruitment of a binding partner in the vicinity of His-147 could initiate blade 4 unfolding, thus presenting buried features to potential interacting partners.
Thus biochemical and dimerization studies single out the two blades, 3 and 4, precisely where the extra densities are observed for complexes that contain activated PKC βII bound to RACK1. This observation strongly suggests that the densities are due to two subdomains of PKC βII recruited to RACK1 and bound to blades 3 and 4. (Fig. 2F, S2C). Evidently, more elaborate biochemical and structural studies will be required to confirm this suggestion and delineate the precise binding configuration.
Other peptide-mapping studies suggest that an additional PKC βII-binding sequence resides in the inner β-strand of blade 6 of RACK1 (Ron and Mochly-Rosen, 1995; Ron et al., 1994, 1995). In our study, we observe no such interaction (Fig. 2B–I, S2B–F). Interestingly, the peptide-mapping studies have been performed on ribosome-free RACK1 and activated PKC βII, which might explain the apparent discrepancy.
4. Orientation analysis in the PKC-40S data set
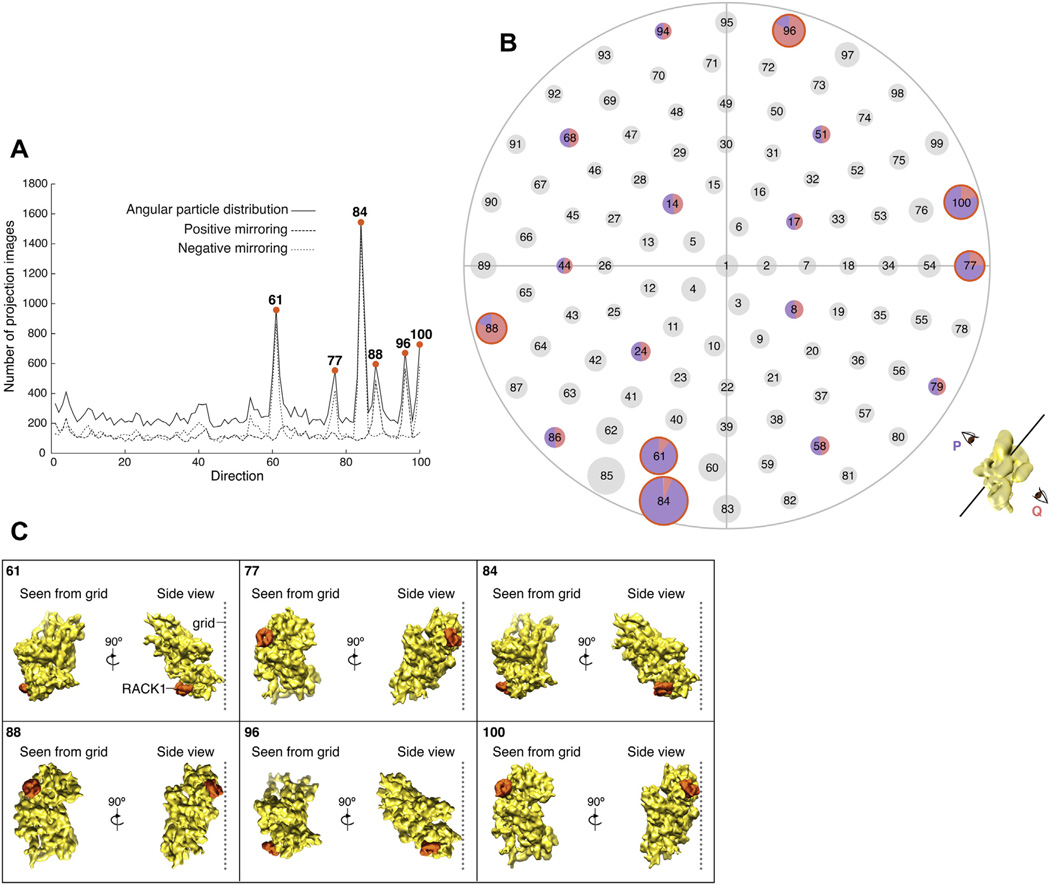
With the use of affinity grids, there is the concern that the chemical linking of the specimen to the lipid monolayer might introduce strongly preferred orientations in cryo-samples. By examining the distribution of projection image orientations obtained in angular refinement, we investigated the existence of such overrepresented views in our PKC-40S data set. We discretized projection images into one hundred bins based on view-directions. Overall, we observed six overrepresented view-directions (Fig. 3A and B, S3). Hence, tethering of the PKC-40S complex to the affinity grid results in a significant preference for the complex to assume certain orientations in the cryo-sample. While the orientation bias is not strong enough to introduce artifacts in the reconstruction, it is in itself informative as it reflects on the position of the ribosome in the supporting ice relative to the lipid layer.
Fig. 3.
Orientation analysis in the PKC-40S data set. (A and B) Angular particle distribution plots for PKC-40S data set sorted in 100 view-directions. (A) The number of particles per direction, and in addition, positive and negative mirroring assigned during alignment are shown. (B) Each view on the starry sky plot is associated with two opposite orientation assignments, which we call P (in purple) and Q (in pink). For most views (circles in black) the same numbers of particles are facing in opposite directions, as indicated by the pie chart in a few randomly chosen examples. In contrast, the particles falling into the six overrepresented views (circled in orange) show a strong preference for one of the two directions. Cartoon at the bottom right explains the two opposite orientations, for an arbitrary view, giving rise to mirrored projections. (C) Relative proximity of RACK1 (in PKC-40S complex) to the cryo-grid in the six overrepresented orientations (as seen from the grid and the side-view, RACK1 is highlighted in orange).
For each view, two orientation assignments are possible, the two assignments being related to each other by a rotation of 180° (i.e., either assigned to one end of the line representing the view direction or to the opposite end). Taking this into account, it is evident that there is a nearly even distribution between the two possible orientations in each view direction for all but the six overrepresented views (Fig. 3A and B). For all six overrepresented views, however, we observe more than 77% of projection images assigned to identical orientations. Keeping this in mind, an analysis of the geometry of the six overrepresented views of the complex relative to the cryo-grid reveals that, as expected, ribosomal subunits are in orientations that bring RACK1 (through which PKC-40S is tethered) in close proximity to the affinity layer of the cryo-grid (Fig. 3C).
In conclusion, we show that applying the affinity grid technique makes it possible to visualize the functional complex consisting of PKC βII and apo-40S (PKC-40S) with high occupancy of the bound ligand. We thereby demonstrate that the affinity grid technique can be utilized to reduce time and increase efficiency in obtaining cryo-EM 3D reconstructions of ligand-bound molecules by using His-tagged ligands. Specifically, our analysis, utilizing cryo-EM in combination with focused alignment and analysis of orientations of PKC-40S in our data set, supports tentative biochemical reports on a direct interaction between PKC βII and RACK1 (Ron et al., 1994). Furthermore, our map shows a novel site of possible interaction between activated PKC βII and RACK1 when bound to the ribosome. Finally, our study puts this PKC βII/RACK1 interaction into the context of the ribosomal 40S subunit, thereby providing a direct link to the reports indicating that the PKC βII-RACK1 interaction is involved in the regulation of protein translation.
Supplementary Material
Acknowledgements
We thank Melissa Thomas for assistance with the illustrations. The work was supported by HHMI and grant R01 GM29169 (to JF).
Footnotes
Appendix A. Supplementary material
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.jsb.2012.11.006.
References
- Adams DR, Ron D, Kiely PA. RACK1, a multifaceted scaffolding protein: structure and function. Cell Commun. Signal. 2011;9:22. doi: 10.1186/1478-811X-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203–1209. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Grosso S, Volta V, Vietri M, Gorrini C, Marchisio PC, et al. Eukaryotic ribosomes host PKC activity. Biochem. Biophys. Res. Commun. 2008a;376:65–69. doi: 10.1016/j.bbrc.2008.08.118. [DOI] [PubMed] [Google Scholar]
- Grosso S, Volta V, Sala LA, Vietri M, Marchisio PC, et al. PKCbetaII modulates translation independently from mTOR and through RACK1. Biochem. J. 2008b;415:77–85. doi: 10.1042/BJ20080463. [DOI] [PubMed] [Google Scholar]
- Kelly DF, Dukovski D, Walz T. Monolayer purification: a rapid method for isolating protein complexes for single-particle electron microscopy. Proc. Natl. Acad. Sci. USA. 2008a;105:4703–4708. doi: 10.1073/pnas.0800867105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly DF, Abeyrathne PD, Dukovski D, Walz T. The affinity grid: a prefabricated EM grid for monolayer purification. J. Mol. Biol. 2008b;382:423–433. doi: 10.1016/j.jmb.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly DF, Dukovski D, Walz T. Strategy for the use of affinity grids to prepare non-His-tagged macromolecular complexes for single-particle electron microscopy. J. Mol. Biol. 2010a;400:675–681. doi: 10.1016/j.jmb.2010.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly DF, Dukovski D, Walz T. A practical guide to the use of monolayer purification and affinity grids. Methods Enzymol. 2010b;481:83–107. doi: 10.1016/S0076-6879(10)81004-3. [DOI] [PubMed] [Google Scholar]
- Leonard TA, Rozycki B, Saidi LF, Hummer G, Hurley JH. Crystal structure and allosteric activation of protein kinase C betaII. Cell. 2011;144:55–66. doi: 10.1016/j.cell.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Roberts R. WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases. Cell Mol. Life Sci. 2001;58:2085–2097. doi: 10.1007/PL00000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YV, Hubbi ME, Pan F, McDonald KR, Mansharamani M, et al. Calcineurin promotes hypoxia-inducible factor 1alpha expression by dephosphorylating RACK1 and blocking RACK1 dimerization. J. Biol. Chem. 2007;282:37064–37073. doi: 10.1074/jbc.M705015200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCahill A, Warwicker J, Bolger GB, Houslay MD, Yarwood SJ. The RACK1 scaffold protein: a dynamic cog in cell response mechanisms. Mol. Pharmacol. 2002;62:1261–1273. doi: 10.1124/mol.62.6.1261. [DOI] [PubMed] [Google Scholar]
- Miluzio A, Beugnet A, Volta V, Biffo S. Eukaryotic initiation factor 6 mediates a continuum between 60S ribosome biogenesis and translation. EMBO Rep. 2009;10:459–465. doi: 10.1038/embor.2009.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson J, Sengupta J, Frank J, Nissen P. Regulation of eukaryotic translation by the RACK1 protein: a platform for signalling molecules on the ribosome. EMBO Rep. 2004;5:1137–1141. doi: 10.1038/sj.embor.7400291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Mochly-Rosen D. An autoregulatory region in protein kinase C: the pseudoanchoring site. Proc. Natl. Acad. Sci. USA. 1995;92:492–496. doi: 10.1073/pnas.92.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Chen CH, Caldwell J, Jamieson L, Orr E, et al. Cloning of an intracellular receptor for protein kinase C: a homolog of the beta subunit of G proteins. Proc. Natl. Acad. Sci. USA. 1994;91:839–843. doi: 10.1073/pnas.91.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Luo J, Mochly-Rosen D. C2 region-derived peptides inhibit translocation and function of beta protein kinase C in vivo. J. Biol. Chem. 1995;270:24180–24187. doi: 10.1074/jbc.270.41.24180. [DOI] [PubMed] [Google Scholar]
- Ron D, Jiang Z, Yao L, Vagts A, Diamond I, et al. Coordinated movement of RACK1 with activated betaIIPKC. J. Biol. Chem. 1999;274:27039–27046. doi: 10.1074/jbc.274.38.27039. [DOI] [PubMed] [Google Scholar]
- Schechtman D, Mochly-Rosen D. Adaptor proteins in protein kinase C-mediated signal transduction. Oncogene. 2001;20:6339–6347. doi: 10.1038/sj.onc.1204778. [DOI] [PubMed] [Google Scholar]
- Sengupta J, Nilsson J, Gursky R, Spahn CM, Nissen P, et al. Identification of the versatile scaffold protein RACK1 on the eukaryotic ribosome by cryo-EM. Nat. Struct. Mol. Biol. 2004;11:957–962. doi: 10.1038/nsmb822. [DOI] [PubMed] [Google Scholar]
- Sondek J, Siderovski DP. Ggamma-like (GGL) domains: new frontiers in G-protein signaling and beta-propeller scaffolding. Biochem. Pharmacol. 2001;61:1329–1337. doi: 10.1016/s0006-2952(01)00633-5. [DOI] [PubMed] [Google Scholar]
- Stebbins EG, Mochly-Rosen D. Binding specificity for RACK1 resides in the V5 region of beta II protein kinase C. J. Biol. Chem. 2001;276:29644–29650. doi: 10.1074/jbc.M101044200. [DOI] [PubMed] [Google Scholar]
- Yatime L, Hein KL, Nilsson J, Nissen P. Structure of the RACK1 dimer from Saccharomyces cerevisiae. J. Mol. Biol. 2011;411:486–498. doi: 10.1016/j.jmb.2011.06.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.