Abstract
With the sequencing and assembly of the rat genome comes the difficult task of assigning functions to genes. Tissue localization of gene expression gives some information about the potential role of a gene in physiology. Various examples of the utility of multiple tissue gene expression data sets are illustrated here. First, we highlight their use in finding genes that might play an important role in a particular tissue on the basis of exclusive expression in that tissue or coexpression with a gene or genes with known function. Second, we show how this data might be used to explain known phenotypic differences between strains. Third, we show how expression patterns of genes in a genomic interval might identify candidate genes in quantitative trait loci (QTL) mapping studies. Lastly, we show how multiple tissue and species data can help researchers prioritize follow up studies to microarray experiments. All of these applications of multiple tissue gene expression data sets will play a role in functionally annotating the rat genome.
The rat is preferred to mice to model some aspects of human physiology and disease due to its larger size, more complex metabolism, and advanced intelligence. Rats were the first mammalian species used for scientific research, and some important genetic discoveries were first made in this species (Jacob and Kwitek 2002). However, mice have been the favored model species for geneticists due to the relative ease in which transgenic and knockout mice can be generated. Researchers who are dependent on rats as experimental subjects have been looking forward to the public release of the rat genome assembly. The promise is that more genetic information about this important model species will speed characterization of gene function by implicating genes in more advanced disease models.
Regional gene expression data can be used in many ways to functionally annotate the genome (Su et. al. 2002). For example, when a researcher finds that an uncharacterized transcript shows an interesting expression pattern from a microarray study, location of expression is one important clue to the possible importance of that transcript in the physiology that the researcher is attempting to model. Likewise, knowledge that the transcript of interest has a human ortholog with a similar expression pattern across tissues is another hint of the significance of that transcript in the disease model. Regional expression data can also be used to find novel genes that are likely to have an important function. For example, genes that are coexpressed in particular tissues, along with genes of well-characterized function, are likely to also play a prominent role in the function of those tissues. Similarly, genes that are coexpressed with known genes of a particular biochemical pathway might also play a role in that pathway (Staudt and Brown 2000). Although this type of guilt-by-association approach has many critics, it has recently been shown to be successful if multiple species are compared (Stuart et. al. 2003). Recently, a similar approach used our public mouse and human gene expression set (http://expression.gnf.org) to identify a disease gene for a human cytochrome c oxidase deficiency on the basis of the mapped chromosomal locus and the pattern of gene expression across multiple tissues (Mootha et al. 2003).
Here, we examine normal physiological expression levels of ∼7000 characterized rat genes and 1000 EST clusters in 11 peripheral and 15 brain regions in three common out-bred rat strains. We describe various applications of this data set including (1) prioritization of microarray follow-up studies, (2) finding genes that likely play an important role in tissue function, (3) finding gene expression differences that might explain phenotypic differences between strains, and (4) narrowing the search for genes that contribute to quantitative trait loci (QTL). Accompanying query tools are available on our Web site at http://expression.gnf.org or http://symatlas.gnf.org.
RESULTS
Analysis of Strain Differences
Sprague Dawley, Wistar, and Wistar Kyoto rats are some of the more common albino strains used to model human disease and physiology. Phenotypic differences that exist between the strains tend to complicate their use as models for human disease. However, these differences will ultimately reveal physiological roles of some genes. Here, we compare gene-expression differences between strains in a brain region mediating a known phenotypic difference.
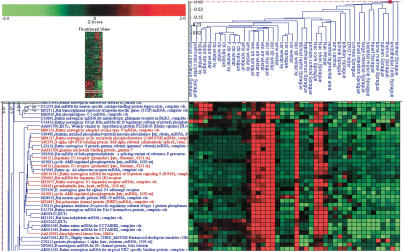
Wistar Kyoto rats show exaggerated responses to a number of stressors, and are thus often used as a model to study the relationship between stress and depression (Rittenhouse et. al. 2002). The first successful antidepression drugs worked to increase catecholamine release in cerebral cortical areas, and many more recent drugs also potentiate cortical catecholamine levels by blocking removal from synapses (Baldessarini 1996). Catechol-O-methyltransferase (COMT) is a key enzyme in the catabolism of catecholamines (Hoffman et. al. 1996). Wistar Kyoto rats show four- to sevenfold higher levels of mRNA for COMT in the cerebral and frontal cortices than Sprague Dawley rats, consistent with a decrease in synaptic levels of catecholamines and a susceptibility to depression (Fig. 1). One report has shown an alteration in stress-induced norepinephrine release in Wistar Kyoto, but not Sprague Dawley rats, but a defect in norepinephrine catabolism was not described (Tejani-Butt et al. 1994). However, another study reported that Wistar Kyoto rats are subsensitive to antidepressants, consistent with higher COMT levels in this strain (Lahmame and Armario 1996).
Figure 1.
Catechol-O-Methyltransferase (COMT) mRNA levels are increased in Wistar Kyoto compared with Sprague Dawley frontal and cerebral cortices. Red represents expression level of a transcript that is above the median expression level across the tissues shown, whereas green represents expression below the median. The intensity of the color corresponds to the magnitude of the change.
Gene Coexpression Patterns
Coexpression of uncharacterized genes with genes of important physiological function can be examined with this data set. Such an approach has been hypothesized to be effective in assigning functions to genes (Staudt and Brown 2000). We used Pearson analysis to find sequences that are coexpressed with any single gene across all tissues in this data set. As an example, we chose the D1 dopamine receptor, as it has an enriched expression pattern in dopamine terminal areas, and plays an important role in the reinforcing behaviors related to drugs of abuse (Self and Stein 1992). Ten sequences have expression correlation coefficients of at least 0.8 to the D1 receptor (Fig. 2). Some of these sequences have previously been shown to be enriched in dopamine terminal areas and play an important role in dopamine signaling and striatal function. One sequence, the rat potassium channel protein RHK1 = Kcna4 (GenBank #M32867_at), has been predominantly studied in heart, but is also expressed throughout the basal ganglia (Chung et al. 2000). But, on the basis of our expression data showing strongest expression in both dorsal and ventral striatum, and especially nucleus accumbens core and shell, it may play a predominant role in both drug addictive and locomotor behaviors (Brundege and Williams 2002).
Figure 2.
A set of transcripts shows a high expression correlation to the D1 dopamine receptor across many rat tissues. Highlighted sequences had Pearson coefficients across tissues of at least 0.8. Red represents expression level of a transcript that is above the median expression level across the tissues shown, whereas green represents expression below the median. The intensity of the color corresponds to the magnitude of the change.
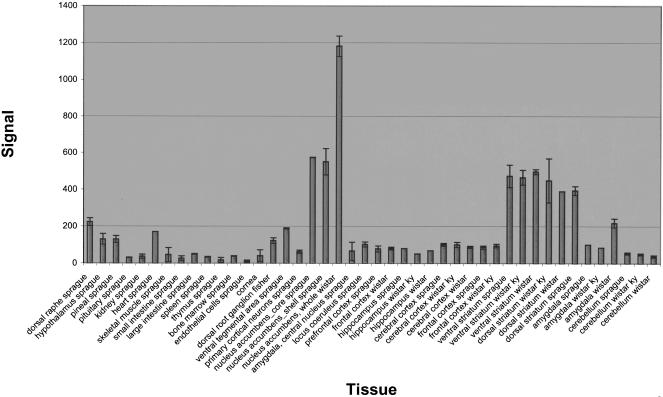
A similar search for correlated expression patterns, using a seed set yields more sequences with the same general expression pattern. We used a function of the Rosetta Resolver algorithm (GROW, see Methods) that searches for genes with a similar expression pattern to a defined set of genes. As an example, several genes with known roles in striatal function were chosen for a seed set (GenBank accession nos. X57659, X56065, M35077, AB019145). Using this method, many additional sequences with known roles in striatal neurotransmitter signaling are found, including type V adenylyl cyclase (M96159), CaM-PDE (M94537), and a striatal-enriched phosphatase (S49400) (Supplemental Table 1 available online at www.genome.org). In addition, a transcribed sequence of unknown function shows a dramatic enrichment in the nucleus accumbens core and shell, whole nucleus accumbens, and dorsal and ventral striatum of all rat strains analyzed (Affymetrix probe set rc_AI639435_at, Accession no. AI639435; Fig. 3). This expression pattern suggests that this unknown sequence may play an important role in striatal function.
Figure 3.
An uncharacterized transcript shows an enriched expression in all striatal (dorsal and ventral striatum, nucleus accumbens) tissues. Expression profile of this transcript, rc_AI639435_at = AI639435, is displayed using Affymetrix MAS5 signal intensity across many tissues. This sequence was found using the GROW function of the Rosetta Resolver software package (see Methods and Supplemental Table 1), and shows a highly correlated expression pattern to known striatal genes. A similar image can be obtained on http://expression.gnf.org/ratlas.
Tissue-Enriched Gene Expression
The Web interface for this data set is useful for finding sequences that are uniquely enriched in expression in particular tissues. The Web site tool allows for selection of one to several tissues, setting relative levels of expression between tissues, and specifying whether or not expression should be exclusive for the chosen tissue or set of tissues. The method simply searches for genes on the basis of specified fold expression levels in a tissue over the median expression level across all tissues. Supplemental Table 2 shows the results of a systematic search for sequences enriched in several brain regions over other tissues. Genes known to be enriched in particular regions, like the dopamine transporter in the ventral tegmental area, validate the utility of this tool. Some EST sequences also possess a restricted tissue distribution. For example, both the pituitary and the pineal contain several ESTs that are enriched in those regions and, given their similar restricted expression pattern to known genes, would indicate that they play an important role in the physiology of these tissues.
Using Gene Expression Information to Find Candidate Genes in Quantitative Trait Loci
QTL mapping is a powerful way to find chromosomal locations that contribute to human disease. Identifying the genes underlying QTL has been difficult, but is occasionally successful (Horikawa et. al. 2000; Hugot et. al. 2001; Ogura et. al. 2001). QTL mapping in rat may be useful to find genes responsible for various aspects of human disease (Jacob and Kwitek 2002). However, one major difficulty of the QTL approach is the rather large genomic regions that define a QTL, as anywhere from tens to hundreds of genes may reside in a locus. Trying to narrow down this QTL region and identify candidate genes has been a stumbling block for QTL researchers. In addition, for a complex disease, many loci may contribute to the phenotype. With additional sources of information about the genes that reside in a genomic interval, such as tissue localization of gene expression, it may be possible to identify potential candidates. As an example of this application of our data set, we chose a QTL from the literature associated with a preference for alcohol intake. RatMap http://ratmap.gen.gu.se/ was used to find QTLs for alcohol preference. A high lod score for alcohol preference is found on chromosome 4 (Bice et. al. 1998; Carr et. al. 1998; Terenina-Rigaldie et. al. 2003). Neuropeptide Y (NPY) was considered as a candidate gene in this study, but sequencing of that gene did not reveal any nucleotide differences (Bice et. al. 1998), and expression differences were not detected (Liang et. al. 2003) between the alcohol-preferring and alcohol-nonpreferring rats.
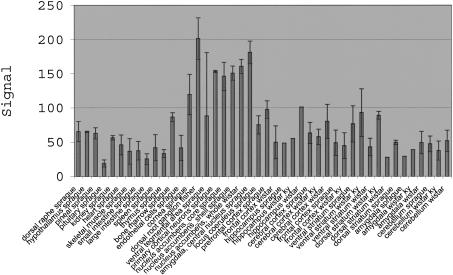
Genes responsible for alcohol preference should be expressed in the nucleus accumbens; especially the nucleus accumbens shell region and/or the central nucleus of the amygdala (Koob 1999; McBride 2002). Using resources from Affymetrix (http://www.affymetrix.com), UCSC (http://genome.ucsc.edu), and our Web site (http://expression.gnf.org/ratlas), all transcripts represented on the RG_U34A chip can be found that flank the marker D4Rat34, the peak of the maximum lod score of 9.2 (Bice et. al. 1998) from position 78 Mb to 92 Mb. This interval was chosen to include the previous candidate gene, NPY, and include D4Rat34 at the center. Forty-two probe sets are included in this interval (Supplemental Table 3). Several transcripts show at least enriched expression in the brain regions of interest (enter all probe sets from Supplemental Table 3 into the dialog box on http://expression.gnf.org/ratlas). One of those transcripts near D4Rat34 is Corticotropin Releasing Hormone (or Factor) Receptor 2 (Accession no. U16253; Fig. 4). Previous studies have shown that corticotropin releasing factor (CRF) plays a role in alcohol intake (Bell et. al. 1998; Olive et. al. 2002). In addition, alcohol-preferring and alcohol-nonpreferring rats show differing responses to CRF (Ehlers et. al. 1992). All of these reasons point to CRHR2 as a potential candidate gene for alcohol preference. This example illustrates the value of tissue-expression information in prioritizing the search for candidate genes from QTL studies.
Figure 4.
Corticotropin release hormone receptor 2 shows enriched expression in brain regions involved in alcohol preference, especially the nucleus accumbens core and shell, whole nucleus accumbens, and central nucleus of the amygdala. Expression profile of this transcript is displayed using Affymetrix MAS5 signal intensity across many tissues. A similar image for this sequence, U16253_at = U16253, can be found at http://expression.gnf.org/ratlas.
Using Gene Expression Information to Prioritize Microarray Follow-Up Experiments
Tissue expression information is a useful guide for the prioritization of follow-up experiments from gene expression studies. An example of the utility of our gene atlas in this role is from a rat model of addiction. Drug addicts and laboratory animals with drug experience, once made abstinent, very often return to consuming the drug once exposed to a previous environment related to drug taking (Stewart 1983). Recent studies have found that extinction training, a form of inhibitory learning, reverses or normalizes many neural alterations in addicted animals. For example, rats receiving extinction training after drug self-administration and abstinence (referred to here as 6EXT) show opposite interests in drug seeking when placed in the drug-taking environment, compared with animals that previously self-administered the drug followed by abstinence without extinction training (referred to here as 6WD). Gene expression differences between these two groups, in a relevant brain region, might reveal additional biochemical alterations responsible for drug craving. The brain region that we examined was the shell portion of the nucleus accumbens, one of the central brain regions in drug addiction research (Koob 1999).
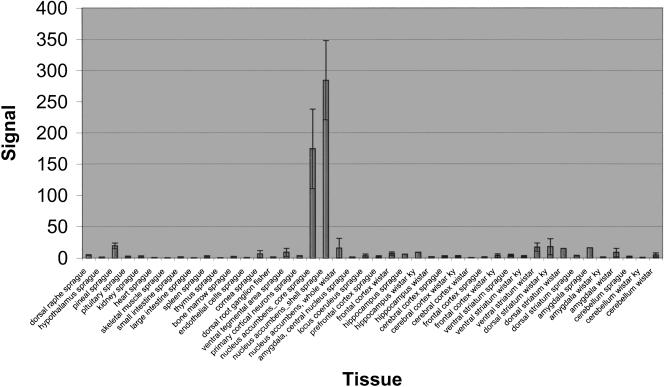
A comparison of the 6WD and 6EXT groups yields several transcripts for follow-up studies on the basis of their known function in particular tissues (Supplemental Table 4). ESTs of unknown function were initially ignored, as we had no further information about them. However, once we examined expression data across a variety of tissues, some ESTs that demonstrated interesting expression patterns could be moved up the priority list for further studies. For example, one sequence (Affymetrix probe set AF055714UTR#1_at, Accession no. AF055714), in addition to being the most down-regulated transcript in the comparison, also shows highly enriched expression in the nucleus accumbens core and shell of Sprague Dawley rats (Fig. 5). In addition to its regulation by the behavioral condition of interest, this tissue-restricted pattern suggests a prominent role in the function of the nucleus accumbens.
Figure 5.
A novel transcript with differential expression in a model of cocaine craving shows a dramatically enriched expression in drug addiction-related brain regions. Expression profile of this transcript, AF055714UTR#1_at = AF055714, is displayed using Affymetrix MAS5 signal intensity across many tissues. A similar image can be obtained on http://expression.gnf.org/ratlas.
Comparison of the expression pattern of rat and human orthologous genes can help prioritize followup experiments from rat gene-expression studies, as well as determine whether or not rats should be used as models when studying the functions of particular genes. We therefore compiled a list of rat to human orthologs using HomoloGene (Supplemental Table 5). We then compared expression data from rat and human across tissues in our data sets. An example of the usefulness of this information is illustrated here. From the cocaine-craving experiment described above, the most differentially expressed gene list (Supplemental Table 4) is compared with a list of genes showing the highest expression correlations between human and rat (from Supplemental Table 5). One of the resulting sequences is BHF-1, also called NeuroD1, a basic helix-loop-helix protein that, upon a preliminary search of the human ortholog in Locus Link (http://www.ncbi.nlm.nih.gov/LocusLink/ locus ID: 4760), seems to be primarily involved in insulin transcription and diabetes. Comparison of transcription patterns across tissues in both species indicates that it is highly expressed, manyfold over any other tissue, in the cerebellum. Examination of the expression pattern of mouse and human orthologs, across more tissues, further highlights its enhanced expression in the cerebellum (see probe set 36768_at for U95A and 92717_at for U74A from http://expression.gnf.org). A more extensive look into the literature has suggested a role for BHF-1 in neuronal development (Liu et al. 2000; Franklin et al. 2001). This finding supports the theory that some type of neuronal remodeling might be occurring during the extinction process (Schmidt et al. 2001; Henry and Garcia 2002) and may be responsible for the loss of interest in cocaine seeking. Using the tissue-expression pattern of this gene in rat and observing that its expression pattern is conserved in humans and enriched in the cerebellum, focuses additional importance on its neuronal function.
DISCUSSION
We describe a data set and accompanying Web site of rat gene expression across multiple tissues in commonly used strains. Multiple examples of how this data set can reveal interesting potential functions of genes are illustrated. Our public human and mouse gene-expression data (http://expression.gnf.org) has been an important resource for functional gene discovery. We believe the present rat data set will accelerate gene discovery in this common model for human physiology.
Careful consideration of the potential experimental variables should be recognized in these large data sets. This is especially true with public data sets that likely contain samples taken from a variety of laboratories. Although standard Affymetrix procedures were used in the two laboratories where these microarray studies were done, some slight laboratory-specific or operator-specific differences may exist. To minimize these potential differences, care was taken in this study to only include arrays and samples of similar quality (percent present scores, background, actin, and GAPDH 3′/5′ ratios). Another potential variable becomes more pronounced when increasingly refined dissections are taken, and when similar dissections are taken from different laboratories and combined into one database. For this reason, detailed descriptions of the dissection procedures should be given when dissection borders are not obvious.
Basal gene expression measurements across multiple tissues can be used as reference data to prioritize follow-up experiments of expression array studies. Multiple simultaneous measurements inherent in expression studies result in many false-positive expression changes. In addition to the false positives, in an organism such as the rat, in which most genes are uncharacterized, some decision has to be made whether or not to pursue further experiments with the uncharacterized transcripts. Researchers often perform sequence analysis to investigate the function of an uncharacterized transcript; however, identifying common domains from sequence only reveals a molecular function and usually reveals no information on a gene's physiological function. An additional source of information is the pattern of gene expression in normal physiological tissues, which often can help the researcher decide whether or not to pursue a particular uncharacterized transcript based on a more complete description of gene function. A scenario can also be imagined where drug targets or diagnostic markers are sought from an expression experiment. Of the desirable expression changes found, some priority should be given to those genes that are expressed in the target or diseased tissue over other organs (Welsh et al. 2003).
Regional expression data from multiple species can add an additional level of analysis to results of a microarray study. One should focus on regional expression patterns that are conserved across species, or at least consistent with human expression patterns. In the rat-to-human comparisons shown here, previously unknown discrepancies in expression patterns between the species can be found with several genes (see Interleukin 18, as U133A probe set 206295_at in Supplemental Table 5). Caution should be taken, however, in making conclusions when a limited set of tissues is compared, as high or low correlations might be due to the absence of a tissue where the key function of a particular gene is performed. For example, the present data set is lacking a couple of key organs, such as liver and lung. But when considered carefully, these types of comparisons can help a researcher discern whether or not to study a particular gene in a rodent model if the goal is to predict human physiology.
This gene expression data set can also aid in the hunt for functions of uncharacterized genes by pairing their expression patterns to known genes. In the few examples shown here, genes with expression patterns restricted to particular tissues likely play an important role in the function of those tissues. Likewise, genes that are coexpressed with members of a biochemical pathway might play a role in that pathway. A similar recent study in the Malarial parasite Plasmodium falciparum, although using various life-cycle stages instead of different tissues, has proven the utility of such an approach (Le Roch et al. 2003).
This data set can also help identify potential candidate genes from published rat QTL studies. The genomic interval given in the example above unveiled several interesting genes based on expression pattern alone (paste probe set identifiers from Supplemental Table 3 into the dialog box on expression.gnf.org/ratlas). One of the genes was α synuclein, which a previous study demonstrated to be differentially expressed between alcohol-preferring and alcohol-nonpreferring rats (Liang et.al. 2003; Supplemental Table 3). That study found that this expression difference was due to a polymorphism in the α synuclein gene, but the authors also suggested that multiple genes in the same QTL might influence alcohol consumption in these rats. CRHR2 should also be considered as a candidate gene influencing alcohol intake and preference. Studies suggest that CRHR2 does not play as prominent a role in the stress response and. therefore, in the major biological effect of its ligand, CRF, as CRHR1 (Rivier et. al. 2003). However, additional lines of evidence suggest that CRHR2 should be considered along with α synuclein. First, CRHR2 is relatively enriched in expression in two brain regions implicated in alcohol intake, the nucleus accumbens and the central nucleus of the amygdala (Fig. 4; Koob 1999; McBride 2002). Second, CRF plays a role in alcohol consumption (Bell et. al. 1998; Olive et. al. 2002). And third, alcohol-preferring and alcohol-nonpreferring rats show a differential response to CRF (Ehlers et. al. 1992). Future studies should determine the role of this specific receptor in alcohol intake, and if any nucleotide differences exist in this gene between preferring and nonpreferring rats. Although this gene could have been found simply through browsing QTLs in Ratmap (http://ratmap.gen.gu.se/) and using various tools to view genes in the relevant genomic intervals, examination of its expression pattern across multiple tissues rapidly identified it as a potential candidate gene.
Lastly, the data set presented here can help to explain some of the phenotypic differences that have been known to exist between common rat strains. In our search for the best model for human disease, we will learn more about the disease by comparing the strains. Some of the phenotypic differences might be predicted from genes of known function. It might also be possible to exploit these strain differences to discover the functions of novel genes, or to discover additional functions of known genes.
The public release of this data set coincides with the release of the rat genome sequence. It won't be long until full-transcriptome rat chips are commercially available and larger data sets than the one presented here are generated. These types of reference data sets will speed the characterization of rat gene function, and ultimately, the function of human genes that the rats are meant to model.
METHODS
Tissue and Microarray Processing
Tissues for expression studies were collected from a variety of sources (Supplemental Table 6). Tissues were homogenized in Trizol (Invitrogen), and total RNA was purified with Rneasy columns (Qiagen). Replicates consisted of either duplicate pools of tissue used for each array, or one tissue from one animal per array (see Supplemental Table 6). Five micrograms total RNA, or 0.2 μg poly(A+) RNA, was used for cDNA synthesis and cRNA amplification, and chips were hybridized to RGU34A arrays (Affymetrix) according to standard Affymetrix protocols (Affymetrix Expression Analysis Technical Manual, http://www.affymetrix.com/support/technical/manuals.affx). Arrays were washed and stained with standard Affymetrix reagents. Arrays were scanned with an Affymetrix scanner (model GA 2500). Data was analyzed both with MAS5 (Affymetrix) by global scaling to a target intensity of 200 and Rosetta Resolver.
Training of Drug-Seeking and Nonseeking Rats
Detailed methods describing the surgery and training of the rats used for the cocaine-craving study can be found in Sutton et. al. (2003). The drug-craving group of rats, 6-wk withdrawal (6WD), self-administered cocaine for 15 d in 6 h/d sessions, then were made abstinent for 6 wk, a time when a parallel group of identically trained animals exhibit profound drug-seeking behavior when returned to the environment where they previously self-administered the drug. Another group, 6-wk extinction (6EXT), self-administered the same amount of cocaine as the 6WD rats, and then also was made abstinent for 6 wk. However, unlike the 6WD group, the 6EXT group underwent extinction training during the final week of the 6-wk abstinence period, which involved returning rats to the previous drug-paired environment and allowing them to attempt to self-administer a drug when it was not available. After this period, the 6EXT group no longer pursued cocaine reinforcement.
Analysis of Differential Gene Expression
For finding gene-expression differences between strains in the frontal and whole cerebral cortices, both MAS5 (global scaling to a value of 200) and Rosetta Resolver analysis (see below) was done, and the intersection between the data sets were reported. Briefly, pairwise comparisons in MAS5 used three filters, t-test P-values of <0.05, absolute fold changes of at least 2, and minimum expression values (group average signals had to exceed a value of at least 100 in at least one group). Rosetta Resolver filters were absolute fold changes of at least 2 and P-values <0.05, using Resolver's ratio ANOVA function. Resolver ANOVA analysis is similar to standard ANOVA, but instead uses two inputs, expression measurement quantity, and estimated error of measurement quantity. This additional input provides more reliable variance measurements, a necessity when the number of replicates is small (Rajagopalan 2003). This error estimate also brings extra degrees of freedom to the analysis, allowing for fewer false positives and false negatives (see http://www.rosettabio.com/publications/default.htm for additional references). For finding genes differentially expressed between cocaine-craving and noncraving rats, only the Resolver data was used.
For display of differential expression between strains, MAS 5- and Resolver-processed data (see above) was further filtered in Rosetta Resolver's clustering algorithm according to the following criteria (detection P < 0.05 and present score required for at least three chips, coefficient of variation across all samples at least 0.5). Only 10 sequences remained after these three filtering steps.
Gene Coexpression/Tissue Enrichment
For finding genes coexpressed with the D1A dopamine receptor, Pearson analysis was performed across all tissues. To display the results, (Fig. 2) all sequences on the RG_U34A chip and all experiments in this data set were clustered by Z score with an agglomerative algorithm. This algorithm used the following parameters: average link, Euclidean distance, and individual probes were error-weighted. Sequences were used if their detection Pvalues were <0.01, they were scored as present for at least three chips across the entire data set, and their coefficients of variation across the set were at least 0.5. Sequences with Pearson correlation coefficients of at least 0.8 to the D1 dopamine receptor across all tissues were projected onto this two-dimensional cluster diagram.
For finding genes coexpressed with a set of striatal-enriched genes, GROW (Rosetta Resolver) was performed using this set as a seed set. GROW is a pattern-finding algorithm that searches for additional genes and experiments with a similar pattern to the seed set, and essentially perform a two-dimensional (across genes and tissues) Pearson correlation.
For finding genes with highly enriched expression in particular tissues, the RAtlas expression pattern interface from the Web site (http://expression.gnf.org/ratlas) was used, using the following filters: tissue of interest greater than 10-fold above the median expression level, and nonspecified tissues no more than threefold above the median. The number of exceptions allowed, whether or not exclusive expression was specified, and the combinations of strains used for the search was varied for each tissue to yield a manageable number of transcripts.
Finding Candidate Genes in QTL
The rat QTL database (http://ratmap.gen.gu.se) was used to find genomic intervals associated with quantitative trait differences found in the literature. From a PubMed (http://www.ncbi.nlm.nih.gov/PubMed/) search using “QTL alcohol rat” as keywords, 13 articles were found, and a cursory viewing of the abstracts showed that chromosome 4 was associated with the highest lod score for alcohol preference. From the QTL portion of the RatMap site (http://ratmap.gen.gu.se/qtler/), chromosome 4 and “Eoh” were chosen from the search options, and the genomic interval associated with the published QTL was shown (Carr et. al. 1998). Markers from this genomic interval were searched in the UCSC site via the Affymetrix link (https://www.affymetrix.com/analysis/netaffx/query_ucsc.affx), and probe sets from the RG_U34A chip were downloaded and pasted into the dialog box on the RAtlas Web site (http://expression.gnf.org/ratlas). Expression patterns were examined for all genes, and CRHR2 was chosen, as it had its highest levels of expression in the nucleus accumbens and central nucleus of the amygdala, and lower expression levels in other tissues.
Ortholog Comparisons
Rat to human ortholog comparisons were made using data from Homologene (http://www.ncbi.nlm.nih.gov/HomoloGene/). Only pairs that contained Locus Link identifiers, and only tissues that were common between our human and rat sets, were used. Human expression data for the orthologs was from HG_U133A chips; tissue sources and the complete data set across many more tissues will be described in an upcoming manuscript (A.I. Su, T. Wiltshire, S. Batalov, H. Lapp, K.A. Ching, D. Block, J. Zhang, R. Soden, M. Hayakawa, G. Kreiman, et al., in prep.). The number of othologs that contained Locus Link identifiers and were present on both the RGU34A and HG_U133A chips was 2219 (Supplemental Table 5). For Pearson analysis, the data set was further filtered to find the most dynamically expressed genes (Su et. al. 2002). Thus, transcripts were chosen only if the minimum MAS5 signal intensity was above a value of 100 in at least one tissue (means of all replicates were taken) across both species, and maximum/median values were at least a value of 3 in both species.
Acknowledgments
We thank Mimi Hayakawa for technical assistance. Also, thanks to Christine Sturchler (Novartis) for data file acquisition and Lisa Tarantino for critically reading the manuscript. We thank Teresa Reyes, Tamas Bartfai, Pietro Sanna, Athina Markou, Trevor Young, and Martin Alda for providing some of the tissues.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2161804.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GEO (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE952.]
References
- Baldessarini, R.J. 1996. Drugs and the treatment of psychiatric disorders. In The pharmacological basis of therapeutics, 9th Ed. (eds. P.B. Molinoff and R.W. Ruddon), pp. 431-459. McGraw-Hill, New York.
- Bell, S.M., Reynolds, J.G., Thiele, T.E., Gan, J., Figlewicz, D.P., and Woods, S.C. 1998. Effects of third intracerebroventricular injections of corticotropin-releasing factor (CRF) on ethanol drinking and food intake. Psychopharmacology 139: 128-135. [DOI] [PubMed] [Google Scholar]
- Bice, P., Foroud, T., Bo, R., Castelluccio, P., Lumeng, L., Li, T.-K., and Carr, L.G. 1998. Genomic screen for QTLs underlying alcohol consumption in the P and NP rat lines. Mamm. Genome 9: 949-955. [DOI] [PubMed] [Google Scholar]
- Brundege, J.M. and Williams, J.T. 2002. Differential modulation of nucleus accumbens synapses. J. Neurophysiol. 2002. 88: 142-151. [DOI] [PubMed] [Google Scholar]
- Carr, L.G., Foroud, T., Bice, P., Gobbett, T., Ivashina, J., Edenberg, H., Lumeng, L., and Li, T.K. 1998. A quantitative trait locus for alcohol consumption in selectively bred rat lines. Alcohol Clin. Exp. Res. 22: 884-887. [PubMed] [Google Scholar]
- Chung, Y.H., Shin, C.M., Kim, M.J., and Cha, C.I. 2000. Immunohistochemical study on the distribution of six members of the Kv1 channel subunits in the rat basal ganglia. Brain Res. 875: 164-170. [DOI] [PubMed] [Google Scholar]
- Ehlers, C.L., Chaplin, R.I., Wall, T.L., Lumeng, L., Li, T.K., Owens, M.J., and Nemeroff, C.B. 1992. Corticotropin releasing factor (CRF): Studies in alcohol preferring and non-preferring rats. Psychopharmacology 106: 359-364. [DOI] [PubMed] [Google Scholar]
- Franklin, A., Kao, A., Tapscott, S., and Unis, A. 2001. NeuroD homologue expression during cortical development in the human brain. J. Child Neurol. 16: 849-853. [DOI] [PubMed] [Google Scholar]
- Henry, C. and Garcia, R. 2002. Prefrontal cortex long-term potentiation, but no long-term depression, is associated with the maintenence of extinction of learned fear in mice. Neuroscience 22: 577-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman, B.B., Lefkowitz, R.J., and Taylor, P. 1996. Neurotransmission. In The pharmacological basis of therapeutics, 9th Ed. (eds. P.B. Molinoff and R.W. Ruddon), pp. 105-139. McGraw-Hill, New York.
- Horikawa, Y., Oda, N., Cox, N.J., Li, X., Orho-Melander, M., Hara, M., Hinokio, Y., Linder, T.H., Mashima, H., Schwarz, P.E., et al. 2000. Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat. Genet. 26: 163-175. [DOI] [PubMed] [Google Scholar]
- Hugot, J., Chamaillard, M., Zouali, H., Lesage, S., Cezard, J.P., Belaiche, J., Almer, S., Tysk, C., O'Morain, C.A., Gassull, M., et al. 2001. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature 411: 599-603. [DOI] [PubMed] [Google Scholar]
- Jacob, H.J. and Kwitek, A.E. 2002. Rat genetics: Attaching physiology and pharmacology to the genome. Nat. Rev. Genet. 3: 33-42. [DOI] [PubMed] [Google Scholar]
- Koob, G.F. 1999. The role of the striatopallidal and extended amygdala systems in drug addiction. Ann. NY Acad. Sci. 877: 445-460. [DOI] [PubMed] [Google Scholar]
- Lahmame, A. and Armario, A. 1996. Differential responsiveness of inbred strains of rats to antidepressants in the forced swimming test: Are Wistar Kyoto rats an animal model of subsensitivity to antidepressants? Psychopharmacology 123: 191-198. [DOI] [PubMed] [Google Scholar]
- Le Roch, K.G., Zhou, Y., Blair, P.L., Grainger, M., Moch, J.K., Haynes, J.D., De La Vega, P., Holder, A.A., Batalov, S., Carucci, D.J., et al. 2003. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science 301: 1503-1508. [DOI] [PubMed] [Google Scholar]
- Liang, T., Spence, J., Liu, L., Strother, W.N., Chang, H.W., Ellison, J.A., Lumeng, L., Li, T.K., Foroud, T., and Carr, L.G. 2003. α-Synuclein maps to a quantitative trait locus for alcohol preference and is differentially expressed in alcohol-preferring and -nonpreferring rats. Proc. Natl. Acad. Sci. 100: 4690-4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, M., Pleasure, S.J., Collins, A.E., Noebels, J.L., Naya, F.J., Tsai, M.J., and Lowenstein, D.H. 2000. Loss of BETA2/NeuroD leads to malformation of the dentate gyrus and epilepsy. Proc. Natl. Acad. Sci. 97: 865-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride, W.J. 2002. Central nucleus of the amygdala and the effects of alcohol and alcohol-drinking behavior in rodents. Pharmacol. Biochem. Behav. 71: 509-515. [DOI] [PubMed] [Google Scholar]
- Mootha, V.K., Lepage, P., Miller, K., Bunkenborg, J., Reich, M., Hjerrild, M., Delmonte, T., Villeneuve, A., Sladek, R., Xu, F., et. al. 2003. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc. Natl. Acad. Sci. 100: 605-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura, Y., Bonin, D.K., Inohara, N., Nicolae, D.L., Chen, F.F., Ramos, R., Britton, H., Moran, T., Karaliuskas, R., Duerr, R.H., et al. 2001. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature 411: 603-606. [DOI] [PubMed] [Google Scholar]
- Olive, M.F., Mehmert, K.K., Koenig, H.N., Camarini, R., Kim, J.A., Nannini, M.A., Ou, C.J., and Hodge, C.W. 2002. A role for corticotropin releasing factor (CRF) in ethanol consumption, sensitivity, and reward as revealed by CRF-deficient mice. Psychopharmacology 165: 181-187. [DOI] [PubMed] [Google Scholar]
- Rajagopalan, D. 2003. A comparison of statistical methods for analysis of high density oligonucleotide array data. Bioinformatics 19: 1469-1476. [DOI] [PubMed] [Google Scholar]
- Rittenhouse, P.A., Lopez-Rubalcava, C., Stanwood, G.D., and Lucki, I. 2002. Amplified behavioral and endocrine responses to forced swim stress in the Wistar-Kyoto rat. Psychoneuroendocrinology 27: 303-318. [DOI] [PubMed] [Google Scholar]
- Rivier, C.L., Grigoriadis, D.E., and Rivier, J.E. 2003. Role of corticotropin-releasing factor receptors type 1 and 2 in modulating the rat adrenocorticotropin response to stressors. Endocrinology 144: 2396-2403. [DOI] [PubMed] [Google Scholar]
- Schmidt, E.F., Sutton, M.A., Schad, C.A., Karanian, D.A., Brodkin, E.S., and Self, D.W. 2001. Extinction training regulates tyrosine hydroxylase during withdrawal from cocaine self-administration. J. Neurosci. 21: RC137:1-RC135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Self, D.W. and Stein, L. 1992. The D1 agonists SKF 82958 and SKF 77434 are self-administered by rats. Brain Res. 582: 349-352. [DOI] [PubMed] [Google Scholar]
- Staudt, L.M. and Brown, P.O. 2000. Genomic views of the immune system. Annu. Rev. Immunol. 18: 829-859. [DOI] [PubMed] [Google Scholar]
- Stewart, J. 1983. Conditioned and unconditioned drug effects in relapse to opiate and stimulant drug self-adminstration. Prog. Neuropsychopharmacol. Biol. Psychiatry 7: 591-597. [DOI] [PubMed] [Google Scholar]
- Stuart, J.M., Segal, E., Koller, D., and Kim, S.K. 2003. A Gene-coexpression network for global discovery of conserved genetic modules. Science 302: 249-255. [DOI] [PubMed] [Google Scholar]
- Su, A.I., Cooke, M.P., Ching, K.A., Hakak, Y., Walker, J.R., Wiltshire, T., Orth, A.P., Vega, R.G., Sapinoso, L.M., Moqrich, A., et. al. 2002. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. 99: 4465-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton, M.A., Schmidt, E.F., Choi, K.H., Schad, C.A., Whisler, K., Simmons, D., Karanian, D.A., Monteggia, L.M., Neve, R.L., and Self, D.W. 2003. Extinction-induced upregulation in AMPA receptors reduces cocaine-seeking behaviour. Nature 421: 70-75. [DOI] [PubMed] [Google Scholar]
- Tejani-Butt, S.M., Pare, W.P., and Yang, J. 1994. Effect of repeated novel stressors on depressive behavior and brain norepinephrine receptor system in Sprague-Dawley and Wistar Kyoto (WKY) rats. 1994. Brain Res. 649: 27-35. [DOI] [PubMed] [Google Scholar]
- Terenina-Rigaldie, E., Moisan, M.P., Colas, A., Beauge, F., Shah, K.V., Jones, B.C., and Mormede, P. 2003. Genetics of behaviour: Phenotypic and molecular study of rats derived from high- and low-alcohol consuming lines. Pharmacogenetics 13: 543-554. [DOI] [PubMed] [Google Scholar]
- Welsh, J.B., Sapinoso, L.M., Kern, S.G., Brown, D.A., Liu, T., Bauskin, A.R., Ward, R.L., Hawkins, N.J., Quinn, D.I., Russell, P., et. al. 2003. Large-scale delineation of secreted protein biomarkers overexpressed in cancer tissue and serum. Proc. Natl. Acad. Sci. 100: 3410-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://expression.gnf.org; GNF GeneAtlas Web site.
- http://expression.gnf.org/ratlas; RAtlas link from GNF GeneAtlas Web site.
- http://www.ncbi.nlm.nih.gov/LocusLink/; Locus Link.
- http://www.affymetrix.com/support/technical/manuals.affx; Affymetrix Web site technical manuals.
- http://www.ncbi.nlm.nih.gov/HomoloGene; Homologene.
- http://www.affymetrix.com; Affymetrix main page.
- http://genome.ucsc.edu; UCSC Genome Bioinformatics Site.
- http://www.rosettabio.com/publications/default.htm; Rosetta Biosoftware, publications.
- http://ratmap.gen.gu.se/; Ratmap, The Rat Genome Database.
- http://www.ncbi.nlm.nih.gov/PubMed/; PubMed.
- http://www.ncbi.nlm.nih.gov/geo/; Gene Expression Omnibus home page.
- http://symatlas.gnf.org; GNF GeneAtlas Web site.