Abstract
Background:
Renal cell tumours (RCTs) are clinically, morphologically and genetically heterogeneous. Accurate identification of renal cell carcinomas (RCCs) and its discrimination from normal tissue and benign tumours is mandatory. We, thus, aimed to define a panel of microRNAs that might aid in the diagnostic workup of RCTs.
Methods:
Fresh-frozen tissues from 120 RCTs (clear-cell RCC, papillary RCC, chromophobe RCC (chRCC) and oncocytomas: 30 cases each), 10 normal renal tissues and 60 cases of ex-vivo fine-needle aspiration biopsies from RCTs (15 of each subtype validation set) were collected. Expression levels of miR-21, miR-141, miR-155, miR-183 and miR-200b were assessed by quantitative reverse transcription–PCR. Receiver operator characteristic curves were constructed and the areas under the curve were calculated to assess diagnostic performance. Disease-specific survival curves and a Cox regression model comprising all significant variables were computed.
Results:
Renal cell tumours displayed significantly lower expression levels of miR-21, miR-141 and miR-200b compared with that of normal tissues, and expression levels of all miRs differed significantly between malignant and benign RCTs. Expression analysis of miR-141 or miR-200b accurately distinguished RCTs from normal renal tissues, oncocytoma from RCC and chRCC from oncocytoma. The diagnostic performance was confirmed in the validation set. Interestingly, miR-21, miR-141 and miR-155 expression levels showed prognostic significance in a univariate analysis.
Conclusion:
The miR-141 or miR-200b panel accurately distinguishes RCC from normal kidney and oncocytoma in tissue samples, discriminating from normal kidney and oncocytoma, whereas miR-21, miR-141 and miR-155 convey prognostic information. This approach is feasible in fine-needle aspiration biopsies and might provide an ancillary tool for routine diagnosis.
Keywords: microRNAs, renal cell tumours, diagnostic tool, fine-needle biopsies
Renal cell tumours (RCTs) account for ∼4% of all adult neoplasms and 90–95% of all tumours arising in the kidney, ranking 14th in incidence worldwide, with an age-standardised mortality rate of 1.6 out of 100 000 (Ferlay et al, 2010). Renal cell tumours are morphologically and genetically heterogeneous (Baldewijns et al, 2008), comprising both malignant tumours (which are subclassified mainly as clear-cell renal cell carcinoma (ccRCC, 70–80% of cases), papillary RCC (pRCC, 10–15% of cases) and chromophobe RCC (chRCC, 5–10% of cases) and benign tumours (among which oncocytoma is the most common subtype; Kovacs et al, 1997).
Because histological subtypes differ in clinical aggressiveness and prognosis (Amin et al, 2002; Ficarra et al, 2006), accurate classification is required for appropriate patient management. Moreover, most RCTs are clinically silent at their earliest stages, and 20–30% are diagnosed when metastatic spread has already occurred (Abrahams et al, 2004). Although widespread use of imaging techniques (mainly ultrasonography) has increased detection of suspicious renal masses, prompting new pre-operative diagnostic challenges as histological diagnosis using needle biopsy material meets with important limitations, hampering an accurate categorisation in many instances (Shen et al, 2012). In this setting, diagnosis relies mainly on morphologic features, which show some overlap among tumour subtypes. The discrimination between chRCC (mainly the eosinophilic variant) and oncocytoma is one of the most critical and, sometimes, difficult differential diagnosis. Although these tumour types share some morphologic characteristics, chRCC is a malignant neoplasm, capable of local invasion and metastatic spread, whereas oncocytoma is a benign tumour just requiring a more conservative management.
Over the years, several attempts have been made to assist morphology in differential diagnosis between chRCC from oncocytoma, including immunohistochemical profiles (Abrahams et al, 2004; Lin et al, 2006; Shen et al, 2012), histochemical stains (Skinnider and Jones, 1999) and gene expression analysis (Lee et al, 2011). However, sensitivity and specificity of those techniques are suboptimal and prompt the need for more accurate biomarkers. Interestingly, some recent studies have attempted to discriminate among RCC subtypes using microRNA (miRNA) expression analysis. Nevertheless, most of those studies have mainly dealt with ccRCC or, when the most relevant histological subtypes were included, only a limited number of samples of each subtype have been analysed, precluding a definitive conclusion about their accuracy (Nakada et al, 2008; Petillo et al, 2009; Jung et al, 2009; Fridman et al, 2010; Juan et al, 2010; Youssef et al, 2011; Zhao et al, 2013). MicroRNAs are small non-coding RNAs (∼22 nucleotides in length), which are involved in several essential biological processes such as cell differentiation, growth, apoptosis and proliferation (Esteller, 2011), and their deregulation has been implicated in tumorigenesis, including that of the kidney (Lu et al, 2005; Jung et al, 2009; Chow et al, 2010; White and Yousef, 2010). In addition to the differential expression patterns of miRNAs among RCT subtypes (Petillo et al, 2009; Jung et al, 2009; Juan et al, 2010; Valera et al, 2011; Youssef et al, 2011), altered miRNA expression might also provide relevant prognostic information (Neal et al, 2010).
In two recent reviews (Henrique et al, 2012; Jeronimo and Henrique, 2011), we found that five miRNAs (miR-21, miR-141, miR-155, miR-183 and miR-200b) had been reported as displaying diagnostic or prognostic value in RCT (Nakada et al, 2008; Petillo et al, 2009; Jung et al, 2009; Juan et al, 2010; Youssef et al, 2011). Thus, we aimed to confirm and extend those findings through expression analysis of a five miRNA panel in a single series of RCT, comprising the four major subtypes. We first assessed the expression levels of selected miRNAs in fresh-frozen tissues, focusing on the discrimination between benign and malignant tumours, as well as between chRCC and oncocytoma. In addition, the prognostic value of each miRNAs was determined. Finally, we validated our findings in a series of ex-vivo fine-needle aspiration biopsies from RCT to assess the feasibility of this approach as an ancillary tool in routine pathology.
Materials and methods
Clinical samples
A total of 130 fresh-frozen tissues were prospectively collected and included in this study, comprising 120 RCTs (30 cases of each of the four major subtypes (ccRCC, pRCC, chRCC and oncocytoma)) and 10 morphologically normal renal tissues (obtained from morphologic normal kidney tissue of patients subjected to nephrectomy due to upper urinary urothelial carcinoma; Table 1). In addition, a validation set comprising 60 ex-vivo fine-needle aspiration biopsies from RCT (15 of each subtype) was included. Samples from RCT patients were procured from patients diagnosed and treated at the Portuguese Oncology Institute – Porto (Portugal), between 2003 and 2007, who underwent partial or total nephrectomy, after obtaining informed consent. For each subtype, cases were consecutively selected until it reached 30 (for tissue samples) or 15 (for ex-vivo fine-needle aspiration biopsies) cases. This strategy was used to maximise the representation of the less common RCT types, thus ensuring that tumour heterogeneity in each subtype would be considered in the molecular analyses. Tumour tissue samples, obtained immediately after surgery, were snap-frozen, stored at −80 °C and subsequently cut in a cryostat for RNA extraction. Ex-vivo fine-needle aspiration biopsies of RCTs were obtained through 4–6 passes of a 23-gauge needle attached to a 10-ml syringe, then washed in PBS and stored at −80 °C until further use.
Table 1. Clinical and pathological features of patients included in this study, including the data for the two sets of samples (fresh-frozen tissues and ex-vivo biopsies).
|
Fresh-frozen tissues |
Ex-vivo
aspiration biopsies |
||
|---|---|---|---|
| Tumour | Normal | Tumour | |
| Number of patients, n |
120 |
10 |
60 |
| Age, median (range) |
62 (30–84) |
65 (20–83) |
60 (30–82) |
|
Gender,
n
(%) | |||
| Male | 71 (59.2) | 7 (70.0) | 35 (58.3) |
| Female |
49 (40.8) |
3 (30.0) |
25 (41.7) |
|
Histological subtype,
n
(%) |
|
NA |
|
| Clear-cell RCC | 30 (25.0) | 15 (25.0) | |
| Papillary RCC | 30 (25.0) | 15 (25.0) | |
| Chromophobe RCC | 30 (25.0) | 15 (25.0) | |
| Oncocytoma |
30 (25.0) |
|
15 (25.0) |
|
Pathological stage,
n
(%) |
|
NA |
|
| pT1 | 46 (38.3) | 25 (41.7) | |
| pT2 | 19 (15.9) | 8 (13.3) | |
| pT3 | 24 (20.0) | 12 (20.0) | |
| pT4 | 1 (0.8) | — | |
| NA (oncocytoma) |
30 (25.0) |
|
15 (25.0) |
|
Furhman grade,
n
(%) |
|
NA |
|
| 1 | 3 (2.5) | 0 (0.0) | |
| 2 | 27 (22.5) | 12 (20.0) | |
| 3 | 44 (36.7) | 20 (33.3) | |
| 4 | 16 (13.3) | 12 (20.0) | |
| NA | 30 (25.0) | 16 (26.7) | |
Abbreviations: NA=not applicable; RCC=renal cell carcinoma.
Routine histopathological assessment of all surgical specimens, in formalin-fixed paraffin-embedded tissue, was performed by an expert uropathologist (RH) and included tumour classification (WHO), grading (Fuhrman) and staging (TNM; Eble et al, 2004). Relevant clinical data were collected from clinical charts. This study, as well as the use of samples and access to clinical data, was approved by the Institutional Review Board (Comissão de Ética para a Saúde) of the Portuguese Oncology Institute – Porto.
RNA extraction
Total RNA was extracted from fresh-frozen tissues and ex-vivo aspiration biopsies using Trizol reagent (Invitrogen, Carlsbad, CA, USA), according to manufacturer's instructions. Briefly, 1500 μl of Trizol reagent was added to each 2 ml tube and samples were homogenised using a rotor shaker. Tubes were incubated for 5–10 min at room temperature and then 300 μl of chloroform (Merck, Darmstadt, Germany) were added. Regarding biopsies the protocol was similar, but the Trizol reagent and chloroform volumes were 500 and 200 μl, respectively. Tubes were vigorously hand-shaked for 15 s and incubated for 3 min at room temperature, followed by a 15-min centrifugation at 12 000 g at 4 °C. Next, the upper phase was collected. RNA was purified using the PureLink RNA Mini Kit (Invitrogen), according to manufacturer's indications. RNA concentration and purity ratios were then evaluated using NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). In addition, RNA quality was checked by electrophoresis in a 2% agarose gel.
Reverse transcription
Reverse transcription (RT) was performed using TaqMan MicroRNA Reverse Transcription Kit and Megaplex RT human pool A (Applied Biosystems, Foster City, CA, USA). The reaction mixture had a final volume of 12 μl and included the following: 3 μl of total RNA (750 ng), 1.6 μl of megaplex RT primers (10 × ), 0.4 μl of dNTPs with dTTP (100 mM), 3 μl of MultiScribe reverse transcriptase (50 U μl−1), 1.6 μl of 10 × RT buffer, 0.2 μl of RNase inhibitor (20 U μl−1) and 0.4 μl of nuclease-free water. Reactions were performed in PCR tubes according to the following conditions: 40 cycles at 16 °C for 2 min, 42 °C for 1 min and 50 °C for 1 s, with a final incubation at 85 °C for 5 min.
Quantitative real-time RT–PCR
Quantitative RT–PCR (qRT–PCR) was performed using TaqMan Small RNA Assays (Applied Biosystems) in a 7500 Real-Time PCR system (Applied Biosystems), according to the recommended protocol. Briefly, for each reaction 0.5 μl of TaqMan Small RNA Assay (20 × ), 0.75 μl of RT product, 5 μl of TaqMan Universal PCR Master Mix II no UNG (2 × ) and 3.75 μl of nuclease-free water were added. According to the manufacturer's instructions, the protocol conditions were: 50 °C for 2 min, 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min. Expression levels of the five selected miRNAs (hsa-miR-21: Tm000397; hsa-miR-141: Tm000483; hsa-miR-155: Tm002626; hsa-miR-183: Tm002269; and hsa-miR-200b: Tm002251) were assessed in triplicate for each sample and two water blanks were added to each plate as negative controls.
Results from the qRT–PCR were analysed using the 7500 Software version 2.0.5 (Applied Biosystems). Levels of miRNA expression were determined using the relative standard curve method (Biosystems, 2004). In each sample, the mean quantity of each miRNA was normalised to the mean quantity for the endogenous controls RNU48 and RNU6B, according to the following formula: miRNA expression=candidate miRNA expression mean quantity/((RNU48 mean quantity+RNU6B mean quantity)/2). Results were then multiplied by 10 000 for easier tabulation.
Statistical analysis
Differences in expression levels of the candidate miRNAs among the different histological subtypes were first analysed using a non-parametric Kruskal–Wallis test, followed by pairwise comparisons using non-parametric Mann–Whitney U-test, when appropriate. The relationship between miRNA expression and clinicopathological variables (gender, Fuhrman grade (recoded into two groups: grades 1–2 vs 3–4) and pathological stage (recoded into two groups: pT1–pT2 vs pT3–pT4)) was evaluated using Mann–Whitney U-test. Spearman's non-parametric correlation tests were additionally carried out to ascertain correlations between age and miRNA expression levels. Receiver operator characteristic (ROC) curves were constructed by plotting the true-positive rate (sensitivity) against the false-positive rate (1−specificity) for each miRNA and for the best combination of miRNAs. The selection of the best miRNA panel was achieved using logistic regression, and the areas under the curve (AUCs) were calculated to assess the panel's diagnostic performance. Disease-specific survival (DSS) curves (Kaplan–Meier with log-rank test) were computed for clinical variables (age, gender, histological subtype, Fuhrman grade and pathological stage) and miRNA expression levels. A Cox regression model comprising all significant variables (multivariate test) was computed to assess the relative contribution of each variable to the follow-up status. For the purpose of survival analyses, all cases were coded based on each miRNA expression levels, using the median value as the cut-off value. Statistical analysis was performed using SPSS for Windows, version 20.0 (SPSS, Chicago, IL, USA) and differences were considered statistically significant when P<0.05. For multiple comparisons, the P-value was adjusted according to Bonferroni's method (i.e., the level of significance was adjusted to P<0.05/n, in which n represents the number of groups under comparison).
Results
MicroRNA expression levels and clinicopathological correlates
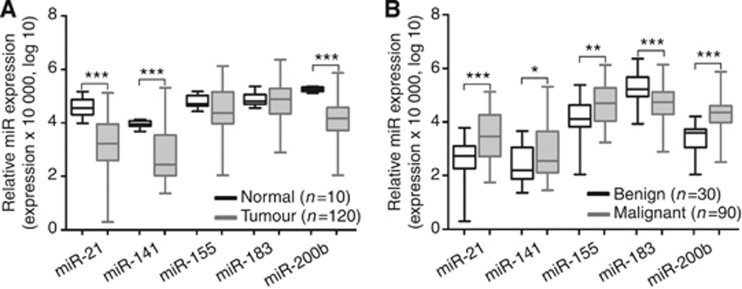
The relative expression levels of miR-21, miR-141, miR-155, miR-183 and miR-200b were determined in fresh-frozen tissues of 120 RCTs and 10 normal renal tissue samples. Relevant clinical and histopathological data are displayed in Table 1. No significant differences in age or gender between RCTs patients and normal tissue donors were apparent. No statistically significant associations were disclosed between miR expression levels and any of the clinicopathological features (age, gender, Fuhrman grade categories or pathological stage). Renal cell tumours showed significantly lower expression levels of miR-21, miR-141 and miR-200b compared with that of normal tissues (P<0.001 for all; Figure 1A and Supplementary Table 1). Moreover, expression levels of all candidate miRNAs differed significantly between benign and malignant RCTs. Oncocytomas displayed lower expression levels for all tested miRs, except miR-183 (Figure 1B and Supplementary Table 2).
Figure 1.
Distribution of miRNA expression levels in kidney tissues. (A) Normal vs tumour tissues. (B) Benign vs malignant tumour tissues. Statistically significant differences are represented as ***P<0.001, **P<0.01 and *P<0.02.
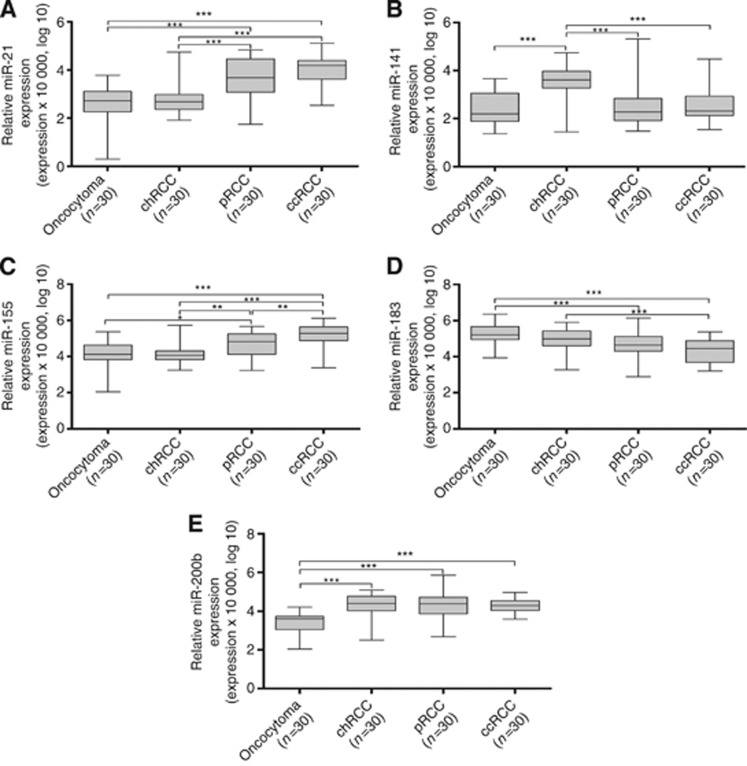
Although there was a wide expression range within the four RCT subtypes, with a significant degree of overlap, expression levels of all miRs differed significantly among them (P<0.001 for all, Kruskal–Wallis test; Table 2). Pairwise comparisons are shown in Table 3 and graphically illustrated in Figure 2. In general, oncocytomas displayed the lower miR expression levels, significantly differing from pRCC or ccRCC regarding four miRs (miR-21, miR-155, miR-183 and miR-200b), and from chRCC in two miRs (miR-141 and miR-200b). Interestingly, ccRCC and pRCC only differed for miR-155 expression levels, whereas chRCC differed from ccRCC and pRCC for miR-21, miR-141 and miR-155 expression levels. In addition, miR-183 expression levels were also different between chRCC and ccRCC (Table 3). Thus, reduced expression of miR-200b surfaced as the most discriminative feature between oncocytomas and RCCs.
Table 2. Distribution of microRNA expression levels among different histological subtypes in fresh-frozen tissues.
| Oncocytoma | chRCC | pRCC | ccRCC | P-value, K–W | |
|---|---|---|---|---|---|
| miR-21 |
5.3 (0.02–60.9) |
4.0 (0.8–560.2) |
47.9 (0.6–689.3) |
155.5 (3.5–1325.8) |
<0.001 |
| miR-141 |
7.9 (0.2–45.9) |
83.5 (0.3–552.2) |
76.8 (0.3–2063.3) |
25.75.7 (0.3–301.2) |
<0.001 |
| miR-155 |
374.9 (1.1–233.7) |
339.6 (14.5–5340.1) |
1054 (17.1–4595.9) |
3148.8 (23.74–13299) |
<0.001 |
| miR-183 |
5034.7 (87.1–23207.1) |
1690.3 (18.8–8013.8) |
1350.1 (7.7–13865) |
512.5 (15.9–2360.7) |
<0.001 |
| miR-200b | 40.3 (1.1–161) | 367.9 (3.3–1244) | 611.6 (4.8–7445.1) | 249.1 (38.1–930.2) | <0.001 |
Abbreviations: ccRCC=clear-cell renal cell carcinoma; chRCC=chromophobe RCC; K–W=Kruskal–Wallis test; pRCC=papillary RCC.
Table 3. Comparison of microRNA expression among renal cell tumour subtypes in fresh-frozen tissues.
|
P-valuea, M–W test |
|||||
|---|---|---|---|---|---|
| miR-21 | miR-141 | miR-155 | miR-183 | miR-200b | |
| Oncotytoma vs ccRCC |
<0.001 |
NS |
<0.001 |
<0.001 |
<0.001 |
| Oncocytoma vs pRCC |
<0.001 |
NS |
0.012 |
<0.001 |
<0.001 |
| Oncotytoma vs chRCC |
NS |
0.001 |
NS |
NS |
0.001 |
| pRCC vs ccRCC |
NS |
NS |
0.003 |
NS |
NS |
| ccRCC vs chRCC |
<0.001 |
0.002 |
<0.001 |
<0.001 |
NS |
| pRCC vs chRCC | <0.001 | <0.001 | 0.002 | NS | NS |
Abbreviations: ccRCC=clear-cell renal cell carcinoma; chRCC=chromophobe RCC; M–W=Mann–Whitney test; NS=not significant; pRCC=papillary RCC.
Statistically significant when P<0.0125, Bonferroni's correction.
Figure 2.
Distribution expression levels of (A) miR-21, (B) miR-141, (C) miR-155, (D) miR-183 and (E) miR-200b according with the histological subtype of RCTs. Statistically significant differences are represented as ***P<0.001, **P<0.003 and *P<0.0125.
Diagnostic performance of miRNA expression levels in tissue samples
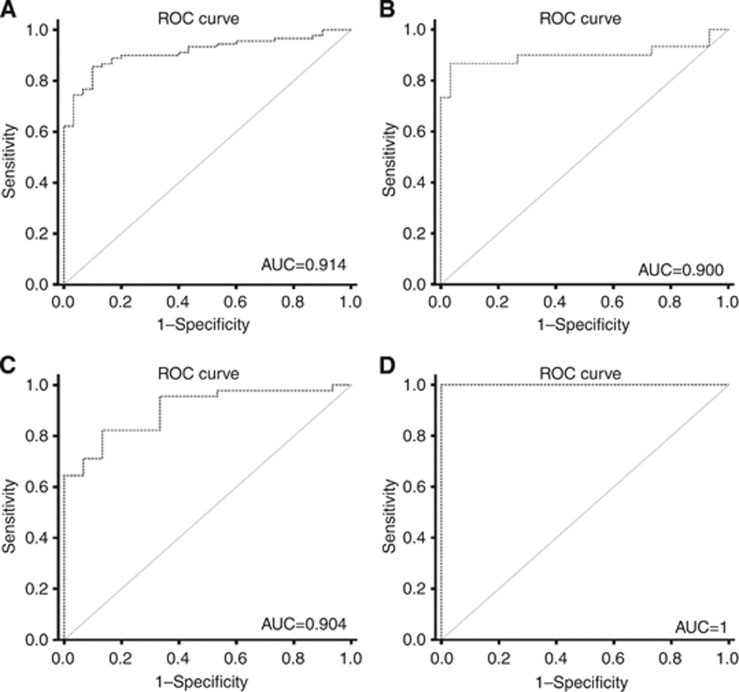
Performance of the five studied miRs was assessed in three different settings: identification of RCTs (vs normal renal tissue), discrimination of malignant from benign tumours and distinction of chRCC from oncocytoma. For that purpose, the cut-off value corresponded to the best performance of each miRNA according to the respective ROC curve analysis. Validity and information estimates for each marker and for the best combination of markers are displayed in Table 4. Receiver operator characteristic curve analysis showed that a panel comprising expressions of miR-141 and miR-200b allowed for the discrimination between RCT and normal renal tissue with 99.2% sensitivity and 100% specificity, corresponding to an AUC of 0.991. In addition, the same panel allowed for the differentiation between benign and malignant tumours with 85.6% sensitivity and 100% specificity, displaying an AUC of 0.912. Furthermore, expression levels of miR-141 and miR-200b also distinguished chRCC from oncocytoma with 90% sensitivity and 100% specificity, corresponding to an AUC of 0.90 (Figure 3A and B).
Table 4. Validity estimates for each tested miR and for the best combination of miRs in each diagnostic setting, in fresh-frozen tissues.
| miR-21 | miR-141 | miR-155 | miR-183 | miR-200b | miR-141 or miR-200b | |
|---|---|---|---|---|---|---|
|
RCT
vs
normal renal tissue | ||||||
| SE | 76.7 | 81.7 | — | — | 97.5 | 99.2 |
| SP | 100 | 100 | — | — | 100 | 100 |
| PPV | 100 | 100 | — | — | 100 | 100 |
| NPV | 26.0 | 31.0 | — | — | 77.0 | 90.9 |
| Accuracy | 78.0 | 83.0 | — | — | 98.0 | 99.2 |
| AUC |
89.9 |
89.7 |
— |
— |
98.7 |
99.1 |
|
RCC
vs
oncocytoma | ||||||
| SE | 48.9 | 25.6 | 50.0 | 72.2 | 96.7 | 85.6 |
| SP | 93.3 | 100 | 83.3 | 73.3 | 90.0 | 100 |
| PPV | 95.7 | 100 | 90.0 | 89.0 | 96.7 | 100 |
| NPV | 37.8 | 13.0 | 35.2 | 46.8 | 67.5 | 69.8 |
| Accuracy | 60.0 | 33.0 | 58.3 | 72.5 | 95.0 | 89.2 |
| AUC |
75.9 |
64.9 |
66.7 |
75.1 |
91.4 |
91.4 |
|
chRCC
vs
oncocytoma | ||||||
| SE | — | 76.7 | — | — | 83.3 | 90.0 |
| SP | — | 86.7 | — | — | 90.0 | 100 |
| PPV | — | 85.2 | — | — | 89.3 | 100 |
| NPV | — | 78.7 | — | — | 84.4 | 90.9 |
| Accuracy | — | 81.6 | — | — | 86.7 | 95.0 |
| AUC | — | 81.9 | — | 89.6 | 90.0 | |
Abbreviations: AUC=area under the curve; chRCC=chromophobe RCC; NPV=negative predictive value; PPV=positive predictive value; RCC=renal cell carcinoma; RCT=renal cell tumour; Se= sensitivity; Sp= specificity
Figure 3.
ROC curves. ROC curves evaluating the performance of the gene panel (miR-141 and miR-200b) as a biomarker for malignant renal tumours (A and C) and as a biomarker of chRCC (B and D). (A and B) Performed in kidney tissue samples; (C and D) performed in ex-vivo aspiration renal biopsies.
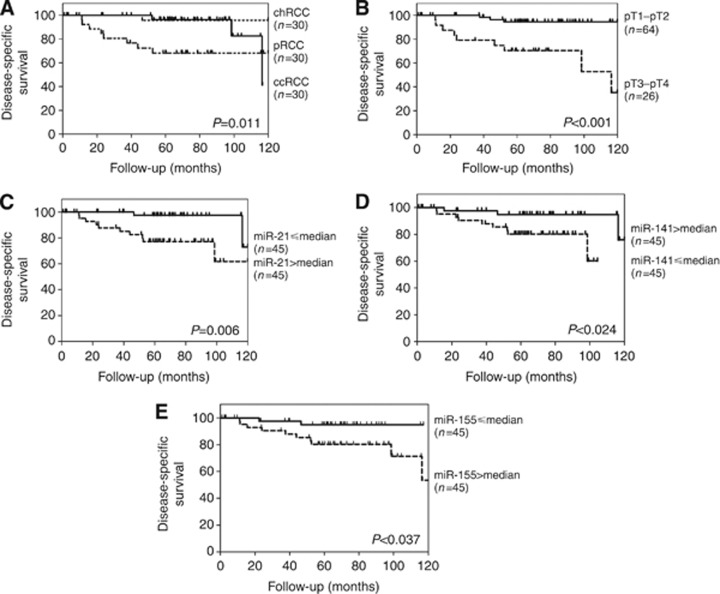
Survival analysis
The median follow-up of this series of RCT patients was 65 months (range: 1–120 months). A total of 12 patients (13.3%) have died from RCC during this period. Disease-specific survival analysis showed that tumour subtype ccRCC or pRCC and higher pathological tumour stage (pT3–T4) were significantly associated with worse outcome (P=0.011 and P<0.001, respectively; Figure 4A and B). Although age at diagnosis over 62 years was associated with worse DSS (P=0.035), gender and Fuhrman grade did not disclose any prognostic value within the available follow-up time. Concerning miRNA expression levels, miR-200b and miR-183 did not exhibit any prognostic value. However, higher expression levels of miR-21 and miR-155, and lower expression levels of miR-141 were associated with worse DSS (P=0.006, P=0.037 and P=0.024, respectively; Figure 4C–E). However, in multivariate analysis only pathological stage independently predicted prognosis, whereas miRNA expression levels did not retain an independent prognostic value (Supplementary Table 3).
Figure 4.
Disease-specific survival according to pathological and molecular parameters. (A) Histopathological classification; (B) pathological stage; (C–E) miR expression levels.
Validation of the miRNA panel in ex-vivo aspiration biopsies
The two best-performing miRNA in tissue samples, miR-141 and miR-200b, were then selected for analysis in ex-vivo samples. This set comprised 60 ex-vivo fine-needle aspiration biopsies. Relevant clinical and histopathological data are summarised in Table 1 and the relative expression levels for each miR are depicted in Supplementary Table 4 and Supplementary Figures S1 and S2.
Remarkably, expression levels of this panel of miRNAs were able not only to distinguish benign from malignant RCT with 73.3% sensitivity and a 100% specificity (AUC of 90.4%), but also oncocytoma from chRCC with 100% sensitivity and 100% specificity (AUC of 100% Figure 3C and D; Table 5).
Table 5. Validity estimates for each tested miR and for the best combination of miRs in each diagnostic setting, in ex-vivo aspiration biopsies.
| (%) | miR-141 | miR-200b | miR-141 or miR-200b | |
|---|---|---|---|---|
|
RCC
vs
oncocytoma | ||||
| SE | 35.5 | 73.3 | 73.3 | |
| SP | 93.3 | 93.3 | 100 | |
| PPV | 94.1 | 97.1 | 100 | |
| NPV | 34.5 | 53.8 | 55.0 | |
| Accuracy | 50.0 | 78.3 | 80.0 | |
| |
AUC |
57.5 |
88.4 |
90.4 |
|
chRCC
vs
oncocytoma | ||||
| SE | 73.3 | 100 | 100 | |
| SP | 93.3 | 100 | 100 | |
| PPV | 91.6 | 100 | 100 | |
| NPV | 77.7 | 100 | 100 | |
| Accuracy | 83.3 | 100 | 100 | |
| AUC | 84.4 | 100 | 100 | |
Abbreviations: AUC=area under the curve; chRCC=chromophobe RCC; NPV=negative predictive value; PPV=positive predictive value; RCC=renal cell carcinoma; Se=sensitivity; Sp=specificity;
Discussion
In this study, we aimed to define a small set of miRs that might allow for accurate identification of RCTs, as well as for discrimination between oncocytoma and RCCs, especially chRCC. This would be of clinical relevance, as diagnostic workup of suspicious renal masses incidentally found by abdominal ultrasonography is increasingly more frequent and demanding. Indeed, each RCT subtype displays quite dissimilar clinical behaviour, ranging from totally benign to overtly malignant, and successful pretherapeutic cytological or histological assessment is limited (Amin et al, 2002; Ficarra et al, 2006). Only a few studies addressed the use of miRNA expression as biomarkers for RCTs detection, and these have been mainly restricted to the ccRCC subtype, or have only analysed a very limited number of samples (Jung et al, 2009; Petillo et al, 2009; Juan et al, 2010; Youssef et al, 2011; Redova et al, 2012; Zhao et al, 2013). After an extensive review of published literature, we selected five miRNAs (miR-21, miR-141, miR-155, miR-183 and miR-200b) with putative diagnostic and prognostic value (Jeronimo and Henrique, 2011; Henrique et al, 2012), and tested them in a relatively large set of tissue samples that comprised the major histological subtypes. To ascertain their clinical and pathological relevance, a validation study was subsequently performed in a set of ex-vivo fine-needle aspiration biopsies.
Of the five miRs tested, three (miR-21, miR-141 and miR-200b) were significantly downregulated in RCTs compared with normal renal tissue. In previous reports, miR-21 was found to be upregulated in RCT (Juan et al, 2010; Faragalla et al, 2012; Zaman et al, 2012), which apparently contradicts our results. However, in those studies, normal renal tissue was obtained from nephrectomy specimens harbouring RCT, which did not occur in our study. This is an important issue, as we have previously shown that morphologically normal renal tissue from kidneys harbouring RCT display epigenetic alterations in line with the respective tumours (Costa et al, 2007). Remarkably, variations in miR-21 expression among RCT subtypes observed in our study matches that reported by Faragalla et al (2012), with ccRCC depicting the highest median levels, followed by pRCC, chRCC and oncocytoma. Indeed, only miR-21 expression levels of ‘normal renal tissue' are notably different between our results and their study (Faragalla et al, 2012). These findings prompt the need for an adequate definition of ‘normal tissue', as the interpretation of results in tumours might be considerably biased.
Concerning miR-141, our results corroborate those of two previous reports (Nakada et al, 2008; Fridman et al, 2010). Thus, higher miR-141 expression levels seem to be a hallmark of chRCC and might constitute a valuable biomarker for discrimination from oncocytoma. Strikingly, a miRNA profiling of ccRCC also identified miR-141 (and 200b) as being downregulated in ccRCC, although with concurrent upregulation of miR-155 (Jung et al, 2009). These results are in line with ours, as we found that the highest miR-155 expression levels in ccRCC and pRCC significantly differed from those of oncocytoma and chRCC. Our findings concerning miR-200b mirror those of Youssef et al (2011), although in a smaller data set. Interestingly, in our larger data set we were able to demonstrate that miR-200b expression levels were significantly lower in oncocytomas compared with all RCC subtypes. Overall, the comparisons of miR expression levels among RCT subtypes also denote the common origin (segment of the nephron) of ccRCC and pRCC on one hand, and of chRCC and oncocytoma on the other hand, emphasising the importance of searching for discriminative biomarkers, which might enable accurate identification of each RCT subtype.
Interestingly, a panel comprising miR-141 and miR-200b demonstrated the best performance in frozen-tissue samples, displaying AUC values ranging from 90.0 to 99.1. Although these results are interesting per se, its clinical usefulness depends on the possibility of using it in diagnostic samples. For that purpose, we further validated this biomarker panel in a set of fine-needle aspiration biopsies performed ex vivo. Although this procedure is not completely equivalent to an imaging-guided diagnostic fine-needle aspiration biopsy performed in a patient (which may yield lower amounts of tumour cells), it is, nonetheless, the best approximation without jeopardising patients' diagnosis. On the other hand, because the nephrectomy specimen is already available its histopathological characterisation is guaranteed, whereas diagnostic biopsies may not be followed by surgical excision, thus precluding accurate tumour classification for comparison purposes. Remarkably, the biomarker panel performance in ex-vivo biopsies was comparable to that of fresh-frozen tissues. To the best of our knowledge, this is the first attempt to demonstrate the feasibility of using miRs as tumour biomarkers in renal tumour biopsies, and may thus constitute a significant step forward in the development of epigenetic-based biomarkers for management of RCC suspects.
The clinical significance of our findings could be extended if miRNA expression levels might convey prognostic information. Thus, we performed DSS analysis using expression levels determined in fresh-frozen-tissue samples. As expected, tumour subtype and pathological stage were of prognostic value in univariate analysis, although only the later showed independent prognostic value in multivariate analysis. Remarkably, miR-21, miR-141 and miR-155 expression levels also displayed prognostic significance in RCC, although only in univariate analysis. A possible explanation for these findings may lie in the association between specific miR expression levels and tumour subtypes. Indeed, whereas for miR-21 and miR-155 the association with poorer DSS was observed for higher (> median) expression levels, the opposite was verified for miR-141. Interestingly, higher miR-21 and miR-155 expression levels and lower miR-141 expression levels were associated with pRCC and ccRCC subtypes, which displayed the worse prognosis compared with that of chRCC. The fact that tumour subtype did not surfaced as independent prognostic parameter for DSS in multivariate analysis is most likely due to the association between tumour subtype and pathological stage, as pT3–4 tumours were mostly of pRCC or ccRCC subtype. Our findings concerning miR-21 and miR-141 are corroborated by previous reports, although with generally smaller patient cohorts (Jung et al, 2009; Faragalla et al, 2012; Zaman et al, 2012). In addition, the prognostic value of miR-155 expression levels has been reported for breast cancer (Song et al, 2012) and non-small cell lung cancer (Yanaihara et al, 2006; Yang et al, 2013), whereas miR-21 and miR-141 expression seem to be of prognostic significance in non-small cell lung cancer (Yanaihara et al, 2006; Yang et al, 2013) and colon cancer (Cheng et al, 2011), respectively.
The aforementioned association of specific miRs altered expression and RCT subtype might also provide clues concerning the cause of miR dysregulation. Renal cell tumour subtypes display characteristic chromosomal aberrations, including whole or partial deletions and duplications (Baldewijns et al, 2008). Strikingly, some of those alterations might explain the altered pattern of miR expression. For instance, miR-200b is mapped at 1p36.33 and loss of 1p or of the whole chromosome 1 is frequently observed in oncocytoma and chRCC. On the other hand, miR-21 and miR-155 are mapped at 17q23.1 and 21q21.2–21.3, which are frequently lost chromosomal regions in chRCC. Conversely, pRCC, which commonly show gain of chromosome 17, are among the RCT subtypes with higher miR-21 expression levels. However, other variations in miR expression might not be explained by chromosomal-level alterations and the respective cause(s) remain to be investigated.
Conclusions
Herein we demonstrate that expression levels of a panel of two miRNAs (miR-141 or miR-200b) allows for accurate distinction of normal kidney from RCT tissue samples, as well as for accurate discrimination among RCT subtypes, including the separation of benign from malignant RCT. Furthermore, the selected miR panel is able to convey prognostic information, although not independent of tumour subtype or pathological stage. Importantly, the same panel displays an impressive performance for accurate detection of RCC in clinical samples obtained from ex-vivo fine-needle aspiration biopsies, demonstrating the feasibility of this approach in routine diagnostic practice.
Acknowledgments
This study was funded by grants from the Research Centre of the Portuguese Oncology Institute – Porto (CI-IPOP-4-2008) and from the European Community's Seventh Framework Programme – Grant number FP7-HEALTH-F5-2009-241783. PC-P is supported by a grant from the Liga Portuguesa Contra o Cancro – Núcleo Regional do Norte. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Abrahams NA, MacLennan GT, Khoury JD, Ormsby AH, Tamboli P, Doglioni C, Schumacher B, Tickoo SK. Chromophobe renal cell carcinoma: a comparative study of histological, immunohistochemical and ultrastructural features using high throughput tissue microarray. Histopathology. 2004;45 (6:593–602. doi: 10.1111/j.1365-2559.2004.02003.x. [DOI] [PubMed] [Google Scholar]
- Amin MB, Tamboli P, Javidan J, Stricker H, de-Peralta Venturina M, Deshpande A, Menon M. Prognostic impact of histologic subtyping of adult renal epithelial neoplasms: an experience of 405 cases. Am J Surg Pathol. 2002;26 (3:281–291. doi: 10.1097/00000478-200203000-00001. [DOI] [PubMed] [Google Scholar]
- Baldewijns MM, van Vlodrop IJ, Schouten LJ, Soetekouw PM, de Bruine AP, van Engeland M. Genetics and epigenetics of renal cell cancer. Biochim Biophys Acta. 2008;1785 (2:133–155. doi: 10.1016/j.bbcan.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Biosystems A.2004. Guide to Performing Relative Quantitation of Gene Expression Using Real-Time Quantitative PCR: Applied Biosystems.
- Cheng H, Zhang L, Cogdell DE, Zheng H, Schetter AJ, Nykter M, Harris CC, Chen K, Hamilton SR, Zhang W. Circulating plasma MiR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PloS One. 2011;6 (3:e17745. doi: 10.1371/journal.pone.0017745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow TF, Youssef YM, Lianidou E, Romaschin AD, Honey RJ, Stewart R, Pace KT, Yousef GM. Differential expression profiling of microRNAs and their potential involvement in renal cell carcinoma pathogenesis. Clin Biochem. 2010;43 (1-2:150–158. doi: 10.1016/j.clinbiochem.2009.07.020. [DOI] [PubMed] [Google Scholar]
- Costa VL, Henrique R, Ribeiro FR, Pinto M, Oliveira J, Lobo F, Teixeira MR, Jeronimo C. Quantitative promoter methylation analysis of multiple cancer-related genes in renal cell tumors. BMC Cancer. 2007;7:133. doi: 10.1186/1471-2407-7-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eble JN, Sauter G, Epstein JI, Sesterhenn IA. World Health Organization Classification of Tumours. IARC: Lyon; 2004. Pathology and genetics of tumours of the urinary system and male genital organs. [Google Scholar]
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12 (12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- Faragalla H, Youssef YM, Scorilas A, Khalil B, White NM, Mejia-Guerrero S, Khella H, Jewett MA, Evans A, Lichner Z, Bjarnason G, Sugar L, Attalah MI, Yousef GM. The clinical utility of miR-21 as a diagnostic and prognostic marker for renal cell carcinoma. J Mol Diagn. 2012;14 (4:385–392. doi: 10.1016/j.jmoldx.2012.02.003. [DOI] [PubMed] [Google Scholar]
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127 (12:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- Ficarra V, Martignoni G, Galfano A, Novara G, Gobbo S, Brunelli M, Pea M, Zattoni F, Artibani W. Prognostic role of the histologic subtypes of renal cell carcinoma after slide revision. Eur Urol. 2006;50 (4:786–793. doi: 10.1016/j.eururo.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Fridman E, Dotan Z, Barshack I, David MB, Dov A, Tabak S, Zion O, Benjamin S, Benjamin H, Kuker H, Avivi C, Rosenblatt K, Polak-Charcon S, Ramon J, Rosenfeld N, Spector Y. Accurate molecular classification of renal tumors using microRNA expression. J Mol Diagn. 2010;12 (5:687–696. doi: 10.2353/jmoldx.2010.090187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrique R, Luis AS, Jeronimo C. The epigenetics of renal cell tumors: from biology to biomarkers. Front Genet. 2012;3:94. doi: 10.3389/fgene.2012.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeronimo C, Henrique R.2011Epigenetic biomarkers in urological tumors: A systematic review Cancer Lette-pub ahead of print 23 December 2011; doi: 10.1016/j.canlet.2011.12.026 [DOI] [PubMed]
- Juan D, Alexe G, Antes T, Liu H, Madabhushi A, Delisi C, Ganesan S, Bhanot G, Liou LS. Identification of a microRNA panel for clear-cell kidney cancer. Urology. 2010;75 (4:835–841. doi: 10.1016/j.urology.2009.10.033. [DOI] [PubMed] [Google Scholar]
- Jung M, Mollenkopf HJ, Grimm C, Wagner I, Albrecht M, Waller T, Pilarsky C, Johannsen M, Stephan C, Lehrach H, Nietfeld W, Rudel T, Jung K, Kristiansen G. MicroRNA profiling of clear cell renal cell cancer identifies a robust signature to define renal malignancy. J Cell Mol Med. 2009;13 (9B:3918–3928. doi: 10.1111/j.1582-4934.2009.00705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs G, Akhtar M, Beckwith BJ, Bugert P, Cooper CS, Delahunt B, Eble JN, Fleming S, Ljungberg B, Medeiros LJ, Moch H, Reuter VE, Ritz E, Roos G, Schmidt D, Srigley JR, Storkel S, van den Berg E, Zbar B. The Heidelberg classification of renal cell tumours. J Pathol. 1997;183 (2:131–133. doi: 10.1002/(SICI)1096-9896(199710)183:2<131::AID-PATH931>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Lee HW, Lee EH, Lee CH, Chang HK, Rha SH. Diagnostic utility of caveolin-1 and MOC-31 in distinguishing chromophobe renal cell carcinoma from renal oncocytoma. Korean J Urol. 2011;52 (2:96–103. doi: 10.4111/kju.2011.52.2.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F, Yang W, Betten M, Teh BT, Yang XJ. Expression of S-100 protein in renal cell neoplasms. Hum Pathol. 2006;37 (4:462–470. doi: 10.1016/j.humpath.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435 (7043:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Nakada C, Matsuura K, Tsukamoto Y, Tanigawa M, Yoshimoto T, Narimatsu T, Nguyen LT, Hijiya N, Uchida T, Sato F, Mimata H, Seto M, Moriyama M. Genome-wide microRNA expression profiling in renal cell carcinoma: significant down-regulation of miR-141 and miR-200 c. J Pathol. 2008;216 (4:418–427. doi: 10.1002/path.2437. [DOI] [PubMed] [Google Scholar]
- Neal CS, Michael MZ, Rawlings LH, Van der Hoek MB, Gleadle JM. The VHL-dependent regulation of microRNAs in renal cancer. BMC Med. 2010;8:64. doi: 10.1186/1741-7015-8-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petillo D, Kort EJ, Anema J, Furge KA, Yang XJ, Teh BT. MicroRNA profiling of human kidney cancer subtypes. Int J Oncol. 2009;35 (1:109–114. doi: 10.3892/ijo_00000318. [DOI] [PubMed] [Google Scholar]
- Redova M, Poprach A, Nekvindova J, Iliev R, Radova L, Lakomy R, Svoboda M, Vyzula R, Slaby O. Circulating miR-378 and miR-451 in serum are potential biomarkers for renal cell carcinoma. J Trans Med. 2012;10:55. doi: 10.1186/1479-5876-10-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen SS, Truong LD, Scarpelli M, Lopez-Beltran A. Role of immunohistochemistry in diagnosing renal neoplasms: when is it really useful. Ach Pathol Lab Med. 2012;136 (4:410–417. doi: 10.5858/arpa.2011-0472-RA. [DOI] [PubMed] [Google Scholar]
- Skinnider BF, Jones EC. Renal oncocytoma and chromophobe renal cell carcinoma. A comparison of colloidal iron staining and electron microscopy. Am J Clin Pathol. 1999;111 (6:796–803. doi: 10.1093/ajcp/111.6.796. [DOI] [PubMed] [Google Scholar]
- Song CG, Wu XY, Fu FM, Han ZH, Wang C, Shao ZM. [Correlation of miR-155 on formalin-fixed paraffin embedded tissues with invasiveness and prognosis of breast cancer] Zhonghua Wai Ke Za Zhi. 2012;50 (11:1011–1014. [PubMed] [Google Scholar]
- Valera VA, Walter BA, Linehan WM, Merino MJ. Regulatory effects of microRNA-92 (miR-92) on VHL gene expression and the hypoxic activation of miR-210 in clear cell renal cell carcinoma. J Cancer. 2011;2:515–526. doi: 10.7150/jca.2.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White NM, Yousef GM. MicroRNAs: exploring a new dimension in the pathogenesis of kidney cancer. BMC Med. 2010;8:65. doi: 10.1186/1741-7015-8-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9 (3:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- Yang M, Shen H, Qiu C, Ni Y, Wang L, Dong W, Liao Y, Du J. High expression of miR-21 and miR-155 predicts recurrence and unfavourable survival in non-small cell lung cancer. Eur J Cancer. 2013;49 (3:604–615. doi: 10.1016/j.ejca.2012.09.031. [DOI] [PubMed] [Google Scholar]
- Youssef YM, White NM, Grigull J, Krizova A, Samy C, Mejia-Guerrero S, Evans A, Yousef GM. Accurate molecular classification of kidney cancer subtypes using microRNA signature. Eur Urol. 2011;59 (5:721–730. doi: 10.1016/j.eururo.2011.01.004. [DOI] [PubMed] [Google Scholar]
- Zaman MS, Shahryari V, Deng G, Thamminana S, Saini S, Majid S, Chang I, Hirata H, Ueno K, Yamamura S, Singh K, Tanaka Y, Tabatabai ZL, Dahiya R. Up-regulation of microRNA-21 correlates with lower kidney cancer survival. PLoS One. 2012;7 (2:e31060. doi: 10.1371/journal.pone.0031060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao A, Li G, Peoc'h M, Genin C, Gigante M. Serum miR-210 as a novel biomarker for molecular diagnosis of clear cell renal cell carcinoma. Exp Mol Pathol. 2013;94 (1:115–120. doi: 10.1016/j.yexmp.2012.10.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.