Abstract
Atlas is a suite of programs developed for assembly of genomes by a “combined approach” that uses DNA sequence reads from both BACs and whole-genome shotgun (WGS) libraries. The BAC clones afford advantages of localized assembly with reduced computational load, and provide a robust method for dealing with repeated sequences. Inclusion of WGS sequences facilitates use of different clone insert sizes and reduces data production costs. A core function of Atlas software is recruitment of WGS sequences into appropriate BACs based on sequence overlaps. Because construction of consensus sequences is from local assembly of these reads, only small (<0.1%) units of the genome are assembled at a time. Once assembled, each BAC is used to derive a genomic layout. This “sequence-based” growth of the genome map has greater precision than with non-sequence-based methods. Use of BACs allows correction of artifacts due to repeats at each stage of the process. This is aided by ancillary data such as BAC fingerprint, other genomic maps, and syntenic relations with other genomes. Atlas was used to assemble a draft DNA sequence of the rat genome; its major components including overlapper and split-scaffold are also being used in pure WGS projects.
The most effective and economical way to provide comprehensive information about an organism is through a whole-genome sequencing project. This perspective and the Human Genome Project (HGP) have advanced DNA sequencing technology so that the raw data can be generated for virtually any genome in a timely and affordable manner. Several different genome assembly strategies have also been developed, to efficiently merge the primary data to construct an ordered and accurate final sequence. Specialized assembly software underlies each of these strategies and defines the precise qualities of the data to be generated.
The current focus of large genome projects is on producing draft sequences. Initial definitions of a “draft” required only relatively low genome sequence coverage (Bouck et al. 1998), as data imperfections in genome assemblies were expected to be cured at a final finishing stage. Subsequently, several projects have been launched without the expectation of production of a finished grade, increasing the importance of the quality of the draft sequence generated in the initial phase. This provides a considerable challenge for large genomes with repeated sequences and high levels of polymorphism. Although there is no formal definition of a “draft,” the current general agreement is that there should be sequence redundancy of at least 6×, covering >90% of the genome at high enough quality to allow genes and other features to be reliably discerned, without the need for additional experimentation.
The whole-genome shotgun (WGS) method, involving sequencing of clones randomly selected from libraries of genomic DNA with different insert sizes, has found widespread acceptance. WGS “reads” are assembled using information from sequence overlaps, pairing of reads from each end of subclone inserts, and distances between paired reads from libraries of different insert sizes. This method produced the first complete sequence of a free-living organism, the bacterium Haemophilus influenzae (Fleischmann et al. 1995), and is now used confidently for sequencing genomes that are not too large or complex. It has also been used for draft sequences of larger genomes such as the human (Venter et al. 2001), mouse (Waterston et al. 2002), Drosophila melanogaster (Adams et al. 2000) and Drosophila pseudoobscura (S. Richards, Y. Liu, B.R. Bettencourt, P. Hradecky, S. Letovsky, R. Nielsen, K. Thornton, M.J. Todd, R. Chen, R.P. Meisel, et al., in prep.), Anopheles gambiae (Holt et al. 2002), Fugu rubripes (Aparicio et al. 2002), and Ciona intestinalis (Dehal et al. 2002), as well as lower-quality draft sequences of rice (Goff et al. 2002; Yu et al. 2002) and dog (Kirkness et al. 2003) genomes. The appeal of the WGS method is that relatively few clone libraries need be constructed and no mapping information for the clones is needed. However, abundant repetitive sequences and large duplications, as found in mammalian genomes, can complicate the final assembly. For example, gaps are formed at regions that are highly repetitive, and large duplications can be collapsed into a single sequence. Several programs have been developed for assembly of large (>100 Mb) genomes including the Celera assembler (Myers et al. 2000), Arachne (Batzoglou et al. 2002; Jaffe et al. 2003), Phusion (Mullikin and Ning 2003), Jazz (Dehal et al. 2002), and PCAP (Huang et al. 2003).
An alternative approach for genome assembly, illustrated by the “map first, sequence later” philosophy of the HGP, is the “clone-by-clone” (CBC) method. A clone map of the genome is first constructed using restriction enzyme digestion fingerprinting and specialized software (Soderlund et al. 1997). A minimal tiling path is derived from the map, and each clone in the tiling path is shotgun-sequenced and assembled (Kent and Haussler 2001; Choi and Farach-Colton 2003). The entire genome is then reconstructed from these local sequence assemblies. This method was used for several genomes including human (Lander et al. 2001), Escherichia coli (Blattner et al. 1997), Mycobacterium tuberculosis (Cole et al. 1998), Arabidopsis (Arabidopsis Genome Initiative 2000), nematode (C. elegans Sequencing Consortium 1998), and yeast (Goffeau et al. 1996; Mewes et al. 1997). It is also the method used for converting the draft WGS Drosophila melanogaster and mouse sequences into high-quality finished sequence (Celniker et al. 2002; Waterston et al. 2002). The main advantage of the CBC method is the reduction of complexity from whole-genome assembly to local assembly of BAC clones, minimizing complications from genome-wide repeated sequences or large (>20 kb) near-identical (>98%) segmental duplications (Bailey et al. 2002; Waterston et al. 2002; Tuzun et al. 2004). The added cost of this method is the need to map the BACs and construct a large number of shotgun libraries. A variety of programs can be used to assemble the sequences from individual BACs, but the Phred and Phrap software that was used in the HGP is the most widespread (Ewing and Green 1998; Ewing et al. 1998).
To exploit the merits of both the WGS and CBC methods, a “combined approach” for genome assembly was developed for the Rat Genome Sequencing Project (RGSP) (Rat Genome Sequencing Project Consortium 2004). In the combined strategy, a BAC map is constructed and each BAC is sequenced to low (∼1×) coverage (“skimming”). WGS sequencing is also performed, and these reads are localized to regions covered by BACs and then coassembled with BAC skim reads. Although this requires elements of the CBC approach, namely, the need to prepare and sequence BACs from a minimal tiling path, the use of WGS reads allows libraries with multiple insert sizes to be prepared, which is important in building accurate assemblies. The WGS reads are, in addition, less expensive to produce than sequencing of BACs, easier to track in a production sequencing facility, and allow gaps in the BAC tiling path to be filled with genomic sequence. However, the use of BACs provides a framework for local assembly, enhancing the accuracy of the final product.
In the current communication, we present the design, implementation, and operation of the Atlas assembly system that enabled the combined approach to genome assembly. We include selected results obtained in the RGSP demonstrating the effectiveness of our approach (Rat Genome Sequencing Project Consortium 2004). The Atlas components are also being used in different configurations for WGS-only assemblies of other organisms at Baylor College of Medicine, including sea urchin and several insects.
RESULTS
The overall scheme of Atlas, summarized in Figure 1 and Table 1, incorporates an upstream phase of data preparation and localized assembly and a downstream phase of large-scale assembly and mapping. The upstream phase produces combined WGS and BAC read assemblies in four steps: (1) data preparation, including quality checks and read trimming; (2) tabulation of k-mers to identify repetitive sequences; (3) computing overlaps between reads; and (4) assembling WGS and BAC reads into “enriched BACs” (eBACs) and scaffolds, usually one per BAC. The down-stream phase rebuilds the enriched BACs into a genome assembly in four additional steps: (5) identifying bactigs as groups of overlapping BACs; (6) reassembling eBACs into new contigs and scaffolds for each bactig; (7) building superbactigs by linking bactigs; and (8) building ultrabactigs and chromosomes.
Figure 1.
Steps in the Atlas Assembly System. (1) Trim off vector and low-quality portions of reads. (2) Count k-mers in WGS reads, saving the overall distribution plus specific counts for k-mers with copy number above a threshold. (3) Align BAC and WGS reads sharing rare k-mers and save overlap edges for high-quality end-to-end alignments. (4) Enrich each BAC read set with overlapping WGS reads and their mates, assemble using Phrap, scaffold and check for consistent assembly. (5) Arrange BACs into contiguous sets (bactigs) and flag problem BACs for closer quality checking. (6) Assemble bactigs in waves designed to limit the number of BACs that are input to Phrap. (7) Treating each bactig scaffold as a unit, rescaffold to produce superbactigs, detecting problem joins and missed merges between bactigs. (8) Link superbactigs together into ultrabactigs based on remaining (single) mate-pair links, fingerprint contigs, markers and synteny with human and mouse genomes. (9) Format chromosome files with contigs separated by strings of Ns representing gaps. Quality-control feedback steps include (10) examining coassembly scores of problem BACs and removing foreign trays of reads; (11) resolving superbactig conflicts by modifying bactigs and possibly flagging BACs for closer checking; and (12) resolving ultrabactig and mapping conflicts in collaboration with research groups that generated FPC and marker information.
Table 1.
Steps in the Atlas Assembly System
| Stage | Program | Action | Comment |
|---|---|---|---|
| 1. Data preparation | Data quality checks | Contamination (reads from other organisms) and mislabeled reads (ie, from another BAC) are identified and corrected if possible. | |
| trim-reads | Remove low-quality bases, so that only highest-quality sequence is used for finding overlaps between reads. | Trimmed reads only used in finding overlaps; full sequences used to assemble the consensus sequence. | |
| 2. Analyze sequence redundancy | k-mer-counter | Build table of the frequency of oligonucleotides (k-mers). | Only WGS reads used to give most complete and random sampling of the genome. |
| 3. Compute read overlap graph | overlapper | Identify candidate overlaps based on shared rare k-mers. | End-to-end criterion scores alignment on entire overlapping portion of reads. |
| Stringently evaluate overlaps by banded alignment. | |||
| Save overlap graph with stringency annotations. | |||
| 4. eBAC assembly | Coassemble WGS and skim reads that have been assigned to the same BAC to produce eBACs. | ||
| binner | Choose WGS reads with best overlaps to skim reads in a BAC; add read pair mates. | ||
| Phrap | Assemble WGS and skim reads. | ||
| split-scaffold | Split misjoined contigs. | ||
| split-scaffold | Build scaffolds with read pairs. | ||
| 5. Build bactigs | Find overlapping eBACs based on shared reads and more. | ||
| BLASTZ | Confirm overlap by aligning eBACs. | ||
| Compare bactigs to other maps for verification. | |||
| 6. Assembly of bactigs | rolling-phrap | Assemble reads in bactigs. | |
| Phrap | Assemble contigs in bactigs. | ||
| split-scaffold | Split misjoins, build scaffolds. | ||
| 7. Build superbactigs | Link bactigs by read pairs and BAC skim read distribution. | ||
| 8. Build ultrabactigs and map to chromosomes | Link superbactigs by map and synteny data. |
Step 1: Trimming Reads
Reads are trimmed to a standard determined by the RGSP. The standard requires identifying windows of 50 bases, scanning in from each end of the read, with no ambiguous or contaminant (e.g., vector) bases and <1.25 expected errors. The passed region is from the beginning of the first such window to the end of the last such window, and bases not located within or between the windows are removed (Fig. 2). After trimming, WGS reads with <100 bases and BAC reads with <50 bases are omitted from the overlap analysis. WGS reads require longer passing sequences so that their assignment to BACs in later stages can be confirmed with a higher confidence. Trimming ensures that only high-quality bases are used in the next phases of the process. Ultimately, when local assemblies of groups of reads are performed using Phrap, the entire sequence called by Phred is used (Ewing and Green 1998; Ewing et al. 1998).
Figure 2.
Read trimming method. A simplified version of the trimming method is shown, with a window size of 4 and a minimum boundary quality of 2. The actual trimming of the rat genome reads used a window size of 50, a minimum boundary quality of 20, and also imposed other requirements on passing windows and regions.
Step 2: Counting k-mers
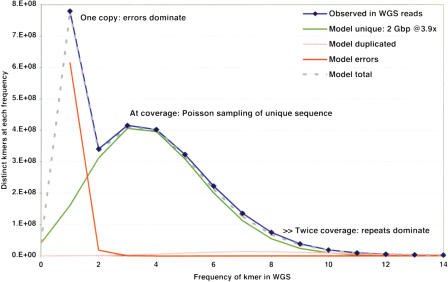
Analysis of genome-wide oligonucleotide frequencies provides estimates of genome size and the true depth of sequence coverage, and guides repeat suppression (Kim and Segre 1999) in the initial assembly stages. We count all subsequences of length k, using the trimmed WGS reads for the most uniform coverage of the genome available (Fig. 3 shows 32-mer occurrences for the RGSP). When there is sufficient coverage of high-quality sequence and the genome has a low amount of heterozygosity, the different ranges of k-mer frequency reflect separate categories of underlying data. Part of the distribution represents k-mers that must result from sequence errors, as each is found only once in the entire set of reads. In contrast, the k-mers that result from frequencies of two to eight occurrences fit closely to a Poisson distribution for unique sequence sampled to a 3.9 × depth of coverage. The total k-mer occurrences in the rat WGS (11.4 billion) can be reduced by the estimated error occurrences (0.65 million), then divided by the estimated coverage to yield an estimated genome size (2.76 billion bases). This method gives robust results for the rat genome, producing estimates that vary little with the length of k-mer used (increasing by 2% for 24-mers) and are consistent with other measurements of the genome size (Rat Genome Sequencing Project Consortium 2004).
Figure 3.
k-mer analysis of WGS reads in the RGSP. The frequency distribution of distinct 32-mer oligonucleotides is shown. The observed distribution is shown as a blue line, and the predicted Poisson distribution of unique 32-mers at 4 × shotgun sequencing coverage is shown as the green line. Models for unique 32-mers resulting from sequencing errors, 32-mers in duplicated regions, and the total 32-mer distribution from these models are shown as orange, pink, and dotted lines.
The table of k-mers and their frequencies is next used to guide overlapper in its selection of pairs of reads to align. To suppress overlaps based on repeated sequence and to avoid testing all O(n2) pairs from n reads, overlapper will only compare reads sharing a “rare k-mer,” defined as having a WGS frequency less than some fixed R. The memory required to store a frequency table, roughly 10 bytes per distinct k-mer (plus 20% hash-table overhead), mandates careful planning. By recording only those k-mers with at least R copies in the WGS, we keep the table small. For the RGSP, using R = 10 gives a table with 59 million distinct k-mers, or 2.1% of the total, and allows us to track an estimated 92% of repetitive copies in the genome. (Setting R = 9 would almost double the size of the table.) When sufficient memory is available for overlapper jobs, we can set R low enough to include borderline k-mers as well as those that are clearly repetitive.
Step 3: Finding Read Overlaps: The overlapper
The Atlas overlapper (see Methods) begins the process of identifying which WGS reads belong to the same genomic regions represented in each BAC. It produces an overlap graph recording high-quality alignments between reads sharing rare sequence. Although overlapper's main requirement of aligning reads is the same as the all-against-all read comparison of WGS-only assembly, some differences are noteworthy. Most significantly, because localized assembly of each BAC will effectively reexamine each read, little detail about each read overlap need be stored.
Key issues for overlapper are run times in limited memory, detecting true genomic overlaps (sensitivity), rejecting alignments because of repeated sequences (specificity), and recording details that will help later resolution of borderline cases. For the rat genome, we aimed for computation on 40 million reads in ∼2 d using a cluster of 50 to 100 Linux processors. We chose to implement a filtration technique (Karp and Rabin 1987; Owolabi and McGregor 1988) in which a fast, sensitive, but nonspecific comparison is performed first to identify candidate overlaps, followed by a slower detailed analysis to validate these candidates. We identify any pair of reads sharing a sufficiently rare k-mer as a candidate overlap, then perform a banded alignment (Chao et al. 1992) to refine the relationship of that pair of reads.
The rarity heuristic based on the saved k-mer table is key to overlapper success. The WGS-based k-mer counts provide a genome-wide view of repetitive sequences that can be loaded into each overlapper job. overlapper parameters for the rarity heuristic include R′, the minimum count kept in its internal k-mer frequency table (no smaller than the R used when creating the saved frequency table) and Y, the maximum count for a k-mer that will seed an overlap. Each pair of reads sharing a k-mer with frequency less than or equal to Y will be aligned, using the rarest k-mer as a seed (k-mers with frequency <R ′ are treated as having the same frequency, and ties are broken arbitrarily). Instead of comparing each of n reads with all others, an O(n2) task overall, or with hundreds of reads that share any (possibly repetitive) k-mer, we compare each read with an average of <Y other reads. For the RGSP, overlapper was able to meet our runtime efficiency goals even with Y set to 100.
Although very efficient, the rarity heuristic cannot by itself be made sufficiently sensitive and specific for following stages. In the RGSP, imposing a seed frequency cutoff Y of 8 would exclude almost 5% of the genomically unique k-mers, limiting sensitivity. But that is still too high for specificity; setting Y even at 6 would let through almost 2% of repeated k-mer occurrences, causing an even larger percentage of false overlaps. The additional banded alignment process allows us to use a larger value for Y for identifying candidate overlaps, yet still reject false overlaps based on the quality of alignment.
overlapper's quality heuristic computes an end-to-end banded alignment of the reads in each candidate overlap (Chao et al. 1992). By limiting the number of consecutive insertions (or deletions) in one read with respect to another, banded alignment is both more efficient and more appropriate to high-stringency comparisons. Evaluating each alignment end to end, over the entire region of overlap implied by the shared k-mer seed, enforces additional stringency.
Step 4: Production of eBACs: The binner and split-scaffold
The WGS reads that fall within each BAC are next selected, based on the best overlaps of each skim read from the BAC. The details are in Methods. All read-pair mates of the chosen WGS reads (even those not passing the trimming phase) were also added. This use of read-pair information also allows the highly repetitive sequences to be included in the assembly, without interfering with the overlap analysis. Thus, a read pair is excluded only if both reads are low-quality or repetitive. As shown in Figure 4, 80%-90% of the expected WGS reads were retrieved as the BAC skim coverage reached 1×, and this increased to >95% recruitment when the BAC coverage reached 2 ×.
Figure 4.
Recruitment of WGS reads into eBACs with increasing BAC skim coverage. Twelve BACs were selected randomly from different chromosomes and used to BAC-Fish WGS reads from a pool representing ∼4 × sequence coverage of the genome. Progressively larger subsets of BAC reads were used to obtain the curves.
A quantitative analysis of yield of WGS reads at each of these steps is presented in Table 2. This summarizes the construction of eBACs for the RGSP. The expected WGS “catch,” considering the number of usable WGS reads, and the average BAC size of 208.8 kb (from FPC data) as a fraction of the total genome size, is 1638 reads. Additional WGS reads, from WGS-WGS overlaps, extending as much as 10 kb off the ends of each BAC, are also recruited. This provides a mechanism to fill gaps in the BAC tiling path with the corresponding genomic sequence. The average catch of 1675 reads is thus very near the expected value for a BAC plus WGS margin, with a standard deviation of ∼20%.
Table 2.
Building eBACs
| Number of | Average (reads) | Std. Dev. (reads) |
|---|---|---|
| Distinct WGS in overlapsa | 2342 | 2011 |
| WGS passing rarityb | 1431 | 352 |
| WGS passing overlap qualb | 2276 | 1947 |
| WGS passing bothb | 1417 | 351 |
| WGS binned with BACc | 1314 | 310 |
| WGS binned + mates | 1757 | 390 |
| WGS in Phrap contigs | 1675 | 368 |
Distinct WGS in all overlaps produced by the overlapper with 95% identity and 100 k-mer copies allowed
Filtering done in Binner based only on overlapper information. Repeat heuristic limits k-mer copies to 12 (three times the coverage). Overlap quality heuristic requires 3 × span/(3 + span-score) ≥ 35, where score is the banded alignment score, and 2 × span/(2 + span-score) would approximate the average distance between discrepancies were there only substitutions (indels have added penalties)
Beyond the k-mer repeat and quality heuristics, only the top six (i.e., coverage × 1.5) WGS overlaps from each end of a BAC read are examined, and they are kept only if strictly better by the quality heuristic than the top discarded overlap
The reads in each BAC bin are next assembled using Phrap, generating an initial set of contigs. A comparison with finished BACs showed that the default options for Phrap gave acceptable results (see Tuning under Methods). However, contigs generated by Phrap are improved by using read-pair information to detect misjoins. Phrap does not use read-pair information, thus we developed the split-scaffold tool to use read pairs to identify misjoins within contigs and split them. After splitting, reads are also removed from contig ends where they conflict with scaffolding and lack a read pair mate inside the contig. Finally, split-scaffold creates scaffolds: contigs that are linked because they each contain one read from a read pair. Two such links are required to consider contigs as reliably joined in a scaffold.
During scaffolding, the contigs/scaffolds at the ends of the BAC insert are identified using several methods, such as searching for BAC end sequence reads, a read pair with one genomic and one vector read, or a chimeric read that is part vector and part genomic (Chen et al. 2004). This information will be used in verifying BAC overlaps (see below). The linearized scaffold is then saved in FASTA format with Ns as placeholders for the sized gaps. It should be noted, however, that this assembly is transient, aimed only at allowing a detailed analysis of BAC overlaps described below. Once the BACs have been accurately laid out, all reads will be reassembled to create the final product.
The BAC-Fisher
An outgrowth of this section of the Atlas assembly pipeline is the BAC-Fisher, a general tool for retrieving WGS reads based on any input DNA sequence. Whereas here this approach is used to “fish” for WGS reads with a BAC clone as “bait,” the bait can be any genomic sequence. This allows identification of reads of interest in a WGS pool with greater specificity, and little loss of selectivity, over performing a simple BLAST search. While BLAST results can be improved by first masking sequences based on a repeated sequence library, for many organisms information about the repeat structure is incomplete or nonexistent, and low-copy duplications cannot be managed by this method in any event. Hence, the Atlas approach of dynamically suppressing repeat-based overlaps provides a useful alternative. An online BAC-Fisher is available for genome projects being conducted at the Baylor College of Medicine Human Genome Sequencing Center (http://www.hgsc.bcm.tmc. edu). This allows researchers to take advantage of WGS data as it is generated, and prior to the complete genome assembly.
Step 5: Generation of Bactigs
Following eBAC and scaffold formation, a two-step strategy is used to generate the global assembly. In bactigging (step 5 in Fig. 1), a clone tiling path is generated using sequence comparisons of the eBAC assemblies and independent mapping data. We call each set of overlapping clones a bactig (note that this usage differs from that in Huson et al. 2001). The bactigs are assembled from eBAC reads using rolling-phrap (step 6 in Fig. 1).
Bactigging and its associated quality checks are designed to reduce misassemblies, such as those caused by genome duplications, which cannot be solved based on reads alone. Because most genome duplications are smaller than a BAC clone, BAC clone pairs with copies of the duplicated region are distinguished owing to divergence in the remainder of their eBAC assemblies. Moreover, unlike the reads alone, each BAC clone has additional information, such as FPC fingerprint pattern and STS marker content. The accuracy of bactigging is assessed by consistency with the FPC and radiation hybrid STS marker maps. Potential errors can be identified and corrected before the bactig assembly. Such comprehensive quality control ensures assembly quality but is more difficult with other WGS assembly methods.
The process of constructing bactigs begins with identifying candidate pairs of overlapping BACs. Rather than conduct all-against-all sequence comparison among enriched BACs, we first identify candidate BAC overlaps based on WGS and FPC information. The eBAC assemblies of overlapping clones will generally share WGS reads; this criterion contributed 33,146 potential BAC overlaps for the RGSP. However, shared WGS reads will be largely suppressed where the shared region is highly repetitive. To avoid missed overlaps, an additional 724 candidate pairs of BACs were identified in the RGSP by linking with two or more read pairs (from WGS, including BAC-end sequences) or by having similar FPC patterns.
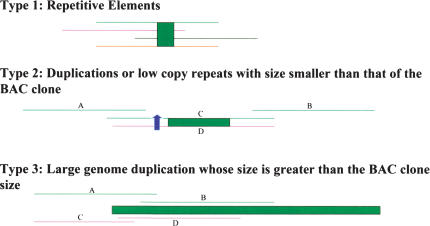
Candidate overlapping pairs are evaluated by alignment using BLASTZ (Schwartz et al. 2003). The linearized sequence from each eBAC scaffold must align end to end for the two BACs to be considered as having a true overlap. False overlaps are excluded by this method except where the shared sequence at the end of each BAC comes from different copies of a duplicated region (Fig. 5, Type 3).
Figure 5.
False clone overlaps caused by repeated sequences. Each thin line represents a BAC clone, and clones with the same color are true overlapping pairs. False overlaps between clones (lines with different colors) are due to highly repeated sequences as well as duplications of small and large regions. Overlaps from highly repetitious sequences are largely dealt with by the overlapper and binner steps and further dealt with at the overlapping BAC detection step. The second and the third situations are dealt with at the bactig linearization step because conflicts can be detected between BACs. For example, in the second case, A/D and B/D clone pairs should overlap based on the clone layout, but this will not be validated.
Two further steps are implemented to eliminate false overlaps caused by largescale, low-copy genome duplications, one of the most difficult types of errors for assembly programs to deal with. First, overlapping BAC pairs are checked for their consistency with other BAC overlaps. Conflicting sets of overlaps are further analyzed for FPC patterns and links between BAC-end reads in the assembly, because these two types of data could potentially extend beyond a duplicated region. An overlap is accepted if it is supported by FPC or BAC end pairs uniquely among potential overlaps. To estimate the specificity of this method, a computer simulation was conducted using 10,000 pairs of BACs that do not overlap to calculate the false-positive rate not excluded by FPC pattern or BAC end read links. Simulations showed that the false-positive error rate is reduced to <0.1% using either FPC or BAC end sequences. Conflicting sets of overlaps that cannot be resolved by these approaches are excluded from subsequent analysis, resulting in gaps in the tiling path. This creates artificial duplications when the overlap is real; these are dealt with later. A bactig is constructed for each remaining set of overlapping BACs. A bactig represents a single contiguous genome region covered by BAC clones, and can often be assembled into a single scaffold.
One remaining problem for bactig construction is the artificial duplications caused by unresolved clone overlaps. Most of these errors are fixed by implementing a feedback loop, to identify true overlapping clones at a later assembly stage, with the aid of long-range information such as RH markers. Previously excluded BAC clone overlaps are considered real if the two clones are located adjacent to each other in a higher order structure, the superbactig that are formed by linking bactigs together through read pairs and confirmed by RH markers (see Step 7 for detail). With this downstream evidence, the bactig is recomputed and subsequent steps are repeated.
A final comprehensive quality control check is implemented to catch rare exceptions that the above filtering misses. The order of BACs in each bactig is compared with independently generated maps. In the case of the RGSP, this included the FPC assembly, the rat radiation hybrid STS map, and the human and mouse genomes, to uncover potential errors such as misjoins. About 60% of the RGSP BACs could be mapped with one or more STS markers, and ∼90% of the BACs could be mapped to consistent locations in the mouse and human genomes. This allowed the quality of each bactig to be evaluated at high resolution. A voting system is implemented in which bactigs (generated based on high-stringency sequence overlaps) are considered verified if supported by any of these maps. Bactigs that are not supported by any map while in conflict with at least two maps are considered potential errors. For the RGSP, seven bactigs (out of >1600) that conflicted with these maps were identified. Close examination of the sequence from these bactigs indicated that all had false overlaps created by duplication regions. These were corrected by breaking the bactigs manually.
In the current rat assembly, 21,689 BACs are grouped into 1607 bactigs, whereas 224 BACs remain as singletons. Among the singletons, 40 are synthetic projects (see Data Quality Assessment in Methods). A quality assessment of RGSP bactigs was made on the bactigs covering two QTL regions, Rf1 and MCS1, for which physical maps of the clone layout had been generated independently. We found only 32 Atlas bactigs in disagreement with the physical map; in consultation with the RGSP Consortium, we determined that the bactigs were more likely to be correct. In sum, very-high-quality bactigs were constructed that were then used as units for the subsequent assembly.
Step 6: Bactig Assembly: Wave Grouping and rolling-phrap
Once the BAC layout into bactigs is determined, the reads within overlapping BACs are assembled into a single sequence for each bactig using a process called rolling-phrap (see Methods). The megabase-sized bactigs can be rather large to assemble en masse with Phrap. To limit the genomic scope of each Phrap run and minimize misassembly caused by repeated sequences, we organize each bactig into sets of overlapping BACs and assemble using rolling-phrap (Fig. 6). The basic idea is to use Phrap to coassemble only small sets of overlapping BACs to keep the assembly size reasonable, outputting contigs and their reads as the process moves through the bactig. The result is an assembly wave that moves through the bactig, assembling a much larger region, with fewer misjoins, than would be possible by pooling all of the reads for a single Phrap run. Arbitrarily choosing one end of a bactig as the left end, the first wave includes all BACs overlapping the leftmost BAC in the layout (Fig. 6). The next wave adds any new BACs overlapping the previous wave. As each wave completes, contigs are removed when their BAC reads come only from BACs that do not overlap the next wave. Using this technique, we assembled bactigs as large as 115 BACs, involving 287,000 reads and with a final assembly size of 12.1 Mb. Much larger assemblies are possible; this is merely the largest we encountered for the RGSP.
Figure 6.
The rolling-phrap process limits the scope of reads presented to Phrap in each wave. All reads for an eBAC are added in the first wave including that eBAC. Contigs are recorded in an .ace file, and the corresponding reads are removed from the Phrap input, until a contig contains (almost) no reads from eBACs overlapping the next wave. (1) First wave containing all eBACs overlapping leftmost eBAC (arrow); (2) emit pure leftmost-eBAC contigs (not overlapping and therefore not merged with any other eBAC); (3) second wave containing all eBACs overlapping next leftmost eBAC contributing new sequences; (4) emit contigs solely from first-wave eBAC regions; (5) third wave containing all eBACs overlapping next leftmost eBAC contributing new sequences; (6) emit new contigs solely from first and second-wave eBAC regions; (7) fourth wave; (8) emit remaining contigs.
The same splitting and scaffolding process is applied as in the eBAC assemblies, with split-scaffold tuned to take better advantage of long insert pairs (50-kb inserts, BAC ends), which are more useful at the megabase scale of bactigs (see Methods). For the RGSP, a total of 6247 scaffolds are obtained for all the bactigs including 2745 single contig scaffolds. On average, 1.9 multicontig scaffolds were obtained for each bactig.
The improvement in overall scaffold size by split-scaffold is significant. When 39 BACs that overlapped finished regions in the genome were analyzed by comparison to the finished sequences, the N50 for scaffolds increased from 974 kb to 1666 kb with the splitting and trimming procedure.
Step 7: Generation of Superbactigs
Higher-order structures called superbactigs are generated by linking bactigs based on read-pair data. Instead of contigs, scaffolds from each bactig are the units for this process. Similar to the construction of scaffolds described above, two or more read pairs are required to link two scaffolds. For the RGSP, we were able to merge 6247 bactig scaffolds into 4584 new scaffolds. Based on these scaffolds, bactigs could be linked to form 824 superbactigs and 93 singletons, 30 of which were synthetic projects. To further reduce the number of scaffolds in each superbactig, the clone tiling path of the superbactigs can be used to determine the order and orientation of the scaffolds. The position and orientation of each scaffold relative to the clone tiling path is calculated first by examining the distribution of BAC skim reads in each scaffold. Scaffolds are considered adjacent to each other if they both contain BAC skim reads from the same BAC clone. These neighbor scaffolds can then be linked and the gap size estimated based on the underlying BAC clone. As a result, one main scaffold can be obtained for each superbactig, which can ultimately be placed on chromosomes (see below). Small scaffolds that could not be merged into the main scaffold are placed into the random chromosome file. For all 137,901 contigs of the RGSP, only 4289 contigs were these unplaced scaffolds, accounting for 1.3% of the sequence. Similar to the bactig, superbactigs were also validated by comparing the clone layout with the rat RH map, FPC assembly, and human/mouse comparative mapping information. Four superbactigs were corrected as a result of this process. Finally, a linearized sequence is generated for each superbactig.
Step 8: Ultrabactigs and Placement on Chromosomes
Additional steps are used to extend the superbactigs to facilitate placement of the sequence on chromosomes. The larger the sequence units are, the more likely they will be mapped by chromosomal anchors. Two types of data were used in the RGSP for this extension process, the rat FPC assembly and single read-pair links. Superbactig pairs that are adjacent to each other on the FPC map were linked together if further supported by a single read-pair link or syntenic consistency with the human and mouse genomes. Similarly, superbactig pairs that were linked by a single read pair were merged together when this merge was further supported by the rat FPC assembly, the RH map, or human/mouse synteny. Superbactigs linked in this manner are called ultrabactigs. The gap size for these links was estimated based on clone fingerprint size or read-pair distances. Like previous steps, the ultrabactigs were compared against the RH markers and human and mouse genomic sequences to ensure accuracy. At the end of the RGSP, the number of ultrabactigs was reduced to 469 pieces with 77 remaining as single BAC clones.
The rat RH map (v3.4) was used to place the ultrabactigs obtained above onto individual chromosomes. To anchor sequence to a chromosome, e-PCR and NCBI-BLASTN are used to map marker primers or sequences onto individual ultrabactigs (see Methods).
To place the ultrabactigs onto chromosomes, the marker locations on the ultrabactigs were converted from the superbactig locations based on their order, orientation, and gap sizes. Dominant windows and slopes were calculated to determine the chromosome location and orientation of each ultrabactig. For those without solid marker information, rat/mouse and rat/human synteny information was used to insert them between or at the ends of marker-ordered ultrabactigs. The final order and orientation were manually adjusted after considering marker quality of the RH map, quality of marker mapping, FPC assembly, and mouse/human comparative information to maximize the synteny.
Adjacent superbactigs on each chromosome were further linked if there were any appropriate read pairs or FPC suggested links. This reduced the RGSP ultrabactigs to 419 pieces with 71 singletons; 291 pieces were placed on chromosomes. Most of the 128 unplaced pieces are either singletons or short superbactigs consisting of only a few clones.
Availability
The components of the Atlas system may be freely downloaded from the BCM-HGSC Web site at http://www.hgsc.bcm.tmc.edu/downloads/software/atlas/.
DISCUSSION
The strategy used to sequence the rat genome was a combined approach using elements of CBC BAC sequencing with WGS sequencing (Rat Genome Sequencing Project Consortium 2004). This contrasts with the human genome, which was constructed entirely out of BAC reads (Lander et al. 2001), and the mouse genome (Waterston et al. 2002), which was constructed entirely out of whole-genome shotgun reads. A mixed approach was also used for the human genome draft produced by Celera Genomics (Huson et al. 2001; Venter et al. 2001), but differs in several respects from the approach described here. For the rat, low coverage (∼1 ×-2×) skims from a 1.6 × clone coverage set of BACs were produced in addition to a 4 ×-5 × coverage of WGS reads. The goal of the BAC skims was to provide localization that could be used to boost the confidence of the assembly, especially in repeat regions, as compared with a WGS-only assembly. Despite the mixed nature of the source data, the assembly of the rat data has more in common with a WGS-only assembly than it does with the CBC approach. The rat assembly required the same all-against-all read comparison that is needed for WGS-only assemblies. This all-against-all read comparison is both a major part of the computational load of the rat assembly as well as a key software component determining its success.
The success of this approach emphasizes the utility of BACs in large-scale sequencing projects. The Atlas assembly system is designed to take both BAC and WGS data as inputs for the genome assembly. This is an important distinction between Atlas and the other popular genome assemblers in use, which are not designed to exploit the unique information associated with BAC clone sequences.
The critical issue in assembling genomes is the method of dealing with repeated sequences. Atlas initially identifies repeats through oligonucleotide frequency analysis and excludes them from the overlap analysis of sequences. Repeated sequences appear in the assembly only as read pairs after the main layout has been derived. However, because of low-frequency repeats, which are not readily distinguished from the redundant sequences from high-coverage sequencing, assemblies can be erroneous. To address this, extensive and repeated checking of assemblies occurs throughout the assembly process, using a variety of methods to detect repeat-induced errors. These include checks of intrinsic properties of assemblies, such as template and read-pair distributions, as well as comparison to external data sets such as FPC and STS maps and syntenic relations with other genomes. These checks are performed at virtually every stage of the process, which is highly iterative to allow feedback from errors that only appear in downstream analysis. All of this produces a robust draft consensus sequence with a high degree of consistency with existing information.
The methods described here clearly highlight the complexity and pitfalls in assembly of large genomes. Current large-scale projects (save the mouse) plan to produce draft (unfinished) genome sequences, further emphasizing the need for high-accuracy assembly procedures. This is not only a software challenge: ancillary data such as FPC maps, STS maps, EST and cDNA, use of BAC based assembly, and sufficient WGS coverage are all essential to get an accurate product. The uses of many of these types of data are clearly illustrated in this report.
METHODS
Overlap Detection
The overlapper performs banded, end-to-end alignments on pairs of reads (Chao et al. 1992), seeded by a shared, rare 32-mer (based on the saved frequency counts). A score of +1 is awarded for each base match, −1 for each substitution, and −2 for insertions or deletions [thus, if there are no indels, (span − score)/2 gives the number of substitutions]. For the RGSP, the settings used were:
maximum seed frequency 12;
bandwidth 3 (total band size of 7, counting main diagonal);
one read set sampled completely, the other taking every sixteenth 32-mer;
maximum mismatch 3% (computed from the span and score as if all mismatches were substitutions, so that <3% indels would be allowed).
Although all WGS-WGS and BAC-WGS overlaps were computed, the Atlas assembly relied primarily on BAC-WGS overlaps. Each overlap is saved as a directed edge that specifies the span, score, left extension, right extension, strand, and global frequency of the seeding k-mer. The span is simply the number of bases the two reads overlap; the score is the banded alignment score in the span region. The left and right extensions are the magnitudes of the nonoverlap regions between the two reads, where a positive extension indicates that the origin read of the edge is longer in that direction, and a negative extension indicates that the sink read of the edge is longer. The strand is indicated by the letter “f” for same-strand and “r” for opposite-strand overlaps.
Sorting WGS Reads Into BAC Bins
Overlap information is used to select the best six overlapping WGS reads. Six was initially chosen as a reasonable threshold value because it was 1.5 times the average WGS coverage of the RGSC project. This value was later verified through the tuning process described below. Overlap information is first sorted according to a modified weight, approximately the number of matching bases per mismatch, with an adjustment to favor longer matches:
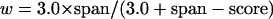 |
This is derived from an approximation for bases between mismatches:
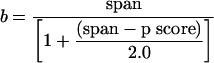 |
For the RGSP, overlap edges with a weight <35 were discarded, and the WGS reads with the best six overlaps at each end of the skim read were kept, provided that their weight was better than the seventh overlap at that end.
Tuning Enriched BAC Assemblies
We tuned parameters of overlapper, binner, and Phrap to provide the best chained alignment between our assemblies on six BACs (Baylor names gapt, gcpq, ggbv, gmez, gymn, and kbas) and finished sequences generated independently at NHGRI from the same strain (generously provided by Eric Green (NHGRI).
Variations that were tested included:
Phrap option settings: defaults, “-shatter_greedy,” or “-trim_qual 16 -penalty -5” (these options had in turn been selected from previous, more extensive searches over the Phrap parameter space).
24-mers versus 32-mers in the k-mer counting and overlapper.
Limits on frequency of k-mer seed for overlaps (12, 20, 100).
-
Limits on maximum overlaps to a BAC read:
- One set of parameters limited the total overlaps to 8, 10, 12, or 14 for each BAC read, and ignored all overlaps on that read if the limit was exceeded.
- Another version took the best N overlaps at each end of a BAC read, with weight better than the (N + 1) overlap, for N = 4, 6, or 8.
Assembled scaffolds were scored both on total finished sequence covered with chains of nonoverlapping exact matches in the correct order and orientation. The TIGR tool Mummer (v2.12; Delcher et al. 2002) was used to compute exact matches, which were trimmed to eliminate overlapping matches and filtered to discard matches disordered with respect to higher-scoring matches.
The percentage coverage ranged from 80%-96% of the actual overlap of the sequences. The best options based on these sequences had:
either 32-mers or 24-mers (no significant difference);
12 as the maximum seed frequency;
35 as the minimum overlap weight;
12 as the limit on total overlaps to each BAC read.
However, manual examination of unfinished assemblies for regions of likely genomic duplication led us to believe that the “limit on total overlaps” heuristic would not do as well as the “best N overlaps” heuristic for those regions. The best settings with that heuristic were used for the enriched BAC preliminary assemblies for the Rnor3.1 release. These were the same as above, using 32-mers, but with the “best N overlaps” heuristic for N = 6. The results were almost identical except that one of the six BACs ended up in two scaffolds instead of one.
rolling-phrap
The leftmost BAC in a bactig is used as the initial source-BAC, and reads from this source-BAC are coassembled with reads from BACS that overlap it in the bactig (Fig. 6). Typically there are a small number of overlapping BACs, called the query-BACs, thus the coassembly involves <10,000 reads. Assemblies of this size are well within the routine capabilities of Phrap. Using Phrap for this small-scale assembly takes advantage of Phrap's ability to use base quality information, as well as the enormous validation that has gone into Phrap over the years. Once this coassembly is performed, the resulting contigs are examined to see which ones contain only source-BAC reads and which are mixtures of source and query reads. Source-only contigs are contigs that do not overlap any query-BACs. These contigs are thus demonstrated to be on the left end of the assembly and will not overlap any contigs further to the right in the rolling-phrap assembly. These contigs are thus complete and can be written out to an “ace” file (which the software constructs, by analogy with Phrap), and the reads associated with these contigs can be removed from the coassembly set of reads. Then, the remaining reads from the query-BAC contigs and the mixed source/query-BAC contigs become the new source-BAC reads. The bactig tiling path is consulted to determine which BACs directly overlap these new source-BACs. These overlapping BACs become the new query-BACs and their reads are coassembled with the new source-BAC reads. The result is another set of contigs, some containing only source-BAC reads, some containing a mixture of source-BAC and query-BAC reads, and some made up only of query-BAC reads. The contigs with only source-BAC reads are written to the growing ace file, the associated reads are removed from further consideration, the query-BACs become the new source-BAC, and the entire process repeats until this wave of coassemblies has moved through the entire bactig. Because there is some noise in the process, and some reads migrate inappropriately, we consider a contig source-only when it is >90% source-only reads (counting bases). Thus, a contig with just a few query reads may be considered not to significantly overlap the query-BACs and be put into the output. For the assembly of the RGSP, 1607 multi-BAC bactigs had to be assembled with rolling-phrap. Using ∼40 nodes of a Linux computer cluster, these assemblies were performed in 3 d.
Atlas-scaffold and split-scaffold
split-scaffold is applied both in the enriched BAC assemblies (after Phrap) and in the bactig assemblies (after rolling-phrap). A greedy algorithm similar to previous published methods is used to generate scaffolds for all the assembled contigs of each bactig (Myers et al. 2000; Batzoglou et al. 2002). All read pairs that link two contigs are first identified, and gap sizes are estimated based on sizes of inserts and linking read locations. To exclude sporadic links between contigs, an appropriate window for gap sizes is used to find the best sets of links between contigs. Paired ends that give gap size estimates within the window size are bundled, and the dominant bundle of links between two contigs is determined. The gap size and its standard deviation between contigs are then calculated by averaging passed read-pair links. Libraries with insert size <10 kb, 50 kb, and 250 kb are used sequentially in this process with the corresponding window size set at 3 kb, 10 kb, and 50 kb. Contig pairs whose estimated gap size are less than −4 standard deviations are discarded. In addition, contigs that linked to multiple overlapping contigs are also discarded in the process. The weight of each link is calculated by the number of read pairs in the bundle. The link with the highest weight is added to the end of a scaffold if it satisfies the positional constraints. Links between the newly formed scaffold and all other contigs and scaffolds are then updated by recalculating the read pairs. This process is iterated until no links with two or more read pairs are left.
Misjoins were corrected in a multistep process. The essence of this process is to use the same read-pairing engine used in scaffolding to discover regions of contigs with inconsistent read pairs. Some inconsistencies were due to small numbers of WGS reads that did not belong in the contig. In these cases, the contig as a whole does not need to be fixed, only the extraneous reads removed. Thus, we could not assume that simply splitting contigs was the proper response to an inconsistency. We used the inconsistent read pairs to identify suspicious regions, which were examined for misjoins. Suspicious read pairs were identified as being one end interior to a contig with the paired end in another contig. Each interior contig point was identified as a checkpoint if the left, the right, or both sides had read-pair evidence linking it to another contig. Based on empirical testing, we identified three rules that determined when to split contigs:
If the depth of read-pair coverage drops to zero (a template crash) near a checkpoint, split the contig in the region.
If the BAC coverage goes to zero for a region >2 kb in a region near a checkpoint, split the contig in the region of the BAC coverage crash.
If the BAC coverage goes to zero for a region <2 kb and there is both a left and right checkpoint in that region, split on the region around the checkpoint.
After splitting problem contigs, split-scaffold checks the ends of contigs, solely for template crashes, and where these are found, and the contig presents a scaffolding conflict, the contig ends are trimmed by removing reads.
After this round of refining the assembly, scaffolds are generated that order and orient as many contigs as possible based on at least two read-pair links. Two scaffold versions are produced for each bactig: one from the contigs with splitting, and another from the contigs with splitting and trimming. The version with the longer scaffold is retained as the final version. The scaffolds >15 kb contain from these bactig assemblies (and eBAC assemblies for single-BAC bactigs) the full set of contigs used in generating the final sequence.
Mapping Ultrabactigs to Chromosomes
BLAST was used to compare sequences of 21,485 RH markers against the linearized sequence of each superbactig without repeat masking. The results were first parsed by removing those hits whose score was below 200, identity <95%, e-value >0.01, or were not in the top five matches. To reduce matches from highly repeated regions, matches (score <1000) were further removed if the number of matches involved in either query or target sequence exceeded a threshold (5 for query, 10 for target). After this step, the location of each marker was determined by the most dominant window that matched most of the marker sequence (>50%). To help resolve inconsistencies from different markers for chromosome placement, the quality of the mapping was scored based on the percentage of marker sequence matching in the window. This resulted in 19,221 markers being mapped to superbactigs. The results of e-PCR were directly used, which mapped 18,458 out of 24,600 RH markers. A total of 22,141 markers were mapped by e-PCR and BLAST to superbactigs.
Data Quality Assessment
Quality assessment of reads and other data occurs throughout an Atlas project as even relatively low numbers of contaminating reads or mixed BAC projects can be problematic. Methods have been described for this process (Stojanovic et al. 2002), but our methods are designed for our environment of low-coverage projects and the combined assembly strategy.
Contigs and scaffolds from each BAC are analyzed for uniform participation of microtiter plates (trays) containing BAC skim sequencing reactions. Reads from trays with unbalanced participation are removed as potentially mixtures of BACs or other contamination, and the eBAC is reassembled. Similarly, if an eBAC assembly size is significantly larger than the FPC size of the BAC or if there are too many scaffolds, the BAC is rejected as a potential mixture. BACs failing any test are excluded from the genome assembly and flagged for purification.
In most cases of mixtures of reads, it is possible to select one read set (reads in a set of contigs) to remain with a BAC and remove all others. For example, in cases in which a clear majority is contradicted by a small minority, we assume the majority to be the correct sequence for the BAC. In other situations, it is possible to assign one read set to a BAC based on BAC end sequences or clones that overlap based on FPC data. In a minority of BACs, neither of these techniques is informative. We resort to splitting such projects in two and noting that the true match to the clone is unknown. We also use the overlapper to detect relatedness between read sets based on k-mer content. This technique has proven particularly useful in the detection of problems with whole-genome shotgun trays, which are among the most difficult to detect with the contig-based purifier.
Reads selected for removal are not discarded. Instead, when a read set is composed of at least four 96-well trays or comprises at least half the reads in a BAC project, we create a “synthetic” project to hold it. Although a synthetic project is not associated with any known clone, it is otherwise assembled and treated exactly as an ordinary BAC project and can contribute to the genome assembly. The final rat genome assembly included 869 synthetic projects containing 359,967 reads that would otherwise have been wasted. In the final analysis, this quality control approach passed 98.7% of eBAC assemblies, and 99.3% of these were included in the final assembly. This included 97.7% of passed BACs that had been purified after previously failing and 93.8% of passed synthetic projects, a slightly lower rate because more synthetic projects created by splitting projects in half were too small to be useful. This high success rate validates the power and correctness of this purification system. Overall, 8.4% of our BAC sequencing projects were subject to purification at one time or another. But purification reduced the number of failed BAC projects from ∼8.9% to 1.3%. The purification system restored 420,894 reads to useful locations either in their assumed projects of origin or in synthetic projects included in the assembly.
In addition to this scrutiny of sequence reads, bactigging produces a list of eBACs that cannot be included in a consistent layout, owing to excessively large numbers of overlaps. These are then re-examined for cross-contamination problems. Similarly, where superbactigging reveals problems with the underlying bactigs, the bactig layouts are re-examined. These and other quality assessment mechanisms are outlined in Table 3.
Table 3.
Quality Assessment Measurements at Various Stages of Atlas Assembly
| Reads | BCM trace-quality (TQ) analysis on all traceruns primary feedback to production group. |
| BCM-cross-repeat scan for wrong-organism repeats (similar to RepeatMasker-primspec). | |
| All reads scanned for first and last 50-base window with no contaminant matches and <1.25 expected errors. | |
| Head and tail beyond the windows trimmed off. Remaining insert required to have 100 bases of Phred quality ≥20 (for WGS) or 50 such bases (for BAC reads). | |
| Trimmed reads used to compute oligo frequencies (32-mers) over all WGS sequence; only oligos with frequency ≤12 (∼3 times the coverage) used to seed overlaps. | |
| Untrimmed BAC and WGS reads used in assembly (masked for contaminant and vector), WGS read must have passed quality or be mate of passed and fished read. | |
| eBAC assembly internal checks | Post-Phrap, paired ends used to split and trim contigs which are inconsistent internally or cannot be consistently scaffolded. |
| BAC purification QC | Each tracerun (96-well group) checked for coassembly against other Traceruns. |
| Coassembly indicated by participation in same contigs (one test) or in same scaffold (for comparison). | |
| Groups of traceruns not coassembling with the bulk of a project are pulled, and if comprising ≥200 passed reads, placed in a “synthetic project.” | |
| (Or relocated to their correct original project if possible, based on both sequence similarity and lab tracking proximity.) | |
| Bactigging QC | Linearized sequence for each enriched BAC scaffold BLASTZ'd against others (rendered efficient by prefilter for shared WGS reads). |
| Enriched BACs with excess overlaps flagged for closer examination in BAC purification. | |
| Bactigs reassembled, scaffolded into superbactigs, laid out by markers. Adjacent bactigs whose terminal BACs had low-confidence overlaps are re-examined for overlaps and joined if confirmed. | |
| Mapping QC | Markers, mouse synteny, human synteny, and FPC all examined simultaneously along with superbactig data primarily driven by BAC ends; feedback between assembly layout of BACs and FPC mapping group. |
| Overall checks | Alignment with finished BACs (and multi-BAC regions from NISC) dot-plotting (BLASTZ and atlas-dot). Alignment and scoring using MUMmer. |
| Large-scale alignment with Mouse, dot-plotting (BLASTZ and atlas-dot); see mapping QC. | |
| Duplications and collapses: | |
| Oligo analysis (24-mers): check for regions overrepresented in assembly (artifactual duplications) especially at bactig and superbactig boundaries-found and corrected small number of cases (<6). | |
| Approximately 4% of unique WGS oligomers missing in final assembly (as compared with 1% in Mouse)-Oligomers with frequency 20-50 underrepresented in Mouse (by 10% to 20%)-oligomer representation in Rnor 3.1 consistently ∼96% beyond frequency = 100. | |
| cDNA and EST alignment consistency. |
Acknowledgments
This project was supported by grant U54 HG02345 from the NHGRI and NHLBI to R.A.G. We thank members of the Rat Genome Sequencing Project Consortium who provided the data for this project and provided feedback as to the quality of the assembly. At the BCM-HGSC we particularly thank Bingshan Li, Yue Liu, Qin Xiang, and Erica Sodergren, who provided invaluable assistance and input to this project; David Wheeler and Zhengdong Zhang also contributed to the quality assessment. We are grateful to John Bouck and Harley Gorrell for making early contributions to the work. We thank Gerard Bouffard and Eric Green (NHGRI) for access to shotgun reads and finished sequences on rat BACs that were used for quality checks of this assembly and Ann Kwitek and Howard Jacob (Medical College of Wisconsin) for assistance with the rat radiation hybrid map.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2264004.
Footnotes
[The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: the Rat Genome Sequencing Project Consortium, Gerard Bouffard, and Eric Green.]
References
- Adams, M.D., Celniker, S.E., Holt, R.A., Evans, C.A., Gocayne, J.D., Amanatides, P.G., Scherer, S.E., Li, P.W., Hoskins, R.A., Galle, R.F., et al. 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185-2195. [DOI] [PubMed] [Google Scholar]
- Aparicio, S., Chapman, J., Stupka, E., Putnam, N., Chia, J.M., Dehal, P., Christoffels, A., Rash, S., Hoon, S., Smit, A., et al. 2002. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297: 1301-1310. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796-815. [DOI] [PubMed] [Google Scholar]
- Bailey, J.A., Gu, Z., Clark, R.A., Reinert, K., Samonte, R.V., Schwartz, S., Adams, M.D., Myers, E.W., Li, P.W., and Eichler, E.E. 2002. Recent segmental duplications in the human genome. Science 297: 1003-1007. [DOI] [PubMed] [Google Scholar]
- Batzoglou, S., Jaffe, D.B., Stanley, K., Butler, J., Gnerre, S., Mauceli, E., Berger, B., Mesirov, J.P., and Lander, E.S. 2002. ARACHNE: A whole-genome shotgun assembler. Genome Res. 12: 177-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner, F.R., Plunkett III, G., Bloch, C.A., Perna, N.T., Burland, V., Riley, M., Collado-Vides, J., Glasner, J.D., Rode, C.K., Mayhew, G.F., et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277: 1453-1474. [DOI] [PubMed] [Google Scholar]
- Bouck, J., Miller, W., Gorrell, J.H., Muzny, D., and Gibbs, R.A. 1998. Analysis of the quality and utility of random shotgun sequencing at low redundancies. Genome Res. 8: 1074-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The C. elegans Sequencing Consortium. 1998. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science 282: 2012-2018. [DOI] [PubMed] [Google Scholar]
- Celniker, S.E., Wheeler, D.A., Kronmiller, B., Carlson, J.W., Halpern, A., Patel, S., Adams, M., Champe, M., Dugan, S.P., Frise, E., et al. 2002. Finishing a whole-genome shotgun: Release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 3: RESEARCH0079, 1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao, K.M., Pearson, W.R., and Miller, W. 1992. Aligning two sequences within a specified diagonal band. Comput. Appl. Biosci. 8: 481-487. [DOI] [PubMed] [Google Scholar]
- Chen, R., Sodergren, E., Weinstock, G.M., and Gibbs, R.A. 2004. Dynamic building of a BAC clone tiling path for the rat genome sequencing project. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Choi, V. and Farach-Colton, M. 2003. Barnacle: An assembly algorithm for clone-based sequences of whole genomes. Gene 320: 165-176. [DOI] [PubMed] [Google Scholar]
- Cole, S.T., Brosch, R., Parkhill, J., Garnier, T., Churcher, C., Harris, D., Gordon, S.V., Eiglmeier, K., Gas, S., Barry III, C.E., et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393: 537-544. [DOI] [PubMed] [Google Scholar]
- Dehal, P., Satou, Y., Campbell, R.K., Chapman, J., Degnan, B., De Tomaso, A., Davidson, B., Di Gregorio, A., Gelpke, M., Goodstein, D.M., et al. 2002. The draft genome of Ciona intestinalis: Insights into chordate and vertebrate origins. Science 298: 2157-2167. [DOI] [PubMed] [Google Scholar]
- Delcher, A.L., Phillippy, A., Carlton, J., and Salzberg, S.L. 2002. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 30: 2478-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing, B. and Green, P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8: 186-194. [PubMed] [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M.C., and Green, P. 1998. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 8: 175-185. [DOI] [PubMed] [Google Scholar]
- Fleischmann, R.D., Adams, M.D., White, O., Clayton, R.A., Kirkness, E.F., Kerlavage, A.R., Bult, C.J., Tomb, J.F., Dougherty, B.A., Merrick, J.M., et al. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269: 496-512. [DOI] [PubMed] [Google Scholar]
- Goff, S.A., Ricke, D., Lan, T.H., Presting, G., Wang, R., Dunn, M., Glazebrook, J., Sessions, A., Oeller, P., Varma, H., et al. 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92-100. [DOI] [PubMed] [Google Scholar]
- Goffeau, A., Barrell, B.G., Bussey, H., Davis, R.W., Dujon, B., Feldmann, H., Galibert, F., Hoheisel, J.D., Jacq, C., Johnston, M., et al. 1996. Life with 6000 genes. Science 274: 546-567. [DOI] [PubMed] [Google Scholar]
- Holt, R.A., Subramanian, G.M., Halpern, A., Sutton, G.G., Charlab, R., Nusskern, D.R., Wincker, P., Clark, A.G., Ribeiro, J.M., Wides, R., et al. 2002. The genome sequence of the malaria mosquito Anopheles gambiae. Science 298: 129-149. [DOI] [PubMed] [Google Scholar]
- Huang, X., Wang, J., Aluru, S., Yang, S.P., and Hillier, L. 2003. PCAP: A whole-genome assembly program. Genome Res. 13: 2164-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson, D.H., Reinert, K., Kravitz, S.A., Remington, K.A., Delcher, A.L., Dew, I.M., Flanigan, M., Halpern, A.L., Lai, Z., Mobarry, C.M., et al. 2001. Design of a compartmentalized shotgun assembler for the human genome. Bioinformatics 17 Suppl 1: S132-S139. [DOI] [PubMed] [Google Scholar]
- Jaffe, D.B., Butler, J., Gnerre, S., Mauceli, E., Lindblad-Toh, K., Mesirov, J.P., Zody, M.C., and Lander, E.S. 2003. Whole-genome sequence assembly for mammalian genomes: Arachne 2. Genome Res. 13: 91-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp, R.M. and Rabin, M.O. 1987. Efficient randomized pattern-matching algorithms. IBM J. Res. Dev. 31: 249-260. [Google Scholar]
- Kent, W.J. and Haussler, D. 2001. Assembly of the working draft of the human genome with GigAssembler. Genome Res. 11: 1541-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S. and Segre, A.M. 1999. AMASS: A structured pattern matching approach to shotgun sequence assembly. J. Comp. Biol. 6: 163-186. [DOI] [PubMed] [Google Scholar]
- Kirkness, E.F., Bafna, V., Halpern, A.L., Levy, S., Remington, K., Rusch, D.B., Delcher, A.L., Pop, M., Wang, W., Fraser, C.M., et al. 2003. The dog genome: Survey sequencing and comparative analysis. Science 301: 1898-1903. [DOI] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Mewes, H.W., Albermann, K., Bahr, M., Frishman, D., Gleissner, A., Hani, J., Heumann, K., Kleine, K., Maierl, A., Oliver, S.G., et al. 1997. Overview of the yeast genome. Nature 387: 7-65. [DOI] [PubMed] [Google Scholar]
- Mullikin, J.C. and Ning, Z. 2003. The phusion assembler. Genome Res. 13: 81-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers, E.W., Sutton, G.G., Delcher, A.L., Dew, I.M., Fasulo, D.P., Flanigan, M.J., Kravitz, S.A., Mobarry, C.M., Reinert, K.H., Remington, K.A., et al. 2000. A whole-genome assembly of Drosophila. Science 287: 2196-2204. [DOI] [PubMed] [Google Scholar]
- Owolabi, O. and McGregor, D.R. 1988. Fast approximate string matching. Software Practice and Experience 18: 387-393. [Google Scholar]
- Rat Genome Sequencing Project Consortium. 2004. Genome sequence of the Brown Norway Rat yields insights into mammalian evolution. Nature (in press). [DOI] [PubMed]
- Schwartz, S., Kent, W.J., Smit, A., Zhang, Z., Baertsch, R., Hardison, R.C., Haussler, D., and Miller, W. 2003. Human-mouse alignments with BLASTZ. Genome Res. 13: 103-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderlund, C., Longden, I., and Mott, R. 1997. FPC: A system for building contigs from restriction fingerprinted clones. Comput. Appl. Biosci. 13: 523-535. [DOI] [PubMed] [Google Scholar]
- Stojanovic, N., Chang, J.L., Lehoczky, J., Zody, M.C., and Dewar, K. 2002. Identification of mixups among DNA sequencing plates. Bioinformatics 18: 1418-1426. [DOI] [PubMed] [Google Scholar]
- Tuzun, E., Bailey, J.A., and Eichler, E.E. 2004. Recent segmental duplications in the working draft assembly of the brown Norway rat. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Venter, J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304-1351. [DOI] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- Yu, J., Hu, S., Wang, J., Wong, G.K., Li, S., Liu, B., Deng, Y., Dai, L., Zhou, Y., Zhang, X., et al. 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296: 79-92. [DOI] [PubMed] [Google Scholar]