Abstract
CLONEPICKER is a software pipeline that integrates sequence data with BAC clone fingerprints to dynamically select a minimal overlapping clone set covering the whole genome. In the Rat Genome Sequencing Project (RGSP), a hybrid strategy of “clone by clone” and “whole genome shotgun” approaches was used to maximize the merits of both approaches. Like the “clone by clone” method, one key challenge for this strategy was to select a low-redundancy clone set that covered the whole genome while the sequencing is in progress. The CLONEPICKER pipeline met this challenge using restriction enzyme fingerprint data, BAC end sequence data, and sequences generated from individual BAC clones as well as WGS reads. In the RGSP, an average of 7.5 clones was identified from each side of a seed clone, and the minimal overlapping clones were reliably selected. Combined with the assembled BAC fingerprint map, a set of BAC clones that covered >97% of the genome was identified and used in the RGSP.
The rapid accumulation of genomic sequence data provides the opportunity for studying biology at the genome scale, leading to identification of the complete transcriptome, understanding gene regulatory networks, or studying evolution at the molecular level.
The most systematic and efficient way to obtain sequence data is through whole genome sequencing projects. Three different strategies were used to generate draft sequences for the human, mouse, and rat genomes, namely, the “clone by clone” (CBC) for human, the whole genome shotgun (WGS) for mouse, and a hybrid strategy for rat (Lander et al. 2001; Waterston et al. 2002; Rat Genome Sequencing Project Consortium 2004). In the CBC approach, individual BAC clones are shotgun-sequenced, the sequence of each BAC clone is generated by assembling the corresponding sequencing reads, and the sequence of the whole genome is obtained by merging overlapping BAC clone sequences. To minimize sequencing the same genomic region multiple times, a set of minimally overlapping clones covering the whole genome is determined beforehand. In contrast, the WGS approach shears the whole genome into small fragments that are sequenced, and all the reads are then assembled simultaneously to generate the consensus sequence for the entire genome. Compared with the CBC method, a small number of DNA libraries are constructed for sequencing, and a predetermined clone tiling path is not required. However, repetitive sequences and large duplications in the genome can make the final assembly less complete and more error prone compared with that obtained by the CBC method. A CBC approach is often needed to generate the finished sequence for a large region sequenced by the WGS method (Waterston et al. 2002).
To combine the merits of both methods, a hybrid strategy was developed for the Rat Genome Sequencing Project (RGSP; Rat Genome Sequencing Project Consortium 2004). Selected BAC clones covering the whole genome were “skim”-sequenced to ∼2× coverage. This was done in parallel with generation of WGS sequences to ∼4× coverage. WGS reads were localized to individual BAC clones using the ATLAS genome assembly software (Havlak et al. 2004). These localized WGS reads were then assembled together with BAC skim reads using a local assembly program, PHRAP (Gordon et al. 1998). The final assembly was generated by merging overlapping BAC clones together (Havlak et al. 2004). Both the high throughput of the WGS approach and important local information provided by the CBC approach were used in this scheme. Like the WGS approach, a majority of the sequence reads were generated from WGS libraries with very high throughput. On the other hand, similar to the CBC strategy, the global assembly is generated by merging of local assemblies. As a result, sequence assembly complexity is greatly reduced and the issue of genome duplications can be better handled (Tuzun et al. 2004).
Similar to the CBC approach, this hybrid strategy requires an optimal BAC clone tiling path. The current method for construction of such tiling paths relies on ordering BAC clones based on their restriction enzyme digestion patterns using the FPC software (Soderlund et al. 1997; McPherson et al. 2001). BAC clones are digested with restriction enzymes, and the resulting fragment sizes are used to determine the similarity and overlap. Clones are then assembled into clone contigs using the FPC program (Soderlund et al. 1997, 2000). Only tens of fragments are generated with each enzyme and, because of this and the low resolution of gel electrophoresis, overlaps between clones can be missed or false overlaps can be obtained. As results, the relative positions of BAC clones in the assembly may not be accurate. One method for improving clone selection is to combine the sequencing information with FPC. By anchoring the FPC assembly to the draft genome sequence contigs through BAC end reads, a more accurate minimal clone tiling path can be identified (Engler et al. 2003). Moreover, for a mammalian genome, initial automatic assembly of BAC clones based on FPC data results in as many as 10,000 contigs. To reduce the number of contigs and increase continuity, laborious manual merging and additional information such as genome sequence are necessary. Thus, there are limitations on the use of FPC for selecting a clone tiling path with low redundancy at the beginning stage of the sequencing project.
RESULTS
With the hybrid strategy used in the RGSP, it was essential to select a set of BAC clones that covered the entire genome for skim sequencing. To reduce clone redundancy, a minimal clone tiling path covering the genome needed to be constructed. As noted earlier, it is difficult to identify BAC clone pairs with small overlaps at high confidence using FPC alone. As a result, the average size of clone contigs assembled based on FPC data is often small. Indeed, the 200,000 rat BAC clones from the RGSP were initially assembled into >10,000 FPC contigs with an average contig size of less than two times the BAC insert size (Schein 2003; M. Krzywinski, C. Fjell, J. Asano, S. Barber, I. Bosdet, M. Brown-John, S. Chan, R. Chiu, S. Chand, A. Cloutier, et al., pers. comm.). Although this level of assembly was useful for seed clone selection, further merging of these contigs into bigger structures was necessary for final selection of a low-redundancy clone path covering the whole genome. Manual merging of these small contigs is time-consuming and could not be conducted efficiently until the later stage of the project, when more DNA sequence information was available. In fact, much of the merging of the rat FPC map was conducted based on the first version of the rat genome sequence assembly. Therefore, it was necessary to develop additional methods to generate an optimal clone tiling path that covers the entire genome with high accuracy.
In addition, it was necessary to build the BAC tiling path and select clones for sequencing before the complete FPC map was available and simultaneous with sequencing of BACs and production of WGS data (Rat Genome Sequencing Project Consortium 2004). Although genome projects are often performed with sequential steps of map construction followed by sequencing, the desire to reduce the duration of projects leads to simultaneous clone mapping and sequencing. This “just in time” methodology was developed for the RGSP, starting with seed BAC clones, which were selected from early FPC contigs so as not to overlap. These clones were sequenced and then used to select other clones for a tiling path, as described in this report. To increase the efficiency, sensitivity, and accuracy of the tiling path building process, a software pipeline was developed (CLONEPICKER; Fig. 1) that automatically identified the minimal overlapping neighbor clone by combining BAC fingerprints with other information, including end sequences of a large collection of BACs and the local assemblies of seed clones.
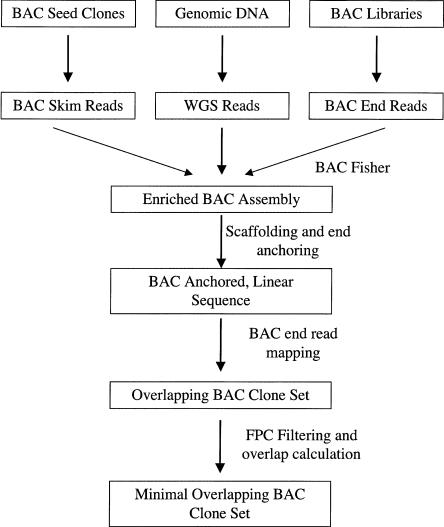
Figure 1.
The CLONEPICKER pipeline. BAC skim reads from seed clones, WGS reads, and BAC end reads were used to establish the enriched BAC seed clone assembly using the ATLAS tools. Candidate BAC clones that overlap with the seed clone were first identified based on BAC end reads that mapped to the enriched BAC assembly. These candidate clones were then assessed by comparing their restriction enzyme digestion patterns. Clones that passed this filtering step were then analyzed for their overlap size with the seed clone. Clones with minimal overlap with the seed clones were selected as new clones for sequencing.
Reads from skimmed BAC seed clones were first used to identify all the WGS reads and BAC end reads that were mapped to this clone using the ATLAS program. These reads were then assembled with the skim reads into sequence contigs of an “enriched” BAC. To determine the BAC clones that extend from the seed clone with minimal overlaps, the order and orientation of the contigs in each enriched BAC were first determined by read mate pairs (two reads from the opposite ends of the same clone template). Two contigs were considered to be adjacent to each other if mate pairs were assembled into these two contigs, thus linking the contigs into a scaffold. Next, contigs that contained the end of the BAC clones were identified by the presence of BAC end reads in the contig, skim reads that contained the cloning junctions, and/or skim read pairs that covered the cloning junctions (Fig. 2A). This allowed anchoring of the scaffold to the clone end. Third, potential overlapping BAC clones were identified from BAC end reads that were assembled into enriched seed BAC contigs. Overlaps of these clones were validated by their fingerprint patterns, which eliminated false positives caused by BAC end read mapping errors. False clones, which had incompatible fingerprint patterns with the rest of clones in the group, were excluded (Fig. 2B). Fourth, BAC clones that had the minimal overlap with the original clone were identified and selected for further sequencing. The overlap size between the candidate BAC clone and the seed clone was calculated from the position within seed BAC contigs where its end reads were mapped. As illustrated in Figure 2B, clone d had the least overlap with the seed clone because its BAC end read mapped to the contig that was closest to the cloning end. Therefore, clone d was considered the optimal clone and added to the tiling path. This process was repeated to construct a tiling path incrementally, allowing BAC clone selection for sequence skims without a delay while the complete BAC clone map was being constructed. This strategy was used to provide ∼1700 rat BAC clones at the rate of 200 per week for sequencing at the later stage of the RGSP when sufficient BAC clones could not be identified by the FPC approach. Together with the FPC method (M. Krzywinski, C. Fjell, J. Asano, S. Barber, I. Bosdet, M. Brown-John, S. Chan, R. Chiu, S. Chand, A. Cloutier, et al., pers. comm.), a clone set that covered >97% of the rat genome was identified for the RGSP.
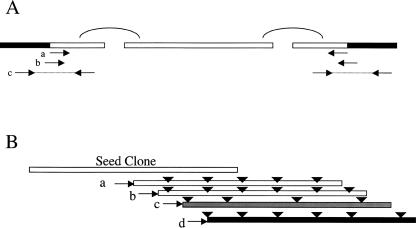
Figure 2.
(A) Anchoring of the scaffold to the cloning end for each BAC clone. Each box represents a sequence contig obtained from the BAC assembly. Solid parts of the box indicate the cloning vector. Contigs were first linked into scaffolds using paired end reads, shown as curve connections in the figure. Three types of reads/clones were used to identify the clone junctions: (a) BAC end reads, (b) insert-vector junction reads, and (c) insert-vector junction clones. (B) Restriction enzyme digestion patterns of the candidate clones were used to filter out false positives caused by mismapping of the BAC end reads. The triangles represent restriction enzyme recognition sites. Clones a, b, and d share common sites, whereas clone c has a very different pattern. As a result, clone c is excluded from further analysis. Clone d is selected as having the minimal overlap.
Obtaining the Sequence of BAC Seed Clones
The key issue in reducing sequence redundancy in the hybrid strategy is to reduce the overlap between BAC clones. The assembled sequence for each skim-sequenced BAC clone was the basis for identification of additional BAC clones that extended into gaps. Accurate assembly for each BAC seed was obtained using the ATLAS tool (Havlak et al. 2004). The assembly of BAC skim reads with added WGS reads resulted in an average of 96% of the BAC sequence being covered in the enriched BAC assembly. Moreover, the paired reads and libraries with different insertion sizes provided by WGS reads allowed the order and orientation to be determined for most contigs in the BAC. In 87% of the BAC clones, all contigs >1 kb could be linked in one scaffold. When finished sequence and other data such as cDNA sequences were compared with the BAC sequences, excellent collinearity was observed with scaffolds. The average ratio of the sum of the length of the scaffolds of the BAC clone to its FPC size was 1.07 (222 kb vs. 208 kb) with a standard deviation of 0.12. As expected, the scaffold size was slightly bigger than the FPC size because contigs that extend beyond the cloning ends of a BAC clone were also included in scaffolds based on WGS read pairs. Therefore, the sequence of the seed clone was accurately represented by the scaffold sequence. Because of this excellent assembly, end reads from BAC clones that overlap the seed clone were readily identified and properly mapped onto scaffolds.
End Anchoring of the Seed BAC Clone Scaffold
To calculate the overlap size between BACs and the seed clone based on their end read mapping positions on the scaffold, it was necessary to determine the end of the insert in the seed clone first. The sequences for each seed clone were anchored to the insertion ends based on three types of data: BAC end reads, cloning junction reads, and clone vector read pairs. We first attempted to map the BAC end reads from the seed clone to its own sequence. If successful, the insert end could be determined based on its end read position and orientation relative to the sequence. However, because ∼40% of the rat genome is highly repetitive (Rat Genome Sequencing Project Consortium 2004) and the EcoRI site used in cloning BACs has bias toward repetitive regions, less than half of the insertion ends could be identified through this approach. To improve the anchoring efficiency, we explored additional types of data for this process. As shown in Figure 2A, BAC skim reads that cross the cloning junctions could also be used to determine the insert end position. Moreover, read pairs from subclones that span the cloning junctions also provided useful information. Scanning though the BAC skim reads, on average 2.9 junction reads were found for each seed clone, consistent with the 1.5× sequence coverage of the skimmed reads. Similarly, an average of 7.1 clones were found that spanned the cloning junctions for each seed clone. Using these two types of data together with the BAC end reads, ∼80% of the insert ends were identified consistently. In addition, for clones whose ends failed to be anchored, the position of the insert ends could be approximately determined if a major scaffold (scaffolds that exceed 90% of the estimated insert size) existed for the clone. By assigning the cloning end to the end of the major scaffold, a maximum of 10–20 kb error was introduced between the real cloning site and the estimated site. In fact, 89% of all seed clones had a major scaffold, and 87% of them only had one scaffold. As a result, the insert ends were determined for >95% of the seed clones. An average extension of 212 kb, similar to the clone size, was found from the insert end and was used to identify additional BACs that extended into gaps.
Mapping BAC End Reads to the Seed Clone
BACs that overlapped the seed clones needed to be identified at high sensitivity and specificity to ensure the quality of subsequent clone selection. One approach was to find overlapping BACs based on FPC assembly. Another approach was through mapping of the BAC end reads to the sequences of seed clones. To compare these two methods, we first mapped the BAC end reads to the sequences from all seed clones. In the RGSP, a total of 306,779 BAC end reads from ∼185,000 BAC clones were generated. Unlike genomes such as human and mouse, the repetitive elements in the rat genome were not fully characterized at the time. To accurately map these BAC end reads, we used the ATLAS tool instead of other sequence database search tools such as BLASTN. In the ATLAS software, potential repetitive sequences are identified by masking highly abundant k-mers, and only read pairs that share low-abundance k-mers and have good alignment are considered true overlaps. Moreover, read pair information is used in ATLAS in the mapping process so that repetitive reads with their pair uniquely mapped can also be mapped (Havlak et al. 2004). Thus, a higher accuracy and sensitivity was achieved in mapping BAC end reads to the sequences. A total of 244,710 unique BAC end reads were mapped to the sequences, with an average of 18.6 BAC end reads from 15.8 BAC clones overlapping each seed clone. Considering that the average size of each BAC clone was ∼220 kb, one overlapping clone was identified every 27.8 kb. To assess the accuracy and sensitivity of this mapping process, we compared the mapping results with the final FPC assembly. We found that 55% (175,152/318,442) of the overlaps suggested by the BAC end read overlap were confirmed in the FPC assembly. Conversely, 50% (175,129/348,543) of the overlaps suggested by the FPC assembly were confirmed by the end read mapping. We believe that this low confirmation rate mainly reflects the inaccuracy of the clone placement in the FPC assembly. When the confirmation rate was examined for overlapping clone pairs identified using the final genome sequence assembly, a low rate of confirmation by the FPC assembly of 42% (10282/24795) was obtained. In contrast, a rate of 72% confirmation was obtained by BAC end read mapping. Therefore, it appears that through BAC end read mapping, BACs that overlap with seed clones can be identified more accurately than through using FPC alone.
Identification of the Minimal Overlapping BAC Clones
To minimize the sequencing redundancy, the overlap size must be accurately estimated for all potential BACs that overlap with a seed clone to choose the BAC with minimal overlap. One approach for calculating clone overlap is to compare their restriction enzyme digestion patterns. However, due to the low resolution offered by the restriction enzyme digestion fingerprint data, estimation of the overlap size between clones is often not accurate. Another approach is to calculate the overlap size based on the map position and orientation of BAC end reads on the anchored sequences. Figure 3 shows the comparison of clone overlap size calculated from the genome assembly to that estimated from mapping BAC end reads to the scaffolds of a seed clone or the FPC assembly. A strong positive correlation of 0.80 was observed comparing the assembly to BAC end reads, significantly higher than the 0.59 obtained using the FPC assembly. Moreover, the average overlap size estimated by end reads was 89 kb, only 1 kb smaller than the number obtained based on the assembled sequence. In contrast, the average clone overlap estimated based on the FPC assembly was 16% smaller than the size obtained from the sequence. Therefore, using BAC end reads as the basis to estimate clone overlap is more accurate than using the FPC assembly.
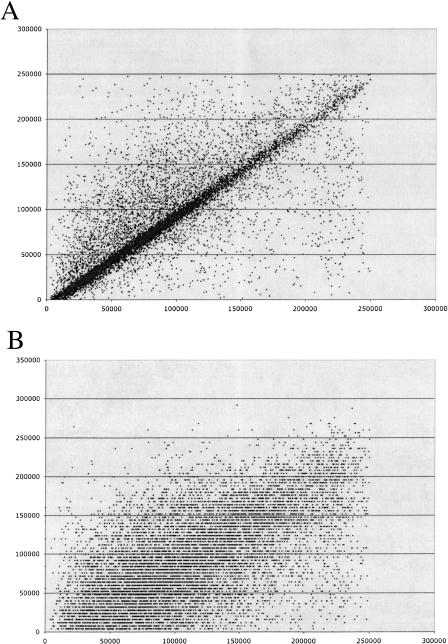
Figure 3.
Scatterplots of the estimated clone overlap against overlap obtained from the sequence of overlapping seed clone pairs. The horizontal axis is the overlap between a seed clone pair based on their final sequence. The vertical axis is the overlap estimated based from (A) BAC end reads or (B) the FPC assembly.
BACs that overlap with the seed clone can be separated into three groups: BACs that are internal or that extend to the left or the right side of the seed clone. BACs with minimal overlaps with the seed clone were selected from the latter two groups to cover new genomic regions. For each seed clone, we found an average of 7.2 BACs extending to either side. To exclude BACs that were introduced through false BAC end read mapping, all the potential BACs from the same group were checked for consistency by their restriction enzyme digestion fingerprints. For BACs that truly extended at the same side of the seed clone, it was very likely that similar restriction fragments will be detected between them. The Sulston score (Sulston et al. 1988; Soderlund et al. 1997) was calculated for each BAC pair within the same group, and BACs that did not have a score lower than 10-10 with any other clones in the group were considered false positives. On average, <0.5 BACs were excluded during this process, consistent with the idea that BAC end read mapping is very accurate. Once the filtering was done, an additional subgrouping step was conducted to deal with genome duplications. It was shown that ∼5% of the human genome is recently duplicated (Bailey et al. 2002). Similarly, recent duplications were found in 2.9% of the rat genome (Tuzun et al. 2004). To ensure that BACs representing all duplicated regions were sequenced, clones passing the previous filtering step were further divided into subgroups based on their FPC patterns. BACs were split into different groups unless they had a Sulston score of <10-10 directly or indirectly (through other clones). Two or more subgroups were found in 389 seed clones, indicating potential duplication regions. For each subgroup/group, the BAC with minimal overlaps with the seed clone was selected as an additional clone for sequencing.
To assess if the CLONEPICKER pipeline was effective in selecting an optimal clone set in the real project, we examined the clone distribution as different stages of the rat sequencing project. The size distributions of continuous genomic blocks covered by seed BAC clones were compared at three stages of the project with 14,000, 17,000, and 19,500 clones sequenced (Fig. 4). Dramatic changes were observed, with the average block size doubling at each stage, indicating that additional seed clones located in regions that were not covered at previous stages were successfully identified. Therefore, using the CLONEPICKER pipeline, we have successfully selected a set of BACs that were evenly distributed across the rat genome.
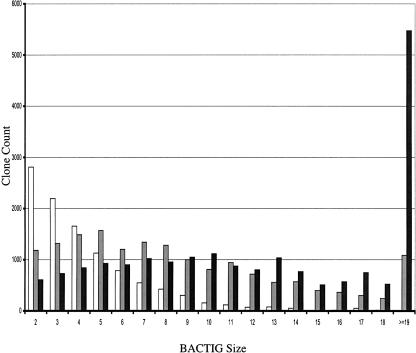
Figure 4.
The distribution of BACTIG size in a series of assemblies. The horizontal axis is the number of overlapping BAC clones in each BACTIG. The vertical axis is the number of each type of BACTIG. Clone distributions from three assemblies with 14,000 (white), 17,000 (gray), and 19,500 (black) BACs are shown.
DISCUSSION
Compared with the pure WGS method, clone-based methods are likely to produce more accurate draft assemblies for mammalian genomes with complicated duplication and repeat structures (Rat Genome Sequencing Project Consortium 2004). A combination of CBC and WGS sequencing is the most cost-effective way for mammalian genome sequencing at the current time. We have developed software that helps to increase the efficiency of such a hybrid sequencing strategy by selecting a set of low-redundancy BACs that cover the whole genome. This software pipeline has been used in the RGSP and yields a clone set that covers >97% of the genome. Most of the redundancy in the current clone set is due to the process adopted in the RGSP. Because of time constraints, FPC, seed clone selection and sequencing, BAC end reads, and WGS read sequencing were all conducted in parallel. Therefore, the tiling path and the minimal overlapping clones were not always ideal because only partial data were available. However, through analyzing results from the CLONEPICKER pipeline on the complete data set of the rat genome, it is clear that the minimal overlapping clones can be reliably identified. Therefore, to achieve the maximum efficiency in the future, it will be desirable to obtain ancillary information first and perform the skim sequencing of individual BACs later.
The key issue for identification of the optimal clone set is to accurately estimate clone overlaps. Sequence-based comparison of overlaps is inherently more accurate than comparing restriction fragments. Because of the low resolution of the restriction enzyme digestion fingerprints, the accuracy of individual clone placement in the FPC assembly is limited. Indeed, even in the final rat FPC assembly, ∼58% of the confirmed overlapping clone pairs from the genome assembly do not overlap in the FPC map. Moreover, the overlap size estimated by the FPC assembly is more inaccurate than the sequence-based method (Fig. 3). Therefore, even with a complete FPC assembly, it is difficult to derive an optimal clone set for sequencing. This situation can be dramatically improved with the integration of BAC end reads and enriched seed clone sequences. Given BAC clone skimmed reads, BAC sequences can be reliably obtained using the ATLAS tools. More than 95% of the time, the insert end can be identified, and the sequence can be anchored accurately. Moreover, the continuity of the sequence is quite good, and an average of 212 kb can be extended inside the clone from the ends. Subsequent mapping of BAC end reads onto these anchored sequences of each seed clone allows identification of potential overlapping BACs. Overlaps calculated between these BACs and the seed clone are more accurate compared with the estimation from the FPC assembly (Fig. 3). The average estimated from the BAC end reads is merely a 1% underestimation of the real size compared with the 16% underestimation by the FPC assembly.
The main limitation of this method is the availability of the sequenced seed clone and the presence of repetitive sequences at the end sequences. This limitation can be overcome by integrating the restriction enzyme digestion and FPC assembly. Clones with very different digestion patterns are likely errors and are excluded from subsequent selection. The FPC assembly can also help to reduce the false-negative rate. BACs can fail to be identified if ends fall into sequence gaps or are repetitive. These BACs can be recovered if suggested by the FPC assembly. Scripts have been developed to incorporate these clones into the seed clone selection process, and a graphical interface was set up for manual inspection, if necessary (data not shown).
Owing to data limitation, it is very difficult to identify the true minimal tiling path covering the whole genome until the final stage of the sequencing project. A different approach may provide a better solution for this problem. Instead of trying to identify the minimal clone set, a clone set with high clone coverage of the genome can be sequenced. Based on the Poisson distribution, >95% of the genome will be covered at least once when clones amounting to threefold coverage are sequenced. To reduce the cost of BAC library production, a pooled array strategy has been proposed (Cai et al. 2001). Instead of making one library for each BAC, BACs are organized into pools for library construction and sequencing, greatly reducing the cost and improving the efficiency. Subsequent deconvolution of these pools allows the preservation of BAC clone information, which can be used during the assembly process. Further development and implementation of this strategy can conceivably eliminate the requirement of map construction, while at the same time preserving the advantage offered by the clone-based sequencing method.
METHODS
Data Sources
The restriction enzyme digestion data and the FPC assembly for rat BAC clones were obtained from ftp://genome.wustl.edu/pub/groups/mapping/rat. The BAC end sequences were downloaded from the TIGR FTP site ftp://tigr.org/pub/data/r_norvegicus/bac_end/bac_end_sequences. Enriched BAC seed clone assemblies were generated using the ATLAS tools. Vector sequences for the cloning vector of the rat BAC library, CHORI-230, were downloaded from http://bacpac.chori.org/vectorsdet.htm.
Data Processing and Software
The FPC assembly file as well as the restriction enzyme digestion pattern for each BAC clone were parsed with scripts and loaded into an ORACLE database for later access. Repeats in BAC end reads were masked using the RepeatMasker program (Smit 1999; http://ftp.genome.washington.edu/RM/RepeatMasker.html). BAC skim reads that contain cloning junctions were identified by the tail-to-tail alignment with the clone vector sequences using banded alignment software developed at HGSC. Similarly, subclones that cover the cloning junctions were identified by the internal alignment of BAC skim reads to the vector sequences. Local assemblies for each BAC seed clone were used to determine the position of BAC end reads, which were then used to calculate clone overlap. Scripts were developed to filter out false overlapping BACs due to end read mapping errors using restriction digestion data. Source code for these scripts is available at http://www.hgsc.bcm.tmc.edu/downloads/software/atlas.
Acknowledgments
This work was supported by grant HG02395 from the NHGRI and NHLBI at the National Institutes of Health. We thank TIGR and the British Columbia Cancer Agency Genome Science Center for sharing the BAC end reads and FPC data.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2171704.
Footnotes
[The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: TIGR and the British Columbia Cancer Agency Genome Science Center.]
References
- Bailey, J.A., Gu, Z., Clark, R.A., Reinert, K., Samonte, R.V., Schwartz, S., Adams, M.D., Myers, E.W., Li, P.W., and Eichler, E.E. 2002. Recent segmental duplications in the human genome. Science 297: 1003-1007. [DOI] [PubMed] [Google Scholar]
- Cai, W.W., Chen, R., Gibbs, R.A., and Bradley, A. 2001. A clone-array pooled shotgun strategy for sequencing large genomes. Genome Res. 11: 1619-1623. [DOI] [PubMed] [Google Scholar]
- Engler, F.W., Hatfield, J., Nelson, W., and Soderlund, C.A. 2003. Locating sequence on FPC maps and selecting a minimal tiling path. Genome Res. 13: 2152-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, D., Abajian, C., and Green, P. 1998. Consed: A graphical tool for sequence finishing. Genome Res. 8: 195-202. [DOI] [PubMed] [Google Scholar]
- Havlak, P., Chen, R., Durbin, K.J., Egan, A., Ren, Y., Song, X.-Z., Weinstock, G.M., and Gibbs, R. 2004. The ATLAS genome assembly system. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- McPherson, J.D., Marra, M., Hillier, L., Waterston, R.H., Chinwalla, A., Wallis, J., Sekhon, M., Wylie, K., Mardis, E.R., Wilson, R.K., et al. 2001. A physical map of the human genome. Nature 409: 934-941. [DOI] [PubMed] [Google Scholar]
- Rat Genome Sequencing Project Consortium. 2004. Genome sequence of the Brown Norway Rat yields insights into mammalian evolution. Nature (in press). [DOI] [PubMed]
- Schein, J.E.A. 2003. High-throughput BAC fingerprinting. In Bacterial artificial chromosomes: Methods and protocols (ed. S. Zhao and M. Stodolsky). Humana Press, Inc., Totowa, NJ.
- Smit, A.F.A. 1999. Interspersed repeats and other mementos of transposable elements in the mammalian genomes. Curr. Opin. Genet. Dev. 9: 657-663. [DOI] [PubMed] [Google Scholar]
- Soderlund, C., Longden, I., and Mott, R. 1997. FPC: A system for building contigs from restriction fingerprinted clones. Comput. Appl. Biosci. 13: 523-535. [DOI] [PubMed] [Google Scholar]
- Soderlund, C., Humphray, S., Dunham, A., and French, L. 2000. Contigs built with fingerprints, markers, and FPC V4.7. Genome Res. 10: 1772-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston, J., Mallett, F., Staden, R., Durbin, R., Horsnell, T., and Coulson, A. 1988. Software for genome mapping by fingerprinting techniques. Comput. Appl. Biosci. 4: 125-132. [DOI] [PubMed] [Google Scholar]
- Tuzun, E., Bailey, J.A., and Eichler, E.E. 2004. Recent segmental duplications in the working draft assembly of the brown Norway rat. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- ftp://genome.wustl.edu/pub/groups/mapping/rat; restriction enzyme digestion data and FPC assembly for rat BAC clones.
- ftp://tigr.org/pub/data/r_norvegicus/bac_end/bac_end_sequences; TIGR FTP site.
- http://bacpac.chori.org/vectorsdet.htm; vector sequences for the cloning vector of the rat BAC library, CHORI-230.
- http://ftp.genome.washington.edu/RM/RepeatMasker.html; RepeatMasker.
- http://www.hgsc.bcm.tmc.edu/downloads/software/atlas; source code for scripts.