Abstract
We define a “threaded blockset,” which is a novel generalization of the classic notion of a multiple alignment. A new computer program called TBA (for “threaded blockset aligner”) builds a threaded blockset under the assumption that all matching segments occur in the same order and orientation in the given sequences; inversions and duplications are not addressed. TBA is designed to be appropriate for aligning many, but by no means all, megabase-sized regions of multiple mammalian genomes. The output of TBA can be projected onto any genome chosen as a reference, thus guaranteeing that different projections present consistent predictions of which genomic positions are orthologous. This capability is illustrated using a new visualization tool to view TBA-generated alignments of vertebrate Hox clusters from both the mammalian and fish perspectives. Experimental evaluation of alignment quality, using a program that simulates evolutionary change in genomic sequences, indicates that TBA is more accurate than earlier programs. To perform the dynamic-programming alignment step, TBA runs a stand-alone program called MULTIZ, which can be used to align highly rearranged or incompletely sequenced genomes. We describe our use of MULTIZ to produce the whole-genome multiple alignments at the Santa Cruz Genome Browser.
The availability of large amounts of genomic sequence data from mammals (Lander et al. 2001; Venter et al. 2001; Waterston et al. 2002; Thomas et al. 2003) and other vertebrates (Aparicio et al. 2002) is transforming major portions of biomedical science (Collins et al. 2003). A crucial step in extracting the wealth of information hidden in these data is to align the sequences, a process that identifies segments that remain similar among the species; these segments may well be under evolutionary constraints and hence are strong candidates for performing an important function. Although some computer programs designed primarily for aligning multiple protein sequences (Thompson et al. 1994; Morgenstern et al. 1999) can be applied to DNA sequence, the belief that DNA presents distinct challenges and opportunities has motivated the development of several multiple alignment programs designed specifically for genomic sequences (Hardison et al. 1994; Bray and Pachter 2003; Brudno et al. 2003; Schwartz et al. 2003b).
To align genomes, the typical approach is to create “reference sequence” alignments, that is, a sequence is fixed as the reference to which all other sequences are compared. Benefits of this approach include simplicity of the aligning programs and of the software that displays alignments. However, a drawback is that regions conserved in a subset of the species, but absent from the reference, are not identified. Another shortcoming of the reference sequence approach is that alignments generated with different reference sequences may be inconsistent. For instance, two genomic positions that are aligned to each other using one reference sequence might be aligned to different positions when another reference sequence is chosen.
To circumvent those deficiencies, we propose that programs produce a set of “blocks” (i.e., local alignments of some or all of the given sequence), in which each position in the given sequences appears precisely once. Any detected match among some or all of the sequences is represented among the blocks, and mutually consistent reference-sequence alignments can be extracted at will. Some vocabulary about alignments will be introduced so that we can more precisely present these informal ideas.
Once the appropriate conceptual underpinnings have been developed, we will describe a new program, called TBA (threaded blockset aligner), which produces such a set of blocks under the assumption that the matching regions occur in the same order and orientation in all species. To illustrate the extraction of several reference-sequence alignments from the same set of blocks, we use a new tool to visualize TBA-generated alignments of vertebrate Hox clusters.
A major component of this paper is our evaluation of software accuracy, which is particularly difficult with programs intended to align noncoding DNA. Here, we use sets of artificial sequences that have been derived through a careful simulation of evolutionary mutations. That way, we “know what the right answer is” and hence can quantify alignment accuracy.
TBA was implemented as a set of independently executing programs. MULTIZ, the component that does the dynamic-programming alignment step, can be used even with sequences that are fragmented or have rearrangements such as inversions and duplications. We use MULTIZ to build whole-genome alignments for the UCSC Genome Browser (Kent et al. 2002), as we describe.
RESULTS
Explanation of Key Terms
Biologists have long been familiar with sequence alignments— rectangular arrays of letters (representing either nucleotides or amino acids) and dashes (indicating insertions or deletions). Moreover, they typically know that sometimes one wants a global alignment (which entirely covers the given sequences), sometimes one prefers a local alignment (which covers only part of the sequences), and frequently the real goal is a set of local alignments. However, even this breakdown is insufficient to describe what TBA does; we need a more refined vocabulary. We use the neutral word “block” in preference to “alignment” for one of the objects computed by our programs; “alignment” means different things to different people.
For what follows, suppose we are given a fixed set of genomic DNA sequences, called the original sequences. A block is a rectangular array of symbols such that removing dashes from any row produces a run of one or more consecutive positions in one of the original sequences or their reverse complements. It is important to note for some of our later discussion that we permit a block to have only one row. However, we require that no column of a block consists entirely of dashes. For brevity, we call a set of such blocks a blockset.
A “ref-blockset” consists of a blockset in which every block has a designated row, all of which come from the same original sequence, called the reference for that ref-blockset. In addition, we require that each position in the reference appear in precisely one block. Without loss of generality, we can assume that the reference row of each block has a positive (not reversed) orientation in the original sequence, because otherwise we can replace each row of the block with its reverse complement. We will use the notation, for example, “human-ref blockset” for a ref-blockset in which a segment of a human chromosome is the reference.
A given sequence, S, is said to “thread” a blockset if every position in S appears precisely once in some block of the blockset. Thus, a ref-blockset is threaded by the reference sequence. If a blockset is threaded by each of the original sequences, we call it a threaded blockset.
Given a threaded blockset, it is straightforward to generate an S-ref blockset for any original sequence S, an operation that we call “projecting onto S.” One merely picks the blocks having a row from S and orders them according to S. In practice, we move the reference row to the top of each block. See Figure 1 for an example. A critical property of projections is that any two ref-blocksets generated by projection from the same threaded blockset are consistent. More precisely, if position x of sequence X aligns to position y of sequence Y in one projection and to position z of Y in another projection, then y = z.
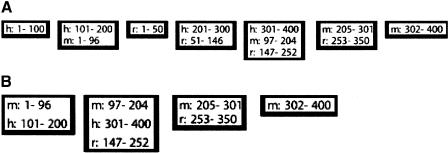
Figure 1.
(A) Blocks (alignments) of a hypothetical threaded blockset for sequences h (400 bp), m (400 bp) and r (350 bp). Only the range of positions in each alignment is given. (B) Projection of the threaded blockset onto m.
The concept of “threaded blockset” readily accommodates complex evolutionary operations such as inversions and duplications. For a concrete example, consider a blockset for the chloroplast genomes of Arabidopsis thaliana and evening primrose (Oenothera elata), shown in Figure 2.
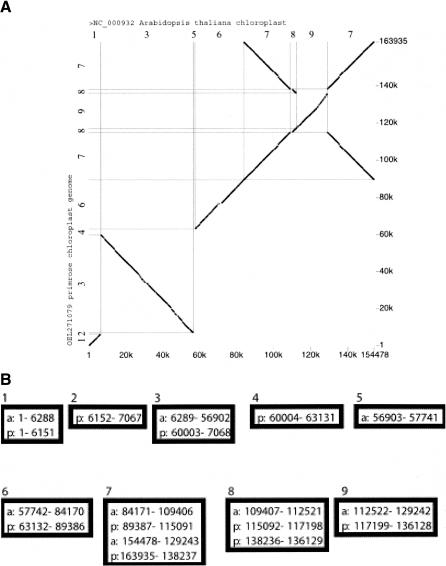
Figure 2.
(A) Alignments between the chloroplast genomes of Arabidopsis thaliana and Oenothera elata (evening primrose). Lines running from lower left to upper right indicate positions of matches on the forward strand (relative to the GenBank entries, NC_000932 and OEL271079, respectively), and lines running from upper left to lower right indicate matches in reverse complement. The alignments were computed and displayed by programs used by the PipMaker Web server (Schwartz et al. 2000). (B) Blocks of a threaded blockset for the chloroplast genomes of Arabidopsis and evening primrose.
The pairwise chloroplast blockset can be represented as a threaded blockset of blocks having 1 to 4 rows, which makes explicit the inversions and duplications. In Figure 2A, horizontal and vertical lines, together with the numbers along the axes, indicate the boundaries of each row in each block. The blockset is shown in Figure 2B with precise start and end positions. Blocks 1, 6, and 9 are the usual pairwise matches in the same orientation. Block 2 has just one row, namely, positions 6152–7067 of the primrose sequence. Block 3 is a pairwise match on the reverse strand (note the decreasing numbers for primrose). Block 7 is a four-way match, involving a multigene inverted repeat (a normal feature of chloroplast genomes) in each species. Block 8 is a threeway match involving the ndhF (NADH-plastoquinone oxidoreductase subunit 5) gene, which is contained in the inverted repeat in primrose but not in Arabidopsis. The projection of these blocks onto each of the species is indicated by the sequence of numbers along the corresponding axis in Figure 2A. Thus, blocks 7 and 8 each appear twice in the projection onto the primrose sequence (once in each orientation); the projection can be thought of as a multiple alignment in which the first row is the primrose sequence (with interspersed gap characters), and which contains between 1 and 4 rows, depending on the region under consideration.
Threaded Blockset Aligner
The current version of the TBA program can automatically produce only a limited kind of threaded blockset. In informal terms, it does not accommodate inversions or duplications, and it is restricted to finding matches that occur in the same order and orientation in the given sequences. (However, a match need not involve every sequence.) A classical global alignment can be obtained by piecing together the blocks of the blockset.
More formally, in addition to being threaded, a blockset currently produced by TBA must have the property that no block in it contains a row on the reverse strand of an original sequence or two rows from the same original sequence, and it must be partially ordered in the following sense. We say that block A “precedes” block B if there is a sequence S such that A and B have rows from S where A's row precedes B's row in S. A threaded blockset is “partially ordered” if the relation “A precedes B” is a (strict) partial order, that is, the blockset does not contain a list B1, B2, B3,..., Bn of one or more blocks where B1 precedes B2, B2 precedes B3, and so on, and Bn precedes B1. Extensions of TBA to automatically produce the richer class of blocksets obtained by dropping these requirements remain as future work.
Applying TBA to Vertebrate Hox Clusters
We applied TBA to approximately the same sequences from the HoxA region that were studied in Santini et al. (2003), four from mammals and four from fish. When the threaded blockset computed by TBA is projected onto tilapia, many noncoding matches with Fugu are apparent, as are a few putatively noncoding matches with mammals (Fig. 3A). We have written an interactive visualization program called Gmaj that facilitates the process of projecting and exploring TBA blocksets, and used it in this analysis. When Gmaj projects the same blockset onto human (Fig. 3B), most of the tilapia–Fugu matches disappear, to be replaced by extensive matches among mammals. Also, the rich annotation of the human sequence is then available, which provides a clue about the match of interest.
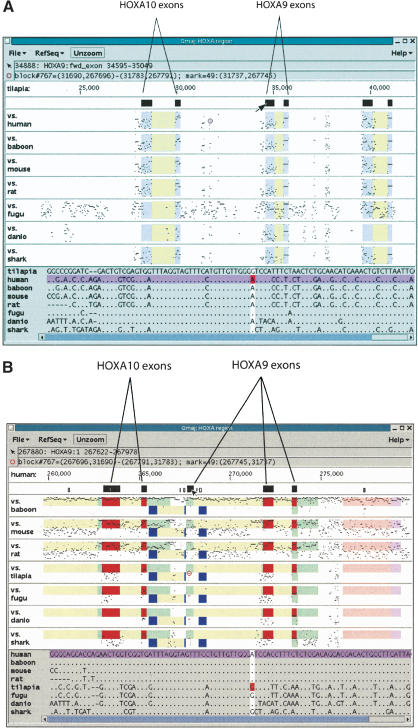
Figure 3.
A threaded blockset for vertebrate HoxA regions, displayed in our interactive blockset viewer Gmaj. (A) The red circle marks a position of interest where the tilapia reference sequence aligns with human. The block containing this position is highlighted in red in all of the alignment panels. Color underlays are blue for exons in the reference sequence and yellow for introns, and the exons are also represented as icons above the alignments. At the top of the Gmaj window, two status lines describe the positions of the mouse pointer and the red circle, respectively. Individual nucleotides for the selected block are displayed in the bottom pane, with the marked position highlighted. (B) The same region projected onto the human sequence. The underlays for human include (green) for EST evidence, (dark blue) for antisense RNA, and (red) for coding sequences. The conserved element from A is part of an alternative 5′-end identified by homology to a human EST from TIGR.
When using Gmaj to switch back and forth between different projections, the user can rest assured that the views, though quite different, are consistent. The matches in a given projection are merely a subset from a comprehensive collection of matches—the threaded blockset.
Evaluation of Alignment Accuracy
The accuracy of an alignment algorithm can be estimated in two ways. First, one can run the aligner on a set of biological sequences in which certain features are known a priori to be orthologous (e.g., coding exons and regulatory modules), and then ask whether these regions have been correctly aligned. This approach has the advantage of evaluating the aligner directly on the type of sequences where it will be put to use. However, it suffers two major drawbacks: (1) most known sets of orthologous features have been identified using some aligner (which implies some circularity), and they are generally so well conserved that most alignment methods will align them correctly; and (2) this approach does not provide any information about the accuracy of the alignment in the regions between these known features.
The alternative approach, which is used in this study, is to use simulations. Here, we simulate sequence evolution, starting with some ancestral sequence and performing mutations along the branches of a predetermined phylogenetic tree until sequences at the leaves of the tree are obtained. These leaf sequences are the ones that are aligned. Because we generated the sequences ourselves, we have access to their phylogenetically correct alignment (that which aligns orthologous bases). We can therefore compare the reconstructed alignment to the true alignment and measure its accuracy. The value of such an approach depends completely on the realism of the simulations; the simulated sequences should look as much as possible like actual biological sequences. We thus took great care to develop a procedure that simulates the evolution of neutral regions of DNA, including context-dependent substitutions (Siepel and Haussler 2003), rates and sizes of insertions and deletions empirically derived from actual data (Kent et al. 2003), and insertion of actual repetitive elements from various families (see Supplemental material). We simulate sequences of length ∼50 kb. It should be noted that the simulation procedure was originally developed by one of us for a completely different purpose than evaluating aligners (M. Blanchette, E. Green, W. Miller, and D. Haussler, in prep.) and that there were no interactions between the development of the MULTIZ and TBA programs and that of the evaluation procedure.
To compare the predicted alignment to the true alignment, we analyze how accurately each pair of species is aligned within the multiple alignment. We start by soft-repeat-masking the simulated sequences using RepeatMasker, and then feed them to each aligner to obtain a global multiple alignment. For species X and Y, we extract the pairwise induced alignments from both the predicted and true multiple alignments, removing positions where both sequences contain a gap. We then compute the agreement score between the two, defined as the fraction of positions of the predicted alignment that agree with the true alignment. There is agreement if the predicted alignment aligns the i-th nucleotide of X to the j-th nucleotide of Y and these two are aligned similarly in the true alignment, or if the i-th (respectively, j-th) nucleotide of species X (respectively, Y) is aligned to a gap in both alignments. Two identical alignments thus obtain an agreement score of 1, whereas two completely different alignments get a score of zero.
We used this methodology to evaluate the accuracy of TBA and to compare it with the latest versions of other multiple alignment programs previously published: CLUSTALW 1.83 (Thompson et al. 1994), DIALIGN 2 (Morgenstern 1999), DIALIGN 2 with CHAOS anchors (Brudno and Morgenstern 2002), MAVID (Bray and Pachter 2003), and MLAGAN (Brudno et al. 2003). Because the DIALIGN and CLUSTALW programs are too slow to be run on our 50-kb-long simulated sequences, we divide the sequences into ∼15 regions, choosing the region boundaries in each species using the true alignment. This should in theory give a small advantage to these two programs because it provides them with a set of anchors derived from the true alignment.
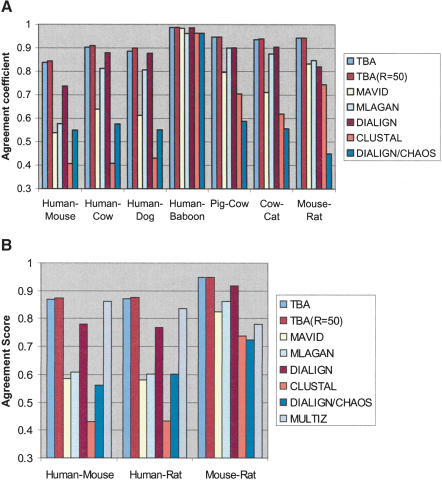
Figure 4A reports the accuracy of the alignment produced by each program for sets of nine orthologous sequences simulating nonfunctional regions of human, chimp, baboon, mouse, rat, cat, dog, cow, and pig sequence (Thomas et al. 2003). As expected, the accuracy of the induced pairwise alignments produced is better for pairs of closely related sequences than for more diverged ones, with human–mouse being the least accurate for most methods. Whereas all methods generally do well from the point of view of the human–primate induced alignments, TBA clearly and uniformly stands out for the more diverged pairs, with the human–rodent alignments being ∼84% more accurate. This represents about one-third of the base-by-base errors made by any of the other programs tested except DIALIGN.
Figure 4.
(A) Accuracy of the multiple alignments produced by different aligners on a set of nine simulated mammalian sequences of length ∼50 kb, as measured on the basis of the pairwise alignments induced by different pairs of species. The scores reported are the average of 50 simulation experiments. See the Methods section (Supplemental material) for an explanation of the R parameter. (B) Accuracy of the multiple alignments produced by different aligners on simulated human, mouse, and rat sequences of length ∼50 kb, as measured on the basis of the pairwise alignments induced by different pairs of species. The scores reported are the average of 50 simulation experiments.
When asked to produce a multiple alignment based only on human, mouse, and rat simulated sequences (Fig. 4B), the accuracy of many aligners on the three pairwise induced alignments seems to increase slightly, indicating that the presence of a larger number of species actually confuses most programs. This is an indication that there is still some room for improvements, as proper use of the information coming from other species should in theory lead to a more accurate alignment. We also used this set of simulations to evaluate the MULTIZ program (see below). MULTIZ produces a set of local multiple alignments that do not necessarily contain all bases of all sequences. To provide a consistent base for evaluation, we complete MULTIZ alignments by assuming that any base missing from the alignment is aligned to a gap in the two other species. This results in an accuracy almost as good as TBA's for human–mouse induced alignments. However, because the MULTIZ alignments are human-referenced, the quality of the induced mouse–rat alignment suffers. The human–rat alignment is also slightly worse than the human–mouse alignment because rat is aligned to human only through mouse.
Finally, it should be noted that a perfect agreement score of 1.0 may often be impossible to achieve, simply because a certain amount of information is lost during sequence evolution and cannot be recovered by any method. Still, obtaining a perfect agreement score is not necessarily required to solve perfectly other problems based on that alignment. For example, errors where a sequence AA should be aligned to A–but was instead aligned to –A are inconsequential when it comes to use the alignment to identify conserved regions (Margulies et al. 2003) or to infer ancestral sequences (M. Blanchette, E. Green, W. Miller, and D. Haussler, in prep.).
The method used so far to measure similarity between induced pairwise alignments is by no means the only one possible. Another intuitive measure would be to compute the sensitivity and specificity of the predicted pairwise alignments (M. Brudno, S. Batzoglou, L. Pachter, pers. comm.), defining the sensitivity as the fraction of aligned bases of the correct alignment that are paired identically in the predicted alignment, and defining the specificity as the fraction of aligned bases of the predicted alignment that are paired identically in the correct one. These scores are particularly useful to determine whether a given program over- or underpredicts aligned bases. Supplemental Figures S2 and S3 show that the sensitivity of TBA, MULTIZ, MLAGAN, and DIALIGN is usually similar to their specificity, with TBA and MULTIZ obtaining sensitivity and specificity slightly higher than the other two programs. On the other hand, MAVID usually obtains better specificities than sensitivities, indicating that it tends to underpredict aligned bases (in the framework of our simulations), whereas the opposite is true for CLUSTALW.
The average running times for each program are given in Table 1. It is immediately clear that the four programs actually designed for aligning large genomic regions (MULTIZ, TBA, MAVID, and MLAGAN) are the only ones able to run fast enough to contemplate whole-genome alignments. Among those, MAVID stands out with a remarkably fast running time. Notice that the running times of DIALIGN and CLUSTALW would be prohibitive if they were required to align the full 50-kb sequences all at once.
Table 1.
Average Running Time to Produce a Multiple Alignment of the Given Sequences
| Program | Running time, human-mouse-rat (sec) | Running time, nine mammals (sec) |
|---|---|---|
| MULTIZ | 7.5 | NA |
| TBA | 7.7 | 70.2 |
| TBA (R = 50) | 8.3 | 82.8 |
| CLUSTALW | 700.4 | 3000.9 |
| MLAGAN | 31.2 | 166.8 |
| MAVID | 6.7 | 38.5 |
| DIALIGN | 2894.2 | 35,954.4 |
| DIALIGN/CHAOS | 764.4 | 13,580.7 |
Each sequence is ∼50 kb. The time for masking repeats using Repeat-Masker is not included. Programs were run on 866-MHz Pentium III processors with 1 Gb of memory.
MULTIZ
TBA is implemented as a suite of independently executing programs. Pairwise alignments between the original sequences are computed by BLASTZ (Schwartz et al. 2003a), and alignments between three or more sequences are computed by a new program called MULTIZ.
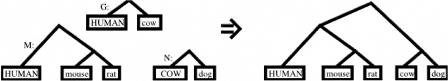
The general problem addressed by MULTIZ is to align two ref-blocksets, say M with reference S and N with reference T, guided by a pairwise S-ref blockset, G, for S and T. The result is an S-ref blockset for the union of the original sequences in M and N. Figure 5 depicts an example. MULTIZ does not require the user to provide M or N if it consists of a single species; in such cases, G supplies the necessary data. G and M are assumed to be sorted by starting position in S, and the output will be similarly sorted.
Figure 5.
Pictorial representation of an application of MULTIZ. M is a human-ref blockset of human, mouse, and rat, whereas N is a cow-ref blockset of cow and dog. MULTIZ uses a pairwise human-ref blockset, G, of human and cow to guide the aligning process. The output is a human-ref blockset of human, mouse, rat, cow, and dog. The reference sequence for each blockset is indicated by capital letters.
The basic computation performed by MULTIZ is to find segments of a block in M and a block in N that are predicted to align to each other according to some block g in G. Those segments are aligned to each other by dynamic programming, using the approximation provided by g to speed up the process. The Methods section (see Supplemental Material) provides more details. There, we also describe a special variant of MULTIZ, which we call HUMOR (HUman-MOuse-Rat), designed specifically to make the human-ref blockset of human–mouse–rat that we analyzed for the Rat Consortium (The Rat Genome Sequencing Project Consortium 2004).
Whole-Genome Alignments on the Santa Cruz Genome Browser
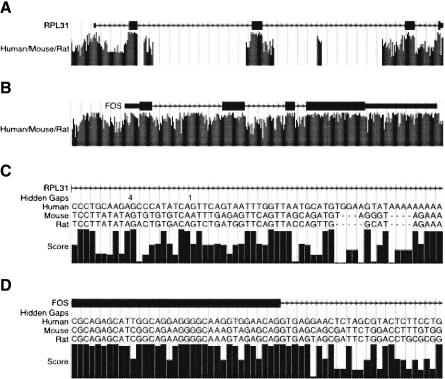
The browser at http://genome.ucsc.edu currently displays MULTIZ and HUMOR alignments between the mouse, rat, and human genomes (Fig. 6). The underlying alignment files are also available in the downloads section of the genome.ucsc.edu site, as is documentation on the MAF (Multiple Alignment Format) that HUMOR, MULTIZ, and TBA use. The browser graphs a local score related to the MULTIZ score, which is described further in the Supplemental material.
Figure 6.
UCSC Genome Browser display of HUMOR alignments. (A) Ribosomal protein RPL31. The human/mouse/rat track shows the MULTIZ score normalized as described in the text. The high conservation of exons relative to introns is typical of many genes. (B) Transcription Factor FOS. In highly regulated genes such as this one, it is not unusual to find extensive conservation outside of protein-coding exons. (C) Closeup of a poorly conserved part of a RPL31 intron. When the display is zoomed in close enough, the base-by-base alignment is displayed as well as the score graph. Because the alignment is projected onto the reference sequence, a “Hidden Gaps” row indicates areas where in the full alignment there would be dashes in the reference sequence row. Clicking on the human/mouse/rat track takes you to a details page that displays the full alignment. (D) Closeup of an exon/intron boundary in FOS. The canonical “GT” 5′ consensus sequence is usually conserved, but then conservation falls off for the rest of the intron.
How TBA Was Built
Figure 7 summarizes TBA's implementation. Given a node of the phylogenetic tree, TBA loops over the BLASTZ blocksets between sequences from each of the two subtrees. For each such pairwise blockset G, TBA uses MULTIZ to align the blocksets X and Y, which contain segments of blocks among sequences from one of the subtrees that haven't yet been aligned to sequences from the other subtree. MULTIZ(G,M,N) signifies the set of new blocks that MULTIZ produces from blocksets M and N, guided by the pairwise blockset G. We use the symbols “+” to denote the union of two blocksets, and, for example, “X - T” to denote the operation of removing all segments of blocks in X that MULTIZ has aligned to the blockset for the other subtree.
Figure 7.
The TBA implementation.
Because TBA's invocations of MULTIZ are independent of one another, one invocation cannot know how the blocks it is producing relate to blocks produced by earlier invocations at the same node of the phylogenetic tree. Consequently, to guarantee the partial order condition, TBA occasionally breaks apart a block (into the block of rows from the left subtree and the block from the right subtree) to re-establish the partial order condition. (The process is described in some detail in the Supplemental material.) Partial order is necessary and sufficient to guarantee that the blocks can be ordered so as to be threaded left-to-right by every original sequence, which we use to create a “multiple alignment” for comparison with the output of other aligners.
DISCUSSION
We have developed new software to align multiple mammalian genomic sequences, used the software to align the human, mouse, and rat genomes, and given the UCSC Browser a capability to let users explore and/or download those alignments. This paper announces the availability of those resources. In addition, the paper contributes to three facets of the art and science of aligning multiple genome sequences, namely, problem formulation, software evaluation, and implementation strategy.
Problem Formulation
Current discussions of the best way to align genome sequences often focus on identifying a useful blend of local and global alignment strategies. We believe that a wider range of notions is needed, and here we propose a conceptual framework that we find useful: threaded blocksets.
Numerous papers extend the classic dynamic-programming method for pairwise sequence alignment to more general classes of bioinformatics problems, such as addressing the types of objects to be aligned and the scoring schemes to be optimized. The most widely used extension is to multiple sequence alignment, based on the observation that the “sequences” being aligned can themselves be alignments. Another observation is that the objects being aligned can deviate somewhat from being totally ordered. That is, under certain conditions, a pair of positions in one of the objects being aligned can be such that neither position comes before the other. Typically, one of the required conditions is that cycles are not allowed, for example, there cannot exist positions x, y, and z, with x before y, y before z, and z before x. For methods of this type, see Sankoff and Kruskal (1983, pp. 265–274), Hein (1989), Myers (1996), and Lee et al. (2002). Our “partially ordered threaded blocksets” fall into this category. What we particularly like about threaded blocksets is that they handle a range of evolutionary operations much broader than merely insertions and deletions.
Software Evaluation
Schwartz et al. (2003a) suggest that the maxim “it is an order of magnitude easier to design two good programs than it is to tell which one is better” applies to pairwise genomic alignment software. We think that the same is true for multiple alignment programs. Generating test data by simulation of evolutionary processes is clearly a useful approach, and it may be the best approach for some purposes. This is the method used recently by Pollard et al. (2004) for evaluating the accuracy of programs computing pairwise alignments. Our simulation procedure is done in the same spirit but differs from theirs on several points: (1) Because we are interested in multiple alignments, our simulations yield as many orthologous sequences as desired. (2) We simulate purely nonfunctional sequence, without interspersed conserved regions, whereas Pollard et al. (2004) simulated unconstrained regions with interspersed constrained segments. (3) Our simulations include realistic insertion of mammalian interspersed repeats, whereas Pollard et al. (2004) used an indel model appropriate for Drosophila.
Of course, our comparisons to programs written by other groups must be interpreted with extreme caution, for several reasons. First, the authors of those programs may have preferred different parameter settings in their programs for these particular experiments, and they may now have more recent versions that would perform better. Also, the properties of the simulation may favor some programs over the others, even if it is designed independently from any of the alignment programs. For instance, because our evolutionary simulations model only nonfunctional sequences, the generated sequences tend not to contain well-conserved regions like coding exons that many aligners (including MULTIZ and TBA) may use as anchors during the alignment process. However, it is unclear which of the programs tested would benefit the most from simulations that are more realistic in this respect. Finally, the accuracies obtained for all aligners may overestimate slightly the accuracies on biological data, because we do not model the presence of low-complexity regions or of repetitive regions that are unknown to RepeatMasker.
Implementation Strategy
Our implementation of TBA/MULTIZ experimented with the strategy of building a multiple aligner as a suite of independently executing programs, which has advantages and disadvantages compared with the more common approach of linking independently compiled procedures into a single executable program. A main advantage is in ease of software maintenance. TBA's components can be tested and modified individually, rather than being difficult-to-isolate parts of a monolithic piece of software. Also, several of the components, including MULTIZ and programs to compute projections and to reorder rows within blocks, are useful in their own right. On the other hand, monolithic programs gain execution-time efficiency by avoiding intermediate input/output operations. More important, limiting intermediate files to blocksets (as we did) means that potentially useful data structures cannot be passed from one component to another; in our case, this required periodic re-enforcement of the partial order condition. On the whole, we feel the experiment verified that the implementation strategy is viable.
Open Problem
Although threaded blocksets appear adequate to describe homology relationships in genomic regions that include complex rearrangements, we do not currently know how to produce such blocksets automatically, accurately, and reliably. Here, we have described an approach for handling genomic regions for which it is sufficient to consider only nucleotide substitutions and inversions/deletions of arbitrary size. An important goal is to develop software that automatically, efficiently, and reliably creates a threaded blockset for entire mammalian genomes.
METHODS
See Supplemental material.
Acknowledgments
We thank Adam Siepel for helpful comments; Claude dePamphilis for suggesting the chloroplast example; Piotr Berman for discussions about the partial order condition; and Lior Pachter, Michael Brudno, and Serafim Batzoglou for many helpful comments about evaluation of alignment programs. We also thank the NISC Comparative Sequencing Program for generating multispecies sequences for some of the analyses performed here. C.R., L.E., and W.M. are supported by NIH grant HG-02238, with additional support to L.E. from HG02325; M.B. and D.H. by the Howard Hughes Medical Institute and NHGRI Grant 1P41HG02371; and W.J.K., K.M.R., R.B., K.R., and H.C. by NHGRI Grant 1P41HG02371. K.M.R. is a Howard Hughes Medical Institute Predoctoral Fellow.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1933104.
Footnotes
[Supplemental material, including the Methods section, is available online at www.genome.org. The multiple alignments produced by MULTIZ can be viewed at the Santa Cruz Genome Browser or downloaded in bulk. TBA, simulated test data, and the Gmaj visualization tool can be downloaded from http://bio.cse.psu.edu/.]
References
- Aparicio, S., Chapman, J., Stupka, E., Putnam, N., Chia, J.M., Dehal, P., Christoffels, A., Rash, S., Hoon, S., Smit, A., et al. 2002. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297: 1301-1310. [DOI] [PubMed] [Google Scholar]
- Bray, N. and Pachter, L. 2003. MAVID multiple alignment server. Nucleic Acids Res. 31: 3525-3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brudno, M. and Morgenstern, B. 2002. Fast and sensitive alignment of large genomic sequences. In Proceedings of the IEEE Computer Society Bioinformatics Conference, pp. 138-150. IEEE Press. [PubMed]
- Brudno, M., Do, C.B., Cooper, G.M., Kim, M.F., Davydov, E., Green, E.D., Sidow, A., Batzoglou, S., and NISC Comparative Sequencing Program. 2003. LAGAN and Multi-LAGAN: Efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 13: 721-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, F.S., Green, E.D., Guttmacher, A.E., and Guyer, M.S. 2003. A vision for the future of genomics research. Nature 422: 835-847. [DOI] [PubMed] [Google Scholar]
- Hardison, R.C., Chao, K.-M., Schwartz, S., Stojanovic, N., Ganetsky, M., and Miller, W. 1994. Globin Gene Server: A prototype e-mail database server featuring extensive multiple alignments and data compilation for electronic genetic analysis. Genomics 21: 344-353. [DOI] [PubMed] [Google Scholar]
- Hein, J.A. 1989. A new method that simultaneously aligns and reconstructs ancestral sequences for any number of homologous sequences, when the phylogeny is given. Mol. Biol. Evol. 6: 649-668. [DOI] [PubMed] [Google Scholar]
- Kent, W.J., Sugnet, C., Furey, T., Roskin, K., Pringle, T., Zahler, A.M., and Haussler, D. 2002. The Human Genome Browser at UCSC. Genome Res. 12: 996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, W.J., Baertsch, R., Hinrichs, A., Miller, W., and Haussler, D. 2003. Evolution's cauldron: Duplication, deletion, and rearrangements in the mouse and human genomes. Proc. Natl. Acad. Sci. 100: 11484-11489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Lee, C., Grasso, C., and Sharlow, M. 2002. Multiple sequence alignment using partial order graphs. Bioinformatics 18: 452-464. [DOI] [PubMed] [Google Scholar]
- Margulies, E.H., Blanchette, M., Haussler, D., Green, E.D., and NISC Comparative Sequencing Program. 2003. Identification and characterization of multi-species conserved sequences. Genome Res. 13: 2507-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern, B. 1999. DIALIGN 2: Improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics 15: 211-218. [DOI] [PubMed] [Google Scholar]
- Myers, E.W. 1996. Approximate matching of network expressions with spacers. J. Comput. Biol. 3: 33-51. [DOI] [PubMed] [Google Scholar]
- Pollard, D.A., Bergman, C., Stoye, J., Celniker, S., and Eisen, M.B. 2004. Benchmarking tools for the alignment of functional noncoding DNA. BMC Bioinformatics 5: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Rat Genome Sequencing Project Consortium. 2004. Genome sequence of the Brown Norway Rat yields insights into mammalian evolution. Nature (in press). [DOI] [PubMed]
- Sankoff, D. and Kruskal, J.B. 1983. Time warps, string edits, and macromolecules: The theory and practice of sequence comparison. Addison-Wesley, Reading, MA.
- Santini, S., Boore, J.L., and Meyer, A. 2003. Evolutionary conservation of regulatory elements in vertebrate Hox gene clusters. Genome Res. 13: 1111-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, S., Zhang, Z., Frazer, K.A., Smit, A., Riemer, C., Bouck, J., Gibbs, R., Hardison, R.C., and Miller, W. 2000. PipMaker—A web server for aligning two genomic DNA sequences. Genome Res. 10: 577-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, S., Kent, W.J., Smit, A., Zhang, Z., Baertsch, R., Hardison, R.C., Haussler, D., and Miller, W. 2003a. Human–mouse alignments with BLASTZ. Genome Res. 13: 103-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, S., Elnitski, L., Li, M., Weirauch, M., Riemer, C., Smit, A., Green, E.D., Hardison, R.C., Miller, W., and NISC Comparative Sequencing Program. 2003b. MultiPipMaker and supporting tools: Alignments and analysis of multiple genomic DNA sequences. Nucleic Acids Res. 31: 3518-3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel, A. and Haussler, D. 2003. Phylogenetic estimation of context-dependent substitution rates by maximum likelihood. Mol. Biol. Evol. (in press). [DOI] [PubMed]
- Thomas, J.W., Touchman, J.W., Blakesley, R.W., Bouffard, G.G., Beckstrom-Sternberg, S.M., Margulies, E.H., Blanchette, M., Siepel, A.C., Thomas, P.J., McDowell, J.C., et al. 2003. Comparative analysis of multi-species sequences from targeted genomic regions. Nature 424: 788-793. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTALW: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter, J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304-1351. [DOI] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alesandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://bio.cse.psu.edu/; TBA, simulated test data, and the Gmaj visualization tool.
- http://genome.ucsc.edu; MULTIZ and HUMOR alignments.