Abstract
The rat is an important animal model for human diseases and is widely used in physiology. In this article we present a new strategy for gene discovery based on the production of ESTs from serially subtracted and normalized cDNA libraries, and we describe its application for the development of a comprehensive nonredundant collection of rat ESTs. Our new strategy appears to yield substantially more EST clusters per ESTs sequenced than do previous approaches that did not use serial subtraction. However, multiple rounds of library subtraction resulted in high frequencies of otherwise rare internally primed cDNAs, defining the limits of this powerful approach. To date, we have generated >200,000 3′ ESTs from >100 cDNA libraries representing a wide range of tissues and developmental stages of the laboratory rat. Most importantly, we have contributed to ∼50,000 rat UniGene clusters. We have identified, arrayed, and derived 5′ ESTs from >30,000 unique rat cDNA clones. Complete information, including radiation hybrid mapping data, is also maintained locally at http://genome.uiowa.edu/clcg.html. All of the sequences described in this article have been submitted to the dbEST division of the NCBI.
Genomic resources have proven to be very useful in both human and mouse genetic studies. For example, the human UniGene set (Schuler 1997) and GeneMap `99 (Deloukas et al. 1998) at National Center for Biotechonology Information (NCBI) were invaluable in the recent identification of two Bardet-Biedl syndrome genes (Nishimura et al. 2001; Mykytyn et al. 2002). The rat also provides several excellent established physiological and biochemical models for the study of genetically complex human diseases, including hypertension (Hilbert et al. 1991; Jacob et al. 1991), renal disease (Brown et al. 1996), behavioral disorders (Moisan et al. 1996), and auto-immune disorders (Jacob et al. 1992). To make efficient use of the rat as a model for human disease and physiology, a first step is to identify a comprehensive set of genes. This process of gene identification is dubbed gene discovery.
Large-scale production of expressed sequence tags (ESTs) from arrayed cDNA clones has proven to be the most efficient and cost-effective strategy for gene discovery. EST-based gene discovery strategies are advantageous for a few reasons, not the least of which is that purely computational methods of gene prediction are notoriously inaccurate in higher eukaryotes, in which only a small fraction of the genome codes for transcribed genes (Guigo et al. 2000). However, it is the coupling of both EST and genomic sequence data that is most desirable as it allows for determination of the structure of each gene. It is also noteworthy that ESTs are invaluable for genome sequence annotation and for identification of orthologous relationships between sequences of different, and often evolutionarily distant, organisms. The latter is of essence in using rat models for the study of human diseases.
Typically, cDNA libraries used for production of ESTs are oligo-dT-primed and directionally cloned. Thus, the sequence obtained from the 3′ end of a cDNA clone, that is, 3′ EST, corresponds to the 3′ end of the mRNA. Depending upon the length of both the 3′ EST and that of the 3′ untranslated sequence of the mRNA, a 3′ EST may contain none or very limited coding sequence information. Because 3′ untranslated sequences are less conserved than are coding regions as a general rule (Makalowski and Boguski 1998) and are relatively long (750-bp average length; Pesole et al. 1999), a 3′ EST can be used as a fingerprint to unequivocally identify a transcript. For this reason, each unique 3′ EST that contains a bona fide polyadenylation signal sequence and tail can be tentatively considered as representing a different mRNA and, except for the cases of alternative splicing and/or differential polyadenylation, a different transcription unit. Conversely, 5′ ESTs are derived from the 5′ ends of the cDNAs and may encompass 5′ untranslated, coding, and/or 3′ untranslated sequences depending upon the length of the EST and whether the cDNA corresponds to a full-length or a truncated copy of the mRNA. Typically, however, 5′ ESTs span coding sequences and thus often enable identification of similarities between evolutionarily conserved sequences from different organisms. The latter is invaluable in identifying orthologous relationships and candidate functions to otherwise unknown transcripts. It is noteworthy that ESTs are single-pass sequences, and as such have an error rate of ∼3% (Hillier et al. 1996). This poses certain challenges to computational methods for identification of nonredundant sets of ESTs, such as NCBI's UniGene collections.
The EST approach to gene discovery has been successfully applied to a number of organisms (Adams et al. 1995; Hillier et al. 1996; Marra et al. 1999; Dimopoulos et al. 2000; Blackshear et al. 2001; Whitfield et al. 2002). It should be acknowledged, however, that despite its advantages, there are certain limitations to this approach, not the least of which is the redundant generation of ESTs derived from the most common transcripts, that is, mitochondrial RNAs, ribosomal RNAs, and mRNAs of the super-prevalent and intermediate frequency classes (Bishop et al. 1974). This is a problem that can significantly impair the overall efficiency of a gene discovery program that relies solely on the generation of ESTs from cDNA clones randomly picked from standard (non-normalized) libraries. Accordingly, the use of normalized cDNA libraries in which all clones are represented at a comparable frequency (Soares et al. 1994; Bonaldo et al. 1996) has proven most advantageous (Hillier et al. 1996; Marra et al. 1999; Dimopoulos et al. 2000; Blackshear et al. 2001; Whitfield et al. 2002). It is noteworthy, however, that the process of normalization only contributes to minimize redundancies within libraries, and it is particularly advantageous to minimize redundant identification of tissue-specific mRNAs. Redundant production of ESTs derived from ubiquitously expressed mRNAs constitutes a major problem at intermediate to advanced phases of gene discovery programs. Hence, we have argued that this problem can be more effectively addressed by the use of subtractive libraries that are progressively enriched for novel ESTs (Bonaldo et al. 1996; Soares 1997). This is the rationale behind our strategy to generate ESTs from serially subtracted normalized libraries.
Serial subtraction of normalized libraries is an iterative process whereby arrayed sets of cDNAs, from which ESTs have been derived, are pooled and used as a driver in a subtractive hybridization with one or a pool of normalized or subtracted libraries. It is noteworthy that our cDNA clones contain library-specific sequence tags to enable computational identification of library and tissue of origin of ESTs obtained from pooled libraries (Gavin et al. 2002). Because the representation of the driver population is significantly reduced in the resulting subtracted library, redundant generation of ESTs is greatly minimized. Hence, every new library of a series is enriched for novel and progressively rarer ESTs. Here we describe the use of this strategy to identify a comprehensive nonredundant collection of rat ESTs with unprecedented efficiency.
RESULTS
Sequencing and Gene Discovery
Within the scope of this project, >100 individually tagged oligo-dT-primed directionally cloned cDNA libraries were constructed, collectively representing a wide range of tissues and stages of development. These are referred to as start libraries. A complete list of the start libraries constructed for this project is presented in Table 1.
Table 1.
Description of Tissue Sources
| Library | Description of mRNA Sources |
|---|---|
| A0 | Adult rat placenta, lung, brain, liver, kidney, heart, spleen, ovary, and muscle |
| E0 | Whole embryo from 8 dpc, 12 dpc, and 18 dpc |
| Y0 | Whole eye (minus lens) |
| G0 | Nodose ganglia, dorsal root ganglia, and trigeminal ganglia |
| AA0 | Atrium, 16.5 dpc |
| AB0 | Ventricle, 16.5 dpc |
| AC0 | Atrioventricular canal, 16.5 dpc |
| AD0 | Atrium, 15 dpc |
| AE0 | Ventricle, 15 dpc |
| AF0 | Atrioventricular canal, 15 dpc |
| AG0 | Ventricle, 13 dpc |
| BO0 | Adult brain subregions: thalamus, cerebellum, hypothalamus, medulla, pons, midbrain, cerebral cortex, corpus striatum, and hippocampus |
| BS0 | Whole embryo, 13 dpc |
| BT0 | Hippocampus, thalamus, midbrain, medulla, corpus striatum, cerebral cortex, and testis |
| BV0p | Whole embryo, 13 dpc |
| BX0 | Whole embryo, 13 dpc |
| CJ0 | Testis, mammary gland, and pregnant uterus |
| CM0 | Adult rat heart (same source as in A0) |
| CS0s | Whole heart: 17 dpc, 19 dpc, 21 dpc, 1 dpb, 12 dpb, 75 dpb, and 200 dpb |
| CT0s | Whole brain: 17 dpc, 19 dpc, 21 dpc, 1 dpb, 12 dpb, 75 dpb, and 200 dpb |
| CU0s | Kidney: 17 dpc, 19 dpc, 21 dpc, 1 dpb, 12 dpb, 75 dpb, and 200 dpb |
| CV0 | Eye |
| CW0s | Aorta: 19 dpc, 21 dpc, 1 dpb, 12 dpb, 75 dpb, and 200 dpb |
| CX0s | Placenta: 17 dpc, 19 dpc, and 21 dpc |
| CY0 | Brown adipose tissue |
| CZ0 | Penis |
| DA0 | Salivary gland |
| DB0 | Bladder |
| DC0 | Seminal vesicle |
| DD0 | Fundus |
| DE0 | Cervix |
| DM0 | Prostate |
| DN0 | Distal colon |
| DO0 | Cell line R3327-5P |
| DQ0 | Cell line R3327-5A |
| DR0 | Osteoblast |
| DS0 | Appendix |
| DY0 | Cartilage |
| DZ0 | Cartilaginous tumor |
| EA0 | Ileum |
| EB0 | Duodenum |
| FJ0 | Embryo |
| FS0 | Swarm rat chondrosarcoma |
| GO0 | Embryo |
| GQ0 | Embryo |
| GR0 | Embryo |
dpc = days post conception.
dpb = days post birth.
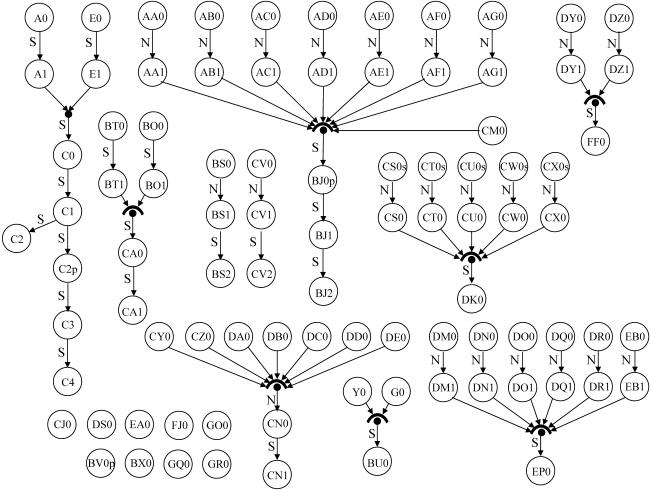
Start libraries were individually normalized, and ESTs were generated from each resulting pair of start and normalized libraries. Several pools of normalized libraries were created, and serially subtracted libraries were derived from each pool. Figure 1 shows the derivation pattern for all of the libraries that were sequenced. Library normalization is indicated by an N-labeled arrow and serial subtraction by an S-labeled arrow. Complete details on the specifics of each particular library are available upon request.
Figure 1.
Summary of libraries created. This figure presents a graphical description of the cDNA libraries created throughout the gene discovery process. The operation of cDNA library normalization is represented by arrows labeled with an “N,” and serial subtraction operations are represented by arrows labeled with an “S.” Pooling of libraries is represented by multiple arrows converging on a single node.
Drawing from cDNA libraries of high sequence complexity collectively representing a large number of tissues and stages of development was essential to maintain a high rate of discovery of novel ESTs throughout the project. This is clearly demonstrated by the brief periods of suppressed novelty when sequencing was performed on single-tissue libraries. From these cDNA libraries, 227,364 ESTs have been generated, and have progressed through the entire sequence analysis pipeline, satisfying all quality criteria. Overall, 90% (205,609) of these ESTs have an identifiable polyadenylation tail. Of those with a polyadenylation tail, 63% (130,331) have one of the two canonical polyadenylation signals (AAUAAA and AUUAAA), and 13.9% (28,564) have an alternative polyadenylation signal (Beaudoing et al. 2000). From those ESTs with a polyadenylation tail, 194,259 (94.5%) have an identifiable tissue tag, allowing determination of tissue of origin even after pooling with other cDNA libraries. This correlation to tissue of origin can only be accomplished in ESTs with a polyadenylation tail because the synthetic library tag is encoded in the oligonucleotide used to prime first-strand cDNA synthesis. The average read-length after trimming for quality and removal of contaminating vector sequence is 429 bases, with an average phred quality value of 34. ESTs were generated from the 5′ end for a nonredundant set of cDNAs. These clones were first rearrayed and then sequenced by using the same sequence processing pipeline used for the 3′ sequences. Every EST (3′ and 5′) generated during this project that was uncontaminated and of sufficient quality was annotated with library information and any other identified feature and submitted to the dbEST database at the NCBI.
Novelty Assessment
By using the UIcluster (version 3.0.5) program, the 3′ EST sequences generated at the University of Iowa were clustered along with 56,000 3′ ESTs from TIGR (derived from the single-tissue normalized libraries pooled to create the A0 and E0 libraries) and a nonredundant set of rat mRNA sequences. This yielded 57,536 clusters from the Iowa-derived ESTs (plus an additional 2673 with the TIGR ESTs), including 28,883 clusters with only a single EST in them. Of these clusters, 51,818 contain ESTs that have an identified polyadenylation tail. Approximately half of these clusters (26,700) were identified as containing one of the two canonical polyadenylation signals. Of note is that the singleton clusters, representing the rarest of observed clones, have a reduced prevalence of detected polyadenylation signal. The prevalence of polyadenylation signal in singleton clusters was 16% lower than for clusters containing two or more sequences.
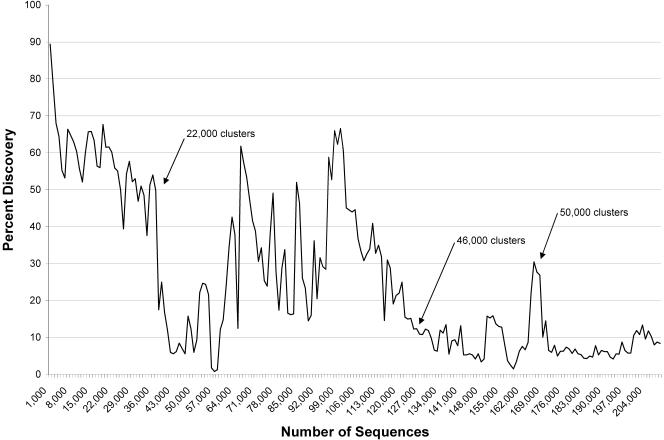
Figure 2 shows the instantaneous novelty in increments of 1000 sequences throughout the EST discovery process. The values indicate the percentage of new clusters defined within each group of 1000 sequences. The initial novelty rate approached 90%, but quickly fell to ∼60%. It is noteworthy that after a single year, and only 40,000 ESTs, there were 22,000 clusters already defined. Next, focused gene discovery in the heart was initiated. Most of the low novelty rate at this point can be attributed to the sequencing of multiple non-normalized, single-tissue libraries from the heart. The more pronounced peaks, between 70,000 and 90,000 sequences, correspond to the sequencing of the normalized rat heart libraries. The subsequent broader peaks represent the discovery obtained from sequencing of the first and second subtracted rat heart libraries. Note that by the time that 120,000 sequences were generated, 46,000 clusters had been identified. Since then, many new single-tissue libraries from previously unsurveyed tissues provided by the Sanger Centre have been constructed and used for production of ESTs. The two peaks at 150,000 and 170,000 sequences demonstrate the relative discovery from the sequencing of the normalized and subtracted versions of a pool of these libraries.
Figure 2.
Incremental discovery throughout gene discovery. This graph tracks the novelty per unit (1000 sequences) for the entire gene discovery project. Peaks in the incremental novelty rate correlate with sequencing of pooled subtracted cDNA libraries. The troughs in the graph correlate with sequencing single tissue non-normalized cDNA libraries.
DISCUSSION
Several analyses were performed on the collected sequence and clustering data. A characterization of the clones and ESTs is presented. Then an assessment of observed alternative polyadenylation is described. Next, a discussion of how clustering results were used to assess both the novelty and quality of the cDNA libraries is presented. The prevalence of alternative polyadenylation was assessed based upon detection of previously described alternative polyadenylation signals. The impact of including mRNA sequences in the clustering procedure is assessed, as well as the correlation between those clusters and those from NCBI's rat UniGene set. The comprehensiveness of the EST collection is then assessed through measuring the representation of known rat mRNAs. Next, cDNA-based gene discovery and full-length sequencing projects are contrasted. The effects of cDNA library normalization and serial subtraction were analyzed to evaluate their impact on the prevalence and length of polyadenylation tails and the use of polyadenylation signal. Finally, the efficiency of gene discovery for this project is compared with that of the human and mouse EST-based gene discovery projects.
Characterization of Clones and Sequences
Insert sizes in the libraries used for rat EST discovery range between 0.4 and 2.5 kb. Thus, one should not expect a significant fraction of the clones in this collection to be full-length. It should be emphasized, however, that the main purpose of this work was to document the feasibility of serial subtraction of normalized libraries as a strategy for rapid development of comprehensive nonredundant collections of ESTs. Because existing subtractive hybridization procedures are not well suited for subtraction of full-length–enriched cDNA libraries, as the presence of a truncated cDNA in a driver population will potentially lead to subtraction of all complementary sequences, i.e., truncated and full-length, the strategy that we devised for EST discovery based on serial subtraction of normalized libraries is best applicable to oligo-dT-primed, directionally cloned cDNA libraries with insert sizes within a narrow range to minimize differential growth of clones with significantly different insert sizes.
It is important to note that the purpose of this sequencing project was to define a comprehensive set of transcribed sequences in the rat. The library normalization and serial subtraction strategy is designed specifically to reduce the redundancy within the set of clones to be sequenced. This reduction of redundancy greatly increases the rate at which novel transcripts are identified. However, this has the additional effect of reducing the ability to validate novel transcripts through the observation of multiple independent cDNA copies. Multiple independent occurrences (ESTs) are commonly used to validate the existence and structure of an otherwise novel transcript. This effect is lessened due to the generation of 5′ EST sequences from a non-redundant set of clones. These sequences often capture significant amounts of coding sequence and splicing, both of which are alternative validation methods. Thus, the synthesis of alignments from paired 3′ and 5′ ESTs serve as validation for most gene structures.
Based upon an analysis of the available rat mRNAs, the average 3′ UTR length observed is 935 bp. This is based upon a set of 4981 nonredundant rat mRNAs with annotated CDS extracted from the rat UniGene build. This figure is significantly longer than those reported in Makalowski and Boguski (1998), in which the average 3′ UTR size was reported to be 411 bp in orthologous human and rodent mRNAs. Only 3′ ESTs with polyadenylation tail and signal were used to validate complete UTR length. This allowed the calculation of UTR length when the mRNA sequence contained a truncated UTR.
To estimate the fraction of cDNAs expected to contain at least partial coding sequence (CDS), we compared 5′ ESTs to the set of rat mRNAs with annotated CDS. This analysis was performed by using a set of 4313 5′ ESTs from cDNA clones derived from normalized libraries with 3′ EST that contained polyadenylation tail and signal. Of the 4313 5′ ESTs, 1193 (28%) matched an mRNA, from which 765 (65%) reached the annotated CDS region.
To estimate the frequency of internal priming, a comparison was made of the alignment of our 3′ ESTs to the known rat mRNAs. Specifically, those 3′ ESTs with polyadenylation tail were assessed based upon the start of alignment. Because of the method used in the creation of the cDNA libraries, the polyadenylation tail is (in general) at least 18 bp long. We used evidence of internal sequence homology that extended significantly into the polyadenylation tail of the cDNA as evidence of internal priming. The assessment was made across a wide diversity of libraries to also assess for variations in the prevalence of internal priming based upon the depth of serial subtraction.
The average rate of internal priming across all of the libraries analyzed was 24.7%. In other words, one fourth of sequences with a polyadenylation tail appear to be caused by an internal priming event. In general, the rate of internal priming increased with increasing numbers of library manipulations. The highest rates of internal priming were observed in the C3 and C4 libraries (37% and 39%, respectively), which are also the most heavily subtracted libraries.
Because of the verification procedures used, we have excellent statistics on the overall rates of clone tracking errors. The clone error rate differed significantly depending on whether a slab-based (ABI 377) or capillary-based (ABI 3700) sequencing platform was used. In the case of the ABI 377 platform, the error rate was 6.1% (912/14,980). In the case of the ABI 3700 platform, the error rate was 3.7% (273/7248). This represents a significant reduction in the overall error rate of the clone collection. It should be noted that because the error may have occurred either in the original sequencing or during verification, the error rate of the clone collection is expected to be half of the rates reported above.
Assessment of Alternative Polyadenylation
One important set of data contained in the clustering analysis is that of polyadenylation signal usage. Previously reported results in humans indicate an 85% utilization of the two canonical polyadenylation signals, with the remaining 15% corresponding to one of 14 alternative polyadenylation signals (Beaudoing et al. 2000). Table 2 shows a distribution of polyadenylation signal usage for those sequences with a polyadenylation tail. Here we see that in good agreement with the previous analysis, 82% (130,331/158,895) of sequences with a polyadenylation signal used one of the two canonical signals (AAUAAA, AUUAAA). A summary of observed alternative polyadenylation signals is presented in Table 3. The relative observed prevalence of the individual signals agrees with previously published results. The most frequently observed alternative (i.e., noncanonical) signals were AGUAAA and UAUAAA, and GGGGCU was the least frequent. The most prominent difference from previous results is the relatively low number of ESTs with an observed AAUAUA polyadenylation signal. In the ESTs analyzed, only 1.1% had an AAUAUA signal, compared with 1.7% in the mRNAs analyzed by Beaudoing et al. (2000).
Table 2.
Observed Distribution of Polyadenylation Signals
| PolyA signal type | Number found in sequences | Number found in clusters |
|---|---|---|
| Canonical | 130,331 | 26,700 |
| Alternative | 28,564 | 8,635 |
| Not detected | 46,714 | 16,483 |
Table 3.
Observed Distribution of Alternative Polyadenylation Signals
| Alternative polyadenylation signal | Prevalence |
|---|---|
| AGUAAA | 4685 |
| UAUAAA | 4140 |
| UUUAAA | 3442 |
| CAUAAA | 2463 |
| AAGAAA | 2401 |
| AAUACA | 2145 |
| GAUAAA | 1718 |
| AAUAUA | 1693 |
| AAAACA | 1623 |
| ACUAAA | 1125 |
| AAUGAA | 1110 |
| AAAAAG | 1106 |
| AAUAGA | 687 |
| GGGGCU | 226 |
It remains to be determined whether the increased representation of 3′ ESTs with alternative polyadenylation signal sequence in serially subtracted libraries is a manifestation of enrichment for rarer transcripts in these libraries. The underlying hypothesis is that polyadenylation of transcripts with weaker alternative signals occurs at lower efficiency, thus resulting in low representation of such transcripts in the A+ population.
Assessment of Library Quality
The ESTs are clustered locally to minimize the delay in providing feedback on library novelty and quality. Rapid assessment of novelty makes selection of libraries for further sequencing possible. Naturally, as more ESTs are derived from a library, the rate of new cluster identification decreases, and it becomes unproductive to continue to generate ESTs from the same library. Hence, the utilization of pooled normalized and/or serially subtracted libraries of high sequence complexity can contribute significantly to increase EST discovery rates. Per-library novelty assessment can also serve as a critical clue in determining contamination problems.
It is important to note that several steps were taken to minimize the incorporation of genomic contaminants within this clone set. Foremost was the treatment of all RNA samples with DNAse to degrade any DNA present after RNA isolation. Because of the utilization of total cellular RNA it is possible that some unprocessed or semiprocessed nuclear transcripts were included. Internal priming in such messages could then produce clones primed from within an intron, thus contributing to genomic contamination. Hence, it is conceivable that a fraction of the 3′ ESTs with tail but no detectable polyadenylation signal sequence may have resulted from internal priming at A-rich stretches within introns of such contaminating nuclear transcripts. Although our analysis indicates the presence of genomic contaminants to be very low, ultimately this question will be best addressed by comparison to a fully annotated rat genomic sequence, once available.
Cluster Merging
Incorporation of the available rat mRNA sequences allows greater accuracy in bringing ESTs derived from the same gene together into the same cluster. Such merging behavior is caused by non-overlapping EST sequences derived from the same gene. Such ESTs may occur for several reasons, the most common being multiple polyadenylation signals, internal cDNA priming, and internal (NotI) cloning sites. Using the mRNA sequences resulted in merging 800 clusters. There were several hundred rat mRNAs to which no corresponding ESTs could be identified. These mRNAs typically lack 3′ UTR sequence, significantly restricting the region to which the ESTs are expected to align. The utilization of genomic sequence to automatically guide the EST clustering process is currently being investigated.
Comparison to NCBI's Rat UniGene
Overall, the set of clusters generated by UIcluster is very similar to the NCBI's UniGene collection. An analysis on cluster composition indicated that only 2.5% of sequences in the same UI cluster appeared in different NCBI UniGene clusters. Similarly, 5.3% of sequences in the same NCBI UniGene cluster appeared in different UI clusters. Of the 53,354 clusters in the current build of the rat NCBI UniGene set, 49,285 (92%) contain at least one EST contributed by this project. In 27,799 (52%) of the clusters, the only ESTs in the cluster are those derived from this project.
Comprehensive Coverage of Rat Transcripts
Of the 5424 nonredundant rat mRNAs available from NCBI's rat UniGene build, 4360 were represented by cDNAs in our clone collection. Of the 1064 mRNAs not represented in our clone collection, 617 are not represented by any EST in the rat UniGene build. Thus, only a small fraction (447; 8%) of rat mRNAs are represented by ESTs other than those described in this manuscript.
Comparison to Full-Length Sequencing Projects
Recent projects involved in identifying novel full-length transcripts are now under way for several species (Stapleton et al. 2002; Strausberg et al. 2002). These projects allow for an interesting comparison to traditional 3′ EST-based gene discovery projects, such as the one described here. Although both projects use cDNA libraries as their fundamental resource, full-length projects use 5′ ESTs to identify those cDNA clones likely to represent full-length cDNAs, whereas traditional gene discovery projects use 3′ ESTs. This is an important difference, as significant effort must be expended either to enrich the cDNA libraries for full-length clones or to sequence sufficient numbers of clones to eventually identify full-length candidates. Both strategies suffer from the large amount of redundancy in the underlying mRNA population. The normalization and subtraction methods described in this article can be used to significantly reduce the redundancy in traditional gene discovery projects. It is noteworthy that specific processes do not currently exist for subtraction of full-length libraries.
Impact of cDNA Library Normalization and Serial Subtraction
In Tables 4 and 5, we present a step-by-step assessment of key EST features across a series of cDNA libraries. These libraries span a spectrum from initial, single tissue libraries to multiply subtracted libraries (e.g., C3 and C4). A trend was observed when the percentage measures of ESTs with polyadenylation tail and signal were viewed jointly. The prevalence of canonical polyadenylation signal declined steadily throughout the series of libraries. Of particular interest was the C4 library. In this library, the alternative polyadenylation signal also began to decrease, likely due to an increase in the frequency of otherwise rare internally primed cDNAs. We have observed that as a general rule, prevalent ESTs without a polyadenylation tail correspond to ribosomal cDNAs, which are at times common in start libraries. These analyses led us to conclude that maximum benefit of this strategy is attained with up to, but no more than, two rounds of subtraction. Additional rounds of subtraction, although successful in facilitating identification of rarer ESTs, are compromised by the increased representation of rare ESTs derived from 3′ end-truncated internally primed cDNAs.
Table 4.
Impact of Serial-Subtraction Procedures on Polyadenylation Tail and Signal
|
Polyadenylation signal
|
||||||
|---|---|---|---|---|---|---|
| Library | Number of 3′ ESTs | Percentage tail | Average tail length | Canonical | Alternative | None detected |
| A0 + E0 | 7118 | 84.29 | 26.4 | 70.18 | 12.31 | 17.50 |
| A1 + E1 | 6861 | 87.96 | 26.5 | 67.64 | 13.14 | 19.22 |
| C0 | 6479 | 85.64 | 26 | 61.49 | 14.71 | 23.81 |
| C1 | 4169 | 82.32 | 24 | 56.50 | 16.38 | 27.13 |
| C2p | 4804 | 79.68 | 23 | 54.57 | 16.82 | 28.61 |
| C3 | 3163 | 77.39 | 22 | 50.00 | 17.08 | 32.92 |
| C4 | 2290 | 75.32 | 22 | 44.64 | 15.36 | 40.00 |
Table 5.
Impact of Normalization and Subtraction on Polyadenylation Tail and Signal
| Polyadenylation signal
|
||||||
|---|---|---|---|---|---|---|
| Library | Number of 3′ ESTs | Percentage tail | Tail length | Canonical | Alternative | None detected |
| Start | 5832 | 77.43 | 27.15 | 84.39 | 7.32 | 8.28 |
| Normalized | 5437 | 80.30 | 25.1 | 77.28 | 10.65 | 12.07 |
| BJ0p | 4116 | 85.45 | 25 | 72.53 | 14.50 | 12.97 |
| BJ1 | 8459 | 82.01 | 24 | 67.20 | 14.23 | 18.57 |
| BJ2 | 5256 | 81.35 | 24 | 67.12 | 14.85 | 18.03 |
Efficiency of Discovery and Comparison to Relative Discovery in Other Projects
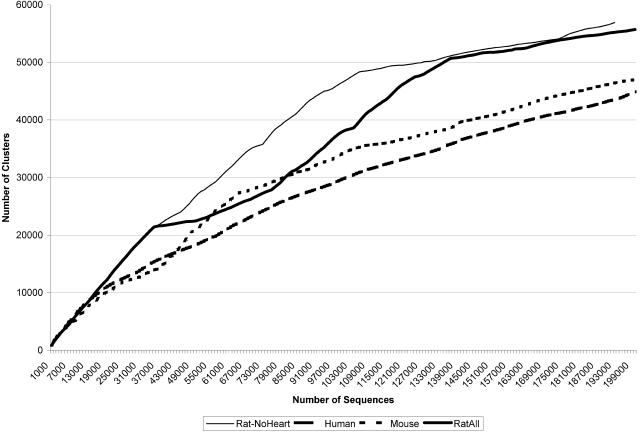
The graph of gene discovery below demonstrates the effectiveness of discovery using serially subtracted cDNA libraries. Figure 3 presents the total number of EST-based clusters identified for a given number of EST sequences. For the first 20,000 sequences, there was relatively little difference between the overall discovery in human, mouse, and rat. However, beyond this point, the rate that novel clusters were identified dropped significantly in both human and mouse. The cause is clearly demonstrated by observing the steady increase in novel cluster discovery. Significantly more clusters were identified before the discovery rate decreased.
Figure 3.
Gene discovery in rat, mouse, and human. Discovery per 1000 sequences is presented to compare the rates of EST-based gene discovery among human, mouse and rat. The rat discovery rate is presented with and without the rat heart libraries to demonstrate the power of focused gene discovery with normalization and serial subtraction of cDNA libraries.
As can be seen in Figure 2, after several critical pooling and subtractions, the discovery rate in rat was significantly increased. This can be seen most dramatically at 65,000 and 100,000 EST sequences. These peaks correspond to the sequencing of several pooled, subtracted libraries derived from microdissected brain regions and previously unsurveyed tissues. To date, the final number of clusters identified in the human and mouse UniGene data set are 121,000 and 88,000, respectively, derived from a total of 1,378,000 human and 1,372,000 mouse 3′ ESTs. Thus, a significant portion of the EST discovery in these projects is not represented in Figure 3. Instead, this figure demonstrates the power of serial subtraction to accelerate gene discovery. This analysis was performed by using only 3′ ESTs to avoid biasing against the human and mouse data sets in which significant 5′ EST sequencing was performed. The ESTs were ordered by accession number to approximate the order in which the sequences were submitted to dbEST (Boguski et al. 1993). In all, the 193,000 locally generated 3′ ESTs (without the heart ESTs) identified 57,000 unique clusters in the rat. It should be emphasized that this corresponds to ∼25% to 30% more clusters identified than in both the human and mouse EST projects after a similar number of sequences had been generated. The heart-derived sequences were removed from this comparison to properly highlight the strategy based upon serial subtraction of normalized libraries. The heart libraries were constructed in parallel as part of this project but were subtracted only for sequences identified in heart libraries. This was done to construct a comprehensive set of heart-derived transcripts for use in cDNA-based microarray studies (J. Laffin, in prep.). When the heart-derived sequences are included in the clustering (see the thick solid line in Fig. 3), the result is a significant reduction in the discovery of new clusters, as the highest and most ubiquitously expressed sequences are reidentified from the heart libraries.
Of interest is that many of the ESTs in the human and mouse projects were derived from normalized cDNA libraries— particularly in the mouse gene discovery project. This is most obvious from 40,000 to 60,000 sequences in the mouse discovery curve in Figure 3. At this point, the mouse discovery rate accelerates faster than that of human. The majority of sequences in that interval are from normalized libraries produced by M. Bento Soares and M. Fatima Bonaldo.
METHODS
cDNA Library Creation
Directionally cloned start (non-normalized), normalized and serially subtracted cDNA libraries were constructed in a plasmid vector (pT7T3-Pac) from DNAse-treated poly(A)+ mRNA isolated from a number of tissues and stages of development of the laboratory rat (Sprague-Dawley), as previously described (Soares 1994; Bonaldo et al. 1996; Soares and Bonaldo 1998). Briefly, first-stranded cDNA was primed with a poly-dT oligonucleotide (TGT TACCATTCTGATGTTGGAGCGGCCGC-N[6-10]-T[18]) that contained a NotI restriction site for directional cloning and a library tag used to identify the tissue of origin (Gavin et al. 2002). Double-stranded cDNA was ligated to EcoRI adaptors (5′-AATTGGCACGAGG-3′, 3′-GCCGTGCTCC-5′), digested with NotI, and directionally cloned into pT7T3-Pac. A complete list of the tissues used is provided in Table 1.
Sequencing
Di-deoxy terminator sequencing was performed in 96-well format by cycle sequencing using rhodamine dye terminator chemistry (ABI). After thermal cycling, sequencing reactions were ethanol precipitated, resuspended in loading buffer containing formamide, denatured, and run on an ABI 377 or an ABI3700 capillary sequencer. A detailed description of our sequencing protocol can be found at http://ratest.eng.uiowa.edu/localdocs/sequencing_protocol.html.
After data capture on the ABI sequencers, the gels were tracked (if necessary) and transferred to a centralized server. From there, the sequences were processed as outlined below and placed into our file-system hierarchy. Nucleotide sequences and perbase quality values were extracted from the ABI-generated chromatograph files (SCF files) by using the phred base-calling program (Ewing et al. 1998).
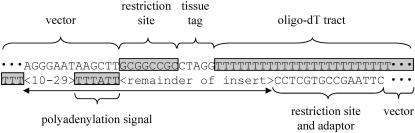
Feature Identification
The set of features that may be identified are presented in Figure 4. These include leading and trailing vector, the cloning restriction sites, library tag, polyadenylation tail, and polyadenylation signal. Every EST should have a cloning restriction site, preceded by vector sequence. The polyadenylation tail and putative polyadenylation signal are both reverse-complemented in this figure because the sequence shown corresponds to that of a 3′ EST. Other potential features that can be detected are repetitive and low-complexity sequences, trailing vector sequence (i.e., reading through the entire insert), and contaminating sequences— bacterial, mitochondrial, and empty vector clones.
Figure 4.
Sequence features of ESTs derived from oligo-dT-primed directionally cloned cDNA libraries. This figure presents the features commonly found in ESTs. The sequence is presented as viewed from the 3′ end, with all expected features shown. These include vector sequence before and after the insert sequence, as well as the flanking restriction sites, inserted library tag, and polyadenylation tail and signal sequence.
The identification of EST features was performed in two stages. First, ESTprep (Scheetz et al. 2003) was used to assess sequence quality and to identify common sequence features, as described above. These features include the initial vector, restriction site, library tag, and polyadenylation tail and signal. ESTprep is configurable with respect to both the set of features to be identified and the level of stringency required. For example, although allofthe 3′ rat EST sequences have a NotI restriction site (used in the cloning process), often basepair errors can accumulate within the NotI recognition sequence (GCGGCCGC); thus, errors are allowed within this sequence. Allowing errors, however, decreases the specificity of the match, thereby increasing the like-lihood of observing such features. To address this problem, detection of leading vector is used to validate detected restriction sites. The end of good-quality sequence is defined as the first stretch of 20 nucleotides in which eight have a phred score <11. This defines the right-most trim site. Only trimmed sequences with (1) at least 100 bases of good quality after trimming, (2) an average per-base phred value >20, and (3) at least 50% of bases greater than phred 20 are propagated through the remainder of the sequence analysis pipeline.
The second stage of feature identification uses the Repeat-Masker program (A.F.A. Smit and P. Green; http://ftp.genome.washington.edu/RM/RepeatMasker.html) to identify ESTs containing contaminating sequence from the bacterial host, the vector, or mitochondrial sequence. ESTs that have significant hits (>85%) to bacterial or mitochondrial sequences or complete vector inserts are removed from further processing. Any trailing vector sequence (due to reading through the cDNA insert) is removed. Sequences containing repetitive elements (such as B1 or LINE elements) or low-complexity (e.g., homopolymeric–AAAAAAA) elements are masked in the ESTs, replacing the nucleotide sequence of the masked region with N's. A complete description of the sequence-processing pipeline can be found in Scheetz and Casavant (2003).
Clustering
Clustering of the 3′ sequences was accomplished by using UIcluster (Trivedi et al. 2002). The input to this process is a set of sequences in the FASTA format and optionally also a list of files containing previously clustered sequences. The sequences used include all rat 3′ ESTs sequenced at The University of Iowa and at The Institute for Genome Research (TIGR; http://www.tigr.org). The clustering was initialized by creating a set of clusters from a non-redundant set of rat mRNA sequences obtained from the NCBI's rat UniGene set.
Default parameters were used when adding the ESTs to the clustering, except as noted below. Options were used to locate the best match to a cluster (rather than the first that matched ≥95%), to search with both the given sequence and its reverse complement, and to select the longest sequence in a cluster as the cluster representative.
The construction of the clustering is a continual process, performed weekly, in which the previous week's output is used as the input for the current week. This is done to minimize rearrangement of clusters from build to build. Manual modifications are made, when necessary, to correct for sequences that are falsely drawn together or when a longer sequence revealed an overlap between clusters.
Acknowledgments
This research was supported in part by NIH-2RO1-HL597898. V.C.S. is an associate investigator of the Howard Hughes Medical Institute. We thank Stephen Brown, Fred Lamb, Hong Lan, Jane B. Lian, the McArdle Laboratory of Cancer Research, Jose Morcuende, Annie Novakovich, Gary S. Stein, Jeff Stevens, Bob Strausberg, and Phillip Wackym for providing samples from several previously unsurveyed tissues.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1414204.
Footnotes
[The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: S. Brown, F. Lamb, H. Lan, J.B. Lian, the McArdle Laboratory of Cancer Research, J. Morcuende, A. Novakovich, G.S. Stein, J. Stevens, B. Strausberg, and P. Wackym.]
References
- Adams, M.D., Kerlavage, A.R., Fleischmann, R.D., Fuldner, R.A., Bult, C.J., Lee, N.H., Kirkness, E.F., Weinstock, K.G., Gocayne, J.D., White, O., et al. 1995. Initial assessment of human gene diversity and expression patterns based upon 83 million nucleotides of cDNA sequence. Nature 377: 3-174. [PubMed] [Google Scholar]
- Beaudoing, E., Freier, S., Wyatt, J.R., Claverie, J-M., and Gautheret, D. 2000. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 10: 1001-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, J.O., Morton, J.G., Rosbash, M., and Richardson, M. 1974. Three abundance classes in HeLa cell messenger RNA. Nature 250: 199-204. [DOI] [PubMed] [Google Scholar]
- Blackshear, P.J., Lai, W.S., Thorn, J.M., Kennington, E.A., Staffa, N.G., Moore, D.T., Bouffard, G.G., Beckstrom-Sternberg, S.M., Touchman, J.W., Bonaldo, M.F., et al. 2001. The NIEHS Xenopus maternal EST project: Interim analysis of the first 13,879 ESTs from unfertilized eggs. Gene 267: 71-87. [DOI] [PubMed] [Google Scholar]
- Boguski, M.S., Lowe, T.M., and Toltoshev, C.M. 1993. dbEST: Database for “expressed sequence tags.” Nat. Genet. 4: 332-333. [DOI] [PubMed] [Google Scholar]
- Bonaldo, M.F., Lennon, G., and Soares, M.B. 1996. Normalization and subtraction: Two approaches to facilitate gene discovery. Genome Res. 6: 791-806. [DOI] [PubMed] [Google Scholar]
- Brown, D.M., Provoost, A.P., Daly, M.J., Lander, E.S., and Jacob, H.J. 1996. Renal disease susceptibility and hypertension are under independent genetic control in the fawn-hooded rat. Nat. Genet. 12: 44-51. [DOI] [PubMed] [Google Scholar]
- Deloukas, P., Schuler, G.D., Gyapay, G., Beasley, E.M., Soderlund, C., Rodriguez-Tome, P., Hui, L., Matise, T.C., McKusick, K.B., Beckmann, J.S., et al. 1998. A physical map of 30,000 human genes. Science 282: 744-746. [DOI] [PubMed] [Google Scholar]
- Dimopoulos, G., Casavant, T.L., Chang, S., Scheetz, T., Roberts, C., Donohue, M., Schultz, J., Benes, V., Bork, P., Ansorge, W., et al. 2000. Anopheles gambiae pilot gene discovery project: Identification of mosquito innate immunity genes from expressed sequence tags generated from immune-competent cell lines. Proc. Natl. Acad. Sci. 97: 6619-6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M.C., and Green, P. 1998. Base-calling of automated sequencer traces using phred, I: Accuracy assessment. Genome Res. 8: 175-185. [DOI] [PubMed] [Google Scholar]
- Gavin, A.J., Scheetz, T.E., Roberts, C.A., O'Leary, B., Braun, T.A., Sheffield, V.C., Soares, M.B., Robinson, J.P., and Casavant, T.L. 2002. Pooled library tissue tags for EST-based gene discovery. Bioinformatics 18: 1162-1166. [DOI] [PubMed] [Google Scholar]
- Guigo, R., Agarwal, P., Abril, J.F., Burset, M., and Fickett, J.W. 2000. An assessment of gene prediction accuracy in large DNA sequences. Genome Res. 10: 1631-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilbert, P., Lindpainter, K., Beckmann, J.S., Serikawa, T., Soubrier, F., Dubay, C., Cartwright, P., Degouyon, B., Julier, C., Takahasi, S., et al. 1991. Chromosomal mapping of 2 genetic-loci associated with blood-pressure regulation in hereditary hypertensive rats. Nature 353: 521-529. [DOI] [PubMed] [Google Scholar]
- Hillier, L., Lennon, G., Becker, M., Bonaldo, M., Chiapelli, B., Chissoe, S., Dietrich, N., DuBuque, T., Favello, A., Gish, W., et al. 1996. Generation and analysis of 280,000 human expressed sequence tags. Genome Res. 6: 807-828. [DOI] [PubMed] [Google Scholar]
- Jacob, H.J., Lindpainter, K., Lincoln, S.E., Kusumi, K., Bunker, R.K., Mao, Y.P., Ganten, D., Dzau, V.J., and Lander, E.S. 1991. Genetic-mapping of a gene causing hypertension in the stroke-prone spontaneously hypertensive rat. Cell 67: 213-224. [DOI] [PubMed] [Google Scholar]
- Jacob, H.J., Pettersson, A., Wilson, D., Mao, Y., Lernmark, A., and Lander, E.S. 1992. Genetic dissection of autoimmune type-I diabetes in the BB rat. Nat. Genet. 2: 56-60. [DOI] [PubMed] [Google Scholar]
- Makalowski, W. and Boguski, M.S. 1998. Evolutionary parameters of the transcribed mammalian genome: An analysis of 2820 orthologous rodent and human sequences. Proc. Natl. Acad. Sci. 95: 9407-9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra, M., Hillier, L., Kucaba, T., Allen, M., Barstead, R., Beck, C., Blistain, A., Bonaldo, M., Bowers, Y., Bowles, L., et al. 1999. An encyclopedia of mouse genes. Nat. Genet. 21: 191-194. [DOI] [PubMed] [Google Scholar]
- Moisan, M.P., Courvoisier, H., Bihoreau, M.T., Gauguier, D., Hendley, E.D., Lathrop, M., James, M.R., and Mormede, P. 1996. A major quantitative trait locus influences hyperactivity in the WKHA rat. Nat. Genet. 14: 471-473. [DOI] [PubMed] [Google Scholar]
- Mykytyn, K., Nishimura, D.Y., Searby, C.C., Shastri, M., Yen, H., Beck, J.S., Braun, T., Streb, L., Cornier, A.S., Cox, G.F., et al. 2002. Identification of the gene (BBS1) most commonly involved in Bardet-Biedl syndrome, a complex human obesity syndrome. Nat. Genet. 31: 435-438. [DOI] [PubMed] [Google Scholar]
- Nishimura, D.Y., Searby, C.C., Carmi, R., Elbedour, K., Van Maldergem, L., Fulton, A.B., Lam, B.L., Powell, B.R., Swiderski, R.E., Bugge, K.E, et al. 2001. Positional cloning of a novel gene on chromosome 16q causing Bardet-Biedel syndrome (BBS2). Hum. Mol. Genet. 10: 865-874. [DOI] [PubMed] [Google Scholar]
- Pesole, G., Liuni, S., Grillo, G., Ippedico, M., Larizza, A., Makalowski, W., and Saccone, C. 1999. UTRdb: A specialized database of 5′ and 3′ untranslated regions of eukaryotic mRNAs. Nucleic Acids Res. 27: 188-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheetz, T.E. and Casavant, T.L. 2004. Informatics for efficient EST-based gene discovery in normalized and subtracted cDNA libraries. In The Practical Bioinformatician (ed. L. Wong). World Scientific Publisher, River Edge, NJ.
- Scheetz, T.E, Trivedi, N., Roberts, C.A., Kucaba, T., Berger, B., Robinson, N.L., Birkett, C.L., Gavin, A.J., O'Leary, B., Braun, T.A., et al. 2003. ESTprep: Preprocessing cDNA sequence reads. Bioinformatics 19: 1318-1324. [DOI] [PubMed] [Google Scholar]
- Schuler, G.D. 1997. Pieces of the puzzle: Expressed sequence tags and the catalog of human genes. J. Mol. Med. 75: 694-698. [DOI] [PubMed] [Google Scholar]
- Soares, M.B. 1994. Construction of directionally cloned cDNA libraries in phagemid vectors. In Automated DNA sequencing and analysis techniques (ed. J.C. Venter), pp. 110-114. Academic Press, London, UK.
- ———. 1997. Identification and cloning and differentially expressed genes. Curr. Opin. Biotechnol. 8: 542-546. [DOI] [PubMed] [Google Scholar]
- Soares, M.B. and Bonaldo, M.F. 1998. Construction and screening of normalized cDNA libraries. In Genome analysis: A laboratory manual, Vol. 2 (eds. B. Birren, et al.), pp. 49-157. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. [Google Scholar]
- Soares, M.B., Bonaldo, M.F., Jelene, P., Su, L., Lawton, L., and Efstratiadis, A. 1994. Construction and characterization of a normalized cDNA library. Proc. Natl. Acad. Sci. 91: 9228-9232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton, M., Liao, G., Brokstein, P., Hong, L., Carninci, P., Shiraki, T., Hayashizaki, Y., Champe, M., Pacleb, J., Wan, K., et al. 2002. The drosophila gene collection: Identification of putative full-length cDNAs for 70% of D. melanogaster genes. Genome Res. 12: 1294-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausberg, R.L., Feingold, E.A., Grouse, L.H., Derge, J.G., Klausner, R.D., Collins, F.S., Wagner, L., Shenmen, C.M., Schuler, G.D., Altschul, S.F., et al. 2002. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl. Acad. Sci. 99: 16899-16903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi, N., Bischof, J., Davis, S., Pedretti, K., Scheetz, T.E., Braun, T.A., Roberts, C.A., Robinson, N.L., Sheffield, V.C., Soares, M.B., et al. 2002. Parallel creation of non-redundant gene indices from partial mRNA transcripts. Future Generation Computer Systems 18: 863-870. [Google Scholar]
- Whitfield, C.W., Band, M.R., Bonaldo, M.F., Kumar, C.G., Liu, L., Pardinas, J.R., Robertson, H.M., Soares, M.B., and Robinson, G.E. 2002. Annotated expressed sequence tags and cDNA microarrays for studies of brain and behavior in the honey bee. Genome Res. 12: 555-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://genome.uiowa.edu/clcg.html; The Coordinated Laboratory for Computational Genomics.
- http://ratest.eng.uiowa.edu/localdocs/sequencing_protocol.html; detailed description of the sequencing protocol.
- http://ftp.genome.washington.edu/RM/RepeatMasker.html; RepeatMasker.
- http://www.tigr.org; The Institute for Genome Research.