Abstract
The laboratory rat is a major model organism for systems biology. To complement the cornucopia of physiological and pharmacological data generated in the rat, a large genomic toolset has been developed, culminating in the release of the rat draft genome sequence. The rat draft sequence used a variety of assembly packages, as well as data from the Radiation Hybrid (RH) map of the rat as part of their validation. As part of the Rat Genome Project, we have been building a high-density RH map to facilitate data integration from multiple maps and now to help validate the genome assembly. By incorporating vectors from our lab and several other labs, we have doubled the number of simple sequence length polymorphisms (SSLPs), genes, expressed sequence tags (ESTs), and sequence-tagged sites (STSs) compared to any other genome-wide rat map, a total of 24,437 elements. During the process, we also identified a novel approach for integrating the RH placement results from multiple maps. This new integrated RH map contains approximately 10 RH-mapped elements per Mb on the genome assembly, enabling the RH maps to serve as a scaffold for a variety of data visualization tools.
Rattus Norvegicus is a major model organism in biomedical research, with over a century of physiological and pharmacological data for baseline biology and pathobiology. The past decade has witnessed a dramatic growth in rat genomic data, resulting in well established genomic resources (Jacob and Kwitek 2002), and culminating in the draft sequence of the whole genome (Rat Genome Sequencing Project Consortium 2004). One such genomic resource, radiation hybrid (RH) maps, are a powerful tool for annotating the rat genome with systems biology information; they have been used to integrate genetic maps from different rat crosses used in QTL mapping, physical and draft genomic maps, and the genomes of different species via comparative mapping (Gosele et al. 2000; Bihoreau et al. 2001; Kwitek et al. 2001; Dobbins et al. 2002; Moujahidine et al. 2002; Chowdhary et al. 2003; Larkin et al. 2003). The VCMap (Kwitek et al. 2001) at the Rat Genome Database (RGD) has chosen the RH maps as the backbone platform for integrating available genomic data as well as for annotating disease QTLs for complex traits and diseases, using the new VCMapView tool (see Twigger et al. 2004). Finally, the maps offer many direct links to the draft genomic sequence and help to validate the assembly of the rat genome sequence.
We, and others, have generated whole-genome RH maps of the rat genome, including simple sequence length polymorphisms (SSLPs), expressed sequence tags (ESTs), and genes, using the T55 rat RH panel (Steen et al. 1999; Watanabe et al. 1999; McCarthy et al. 2000; Scheetz et al. 2001). However, since that time, many more sequence-tagged site (STS) vectors have been generated as a result of the Rat EST Project. Therefore, we improved the RH maps, generating a high-density RH map that integrated markers from all sources with the highest possible resolution and validation. We also improved upon the previous algorithm for building RH maps using the RHMAPPER computer package, resulting in an even higher number of markers placed. As a result, we have built new framework maps for all of the rat chromosomes and were able to place an additional 23,172 markers (SSLPs, genes, ESTs), more than doubling the map density of any other rat whole-genome RH map to date (Steen et al. 1999; Watanabe et al. 1999; Scheetz et al. 2001).
Because these maps were to help to validate the assembly of the genomic sequence, their accuracy is of utmost importance. Therefore, we performed extensive comparisons of the newly generated maps to other maps as well as multiple tests to evaluate potential errors in the maps. These efforts have resulted in high-density RH maps with extensive curation, which can be used for multiple applications in the discovery and understanding of complex disease pathology. Perhaps most importantly, this new RH map provides “sequence hooks” at nearly 10 STSs per Mb of genomic DNA, providing many integration points between various Rat maps as well as interspecies alignments, as illustrated in the VCMapView tool described in the companion paper within this issue (Twigger et al. 2004).
RESULTS
Framework Maps
The framework maps consist of SSLPs ordered with a LOD 3 confidence, meaning approximately 1000:1 odds of accurate order on the chromosome. Only SSLP markers were used to build the framework maps, because they have been previously mapped to a chromosome by genetic mapping, providing independent validation of chromosome assignment, and because they offer a powerful means to integrate genetic maps from different rat crosses.
The RHMAPPER computer package was used to build the framework maps (Stein et al. 1995) using vector data generated by us and from public data submitted to the RH database (RHdb; http://corba.ebi.ac.uk/RHdb/). Only high-quality consensus vectors (see Methods and Steen et al. 1999) were chosen for building the framework maps. The final framework maps (version 3.0) for all 20 rat autosomes and the X chromosome contained a total of 1265 SSLPs ordered at a LOD of 3 versus any other order. These maps can be viewed online at RGD through the VCMapView at RGD (http://rgd.mcw.edu/VCMAP/mapview.shtml). A summary of the maps is shown in Table 1. The overall length of the new framework maps has not significantly changed from our previous framework map, 20,991 cR vs. 19,368 cR, respectively (Steen et al. 1999). The density of framework markers is increased by 29% (1265 vs. 983), with an average bin size of 17.1 cR, compared to 19.7 cR previously (Steen et al. 1999). The lack of significant map length expansion, while increasing the number of elements mapped, is indicative of our efforts to error check, and increases the likelihood that the maps are accurate.
Table 1.
Summary of Rat RH V3.4 Maps
| Chromosome | Framework length (cR3000) | # Framework markers | Placement length (cR3000) | # Total markers | Genome sequence lengtha(Mb) | # STSs per cR3000 | cR3000/Mb | # STSs per Mb | Av. bin size (cR3000) | Av. bin size (Mb) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1681 | 98 | 1722 | 2951 | 268.12 | 1.7 | 6.42 | 11.0 | 17.6 | 2.74 |
| 2 | 1620 | 119 | 1663 | 1960 | 258.22 | 1.2 | 6.44 | 7.6 | 14.0 | 2.17 |
| 3 | 1519 | 89 | 1565 | 1639 | 170.97 | 1.0 | 9.15 | 9.6 | 17.6 | 1.92 |
| 4 | 1094 | 74 | 1134 | 1690 | 187.37 | 1.5 | 6.05 | 9.0 | 15.3 | 2.53 |
| 5 | 1182 | 81 | 1206 | 1823 | 173.11 | 1.5 | 6.97 | 10.5 | 14.9 | 2.14 |
| 6 | 924 | 57 | 954 | 1277 | 147.64 | 1.3 | 6.46 | 8.6 | 16.7 | 2.59 |
| 7 | 1090 | 74 | 1136 | 1591 | 143.08 | 1.4 | 7.94 | 11.1 | 15.4 | 1.93 |
| 8 | 1358 | 82 | 1358 | 1622 | 129.06 | 1.2 | 10.52 | 12.6 | 16.6 | 1.57 |
| 9 | 880 | 57 | 908 | 897 | 113.65 | 1.0 | 7.99 | 7.9 | 15.9 | 1.99 |
| 10 | 1214 | 67 | 1214 | 1190 | 110.73 | 1.0 | 10.96 | 10.7 | 18.1 | 1.65 |
| 11 | 704 | 40 | 733 | 706 | 87.80 | 1.0 | 8.35 | 8.0 | 18.3 | 2.20 |
| 12 | 794 | 48 | 823 | 789 | 46.65 | 1.0 | 17.64 | 16.9 | 17.1 | 0.97 |
| 13 | 770 | 38 | 800 | 831 | 111.35 | 1.0 | 7.18 | 7.5 | 21.1 | 2.93 |
| 14 | 782 | 41 | 826 | 682 | 112.22 | 0.8 | 7.36 | 6.1 | 20.1 | 2.74 |
| 15 | 743 | 48 | 791 | 721 | 109.77 | 0.9 | 7.21 | 6.6 | 16.5 | 2.29 |
| 16 | 789 | 45 | 820 | 850 | 90.22 | 1.0 | 9.09 | 9.4 | 18.2 | 2.00 |
| 17 | 833 | 51 | 879 | 901 | 97.31 | 1.0 | 9.03 | 9.3 | 17.2 | 1.91 |
| 18 | 832 | 51 | 852 | 775 | 87.34 | 0.9 | 9.76 | 8.9 | 16.7 | 1.71 |
| 19 | 726 | 41 | 754 | 627 | 59.22 | 0.8 | 12.73 | 10.6 | 18.4 | 1.44 |
| 20 | 517 | 22 | 562 | 594 | 55.30 | 1.1 | 10.17 | 10.7 | 25.6 | 2.51 |
| X | 941 | 42 | 990 | 321 | 160.78 | 0.3 | 6.16 | 2.0 | 23.6 | 3.83 |
| Total | 20991 | 1265 | 21688 | 24437 | 2719.92 | - | - | - | - | - |
| Average (Autosome) | - | - | - | - | - | 1.1 | 8.87 | 9.6 | 17.6 | 2.10 |
Genome lengths according to Release 3.1 (RGSPC)
Integrated Placement Maps
These new maps offer a better framework onto which additional STSs can be placed. Our placement vector data set consisted of 28,096 vectors, from 22,897 ESTs, 4583 SSLPs, 597 genes, and 19 STSs. These vectors were generated by us and by several other groups and submitted to RHdb (http://corba.ebi.ac.uk/RHdb/), a public repository for RH vector data (Rodriguez-Tome and Lijnzaad 2001). The placement maps were generated using the `place_ markers.pl' script provided with the RHMAPPER package (Stein et al. 1995), but with modifications to allow for integration of markers placed at differing LOD thresholds (described below). In generating and reviewing the placement maps, we found that markers mapping upstream of the first framework marker were assigned a position equivalent to that of the first framework markers, a feature of the placement algorithm within RHMAPPER. Therefore, we modified the RHMAPPER algorithm to correct this problem so that `0 cR' is the designation of the first placed marker and the absolution position of all consecutive markers is calculated.
Traditionally, when using the RHMAPPER program to place markers onto a framework reference map, a single LOD threshold is chosen for the initial two-point analysis to assign a vector to a single chromosome, after which multipoint analyses place the marker with respect to the framework map for that unique chromosome. If markers are not linked at that single threshold, or alternatively are linked to markers on multiple chromosomes, they will not be placed on the map. However, those markers might map uniquely at an alternative LOD threshold. Previously we determined the most markers mapped using an LOD 10 threshold. In order to map additional markers not placed at this empirically chosen threshold, we developed an algorithm for integrating markers mapped at multiple LOD thresholds. Although about 88% of the markers can be placed with a single, two-point linkage LOD score of 10, many markers were either not linked or were linked to markers on more than one chromosome at that single LOD threshold.
Place Marker Function
In order to increase the amount of markers placed in RHMAPPER, we created a novel algorithm to integrate markers placed across a range of two-point LOD thresholds. For the integrated approach, we generated multiple placement maps for each chromosome, each with a different two-point LOD threshold, and then merged them to generate a single integrated placement map for each chromosome. To test the feasibility of our integration algorithm we placed markers from 1500 vectors, randomly chosen from the placement data set onto chromosome 1 (∼10% of the genome), at LODs 8, 9, 10, 11, 12, 13, 14, and 15, and then compared how much variation in marker position, from the top of the chromosome, there was for markers in common in more than one placement map (data not shown). The results indicated that the variation, that is, the difference in the absolute distance on the map, is very small; in fact, the difference in placement never exceeded 0.8 cR, and the order of markers in common between the maps did not change.
Based on these results, we concluded that integrating the different maps might be feasible for the whole genome. Therefore, we constructed placement maps at incremental LOD thresholds from 8–15 for all rat chromosomes and integrated them into a single placement map. Our approach for the integration is out-lined in Figure 1. For each marker, an absolute chromosome distance was calculated, with the first framework marker denoted as `0' cR and markers mapped upstream of that first framework denoted as the negative distance from that framework marker. It is important to note that the same framework map is used for each LOD placement. Therefore, although the raw placement maps have slightly varying lengths, we could normalize the bin distance (distance between framework markers) for all maps to that of a single map (we chose the LOD 15), and proportionately adjust the position of each placement marker within the framework bin. Once all of the placement maps were generated, they were merged into a single integrated map. Re-evaluation of RNO1 with all markers placed (2903 STSs) identified only 66 cases of marker order discrepancy between any of the placement maps and the final integrated map; all order discrepancies were negligible, involving less than 1 cR (average = 0.2 cR; maximum = 0.8 cR), indicating the vigor of the integration algorithm. The integrated map was 21,733 cR in length, and contained 25,026 STSs. By integrating maps generated at multiple LOD thresholds, the density increased over 12% compared to the single LOD 10 map (Table 2).
Figure 1.
Placement map integration algorithm. Placement maps resulting from incremental LOD thresholds between 8 and 15 were aligned. The framework bins for all maps were normalized to that of the LOD 15 map so that the bin sizes were identical across maps (left panel). The position of markers placed within the bins on all maps were accordingly adjusted to be proportional to the normalized bin size (middle panel). The normalized position from the map with the highest LOD threshold for each marker was added to the integrated map, with markers placing upstream of the first framework designated as a negative distance from that framework marker (middle panel, integrated). Finally, the absolute positions of the markers were determined with the first placed marker at 0 cR (right panel, final integrated).
Table 2.
Number of Mapped STSs Before and After Integration of Placement Maps
| Chrom. | # Total markersa | # Mapped L10b | % Total mapped at L10 | % Increase in integrated map |
|---|---|---|---|---|
| 1 | 3001 | 2350 | 78.3% | 21.7% |
| 2 | 2038 | 1515 | 74.3% | 25.7% |
| 3 | 1663 | 1390 | 83.6% | 16.4% |
| 4 | 1718 | 1168 | 68.0% | 32.0% |
| 5 | 1901 | 1600 | 84.2% | 15.8% |
| 6 | 1287 | 1162 | 90.3% | 9.7% |
| 7 | 1616 | 1456 | 90.1% | 9.9% |
| 8 | 1744 | 1524 | 87.4% | 12.6% |
| 9 | 906 | 838 | 92.5% | 7.5% |
| 10 | 1205 | 1156 | 95.9% | 4.1% |
| 11 | 713 | 579 | 81.2% | 18.8% |
| 12 | 801 | 762 | 95.1% | 4.9% |
| 13 | 893 | 825 | 92.4% | 7.6% |
| 14 | 695 | 638 | 91.8% | 8.2% |
| 15 | 731 | 591 | 80.8% | 19.2% |
| 16 | 859 | 787 | 91.6% | 8.4% |
| 17 | 914 | 875 | 95.7% | 4.3% |
| 18 | 778 | 745 | 95.8% | 4.2% |
| 19 | 635 | 577 | 90.9% | 9.1% |
| 20 | 600 | 566 | 94.3% | 5.7% |
| X | 328 | 304 | 92.7% | 7.3% |
| Total | 25026 | 21408 | - | - |
| Average | - | - | 87.9% | 12.1% |
Total numbers reflect numbers placed before curation of the integrated maps
L10 = two-point LOD threshold of 10
Error Checking and Validation of RH Maps
To ensure the most accurate RH maps possible, we performed several validation tests and extensive curation of the integrated placement maps. When a placement map is generated, a two-point LOD threshold is first chosen; markers mapping to a unique chromosome at that threshold are then subject to a multipoint analysis to place that marker relative to the chromosome framework map. However, a marker may map to an additional chromosome (multiple chromosome linkage) at any two-point LOD below the threshold. In order to evaluate a confidence level for the placement of STSs onto a unique chromosome versus multiple chromosomes, we calculated whether a marker remained uniquely assigned to the chromosome given a LOD 2 decrease in threshold (LOD score for a particular placement map that assigned the marker to its position). If the marker remained uniquely linked to that chromosome, its assignment is at least 100 times more likely to map to that chromosome than to another. Only eight of the placed markers failed the LOD 2 confidence test (D7Hmgc1, D9Hmgc1, D10Hmgc1, Dgkg, D12Hmgc2, D14Hmgc2, D17Hmgc2, Tnf). In fact, on average, each marker placed to a single chromosome with approximately 108 higher likelihood than to any other chromosome, indicating that the vast majority of STSs were placed on the correct chromosome.
It is important to note that all maps have some errors in them and that consistency between independent maps is a powerful validation of accuracy. Having multiple maps, such as genetic, RH, and genome maps allows for these consistency checks, given that they are generated essentially independently of one another. To visualize the consistency of the RH maps with other available maps, the new VCMapView tool (http://rgd.mcw.edu/VCMAP/mapview.shtml; see Twigger et al. 2004) was used to align the RH map with two reference genetic maps (SHRSP × BN and FHH × ACI), the RH map generated by Watanabe et al. (1999), and the genome assembly (R3.1; Fig. 2). The VCMapView tool allows dynamic alignment in any chosen order of maps and is a powerful platform for both intra- and interspecies genome comparisons. By these global alignments, it was clear that the order of the RH framework markers is largely consistent with the order of both the genetic maps and the rat genome assembly (R3.1; Fig. 2). These visual comparisons did indicate regions of inconsistencies within the maps, warranting further investigation to determine the source of the inconsistencies. The next method of validation for the RH maps was to identify SSLPs that were previously assigned to a different chromosome in the SHRSP × BN map reported by Steen (1999). We found 116 markers having inconsistent chromosome designation, corresponding to nearly 98% consistency between maps generated by independent means (Table 3). Because the density of SSLPs in common between the genetic and RH maps was relatively low (5012) compared to the number of STSs in common between the RH map and the genome assembly, a more detailed evaluation was performed by determining the interchromosomal inconsistencies between the RH maps and the genome sequence assembly R3.1. The chromosome designation was compared between the RH and genome assembly; markers that are inconsistent between the RH and both the genetic and genome maps help to confirm that they are errors due to the RH map. There were 21,603 STSs mapped in both the RH and the genome assembly, of which 96.2% were consistent between the maps. In four chromosomes, RNO2, RNO5, and RNO8, and RNO13, there were blocks of markers mapped to different chromosomes in both the genetic and genome maps, compared to the RH map, leading to further curation of these chromosomes, including evaluation of both the genetic and genome maps as well as conservation with mouse and human. As a result, 52, 58, 99, and 48 markers were removed from the RH maps for the RNO2, RNO5, RNO8, and RNO13 maps, respectively. Removing these markers brought the overall consistency in chromosome designation to 96.9% (Table 3); this reduced the number of total markers on the map by 257.
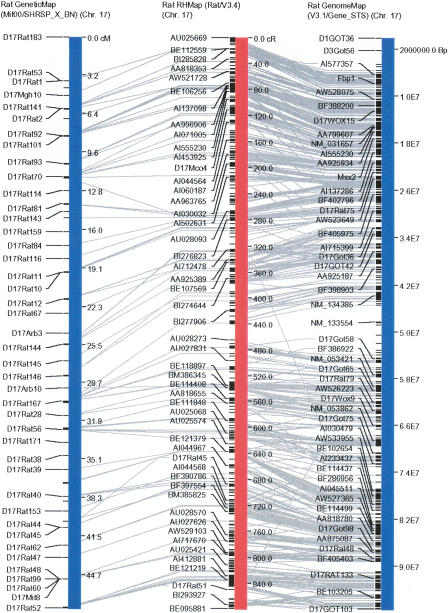
Figure 2.
Screenshot from VCMapView (http://rgd.mcw.edu/VCMAP/mapview.shtml) showing visual alignment between genetic maps (SHRSP × BN), RH map (Version 3.4), and rat genome assembly (Release 3.1) for RNO17. Lines between the maps indicate common STSs between the maps. Please see Twigger et al. 2004, for details on the VCMapView visualization.
Table 3.
Consistency Between RH V3.4, SHRSP × BN, and the Rat Genome Assembly R3.1 (RGSPC)
| Chromosome | # STSs on RH and genetic map | # Discrepant chromosome designation | % Consistent chromosome designation | # STS in RH and genome assembly | # Discrepant chromosome designation | % Consistent chromosome designation | # Binned STSs and RH and genome map | # Discrepant chromosome position | % Consistent chromosome position |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 544 | 8 | 98.5% | 2547 | 88 | 96.5% | 2163 | 213 | 90.15% |
| 2 | 425 | 20 | 95.3% | 1639 | 61 | 96.3% | 1387 | 99 | 92.86% |
| 3 | 331 | 3 | 99.1% | 1371 | 33 | 97.6% | 1086 | 37 | 96.59% |
| 4 | 353 | 10 | 97.2% | 1412 | 84 | 94.1% | 1241 | 113 | 90.89% |
| 5 | 343 | 15 | 95.6% | 1552 | 70 | 95.5% | 1282 | 135 | 89.47% |
| 6 | 300 | 4 | 98.7% | 1088 | 26 | 97.6% | 1098 | 58 | 94.72% |
| 7 | 302 | 3 | 99.0% | 1360 | 63 | 95.4% | 1144 | 68 | 94.06% |
| 8 | 352 | 15 | 95.7% | 1346 | 35 | 97.4% | 1063 | 33 | 96.90% |
| 9 | 193 | 4 | 97.9% | 744 | 36 | 95.2% | 474 | 28 | 94.09% |
| 10 | 224 | 0 | 100.0% | 995 | 22 | 97.8% | 876 | 32 | 96.35% |
| 11 | 144 | 4 | 97.2% | 590 | 15 | 97.5% | 490 | 28 | 94.29% |
| 12 | 169 | 2 | 98.8% | 647 | 15 | 97.7% | 503 | 24 | 95.23% |
| 13 | 175 | 5 | 97.1% | 690 | 10 | 98.6% | 619 | 12 | 98.06% |
| 14 | 136 | 1 | 99.3% | 568 | 17 | 97.0% | 480 | 21 | 95.63% |
| 15 | 155 | 1 | 99.4% | 595 | 22 | 96.3% | 492 | 77 | 84.35% |
| 16 | 165 | 6 | 96.4% | 720 | 18 | 97.5% | 586 | 24 | 95.90% |
| 17 | 224 | 3 | 98.7% | 745 | 15 | 98.0% | 709 | 25 | 96.47% |
| 18 | 167 | 5 | 97.0% | 636 | 23 | 96.4% | 505 | 23 | 95.45% |
| 19 | 121 | 2 | 98.3% | 512 | 16 | 96.9% | 394 | 17 | 95.69% |
| 20 | 76 | 3 | 96.1% | 489 | 7 | 98.6% | 265 | 4 | 98.49% |
| X | 113 | 2 | 98.2% | 244 | 9 | 96.3% | 205 | 9 | 95.61% |
| Total | 5012 | 116 | 20490 | 685 | 17062 | 1080 | - | ||
| Average | 97.8% | - | - | 96.9% | - | - | 94.34% |
The final evaluation was to compare the marker order consistency between the RH and genome maps. The RH map position was compared to the sequence position within each chromosome to identify marker order consistency. Because placement of markers using RHMAPPER is relative only to the framework maps rather than to other placed markers, the fine-level order accuracy is lower than that of the genome sequence. Therefore, each STS on the RH map was first assigned to a bin that included the framework markers between which the STS was placed; given these criteria, the consistency in marker order was 53.8% between the two maps. However, when the bin size was extended to include a bin on either side of the flanking markers, the consistency in mapping between the RH and the genome assembly increased to 94.6 %, reflecting the potential limits of the resolution of RH mapping using the T55 panel (3000Rad). The approximately 5% inconsistency in marker order given a ± one-bin interval is likely due to combined inaccuracies in the RH map and the genome assembly. Having this data facilitates further curation and rectification of either map. The results of the comparisons are summarized in the Table 3 for each chromosome.
Curation of RH Vectors
Some of the RH vectors in our data set came from public sources, and several markers had vectors generated by different groups. Vectors in RHdb are designated with a submitter, an identifying name, primer sequences used to generate the vector, and external sources of information on the template from which primers were chosen; we tracked the source for all mapped vectors. (Rodriguez-Tome and Lijnzaad 2001). However, the template sequence from which primers originated was not included. Furthermore, many vectors in RHdb had no expected size. Therefore, the final step in our validation process was extensive curation, using the data and expertise of the Rat Genome Database. The 24,962 vectors were given a marker type designation, SSLP, EST, or gene, and each type was analyzed separately.
To ensure the highest confidence of vector accuracy, we analyzed vectors to determine whether the primers matched the template for which the assay was named. The 19,431 ESTs were analyzed by comparing the accession IDs against NCBI's dbEST rat records. This analysis showed that 42 ESTs had been retired by NCBI, and these records were removed from the RH map. An additional 190 ESTs were removed from the map, because the primer sequences did not match the appropriate template sequence, resulting in 19,241 validated EST markers. All 5035 SSLPs were checked by comparing the primer pair sequences to RGD's primer pair sequences. The method used was an exact sequence match for both primer pairs and resulted in 5031 validated SSLP markers. The 497 genes were validated using electronic polymerase chain reaction (ePCR) against their template sequences; 75 unverified genes were removed from the map.
After removal of the 268 markers resulting from vector curation and 257 markers resulting from error checking, the final RH map consists of 24,437 markers (Table 1). These maps can be visualized using the VCMapView tool (http://rgd.mcw.edu/VCMAP/mapview.shtml). The map files are available at http://rgd.mcw.edu/pub/rhmap/3.4, and include all detailed curated information, including final vector name, original vector name, primer sequences, chromosome, position, LOD threshold of the placement map from which it was placed, framework or placement flags, expected size, vector, template sequence resulting from our curation, and associated GenBank ID. It is important to note that all curation is linked back to the original vector source to track any changes in nomenclature in the final maps.
DISCUSSION
The rat RH map has become a tool of great importance over the last years, particularly in studies involving complex genetic disease (Dobbins et al. 2002; Moujahidine et al. 2002; Tseng et al. 2002; Wallace et al. 2002). The RHMAP Server at the RGD has been accessed over 79,000 times since June 2000; this public server maps RH vectors using our previous version of the RH map (V2). The RH maps allow the positioning of ESTs, genes, SSLPs, and STSs without the need for polymorphisms and can be used in many ways to annotate the genome with systems biology. Similarly, QTLs from different rat crosses can be integrated using all SSLPs mapped across independent genetic maps. Previously unmapped genes and ESTs can be placed on the RH maps to identify potential candidate genes within a QTL. The RH maps can also facilitate comparisons across species for identification of conserved syntenic regions (Watanabe et al. 1999; Kwitek et al. 2001; Scheetz et al. 2001; Chowdhary et al. 2003; Larkin et al. 2003). Although the rat genome sequence has been released (Rat Genome Sequencing Project Consortium 2004), the sequence is not yet considered `finished'; therefore the RH maps retain their utility in regions of the genome with lower sequence coverage. Furthermore, markers from the RH maps presented here assisted in the validation of the genome assembly by providing confidence in chromosome designation of assembled bactigs.
In this new version (3.4) of the MCW (Medical College of Wisconsin) map, we have generated the highest-density framework placement maps to date, double that of any other described whole-genome rat RH map. We developed a novel integration algorithm that allows for placement at multiple LOD thresholds (Table 1), which increased the density of the maps by 12%. The lack of significant length expansion from our original maps reflects the extensive error checking and curation of the vectors placed. Given the size of the genome assembly (R3.1), the high-density maps result in a resolution of approximately 9 cR(3000) per Mb, and an STS density of ∼1 marker every 100 kb.
Evaluating the number of ESTs mapped to each chromosome determined a lower than expected number (208) on the X chromosome (Table 1), given the chromosome length and the density of over 500 known genes in the mouse. This trend was also noted in a previous rat RH map (Scheetz et al. 2001). The reason for this paucity is not yet clear, but one explanation might be a paucity in genes from the X chromosome in the cDNA libraries from which the ESTs were generated. The genome assembly should help to identify additional genes that should be placed on the X chromosome. These could then be mapped in the RH panel to determine whether there are indeed fewer ESTs from the X chromosome or whether a region of the X chromosome is missing from the RH map.
The resolution of radiation hybrid mapping is determined by the dosage of X irradiation to the cell line from which the panel is generated (Cox et al. 1990). The T55 panel is a 3000-rad panel (McCarthy et al. 2000). The mapping resolution of this panel is ∼17 cR, as indicated in Table 1. By comparing consistency between the STS order on the RH map and genome assembly within a single framework bin versus framework ± one bin, the consistency of mapping increased from 54% to 95%. It is important to note that STSs are placed on the map only with respect to the framework markers, not other placed markers; therefore, the >40% increase in consistency is not completely unexpected with the ± one-bin window. Certainly, there are inaccuracies in all maps, and certainly the 5% inconsistencies reported here are due to errors in both the genome assembly and the RH maps. The importance is identifying these regions of consistency so that an investigator interested in a specific genome region can experimentally confirm the true result. For instance, if fine-mapping below the RH resolution is needed, one might use other means such as in silico, from the genome assembly, and validation by physical mapping using BAC clones spanning this region.
In building this new version of the MCW RH map we sought to provide accurate, detailed and reliable information in what is likely to be the last whole-genome rat RH map built. All other RH maps in the future are likely to be local and will be used to check the sequence assembly, making it essential that the information content of this map be highly detailed and accurate, a feat we believe we have accomplished in this new version of the MCW RH map for the rat genome. The maps presented here offer a powerful complement to the available genetic maps and the genome assembly. They have helped to integrate QTL within rat as well as across species (Jacob and Kwitek 2002), and will continue to be useful for confirmation and validation of the draft genome assembly.
METHODS
RH Vector Data Set
We used vectors produced in our laboratory as part of the NHLBI Rat EST Project (as described by Steen et al. 1999) as well as those submitted to the RHdb (http://corba.ebi.ac.uk/RHdb), primarily by the University of Iowa (also investigators in the Rat EST project) and a collaboration between the Wellcome Trust Centre for Human Genetics and Otsuka Pharmaceutical Company (Watanabe et al. 1999), to create our initial mapping data set consisting of 33,568 vectors including SSLPs, genes, and ESTs. Vectors generated in our laboratory were according to previously described methodology (Steen et al. 1999). Each assay was run in duplicate to minimize false negative results (due to PCR failure), and a consensus vector was generated. This data set was then processed as follows: Distinct vectors referring to the same SSLP or EST, regardless of the vector source, were screened. The vectors with the best quality, judged by the least amount of discordant scores (2's), were kept independent of source. Markers that had more than 10 discordant scores (2's), less than 10 or greater than 39 positive scores (1's) were discarded from the vector data set (with the exception of framework vectors for chromosome 10 (see Framework Map Construction below). These criteria were chosen based on the range of retention, averaged across all chromosomes, for vectors that successfully map to a unique position. The average retention was 26.5%, the minimum was 15%, and the maximum was 39%; we extended the criteria for acceptable vectors to 11% minimum and 42% maximum retention. Following processing, the final data set consisted of 29,353 vectors (framework and placement vectors) from which the RH maps were generated. This data set is available by ftp at http://rgd.mcw.edu/pub/rhmap/3.4.
Framework Map Construction
For generating the framework maps, we selected only vectors with discordant scores ≤4, and with a number of positive scores between (and including) 10 and 39. The only exception was Chr10, as the TK (thimidine kinase) gene at the q telomeric end, used for selection in the hybrid panel construction, resulted in a higher retention rate; therefore, for chromosome 10, all markers with more than 39 positive scores were included in the pool of markers from which the framework map was generated.
The RHMAPPER computer package was used for all framework and placement map generation as described (Steen et al. 1999). We first identified small segments containing vectors that linked with a LOD score of at least 4, by local order permutation. The resulting segments were then merged by identifying vectors that spanned gaps between them and maintained a local order confidence of LOD 4. At this point we also compared our results to the reference genetic maps found at RGD (SHRSP × BN and FHH × ACI). Once an initial RH scaffold was completed, consisting of vectors spanning the chromosome with local permutations (4-marker sliding window) of LOD 4, we used the `grow_ framework' command in the RHMAPPER package to increase the density of the framework maps, with a subsequent manual checking of the markers the program added. Markers that created large gaps, compared to the map before their addition, were removed from the map. We first ran the `grow_framework' command with an LOD score threshold of 4, and subsequently with an LOD score threshold of 3. The only map that wasn't added to by the `grow_framework' command was chromosome 20, as all of the additions created large gaps and undue expansions.
Modifications to RHMAPPER
A feature within RHMAPPER is that markers mapping upstream of the first framework marker are assigned a position equivalent to that of the first framework markers, To correctly place these markers, the true cR distance of all of the markers placed upstream of the topmost framework marker (considered to be at 0 cR) were calculated, and their absolute position on the chromosome was annotated as a negative position from the first framework marker. For example, a marker placed 3.2 cR upstream of the first framework marker is annotated as -3.2 cR. In the final integrated placement map, the first placement marker on the map was converted to 0 cR and all other positions adjusted accordingly; in the example above, the marker placed at -3.2 cR would become 0 cR and the first framework marker would become 3.2 cR.
Placement Map Construction and Integration
We set a `placement too far' threshold of 30 cR for placing additional markers; reducing this parameter from 50 cR (default) minimized map expansion and spurious markers placed at the far ends of the chromosomes. Ten placement maps were independently generated (from the same framework map), with two-point LOD score thresholds ranging from 6 to 15, using the `place_markers' scripts from the RHMAPPER computer package. The RHMAPPER map outputs for LODs 8–15 were used as the base for the final integrated maps (Fig. 1). Although the raw placement maps have slightly varying lengths depending on the two-point LOD threshold chosen, we could normalize the bin distance (distance between framework markers) for all maps to that of a single map (we chose the LOD 15), and proportionately adjust the position of each placement marker within the framework bin. For each placed marker, we identified the normalized position from the map having the highest two-point LOD threshold. For example, a marker might be placed on maps generated at LODs 10, 11, and 12; we would use the highest LOD for the integrated placement, in this example from the LOD 12 map. By this method, every marker has a single integrated position relative to the same framework bin. As a final step, we recalculated the absolute position from the top of each integrated chromosome, with the first placed marker identified as 0 cR.
Map Comparisons
Map discrepancies were determined by comparing the chromosome designations and positions as reported in the SHRSP × BN genetic map, a reference genetic map publicly available at RGD (http://rgd.mcw.edu/pub/publications/1999/steen_genome_research/) and the genome assembly (Release 3.1; http://genome.cse.ucsc.edu/cgi-bin/hgGateway), with the Version 3.4 RH maps.
Curation of RH Vectors
ESTs were first validated by ePCR (Schuler 1997). Using this method, 847 primer pairs did not hit the sequence for the associated GenBank ID. However, the ePCR algorithm requires an expected size for a PCR product, which many of these records did not have. Therefore, these markers were further analyzed by comparison to the rat Version 3.1 genome assembly: 266 ESTs hit the expected chromosome and either hit a sequence that overlapped with the coordinates of the given associated GenBank ID or hit a cluster of ESTs. The longest EST was then chosen for the new representative EST. Because not all ESTs were annotated on the genome, the 581 remaining ESTs were analyzed using RGD's sequence analysis pipeline, which is capable of validating that a given primer pair hits its template sequence without using an expected size for a PCR product. The RGD pipeline does seven independent BLAST comparisons to evaluate primer pairs and template sequences, and collates the results.
The 5035 SSLPs were analyzed using two different methods. First the RH map primers were validated using ePCR against the template sequences for the 3984 SSLPs with available templates. The templates used were a combined set of sequences including RGD SSLP sequences and the sequences for the associated GenBank IDs. Second, all SSLPs were checked by comparing the primer pair sequences to RGD's primer pair sequences. This analysis indicated that 19 primer pairs varied from RGD's primer sequences. However, these were kept in the map based on valid experimental results. The method used was an exact sequence match for both primer pairs.
The source of the gene template sequences was a combination of RGD/NCBI gene sequences and the sequences for the associated GenBank IDs. The conflicts were analyzed manually using BLAST and BLAT algorithms (Altschul et al. 1990; Kent 2002).
Acknowledgments
We thank the RGD curation team for their contributions to the curation and validation of the maps, the Oxford and Otsuka groups for their contribution of public vectors to the RHdb, and past and present members of the H.J.J. and V.C.S. laboratories for their technical assistance. This work has been supported by RO1s HL59826 (H.J.J.), HL64541 (P.J.T.), and HL59789 (V.C.S.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1968704.
References
- Altschul, S., Gish, W., Miller, W., Myers, E., and Lipman, D. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403-410. [DOI] [PubMed] [Google Scholar]
- Bihoreau, M., Sebag-Montefiore, L., Godfrey, R., Wallis, R., Brown, J., Danoy, P., Collins, S., Rouhard, M., Kaisaki, P., Lathrop, M., et al. 2001. A high-resolution consensus linkage map of the rat, integrating radiation hybrid and genetic maps. Genomics 75: 57-69. [DOI] [PubMed] [Google Scholar]
- Chowdhary, B., Raudsepp, T., Kata, S., Goh, G., Millon, L., Allan, V., Piumi, F., Guerin, G., Swinburne, J., Binns, M., et al. 2003. The first-generation whole-genome radiation hybrid map in the horse identifies conserved segments in human and mouse genomes. Genome Res. 13: 742-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, D.R., Burmeister, M., Priece, E.R., Kim, S., and Myers, R.M. 1990. Radiation hybrid mapping: A somatic cell genetic method for constructing high-resolution maps of mammalian chromosomes. Science 250: 245-250. [DOI] [PubMed] [Google Scholar]
- Dobbins, D., Joe, B., Hashiramoto, A., Salstrom, J., Dracheva, S., Ge, L., Wilder, R., and Remmers, E. 2002. Localization of the mutation responsible for osteopetrosis in the op rat to a 1.5-cM genetic interval on rat chromosome 10: Identification of positional candidate genes by radiation hybrid mapping. J. Bone Miner. Res. 17: 1761-1767. [DOI] [PubMed] [Google Scholar]
- Gosele, C., Hong, L., Kreitler, T., Rossmann, M., Hieke, B., Gross, U., Kramer, M., Himmelbauer, H., Bihoreau, M.T., Kwitek-Black, A.E., et al. 2000. High-throughput scanning of the rat genome using interspersed repetitive sequence-PCR markers. Genomics 69: 287-294. [DOI] [PubMed] [Google Scholar]
- Jacob, H.J. and Kwitek, A.E. 2002. Rat genetics: Attaching physiology and pharmacology to the genome. Nat. Rev. Genet. 3: 33-42. [DOI] [PubMed] [Google Scholar]
- Kent, W. 2002. BLAT—The BLAST-like alignment tool. Genome Res. 12: 656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwitek, A.E., Tonellato, P.J., Chen, D., Gullings-Handley, J., Cheng, Y.S., Twigger, S., Scheetz, T.E., Casavant, T.L., Stoll, M., Nobrega, M.A., et al. 2001. Automated construction of high-density comparative maps between rat, human, and mouse. Genome Res. 11: 1935-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin, D., Wind, A.E.-v.d., Rebeiz, M., Schweitzer, P., Bachman, S., Green, C., Wright, C., Campos, E., Benson, L., Edwards, J., et al. 2003. A cattle–human comparative map built with cattle BAC-ends and human genome sequence. Genome Res. 13: 1966-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy, L., Bihoreau, M.T., Kugawa, S.L., Browne, J., Watanabe, T.K., Hishigaki, H., Tsuji, A., Kiel, S., Webber, C., Davis, M.E., et al. 2000. A whole-genome radiation hybrid panel and framework map of the rat genome. Mamm. Genome 11: 791-795. [DOI] [PubMed] [Google Scholar]
- Moujahidine, M., Dutil, J., Hamet, P., and Deng, A. 2002. Congenic mapping of a blood pressure QTL on chromosome 16 of Dahl rats. Mamm. Genome 13: 153-156. [DOI] [PubMed] [Google Scholar]
- Rat Genome Sequencing Project Consortium. 2004. Genome sequence of the Brown Norway Rat yields insights into mammalian evolution. Nature (in press). [DOI] [PubMed]
- Rodriguez-Tome, P. and Lijnzaad, P. 2001. RHdb: The Radiation Hybrid database. Nucleic Acids Res. 29: 165-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheetz, T.E., Raymond, M.R., Nishimura, D.Y., McClain, A., Roberts, C., Birkett, C., Gardiner, J., Zhang, J., Butters, N., Sun, C., et al. 2001. Generation of a high-density rat EST map. Genome Res. 11: 497-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler, G. 1997. Sequence mapping by electronic PCR. Genome Res. 7: 541-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen, R.G., Kwitek-Black, A.E., Glenn, C., Gullings-Handley, J., Van Etten, W., Atkinson, O.S., Appel, D., Twigger, S., Muir, M., Mull, T., et al. 1999. A high-density integrated genetic linkage and radiation hybrid map of the laboratory rat. Genome Res. 9: AP1-8 (insert). [PubMed] [Google Scholar]
- Stein, L., Kruglyak, L., Slonim, D., and Lander, E. 1995. RHMAPPER, installation and user's guide.
- Tseng, J., Erbe, C.B., Kwitek, A.E., Jacob, H.J., Popper, P., and Wackym, P.A. 2002. Radiation hybrid mapping of five muscarinic acetylcholine receptor subtype genes in Rattus norvegicus. Hear. Res. 174: 86-92. [DOI] [PubMed] [Google Scholar]
- Twigger, S.N., Nie, J., Ruotti, V., Yu, J., Chen, D., Li, D., Mathis, J., Narayanasamy, V., Gopinath, G.R., Pasko, D., et al. 2004. Integrative genomics: In silico coupling of rat physiology and complex traits with mouse and human data. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Wallace, C.A., Ali, S., Glazier, A.M., Norsworthy, P.J., Carlos, D.C., Scott, J., Freeman, T.C., Stanton, L.W., Kwitek, A.E., and Aitman, T.J. 2002. Radiation hybrid mapping of 70 rat genes from a data set of differentially expressed genes. Mamm. Genome 13: 194-197. [DOI] [PubMed] [Google Scholar]
- Watanabe, T.K., Bihoreau, M.T., McCarthy, L.C., Kiguwa, S.L., Hishigaki, H., Tsuju, A., Browne, J., Yamasaki, Y., Mizoguchi-Miyakita, A., Oga, K., et al. 1999. A radiation hybrid map of the rat genome containing 5,255 markers. Nat. Genet. 22: 27-36. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://corba.ebi.ac.uk/RHdb/; a repository of radiation hybrid vector data.
- http://rgd.mcw.edu/VCMAP/mapview.shtml; a tool at the Rat Genome Database for dynamic integration of various genome maps (genetic, RH, genome, QTL) and cross-species comparative maps between rat, human, and mouse.
- http://rgd.mcw.edu/pub/rhmap/3.4; ftp site to obtain vector and map information for the rat v3.4 RH maps.
- http://rgd.mcw.edu/pub/publications/1999/steen_genome_research/; ftp site to obtain reference rat genetic maps and our previous version of the rat RH maps as published in Steen et al. 1999.
- http://genome.cse.ucsc.edu/cgi-bin/hgGateway; Genome browser at the University of California at Santa Cruz providing sequence and annotation of the rat genomic sequence assemblies.