Abstract
We have built a whole-genome multiple alignment of the three currently available mammalian genomes using a fully automated pipeline that combines the local/global approach of the Berkeley Genome Pipeline and the LAGAN program. The strategy is based on progressive alignment and consists of two main steps: (1) alignment of the mouse and rat genomes, and (2) alignment of human to either the mouse-rat alignments from step 1, or the remaining unaligned mouse and rat sequences. The resulting alignments demonstrate high sensitivity, with 87% of all human gene-coding areas aligned in both mouse and rat. The specificity is also high: <7% of the rat contigs are aligned to multiple places in human, and 97% of all alignments with human sequence >100 kb agree with a three-way synteny map built independently, using predicted exons in the three genomes. At the nucleotide level <1% of the rat nucleotides are mapped to multiple places in the human sequence in the alignment, and 96.5% of human nucleotides within all alignments agree with the synteny map. The alignments are publicly available online, with visualization through the novel Multi-VISTA browser that we also present.
Multiple sequence alignments represent the fundamental basis for comparative analysis aimed at identification and characterization of functional elements. For example, similarity across large evolutionary distances, detected by a multiple alignment of homologous sequences from several species, usually reveals conserved, and by inference, important, biological features (Gottgens et al. 2002; Boffelli et al. 2003; Kellis et al. 2003; Thomas et al. 2003). Similarly, estimates of local rates of evolution on the basis of multiple alignments give quantitative measures of the strength of evolutionary constraints and the importance of functional elements (Sumiyama et al. 2001; Simon et al. 2002; Cooper et al. 2003). It is with these applications in mind that we embarked on a multiple alignment of the human, mouse, and rat genomes. Accordingly, our alignment formed the basis for global estimates of evolutionary rates and other parameters in these mammalian genomes, as well as for the identification of constrained elements in the human genome (Cooper et al. 2004; Rat Genome Sequencing Project Consortium 2004).
Although there have been several recent efforts to build multiple alignments of smaller bacterial (Hohl et al. 2002) and yeast (Kellis et al. 2003) genomes, the availability of the rat genome presents for the first time the challenge and unparalleled opportunity of building a multiple alignment of several mammalian genomes. Several strategies for pairwise genome alignments were successfully developed for comparing the human and mouse genomes (Waterston et al. 2002). These approaches were based either on local alignment (Ma et al. 2002; Schwartz et al. 2003) or on a local/global technique, in which the mouse contigs are mapped on the human genome by a local aligner initially, and then the homology is confirmed and refined by a global aligner (Couronne et al. 2003). Comparing more than two large and structurally complex genomes presents several new challenges: obtaining a consistent map between several genomes, performing large-scale multiple alignment, and visualizing and interpreting the results.
In this article, we present a multiple alignment of the human, mouse, and rat genomes built using a novel method that expands on the local/global approach of Couronne and colleagues (2003). Our technique is fully automated and efficient: It does not require a prebuilt synteny map, and it is able to align the three mammalian genomes in <1 day on a 24-node computer cluster. Analysis of the alignment indicates high levels of sensitivity and specificity, in that this technique aligns the known functional elements in orthologous regions rather than repeats or spurious hits.
Our multiple alignments of the three genomes have presented novel opportunities for generating biological insights. For example, sites that are present in mouse and rat but absent in human, as judged by the multiple alignment, constitute a novel type of data set for genome-wide estimates of neutral rates of evolution at high local resolution (Cooper et al. 2004; Rat Genome Sequencing Project Consortium 2004); this data set complements the annotation-dependent sites that have traditionally been used for such estimates; namely, ancient repeats and synonymous sites. From the complementary data set, sites present and aligned in all three genomes, genome-wide estimates of the prevalence and rate of evolution of constrained, and presumably functional, elements were obtained.
For exploration of such conserved regions among the three genomes (and additional future genomes), we have developed the Multi-VISTA browser, a user-friendly visualization approach for exploring conserved regions among multiple genomes. This browser provides its users with an interactive environment for analyzing the alignments and patterns of conservation of the three genomes together with related annotation. The browser should be a valuable resource for biologists interested in whole-genome analysis as well as for those interested in the investigation of particular genes or genomic regions.
RESULTS
Overview of Strategy for Multiple Alignment of the Human, Mouse, and Rat Genomes
In our multiple-genome alignment pipeline, we combine the pairwise genome alignment method of Couronne et al. (2003) with the progressive alignment technique that is usually employed by multiple-alignment methods (Thompson et al. 1994). We use LAGAN (Brudno et al. 2003a) as our global aligner, with species-specific parameters.
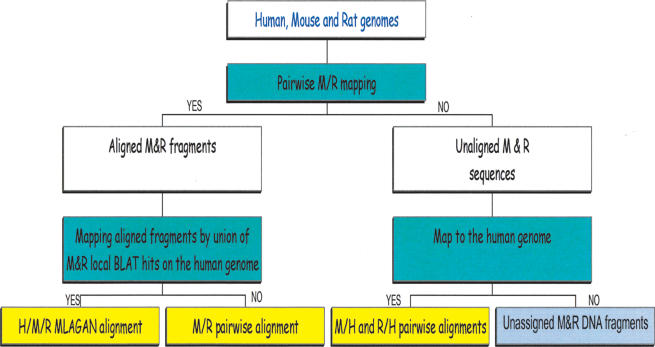
First, the mouse and rat genomes are aligned using the BLAT program (Kent 2002) for approximate mapping, followed by LAGAN global alignment of selected regions (Fig. 1). This step results in a set of mouse-rat “multi-contigs” (global alignments of rat contigs and mouse genomic sequence) as well as the remaining unaligned sequences. Second, the multi-contigs are aligned to human, using the union of all available BLAT local alignments from mouse to human and from rat to human; mouse or rat sequences that could not be aligned to the other rodent are also aligned to human. Using both mouse and rat BLAT alignments to align mouse-rat multi-contigs to human allows us to predict more accurately the ortholog of each multi-contig in the human genome: Only 0.8% (∼2 Mbp) of the rat genome and 7% of the rat contigs were mapped to multiple areas in the human genome, compared with 4.4% of the genome and 32% of the contigs in the pairwise rat/human alignment using the original technique of Couronne et al. (2003).
Figure 1.
General scheme of the method. White boxes show original and intermediate data; green boxes, mapping/alignment steps; and yellow and grey boxes, resulting data.
Because of the importance of alignment parameters to the final quality of the alignment, we have modified LAGAN to use substitution matrices derived specifically for the human, mouse, and rat genomes (Chiaramonte et al. 2002; Blanchette et al. 2004). Because no systematic method of estimating gap penalties for particular genomic sequences is known, these penalties are usually generated empirically (Vingron and Waterman 1994). We analyzed the distribution of insertions and deletions between the human and rodent lineages and selected gap penalties that offered the best tradeoff between accurate alignment of areas with microinsertions and microdeletions and areas in which transposable elements were inserted into one of the genomes.
Using the above method, we generated 11,235 areas of three-way alignments, 74% of which are longer then 200 Kbp in the human sequence. We have verified the quality of the alignments using two different methods. First, we determined the percentage of whole genomes and protein-coding exons that is covered by high-scoring subalignments in our three-way alignments. Second, we compared our alignments with a syntenic map that we generated independently, based on gene predictions, to verify that the alignments correctly map orthologs and that there are few extraneous hits.
Exon-Based Map of Conserved Synteny Among the Three Genomes
Because most gene-prediction programs demonstrate higher accuracy in predicting exons than in predicting entire genes, we built a three-way synteny map based on chains of Fgenesh++-predicted (Solovyev 2002) exons, rather than whole genes. We initially built human/mouse and human/rat pairwise maps, and then resolved them into a single three-way map for human, mouse, and rat. During the construction of pairwise maps, chains of exons are defined as sets of not less than 10 predicted exons, in the same order in each of the two genomes, where at least 70% of the exons have homologs in the other genome (found with BLASTP [Altschul et al. 1997] program). This method requires just a sequential order of similar exons and is expected to be robust with respect to misprediction of gene boundaries, absence of some exons, and misprediction of exon ends. Pairwise syntenic maps are merged into a three-way synteny map by selecting a single genome as a base and merging overlapping parts of the pairwise maps.
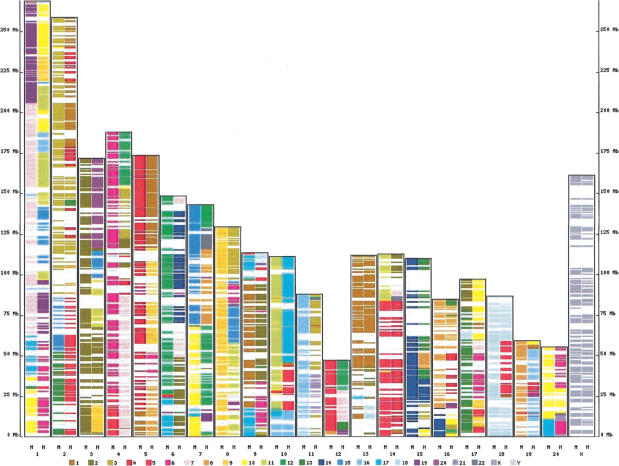
The resulting map has a total of 4497 three-way synteny segments. The rat-based view of the three-way synteny map is presented in Figure 2. Among the 4497 segments, the mouse segment is absent in 191 cases (4.2%), and the rat segment in 315 cases (7%). The total length of three-way synteny segments in the human genome was 674 Mb, with average segment length of 150 kb. These segments are further extended into larger blocks by merging those that are within 5 Mb of each other in every genome. Using this procedure, we find 494 synteny blocks shared among all three genomes (the mouse was absent in six blocks and the rat was absent in seven blocks). The total length of three-way (human-mouse-rat) synteny blocks was 2351 Mb, with an average block length of 4.76Mb.
Figure 2.
Exon-based map of conserved synteny between the rat, human, and mouse genomes. Each rat chromosome (presented along the x-axis) contains two columns, colored according to conserved synteny with chromosomes of the human and mouse genomes. Chromosome color scheme is shown at the bottom.
Evaluation of the Quality of Three-Way Alignments
Agreement Between Alignments and the Exon-Based Map
The multiple alignment of the three genomes and the predicted exon-based synteny map produce complementary, independent data sets that can be used to evaluate the accuracy of both methods. High correlation between these results indicates that overall, the syntenic maps are accurate. To test for this we compared the alignments generated by the automatic alignment pipeline with the exon-based synteny map. A syntenic block and an alignment were considered matching if they overlapped, regardless of a strand or percentage overlap, beause of the presence of small local rearrangements (Brudno et al. 2003b) in the genomes.
The longer alignments (>100 kb in human) exhibited greater than 97% agreement between the two maps, but for very short alignments (1-10 kb), the correlation dropped to 13%. Overall, 87.4% of all alignments and 96.5% of the human nucleotides within alignments lie in regions in which the multiple alignment and the predicted exon-based synteny map agree (see Table 1). Short alignments tend to agree less often with the exon-based map, reflecting our inability to accurately assign short alignments to corresponding synteny regions. This can be caused by several reasons, including the occurrence of multiple gene families and segmental duplications in eukaryotic genomes. In addition, 1636 alignments of total length 305 Mb in human did not overlap a syntenic block in any of the three genomes. This is largely to the result of the coarse nature of the exon-based synteny map, as it does not cover either areas that fall on the borders of regions of conserved synteny or areas that correspond to gene deserts.
Table 1.
Comparison of the Exon-Based Synteny Map and Three-Way Alignments
| Length of alignments (bp) | Total number of alignments | Number of alignments overlapping synteny maps | Agreement between methods (%) | Cumulative agreement (%)a | Total size (Mb) | Total size of disagreement (Mb) |
|---|---|---|---|---|---|---|
| >100 | 8080 | 7848 | 97.1 | 97.1 | 2243 | 59.0 |
| 50—100 | 432 | 301 | 69.7 | 95.7 | 33 | 9.4 |
| 10—50 | 474 | 157 | 33.1 | 92.4 | 12 | 7.3 |
| 1—10 | 613 | 80 | 13.1 | 87.4 | 1 | 0.9 |
| Total | 9599 | 8386 | 87.4 | n.a. | 2289 | 77 (3.4%) |
Cumulative numbers show percentage alignments in agreement with the synteny map for a particular range summed up with higher-length ranges, for example 92.4% agreement was achieved for all alignments longer than 10 kb
Genome Coverage by Three-Way Alignments
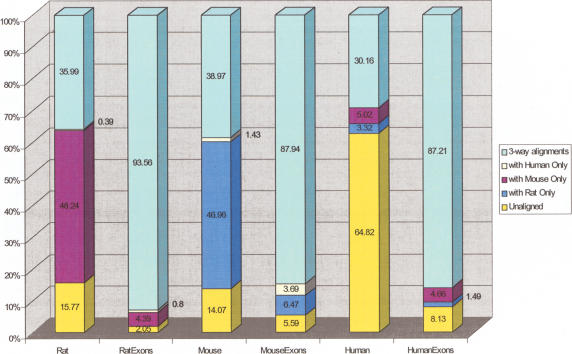
One way to evaluate alignment sensitivity is to compute the percentage of the base pairs of all genomes that are reliably aligned. We used the scoring techniques developed for comparison of the human and mouse genomes (Waterston et al. 2002; Schwartz et al. 2003). We computed overall coverage, as well as coverage of RefSeq exons. The results are summarized in Table 2 and Figure 3. Whereas the overall coverage for the human genome by the mouse (35.2%) is slightly lower then the result achieved using pairwise alignment with LAGAN (36.5%; Brudno et al. 2003b, Table 2, Column 1) it is noteworthy that this coverage was achieved with roughly four times fewer alignments (distinct syntenic segments) and 10% fewer bases aligned than provided by the pairwise method (11,235 vs. 39,163 alignments, 2.7 billion vs. 3 billion human nucleotides), indicating a higher specificity and a better syntenic map. The reduction in coverage is likely the result of both a slightly lower sensitivity of the three-way alignments as compared to the pairwise method and a decrease in nonhomologous, coincidental matches between the genomes.
Table 2.
Coverage of Various Genomic Features of the Human Genome by High-Scoring Subalignments From Within Our Global Alignments of Mouse and Rata
| Category | Mouse with pairwise LAGANb | Mouse from three-way alignments | Rat from three-way alignments | Mouse and rat from three-way alignments |
|---|---|---|---|---|
| Overall | 36.5 | 35.2 | 33.5 | 30.2 |
| Coding (CDX) | 93.2 | 91.9 | 88.7 | 87.2 |
| UTRs | 82.5 | 80.2 | 77.3 | 74.6 |
| Upstream 200 | 77.7 | 77.3 | 74.2 | 70.7 |
Using the criteria of Schwartz et al. (2003)
From Brudno et al. 2003b
Figure 3.
The chart shows the coverage of the three genomes and the RefSeq coding exons on the three genomes in our alignments using the thresholds from the Mouse genome comparisons (Waterston et al. 2002; Schwartz et al. 2003). The chart makes clear that although the bulk of the rat and mouse genomes can be confidently aligned to the other rodent, only a minority (35%-40%) is alignable to human. The percentage of each rodent genome that is aligned to human but not to the other rodent (0.4% of rat, 1.4% of mouse) is reflective of the fraction of the sequence missing in the other rodent.
The coverage of the human genome by both rodents was 1.5%-3.5% lower than that of the rat and 4.7%-6.6% lower than that of the mouse, depending on the category. Overall, the discrepancy between coverage numbers with one versus both rodents can be attributed to the fact that different areas of the two rodent genomes remain unsequenced, but a small percentage of the difference may be caused by regions that are undergoing faster evolution in one rodent than in the other (Yap and Pachter 2004). The difference is smallest for gene-coding regions, in which paralogous genes from the same syntenic area can be aligned instead of unsequenced areas of the genome. It is also possible to compute the fraction of each rodent genome that is missing in the other (because it was unsequenced or because of deletion of large segments) by comparing the fraction of each rodent genome aligned to the outgroup (human) but not to the other rodent (Fig. 3). One can observe that 36% of the rat and 40% of the mouse genomes are aligned to human. As 0.4% of rat is aligned to human and not mouse and 1.4% of mouse is aligned to human and not rat, it is possible to conclude that ∼1.1% (0.4 × [100/36]) of the mouse genome and ∼3.5% of the rat genome are missing in the other rodent.
Multi-VISTA Browser
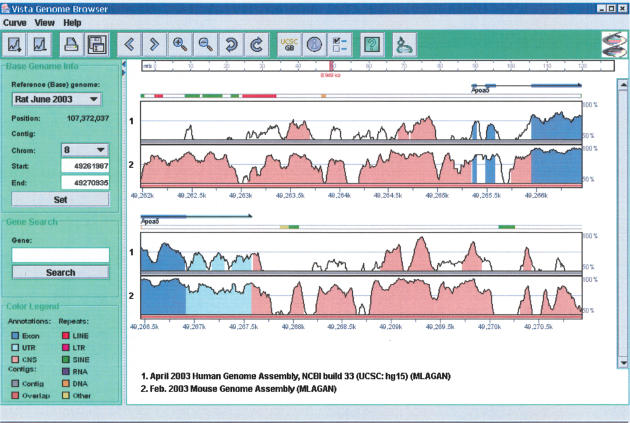
To visualize the results of comparative sequence analysis of multiple genomes in the VISTA format (Dubchak et al. 2000; Mayor et al. 2000), we have developed the Multiple VISTA Browser, a new tool that presents a logical extension of the VISTA browser (Couronne et al. 2003). It can be accessed at http://pipeline.lbl.gov. The Multi-VISTA Browser displays human-mouse-rat multiple alignments on the scale of whole chromosomes, along with annotations. The user may select any of the three genomes as the reference and display the level of conservation between this reference and the sequences of the other two species in a particular interval. The user also has the option of browsing and retrieving alignments, annotation, and pattern of conservation for a specific region of interest. Figure 4 shows the genomic region containing the APOA5 gene on rat chromosome 8. It is clearly seen that this region contains significant areas of rat/human conservation both upstream and downstream from APOA5. Rat and mouse sequences are highly conserved in exon, untranslated region, and intergenic intervals.
Figure 4.
APOA5 region (chr8:49261987-49270935) on the Rat Genome (June 2003, RGSC version 3.1, University of California, Santa Cruz, version rn3) displayed by Multi-VISTA Genome Browser (VGB2.0) accessible through the gateway at http://pipeline.lbl.gov. Conservation plots for human/rat (top plot) and mouse/rat (bottom plot) are displayed on the scale of the rat sequence. Conserved regions above the level of 70%/100 bp are highlighted under the curve, with red indicating a conserved noncoding region; blue, a conserved exon; and turquoise, untranslated region.
DISCUSSION
In this study we aligned the human, mouse, and rat genomes using a progressive local/global technique with the LAGAN multiple alignment program. The computational complexity of whole-genome multiple alignment makes this a computationally interesting problem, whereas the availability of a high-quality alignment between the three genomes should be an invaluable resource for biologists interested in evolution, regulation, and many other aspects of genetics.
Our results indicate that the alignment has high sensitivity and specificity. By comparing our alignment to an independently generated map of protein synteny between the genomes, we conclude that 97% of alignments with a human sequence >100 kbp, and 87% of all alignments, agree with the map. The difference between these numbers can be explained by the lower accuracy of both alignment and synteny map generation when dealing with very short regions of conserved synteny. However, the fact that only 3.4% of the human base pairs in the whole genome alignment are within such nonmatching regions indicates a high overall quality of the synteny map inferred from the alignment. High level of coverage of RefSeq coding exons in all three genomes presents a proof of sensitivity of the method, whereas unambiguous mapping of the vast majority of the rat and mouse contigs shows its specificity. Multiple alignment between human and rodents has increased specificity compared with pairwise alignment between human and a single rodent. Our alignment has allowed for novel biological analyses of the three genomes (Cooper et al. 2004), and we are hopeful that it will become a valuable resource for other researchers.
One drawback of global alignments is their inability to deal with small rearrangement events. A previous study has suggested that as much as 2% of the gene-coding regions of the human genome may have undergone some local rearrangement events since the divergence between human and the rodents (Brudno et al. 2003b), and the local/global approach often is not able to cope with these events. Glocal alignment approaches (Brudno et al. 2003b; Kent et al. 2003) are novel methods for alignment of sequences that have undergone these events while filtering out the spurious matches that are common when employing local aligners. Multiple glocal alignment is a promising area of future research that should allow us to further improve the quality of alignments created by the local/global technique.
Finally, we want to emphasize that additional genomes will help to verify the quality of existing alignments and provide the biologists with additional comparative information with which to judge the evolutionary importance of a region. Recent efforts to analyze multiple alignments and determine the most valuable genomes to sequence in order to improve our ability to determine constrained elements (Cooper et al. 2003) demonstrate that adding several other mammalian genomes will possibly allow us to locate constraints at the individual base pair level. The availability of these genomes would make possible the use of comparative sequence analysis in new areas, such as the determination of individual binding sites.
METHODS
Sequence Data
For the alignment, we used the following versions of genome assemblies: April 2003 Human (National Center for Biotechnology Information build 33, University of California, Santa Cruz [UCSC] version hg15), Februrary 2003 Mouse (UCSC version mm3), and June 2003 Rat Genome Sequencing Project Consortium release 3.1 (UCSC version rn3). All assemblies with associated tracks were downloaded from the UCSC Web site (http://genome.ucsc.edu/). RepeatMasker tables were used during the alignment stage, and RefSeq tables were used for subsequent analysis and visualization.
Progressive Alignment Strategy
The supercontigs comprising the rat genome are divided into regions roughly 250 kb in size in such a manner as not to split contigs of the assembly. These regions are mapped to the mouse, using BLAT. Each resulting local alignment receives a Needleman-Wunsch score (match = +100, mismatch = -70, gap open = -400, gap continue = -20 × log [length gap]). All BLAT local alignments, at most L bases apart (where L is the length of the contig), are grouped together. For groups shorter than L/4, the regions are then extended out by min(50 kb, L/2 - G), where G is the length of the group. For groups with G greater than L/4, the regions are extended out by min(50 kb, L/4). The score of each group is the sum of scores of all local alignments in it. In this manner, each region from rat is mapped to zero or more regions in the mouse. Groups are filtered out if they had a score <70,000, or if <70% of the maximum score of any group associated to the same rat region. The remaining groups define areas of potential synteny that are aligned with LAGAN. We use species-specific substitution matrices (Chiaramonte et al. 2002; Blanchette et al. 2004), with empirically derived gap penalties of -500 for mouse/rat and -800 for human/rodent. LAGAN is run with the fastreject option. This option requires that at least one high-scoring local alignment is found between the two sequences, and clips from each sequence the beginning and ending portions that are more than a cutoff away from an anchored local alignment. This cutoff depends on the quality of the anchor.
The alignments are then clipped on the sides by the scorealign tool included with LAGAN. The resulting alignments are stored as multi-contigs. These multi-contigs, as well as any sequence in the mouse or rat genomes that was not aligned to the other rodent, are mapped to the human genome using the BLAT hits from all available rodent sequences. Here, we use the same thresholds as for the mouse/rat pairwise alignment. The human region is aligned to the mouse/rat contig using LAGAN, and clipped using scorealign (see below).
It is worth noting that the original pipeline used much looser thresholds for BLAT placement between the mouse and rat genomes (groups with score >30% of the maximum were kept, without an absolute threshold). Because the mouse and rat genomes are much closer in evolutionary distance, we are able to use the tighter thresholds in the initial pairwise step without a significant drop in sensitivity, whereas the use of both mouse and rat BLAT hits facilitates placement of the multi-contigs on the human genome, likewise enabling higher thresholds. The tightening of the parameters also speeds up the alignment pipeline by a factor of 10 (15 hours instead of 6 days) and halves the size of the resulting alignments (7 gbytes instead of 14 gbytes) without a noticeable reduction of coverage of genomic features.
The scorealign tool is based on a Hidden Markov Model for finding conserved regions within an alignment without an arbitrary cutoff of percentage similarity within a fixed window size. Given a pairwise alignment and conservation cutoff k%, scorealign returns exactly those regions that are more likely to have resulted from a k% conservation model rather than the background (25%) conservation model. Given a multiple alignment, scorealign performs all pairwise analyses and returns the intersection (or, optionally, the union) of detected pairwise regions. Scorealign can also clip an alignment by returning only the portion that falls between the first and last conserved regions.
Gene Annotation Using Fgenesh++
Synteny maps between the genomes are based on gene predictions for human, mouse, and rat genomes built by Fgenesh++ software developed by Softberry Inc. (Solovyev 2002). Fgenesh++ is among the most accurate gene finders (Solovyev 2002) and is run in a fully automated genome annotation pipeline that includes the following steps:
RefSeq mRNAs are mapped onto the genome by the EST_MAP program. Genomic sequences with mapped mRNAs are excluded from further gene prediction.
Ab initio Fgenesh gene prediction is run on the rest of genome.
Protein homologs of all predicted genes are searched for in the NR database with BLAST (Altschul et al. 1997).
Fgenesh+ gene prediction is conducted on sequences with protein homology.
A second run of ab initio gene prediction is run in regions without predictions from stages 1 and 4.
Fgenesh gene predictions are run in large introns of known and predicted genes.
The Fgenesh++ software consists of a set of Perl scripts and three basic programs: (1) Fgenesh, a Hidden Markov Model-based ab initio gene prediction program; (2) Fgenesh+, which combines protein homology with ab initio prediction; and (3) EST_MAP, which rapidly maps a set of mRNAs/expressed sequence tags to genomic sequence, taking into account statistical features of splice sites. Fgenesh++ was applied to the three genomes after masking interspersed (but not low-complexity) repeats.
Finding Genomic Synteny, Using Chains of Coding Exons
To find chains of exons with conserved synteny between two genomes, we apply the following algorithm:
Compile a set of nonredundant, nonoverlapping exons with at least 10 amino acids in ascending order along each chromosome.
Determine the similarity for each exon in a chromosome of one organism against a set of exons from a chromosome of the compared organism by alignment with the BLASTP aligner (Altschul et al. 1997). The closest homolog for each exon is retained. Subdivide this data into sets of homologous exon chains, where each chain consists of at least of 10 exons, with 70% of all exons in a chain having a homolog on the chromosome of the compared organism.
Two chains of exons are defined to be a conserved syntenic segment if they share at least five pairs of exons with bidirectional hits.
The synteny segments are extended into synteny blocks by concatenating adjacent segments whenever the distances are smaller than a threshold length. We tested thresholds between 1 and 10 Mbp and found that the results are robust to this parameter. The reported results are for the cutoff of 5 Mbp. Some statistics about the three-way synteny map can be found in Table 3. More details of this method, as well as the two- and three-organism synteny maps, can be found at http://www.softberry.com/berry.phtml?topic=human-mouse-rat.
Table 3.
Summary of the Three-Way Synteny Map Based on FGENESH++ Gene Model Predictions in the Three Genomes
| Human | Mouse | Rat | |
|---|---|---|---|
| Total number of gene models | 39,788 | 42,043 | 40,347 |
| Total number of exons | 182,487 | 189,664 | 197,983 |
| Exons in three-way synteny map | 163,345 | 170,760 | 174,812 |
| Percentage of exons in three-way map | 89.5 | 90.0 | 88.3 |
Implementation
Database
The automated pipeline is built on a MySQL database platform selected for its compatibility with major sources of annotation data, such as ENSEMBL (Hubbard et al. 2002) and the UCSC Human Genome Browser (Kent et al. 2002). The database contains the human, mouse, and rat sequences; their annotation; and the alignment data.
Software
The pipeline software is a combination of Perl and C programs. The scheduler gets control data from the database, builds a queue of jobs, and dispatches them to a PC cluster for execution. The main program, running on each node of the cluster, processes individual sequences. A Perl library acts as an interface between the database and the above programs. The use of a separate library allows the programs to function independent of the database schema. The library also improves on the standard Perl MySQL database interface package by providing auto-reconnect functionality and improved error handling.
Data Visualization and Availability
Multi-VISTA Browser, accessible at http://pipeline.lbl.gov, is a Java 2 applet that can display multiple human-mouse-rat alignments, along with genome annotations, using any of the three species as a reference (coordinate) sequence. Its graphical user interface allows for selecting a region to display, scrolling back and forth along the chromosomes of the reference genome, zooming in and out of the region, searching for genes, defining cutoffs to color regions of high conservation, and many other functions. The program is linked with a text browser that provides additional information such as the underlying sequences, alignments, exact location of conserved elements on each genome, and other. The alignments of the three genomes can be downloaded in the eXtended Multi-FastA format from the main pipeline Web site at http://pipeline.lbl.gov/downloads.shtml.
Acknowledgments
We thank the Rat Genome Sequencing Project Consortium for the opportunity to work with the rat genome during the sequencing phases and in the subsequent analysis phase. The analysis group (especially Webb Miller, Ross Hardison, Lior Pachter, David Haussler, and members of their teams) deserves special thanks for crucial suggestions and fruitful discussions. We are grateful to Olivier Couronne for his work on the Berkeley Genome Pipeline; to Lila Tretikov, Michael Teplitsky, and Dmitriy Ryaboy for the development of VISTA Browser for multiple alignments; to Tigran Ishkhanov for evaluating the alignments; and to Chuong Do for designing scorealign and giving helpful suggestions on the manuscript. We are also grateful to the two anonymous reviewers, who made a number of helpful suggestions. The project was partially supported by the Program for Genomic Applications grant from the National Heart Lung and Blood Institute. This work was supported by the Director, Office of Science, of the U.S. Department of Energy under Contract No. DE-AC03-76SF00098. M.B. was supported by a National Science Foundation Graduate Fellowship, and G.C. is a Howard Hughes Medical Institute predoctoral fellow.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2067704.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette, M., Kent, W.J., Riemer, C., Elnitski, L., Smit, A.F.A., Roskin, K.M., Baertsch, R., Rosenbloom, K., Clawson, H., Green, E.D., et al. 2004. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Boffelli, D., McAuliffe, J., Ovcharenko, D., Lewis, K.D., Ovcharenko, I., Pachter, L., and Rubin, E.M. 2003. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science 299: 1391-1394. [DOI] [PubMed] [Google Scholar]
- Brudno, M., Do, C.B., Cooper, G.M., Kim, M.F., Davydov, E., Green, E.D., Sidow, A., Batzoglou, S., and NISC Comparative Sequencing Program. 2003a. LAGAN and Multi-LAGAN: Efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 13: 721-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brudno, M., Malde, S., Poliakov, A., Do, C.B., Couronne, O., Dubchak, I., and Batzoglou, S. 2003b. Glocal alignment: Finding rearrangements during alignment. Bioinformatics 19: 54i-62i. [DOI] [PubMed] [Google Scholar]
- Chiaramonte, F., Yap, V.B., and Miller, W. 2002. Scoring pairwise genomic sequence alignments. Pac. Symp. Biocomput. 115-126. [DOI] [PubMed]
- Cooper, G.M., Brudno, M., Green, E.D., Batzoglou, S., Sidow, A., and NISC Comparative Sequencing Program. 2003. Quantitative estimates of sequence divergence for comparative analyses of mammalian genomes. Genome Res. 13: 813-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, G.M., Brudno, M., Stone, E.A., Dubchak, I., Batzoglou, S., and Sidow, A. 2004. Characterization of evolutionary rates and constraints in three mammalian genomes Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Couronne, O., Poliakov, A., Bray, N., Ishkhanov, T., Ryaboy, D., Rubin, E.M., Pachter, L., and Dubchak, I. 2003. Strategies and tools for whole genome alignments. Genome Res. 13: 73-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubchak, I., Brudno, M., Pachter, L.S., Loots, G.G., Mayor, C., Rubin, E.M., and Frazer, K.A. 2000. Active conservation of noncoding sequences revealed by 3-way species comparisons. Genome Res. 10: 1304-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottgens, B., Barton, L.M., Chapman, M.A., Sinclair, A.M., Knudsen, B., Grafham, D., Gilbert, J.G., Rogers, J., Bentley, D.R., and Green, A.R. 2002. Transcriptional regulation of the stem cell leukemia gene (SCL)-comparative analysis of five vertebrate SCL loci. Genome Res. 12: 749-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohl, M., Kurtz, S., and Ohlebusch, E. 2002. Efficient multiple genome alignment. Bioinformatics 18: S312-S320. [DOI] [PubMed] [Google Scholar]
- Hubbard, T., Barker, D., Birney, E., Cameron, G., Chen, Y., Clark, L., Cox, T., Cuff, J., Curwen, V., Down, T., et al. 2002. The Ensembl genome database project. Nucleic Acids Res. 30: 38-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis, M., Patterson, N., Endrizzi, M., Birren, B. and Lander, E.S. 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423: 241-254. [DOI] [PubMed] [Google Scholar]
- Kent, W.J. 2002. BLAT—The BLAST-like alignment tool. Genome Res. 12: 656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, W.J., Sugnet, C.W., Furey, T.S., Roskin, K.M., Pringle, T.H., Alan, M., Zahler, A.M., and Haussler, D. 2002. The Human Genome Browser at UCSC. Genome Res. 6: 996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, W.J., Baertsch, R., Hinrichs, A., Miller, W., and Haussler, D. 2003. Evolution's cauldron: Duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl. Acad. Sci. 100: 11484-11489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, B., Tromp, J., and Li, M. 2002. PatternHunter: Faster and more sensitive homology search. Bioinformatics 18: 440-445. [DOI] [PubMed] [Google Scholar]
- Mayor, C., Brudno, M., Schwartz, J.R., Poliakov, A., Rubin, E.M., Frazer, K.A., Pachter, L., and Dubchak, I. 2000. VISTA: Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16: 1046-1047. [DOI] [PubMed] [Google Scholar]
- Rat Genome Sequencing Project Consortium. 2004. Genome sequence of the Brown Norway Rat yields insights into mammalian evolution. Nature (in press). [DOI] [PubMed]
- Schwartz, S., Kent, W.J., Smit, A., Zhang, Z., Baertsch, R., Hardison, R.C., Haussler, D., and Miller, W. 2003. Human-mouse alignments with BLASTZ. Genome Res. 13: 103-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, A.L., Stone, E.A., and Sidow, A. 2002. Inference of functional regions in proteins by quantification of evolutionary constraints. Proc. Natl. Acad. Sci. 99: 2912-2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovyev, V.V. 2002. Finding genes by computer: Probabilistic and discriminative approaches. In Current topics in computational biology (eds. T. Jiang et al.), pp. 365-401. MIT Press, Cambridge, Massachusetts.
- Sumiyama, K., Kim, C.B., and Ruddle, F.H. 2001. An efficient cis-element discovery method using multiple sequence comparisons based on evolutionary relationships. Genomics 71: 260-262. [DOI] [PubMed] [Google Scholar]
- Thomas, J.W., Touchman, J.W., Blakesley, R.W., Bouffard, G.G., Beckstrom-Sternberg, S.M., Margulies, E.H., Blanchette, M., Siepel, A.C., Thomas, P.J., McDowell, J.C., et al. 2003. Comparative analyses of multi-species sequences from targeted genomic regions. Nature 424: 788-793. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vingron, M. and Waterman, M.S. 1994. Sequence alignment and penalty choice. Review of concepts, case studies and implications. J. Mol. Biol. 235: 1-12. [DOI] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- Yap, V.B. and Pachter, L. 2004. Identification of evolutionary hotspots in the rodent genomes. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
WEB SITE REFERENCES
- http://pipeline.lbl.gov/; Comparative analysis pipeline gateway at Lawrence Berkeley National Laboratory.
- http://lagan.stanford.edu/; LAGAN Toolkit Web site.
- http://genome.ucsc.edu/; University of California, Santa Cruz, Web site from which the human, mouse, and rat genome assemblies used in this study were downloaded.
- http://pipeline.lbl.gov/downloads.shtml; Download page for whole genome alignments and other materials.
- http://www.softberry.com/berry.phtml?topic=human-mouse-rat; Three-way gene-based synteny.