Abstract
As part of the effort to sequence the genome of Rattus norvegicus, we constructed a physical map comprised of fingerprinted bacterial artificial chromosome (BAC) clones from the CHORI-230 BAC library. These BAC clones provide ∼13-fold redundant coverage of the genome and have been assembled into 376 fingerprint contigs. A yeast artificial chromosome (YAC) map was also constructed and aligned with the BAC map via fingerprinted BAC and P1 artificial chromosome clones (PACs) sharing interspersed repetitive sequence markers with the YAC-based physical map. We have annotated 95% of the fingerprint map clones in contigs with coordinates on the version 3.1 rat genome sequence assembly, using BAC-end sequences and in silico mapping methods. These coordinates have allowed anchoring 358 of the 376 fingerprint map contigs onto the sequence assembly. Of these, 324 contigs are anchored to rat genome sequences localized to chromosomes, and 34 contigs are anchored to unlocalized portions of the rat sequence assembly. The remaining 18 contigs, containing 54 clones, still require placement. The fingerprint map is a high-resolution integrative data resource that provides genome-ordered associations among BAC, YAC, and PAC clones and the assembled sequence of the rat genome.
The rat has historically been an important model organism for physiological, pharmacological, and biochemical studies. Building on the wealth of experimental data and methodology, the rat has become a preferred model for systems biology and the study of many complex diseases (James and Lindpaintner 1997; Aitman et al. 1999; Stoll et al. 2001; Jacob and Kwitek 2002; Yokoi et al. 2002; Olofsson et al. 2003). Here, we describe the creation of major new resources for genomic studies in the rat: (1) the construction of a bacterial artificial chromosome (BAC) fingerprint map spanning the rat genome; (2) the construction of a yeast artificial chromosome (YAC) map covering the rat genome; and (3) integration of both maps with each other and with the assembled rat genomic sequence. The integrated resources provide deep clone coverage and long-range clone continuity for most of the rat genome. This has important implications for studying regions that are not represented by finished sequence and functional genomic approaches that rely on the availability of sequence-anchored clones.
We undertook the construction of the Rattus norvegicus BAC map as part of the international effort to sequence the entire genome (Rat Genome Sequencing Project Consortium 2004). The BAC map was constructed to provide a resource from which clones could be selected for sequencing in a manner similar to that for other large-scale sequencing projects, such as those for human (Lander et al. 2001; McPherson et al. 2001), mouse (Waterston et al. 2002), Drosophila melanogaster (Adams et al. 2000; Hoskins et al. 2000), rice (Barry 2001), and Arabidopsis thaliana (The Arabidopsis Genome Initiative 2000). To generate the BAC map, we used BAC fingerprinting-based methods (Marra et al. 1997; Schein et al. 2004) by which other maps had been constructed, such as those for human (McPherson et al. 2001), mouse (Gregory et al. 2002), and Arabidopsis (Marra et al. 1999; Mozo et al. 1999). Fingerprint maps provide information about the relationships between individual BAC clones in the physical map, and this information can be exploited to select clones for sequencing in a manner that limits sequence redundancy while ensuring coverage of the genome.
The rat sequence assembly (Rnor3.1; http://www.hgsc.bcm.tmc.edu/projects/rat/assembly.html) is 2.75 Gb in size and was generated using a hybrid whole-genome shotgun (WGS) and BAC-based approach (Rat Genome Sequencing Project Consortium 2004). The genome is sequenced to approximately seven-fold genome coverage, with 40% of read coverage generated from ∼21,000 BAC clones. Fingerprint map-based selection of rat BAC clones for sequencing was initiated essentially concurrently with the fingerprinting efforts. Several hundred BAC clones were selected from the map on a weekly basis as fingerprinting progressed and the map evolved. This differs from the hybrid WGS/BAC-based approach used for the mouse (Waterston et al. 2002), in which fingerprinting was substantially complete prior to selection of BAC clones for sequencing. The rat sequence is slightly larger than that of mouse, which is currently estimated to be 2.6 Gb (http://www.ncbi.nih.gov/genome/guide/mouse).
For construction of the YAC-based physical map, we made use of interspersed nuclear elements that are found in the genomes of a wide variety of mammals (Deininger 1989). The most prevalent interspersed repetitive sequence element in the rat genome is the so-called identifier (ID) element (Kim and Deininger 1996). ID elements are members of a family of SINEs found in the rodent genome (Deininger 1989). ID elements consist of a core domain with an average length of 75 bp containing an internal RNA polymerase III promotor, a 10-40-bp poly(A) region, and 5′- and 3′-flanking regions (Deininger 1989; Kim et al. 1994; Kass et al. 1996). The core region of ID elements is considered to be ancestrally derived from alanine tRNA (Daniels and Deininger 1985), and the copy numbers of ID elements are markedly different between species (Sapienza and St Jacques 1986; Anzai et al. 1987; Kass et al. 1996). Among rodent species, the rat has the highest copy number of ID elements, which is estimated to be five times the number found in the mouse genome (Deininger 1989; Kass et al. 1996; Ono et al. 2001), suggesting that the rat ID elements were rapidly amplified after the rat diverged from a common ancestral rodent. We used interspersed repetitive sequence (IRS) PCR technology to generate markers for physical mapping of the rat genome. A single primer was used to amplify sequences that are flanked by ID repeat elements in the rat. We solely used IRS-PCR on low complexity probes, that is, individual BAC or PAC clones. PCR products generated this way can directly be used as markers, that is, probes that can be hybridized to Southern blots. Moreover, each mapped marker at the same time anchors a specific BAC or PAC to the genome from which the individual probe was derived. The generation of large numbers of IRS markers in this way is rapid and cheap, because there is no requirement to sequence markers or to design locus-specific primers.
We have integrated the two physical maps by including into the BAC fingerprint map BAC and PAC clones linked by IRS-PCR markers to the YAC map. Both the BAC map and the YAC map have been anchored to version 3.1 of the rat genome sequence assembly using end sequences for fingerprinted BAC clones. The anchored BAC clones provide an ordered, high-resolution, redundant clone set spanning the sequence assembly, providing the research community with easy identification and access to BAC clones spanning regions of interest in the rat genome.
RESULTS
Generation of IRS-PCR Amplicons for the YAC-Based Map
IRS-PCR amplicons were generated using a single primer complementary to the 5′-sequence of rat ID-consensus sequence from individual RPCI-32 BACs and RPCI-31 PACs. In total, 30,144 BAC clones and 27,648 PAC clones were randomly selected for IRS-PCR amplification. This number of clones represents approximately onefold genome coverage for each library, respectively. We obtained 9378 positive IRS-PCR products for BAC clones and 7601 for the PAC clones. From these, we randomly chose 8397 IRS markers tagging individual BACs (4311) and PACs (4086), which were subsequently used for radiation hybrid mapping and for the identification of YAC clones with overlapping DNA content. We combined two mapping methods to gain information about the proximity of marker loci within the rat genome.
Radiation Hybrid Mapping
Individual IRS-PCR markers were screened against the rat T55 whole-genome radiation hybrid (RH) panel, consisting of 106 rat-on-hamster somatic hybrid cell lines (Watanabe et al. 1999). The observed average marker retention frequency of 28.9% is consistent with previously published results for the T55 panel (Steen et al. 1999; Watanabe et al. 1999; Scheetz et al. 2001). We produced comprehensive BAC and PAC placement maps by mapping the derived markers against the framework-map intervals (Steen et al. 1999; Watanabe et al. 1999) using multipoint maximum likelihood analysis. This resulted in a placement map of 5301 BAC- and PAC-derived markers. Moreover, we produced an independent radiation hybrid framework map using the traveling-salesman problem (TSP) approach (Applegate et al. 1998; Agarwala et al. 2000). The radiation hybrid framework maps produced with the two approaches, however, were highly consistent, indicating that data quality and not algorithmic approach is the critical factor in producing high-quality maps. Framework markers of the TSP map that were placed with odds higher then 1:1000 on the maximum likelihood map, show in 99.2% the same relative order on both maps. Adjacent framework markers were spaced at an average interval of ∼23 centiRay (cR) (2.4 Mb; Steen et al. 1999; Watanabe et al. 1999). In all, markers derived from 2739 BACs and 2562 PACs could be localized with high confidence onto the rat genome by radiation hybrid mapping. Thus, the location of 5301 PACs and BACs can be inferred from the radiation hybrid map.
Marker Content Mapping
For each localized BAC and PAC clone, we additionally identified overlapping YAC clusters by screening two YAC libraries (Cai et al. 1997; Haldi et al. 1997). Each of the 8397 BAC- and PAC-derived markers was individually hybridized against filters containing IRS-PCR-amplified three-dimensional YAC pools. The spotting of IRS-PCR amplified YAC pools and subsequent hybridization allowed an efficient filter-based screening approach of the ∼92,000 clones represented in the two YAC libraries. YAC-based marker content mapping with each of the markers would otherwise amount to >7 × 108 individual assays. About two-thirds of all hybridizations (5803) were successful. Unsuccessful hybridizations were largely caused by experimental reasons related to the hybridization process. We observed on average 8.8 positive YAC hybridization signals in an individual hybridization experiment. This number is significantly lower than expected considering that both libraries combined cover the rat genome ∼20-fold (Cai et al. 1997; Haldi et al. 1997). This phenomenon has previously been observed (Schalkwyk et al. 2001) and was noticed for both YAC libraries screened here and is largely caused by the introduced complexity applying a three-dimensional YAC library pooling strategy. In total, the physical map provides access to 51,323 YAC clones that are directly linked to 5803 BACs and PACs (3266 and 2583, respectively).
YAC Map Construction
The IRS markers were generated from individual BAC and PAC clones representing low-complexity templates. YAC libraries were screened by hybridization-based assays to identify clones containing a given locus. Nearby loci tend to be present in many of the same clones, allowing proximity to be inferred. Marker-content linkage can be detected over distances of ∼800 kb, given the average insert size of the YAC library used here. Hybrid cell lines, each containing many chromosomal fragments produced by radiation breakage, are screened to identify those hybrids that have retained a given locus. Nearby loci tend to show similar retention patterns, allowing proximity to be inferred. Radiation hybrid linkage can be detected for distances of ∼2-3 Mb, given the average fragment size of the RH panel used here. The two methods were used to produce independent maps and were subsequently combined to produce an integrated map. Because RH mapping can detect linkage over large regions, comprehensive RH maps spanning all chromosomes can be assembled with a few thousand loci. The order of the loci can be inferred from the extent of correlation in the retention patterns, although estimates on fine-structure order are not precise. These methods can thus provide “top-down” information about global position in the genome. In contrast, marker-content mapping provides “bottom-up” information. It reveals tight linkage among loci but is useful only over short distances and does not provide extensive long-range connectivity across chromosomes.
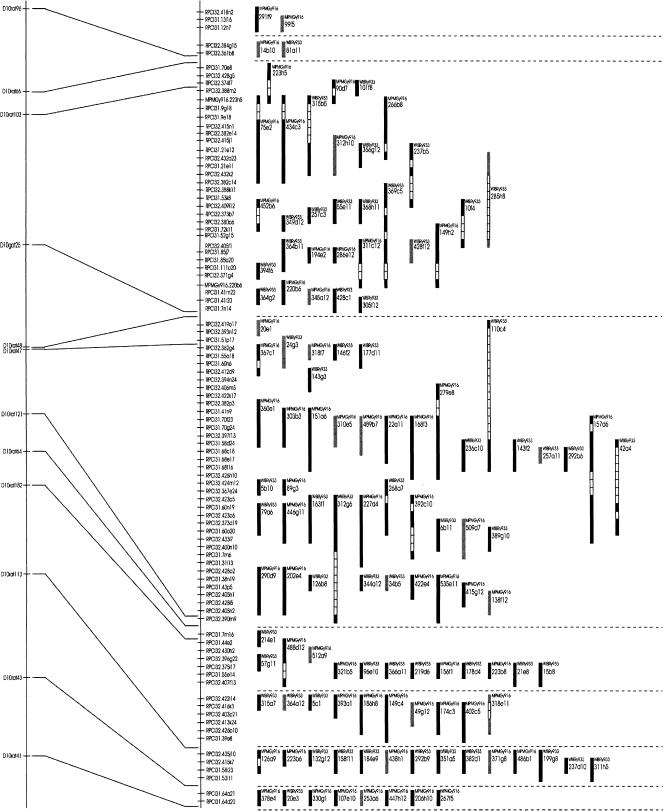
For the construction of a physical map and assembly of contigs, 51,323 YAC clones that gave a positive hybridization signal were considered. This number was successively pruned with considerable care toward chimeric clones, an inherent problem with any YAC library (Green et al. 1999), leaving 31,757 clones (see Methods). We constructed a map of each chromosome by integrating the YAC-linkage information with the known radiation hybrid map positions of the IRS markers using the co2 software package (Hudson et al. 1995). Doubly linked contigs were identified, and then single-linkage information was used to join doubly linked contigs known to lie nearby. The maps were closely inspected to identify apparent conflicts in order between radiation hybrid and YAC-based maps. The final map contains 5803 loci distributed across the 20 autosomes and the X-chromosome. The lack of coverage for the Y-chromosome is due to the fact that the T55 RH panel has been derived from a female donor. The final map was binned into 605 contigs. The markers and chromosomes are described (Table 1), and a representative map from a portion of Chromosome 10 is shown (Fig. 1). All data (including representations of the contigs on each chromosome, tables containing all data) are freely available (http://www.mdc-berlin.de/ratgenome or http://www.molgen.mpg.de/~ratgenome). With an average insert size of 150 kb for each BAC (Osoegawa et al. 2004) and PAC (Woon et al. 1998; http://bacpac.chori.org) clone, this data set spans 835 Mb of genomic sequence, corresponding to roughly 27% of the rat genome cloned in RPCI-32 BACs and RPCI-31 PACs. Assuming an average YAC size of ∼0.8 Mb (Cai et al. 1997; Haldi et al. 1997), this map corresponds to more than eightfold coverage of the rat genome, providing longrange connectivity and coverage for the vast majority of the rat genome.
Table 1.
Distribution of Loci and Contigs on Genome-Wide YAC Map
| Chromosome | Physical length (Mb) | Average spacing (kb) | Loci on RH map | Loci on marker-content map (total loci) | Assembled contigs |
|---|---|---|---|---|---|
| 1 | 268.1 | 402 | 609 | 666 | 65 |
| 2 | 258.2 | 733 | 326 | 352 | 37 |
| 3 | 171.0 | 481 | 304 | 355 | 39 |
| 4 | 187.4 | 568 | 324 | 330 | 32 |
| 5 | 173.1 | 439 | 355 | 394 | 50 |
| 6 | 147.6 | 553 | 273 | 267 | 33 |
| 7 | 143.1 | 409 | 304 | 350 | 38 |
| 8 | 129.1 | 378 | 326 | 341 | 45 |
| 9 | 113.6 | 437 | 227 | 260 | 22 |
| 10 | 110.7 | 260 | 383 | 425 | 31 |
| 11 | 87.8 | 535 | 163 | 164 | 21 |
| 12 | 46.6 | 165 | 259 | 282 | 24 |
| 13 | 111.3 | 435 | 219 | 256 | 23 |
| 14 | 112.2 | 503 | 183 | 223 | 21 |
| 15 | 109.8 | 572 | 171 | 192 | 23 |
| 16 | 90.2 | 458 | 178 | 196 | 20 |
| 17 | 97.3 | 470 | 183 | 207 | 21 |
| 18 | 87.3 | 487 | 165 | 179 | 24 |
| 19 | 59.2 | 438 | 127 | 135 | 15 |
| 20 | 55.3 | 339 | 154 | 163 | 21 |
| X | 160.8 | 2429 | 68 | 66 | 8 |
| Total | 2719.7 | 468 | 5301 | 5803 | 605 |
Figure 1.
View of the YAC-based physical map in a region of rat Chromosome 10.
Clone Fingerprinting
Clones for the BAC-based map were fingerprinted using an agarose gel-based methodology (Marra et al. 1997; Schein et al. 2004) and the restriction enzyme HindIII. The CHORI-230 BAC library was the primary source of clones for construction of the fingerprint map. Fingerprints were attempted on all CHORI-230 clones, and 185,438 (91%) were successful (Table 2). Of the clones that failed to produce fingerprints, 2264 were attributed to empty wells in the library plates, 2880 exhibited poor growth characteristics resulting in insufficient quantities of BAC DNA for fingerprinting, three had their fingerprints manually removed because of poor primary data quality, and 12,167 were rejected by our automated band calling software, BandLeader (Fuhrmann et al. 2003). The latter group included nonrecombinant clones. Automated quality checks (see Supplemental material available online at www.genome.org) on the fingerprint gels detected two duplicated gels (0.1%) and one gel (0.05%) with duplicate lanes. Fingerprinting of the clones on the affected gels was repeated to correct the errors. All successful fingerprints were imported into FPC (Soderlund et al. 1997, 2000; Ness et al. 2002) for assembly and further analysis. The average insert size for CHORI-230 Segment 1 clones was 213 kb and 165 kb for Segment 2 clones (Table 2), matching closely that determined by pulsed field gel electrophoresis (Osoegawa et al. 2004). The distribution of clone insert sizes derived from the fingerprint size data indicated that 99% of all CHORI-230 clones were <280 kb. We manually reviewed the fingerprints of 2141 outlier clones >280 kb. A total of 999 of these were identified as problematic, likely resulting either from well-to-well cross-contamination or from partial HindIII digestion, and were removed from the data set. In total, 184,439 CHORI-230 clones (91%) passed all of our laboratory and data quality checks and were incorporated into the fingerprint database.
Table 2.
Fingerprinting Details
| Library | Attempted fingerprints | Fingerprints in map | Enzyme used to construct library | Avg. insert sizea,b(kb) | Avg. number of HindIII fragmentsb | Estimated genome coveragec |
|---|---|---|---|---|---|---|
| CHORI-230 | ||||||
| Segment 1 | 92,160 | 84,993 | EcoRI | 213 | 57 | 6.7× |
| Segment 2 | 110,592 | 99,446 | MboI | 165 | 45 | 6.0× |
| RPCI-31 | 2541 | 2476 | MboI | 131 | 34 | 0.1× |
| RPCI-32 | 2820 | 2774 | EcoRI | 163 | 42 | 0.2× |
| Total | 208,113 | 189,689 | 186 | 50 | 13.1× |
As determined by fingerprint data
For clones in the fingerprint map
Computed by multiplying the number of clones from each library (or library segment) by the average insert size of the clones, and dividing by the total sequence assembly size (2.75 Gb)
Fingerprints were subsequently attempted on clones from the RPCI-32 BAC and RPCI-31 PAC libraries sharing IRS marker content with clones in the YAC map. Successful fingerprints were obtained for 2774 (98%) and 2476 (97%) of clones from these libraries, respectively (Table 2). These clones were fingerprinted to provide a basis for integration of the YAC map with the fingerprint map. The final number of fingerprinted clones deposited into the fingerprint database was 189,689.
Automated Fingerprint Assembly
Automated fingerprint assemblies were performed using FPC software (Soderlund et al. 1997, 2000; Ness et al. 2002). Default parameters were used with the exception of the cutoff value for the Sulston score (Sulston et al. 1988), which was selected to avoid false-positive clone overlaps and thus minimize the assembly of contigs containing clones from more than one genomic region. This resulted in conservative assemblies consisting of many contigs, each containing highly related clones. The initial automated fingerprint assembly was performed when less than onefold genome coverage had been collected. Thereafter, full assemblies were performed on a weekly basis as fingerprinting progressed. The resultant assemblies, containing contigs of highly related clones, were used to select BAC clones to be sequenced at the Baylor College of Medicine Human Genome Sequencing Center (BCM HGSC) as part of the rat genome sequencing effort (Rat Genome Sequencing Project Consortium 2004). The final automated build performed with the complete set of 184,439 CHORI-230 fingerprints resulted in the assembly of 11,274 contigs (Table 3).
Table 3.
Fingerprint Map Statistics for the Automated Assembly, Manually Edited Map, and Final Merged Map
| Map | Contigs | Clones in contigs | Singletons | Mean contig sizea | Median contig sizea | Largest contigb |
|---|---|---|---|---|---|---|
| Automated assemblyb | 11,274 | 171,297 | 13,142 | 15 | 10 | 425 |
| Manually edited | 634 | 176,171 | 13,518 | 278 (4.6 Mb) | 133 (2.7 Mb) | 3990 (57.2 Mb) |
| Final merged | 376 | 179,794 | 9895 | 453 (8.4 Mb) | 172 (4.4 Mb) | 4179 (60.9 Mb) |
As determined by the number of clones in the contig. The length of the contig is given in parentheses
This assembly contained only CHORI-230 clones
Editing the CHORI-230 Automated Fingerprint Assembly
The CHORI-230 map assembly was subjected to manual review and editing to identify and correct errors within the automated assembly. Editing was performed using tools within FPC, assisted by externally scripted tools. Clone order within contigs was refined, chimeric contigs were identified and misassemblies were corrected. Fingerprint comparisons using only clones at the ends of contigs were performed using higher (less-stringent) cutoff scores to identify singleton clones that extended contigs and to identify potential contig merges. Potential contig merges were examined to evaluate consistency of the fingerprint data at the merge point. Merges were made where supported by the fingerprint data.
To assist with the contig merging process we leveraged the sequence similarity between the mouse and rat genomes, a similar approach to that used by Gregory et al. (2002), in which the human sequence assembly was used to inform contig merges in the mouse fingerprint map. We used BLAST (W. Gish; http://blast.wustl.edu) to compare BAC-end sequences derived from CHORI-230 clones (see Methods) to the MGSC Version 3 mouse genome sequence assembly (http://www.ncbi.nih.gov/genome/guide/mouse/). The resultant coordinates derived for rat BAC ends on the mouse sequence assembly were used to align the fingerprint map contigs to the mouse genome. Manual editing had reduced the number of contigs in the map to 7943, and 7844 of these could be assigned to mouse chromosomes. The contigs were then ordered by their midpoints on the assembly. Contigs overlapping by sequence coordinates were merged into a single contig. Based on these sequence coordinate criteria, a total of 7256 contigs were merged to form 846 contigs. A small, artificial gap was introduced between adjacent clone groups, now called “subcontigs,” comprising previously independent contigs. The subcontigs were ordered within each of the 846 contigs according to their mouse chromosome assignments. This process reduced the overall number of contigs in the map to 1533. We expected that differences in genomic organization between the mouse and rat genomes would result in the joining of some contigs that did not represent adjacent regions in the rat genome, but anticipated that these would be identified and resolved once the rat genome sequence assembly was available for comparison.
Manual review and editing continued subsequent to the mouse assembly-based merges. Within the mouse assembly-merged contigs, adjacent subcontigs were examined to determine if fingerprint overlaps could be detected. Where supported by the fingerprint data, gaps between adjacent subcontigs were removed and the clone groups joined. Additional merges between contig ends were also made based on fingerprint comparisons. This process further reduced the overall number of contigs in the map to 634 (Table 3).
Automated Insertion of RPCI-31 and RPCI-32 Clones Into Edited Fingerprint Map Contigs
To maximize the number of anchors between the YAC map and the fingerprint map, we incorporated into edited fingerprint contigs as many of the YAC-associated RPCI-31 and RPCI-32 fingerprinted clones as possible. The RPCI-31 PAC clone fingerprints were added to the fingerprint database while the editing process was in progress, and a total of 1597 of these remained as singletons in the edited fingerprint map. The RPCI-32 clone fingerprints were added as singletons to the fingerprint database subsequent to completion of manual editing. We used a computational approach to place RPCI-31 and RPCI-32 singletons into edited map contigs, resulting in the placement of 1009 RPCI-31 and 2614 RPCI-32 fingerprints into the edited map contigs. Fingerprints of clones that remained as singletons contained too few fragments to meet the required criteria for accurate placement into contigs. In total, 4502 (86%) of the RPCI-31 and RPCI-32 fingerprints were localized to contigs in the edited map.
Anchoring Fingerprint Map Contigs to the Rat Sequence Assembly
The availability of both a fingerprint map and sequence assembly for the rat genome provides the opportunity to derive a direct link from BAC clones to specific sequence regions, and vice versa. This linkage is useful in several applications, including identification of clones for use in functional studies of genes identified within the sequence, determination of the sequence content of BAC clones of interest identified through other means, such as BAC filter hybridizations, and access to sequencing substrates representing regions of interest in the genome where current sequence coverage or quality is insufficient for analysis. Examination of the linkage between the map and the sequence is likely to identify BAC clones spanning gaps in the sequence assembly. Additionally, map contig overlaps can be identified by sequence where the extent of overlap is insufficient to detect with confidence using fingerprint similarity alone. Furthermore, any discrepancies in the fingerprint map and the sequence assembly locations would identify potential misassemblies in either the map or the sequence. Analysis and resolution of these discrepancies would serve to improve the quality of both the sequence assembly and the fingerprint map. Given that we anticipated some errors in map assembly caused by use of the mouse sequence to identify map contig merges, this latter application was of specific interest to us.
We therefore undertook the correlation of the fingerprint map with the rat genome sequence assembly once it became available, with the aim of maximizing the resolution of the correlation by determining sequence coordinates for as many map clones as possible. We used a combination of BAC-end sequence coordinates and in silico mapping methods to align BAC clones on the genome sequence assembly and used these clone alignments to localize map contigs onto the sequence.
In Silico Mapping Coordinates
The in silico mapping approach used both fingerprint map data and BAC-end sequence alignment data to position map clones onto the sequence assembly. The first step in the in silico mapping process was identification of a sequence region in which a clone was likely to be located (a sequence “neighborhood”) based on BAC-end sequence coordinates (see Methods) of flanking clones in the same map region (see Supplemental material for details on the derivation of sequence neighborhoods). The neighborhoods provide a low-resolution estimation of the position of fingerprint map clones on the genomic sequence.
In the second step of the process, more precise localization of the clones within their sequence neighborhoods was obtained by aligning clone fingerprints with sequence-derived (in silico) restriction maps formed from the neighborhoods (see Supplemental material). We filtered these alignments to remove poor-quality in silico coordinates, using various criteria including the fraction of matched and unmatched fragments between the in silico anchor and the fingerprint. High-quality in silico alignments were identified for 157,527 clones (Supplemental Table 1).
Comparison of paired end sequence coordinates to those generated by the in silico method showed that in 3% of the cases these coordinates did not overlap (Table 4). Mismatches in chromosome assignment accounted for two-thirds of these discrepancies. Comparison of coordinates derived from single end-sequence coordinates to in silico coordinates showed a discrepancy of 60%. The majority of these errors were due to nonoverlapping coordinates on the same chromosome, and only 5% were due to chromosome assignment mismatch. The reduced overlap concordance between the single end sequence coordinates and the in silico coordinates is primarily due to the comparatively much smaller size of the single end coordinates. This is illustrated by the fact that 89% of the nonoverlapping single end coordinates were within 100 kb (less than the average length of a BAC) of the in silico coordinates.
Table 4.
Comparison Between BAC-End Sequence Coordinates and In Silico or Neighborhood
|
Paired end sequence 82,813
|
Single end sequence 54,293
|
None 52,583
|
|||
|---|---|---|---|---|---|
| Coordinate type | Total | Correlationa | Total | Correlationa,b | Total |
| In silico anchor | |||||
| 157,473 | 73,305 | 70,944/900/1461 (97%/1%/2%) | 45,102 | 17,850/25,133/2119 (40%/55%/5%) | 39,066 |
| Neighborhood only | |||||
| 21,976 | 6257 | 5831/50/376 (93%/1%/6%) | 6876 | 5862/379/635 (85%/6%/9%) | 8843 |
| None | |||||
| 10,240c | 3251 | 2315 | 4674 | ||
Only clones in the map are listed here. Paired end sequence coordinates located on different chromosomes or >500 kb apart on the same chromosome have been removed.
Categories are the number of clones with overlapping coordinates/number clones with nonoverlapping coordinates/number of clones with coordinates on different chromosomes. The percentages for each category, calculated as end sequence versus in silico mapping or end sequence versus neighborhood, are given in parentheses
An in silico anchor was considered to be nonoverlapping with a single end sequence coordinate if it fell outside of the bounds of the in silico anchor
9895 of these are singletons
The in silico mapping process is prone to greater positional error in unfinished portions of the assembly than in regions of finished sequence. This is because fingerprint fragments may fail to match in silico digest fragments that contain sequence errors or undetermined base pairs and therefore do not faithfully represent HindIII restriction site sequences or actual restriction fragment sizes. Thus, we expect that in silico coordinates derived from regions of the assembly containing gaps would have a negative impact on the concordance between BAC-end sequence coordinates and in silico coordinates. We also expect that end sequence alignment errors and laboratory tracking errors contribute to the identified discrepancies between end sequence and in silico coordinates; however, the accuracy of the in silico mapping methodology itself needs to be considered.
Assessing the Accuracy of In Silico Mapping Coordinates
To investigate the accuracy of our in silico mapping algorithm, we examined the mapping of a subset of BAC clones that had both in silico and paired end sequence-based coordinates. This control set of 19,223 clones met our criteria for sequence quality and clone size. We found that the positional uncertainty of the in silico coordinates was on the order of two to three HindIII fragments at the end of each clone. The median difference in the left position was -4 kb and in the right position 9 kb. The negative left-end difference and positive right-end difference reflect the fact that the in silico mapping algorithm was designed conservatively to avoid overestimating the left or right end of the clone, and therefore was generally internal to the BAC-end sequence coordinates. The in silico mapping accuracy is therefore good when anchoring to high-quality sequence, but is expected to have lower accuracy in regions of poor sequence quality.
Determination of Fingerprint Map Contig Coordinates
The BAC-end and in silico coordinates were used together to derive locations for fingerprint map clones on the genome sequence to localize the map contigs to the assembly. We determined sequence coordinates for 93% of all map clones. Of all clone coordinates, 55% were based on end sequence coordinates, and the remaining 45% were based on in silico anchors (see Methods). The sequence coordinates were used to identify groups of contiguously overlapping clones aligned to the sequence. These regions of contiguous clone alignments linked the corresponding map contigs to locations on the sequence assembly. Using this approach, 616 of the 634 edited fingerprint map contigs could be aligned to regions on the sequence assembly (see Methods). The remaining unanchored 18 contigs contained a total of 54 clones, with most having two to five clones.
Inconsistencies Between the Fingerprint Map and Sequence Assembly
To identify and investigate differences between clone order in the fingerprint map and on the assembled sequence, we aligned these two data sets using the contig anchor positions. Contigs with alignments to a single contiguous region of the sequence assembly were considered to have consistent map and sequence localizations. We included in this category contigs with a single alignment to assembled chromosomes (1-20, X) plus additional alignments to portions of the assembly without chromosome locations (chrUn). We did not categorize alignments to chrUn sequences as an indication of a potential misassembly because these sequences represent regions of the genome assembly that were not localized to chromosomes, rather than sequences that had been incorrectly assembled. We found that 554 of the 616 anchored contigs, representing 2.37 Gb of the sequence assembly, aligned to single regions of the assembled chromosomes (1-20 and X). Of these 554 contigs, 19 have additional alignment to chrUn sequences, which suggests chromosome locations for these unlocalized regions of the assembly. An additional 24 contigs aligned exclusively to 11 Mb of chrUn sequence.
We found 38 contigs with alignments to at least two disjoint sequence regions on the assembled chromosomes, with 12 of these contigs having additional localization to chrUn sequences. To ascertain the nature of these inconsistencies, we visually examined the affected regions of each of the 38 contigs in FPC and evaluated the strength of the clone overlap using the fingerprints. Of these inconsistencies, we found 19 to be caused by incorrect contig joins that were made based on the mouse sequence assembly. In these cases, the segmentation of the contig on the assembly was between map subcontigs, indicating a join that had been made based only on the sequence but could not be supported by the fingerprint data. These were resolved by splitting the contigs along their sequence region boundaries. In 9 of the 38 contigs, either some of the segments in question contained very few clones (<10 clones) or the entire contig itself was very small (<10 clones), and we ascribe the segmentation to incorrect BAC-end sequence coordinates or incorrectly mapped contig clones. We found 10 of 616 anchored map contigs that appeared to be correctly constructed but were segmented on the sequence assembly. These inconsistencies are detailed in Supplemental Table 2, and their segment locations are also indicated. The total size of fingerprint contigs with visually verified inconsistencies with the rat genome assembly was 95 Mb, or ∼3.5% of the assembled genome. Resolution of these inconsistencies will require further examination, including analysis of the sequence assembly in the affected regions.
Merging Fingerprint Map Contigs Using Sequence Information
The sequence localizations determined for the contigs and singletons in the fingerprint map suggested contig locations for singleton clones and potential merges between closely adjacent map contigs. We used the sequence localizations and fingerprint data to join contigs that were adjacent and for which overlap was discernable. Local clone order was guided by the clone order in the edited fingerprint map and global subcontig, and contig order was guided by the sequence region localizations. Edited map contigs were merged using sequence information in 172 instances in which sequence overlap between edge clones was detected. In an additional 96 cases, we found merges could be made by fingerprint data supported by sequence overlap. Overall, the median sequence overlap between two map contigs in these cases was 70 kb. The result was a merged fingerprint map with 376 contigs, with a mean contig size of 8.4 Mb (Table 3). Sequence coordinates additionally provided contig locations for 3623 singleton clones.
Of the 376 map contigs, 324 were anchored to the chromosome assemblies and provide coverage for 2.69 Gb (99%) of the assembled chromosomes. The contigs are separated on average by 110 kb (Fig. 2; Supplemental Fig. 1). Some of these contigs also contain regions that are anchored to chrUn sequences, totaling 9 Mb. The contig assignment of these clones was maintained with the anchored contigs to which they belonged, thereby assigning a chromosome location to the associated chrUn sequences. A further 34 contigs provide 51 Mb of coverage of the unlocalized assembly. The remaining 18 contigs were unanchored, and their original structure was preserved. Because there may be genomic regions that are not represented by BACs, such as telomeric or centromeric portions that are difficult to clone or maintain in Escherichia coli or regions with an unusual distribution of HindIII sites, our coverage estimate will likely require refinement as the precise size and position of sequence regions become identified.
Figure 2.
Representation of sequence positions of map contigs in the manually edited fingerprint map (green) and merged map (blue) on the first 100 Mb of Chromosomes 1-5. Contigs colored green are localized to a unique contiguous chromosomal interval. The orange color indicates that the contig was incorrectly joined in the fingerprint map and was subsequently split to form two contigs. If a contig is localized to disparate regions of the sequence assembly and appears to be correctly constructed, its glyph is red. To the left of the contig tracks is shown a histogram of the density of anchored clones, cytogenetic band positions, and the sequence scale.
Anchoring YAC Map Contigs to the Fingerprint Map and the Rat Sequence Assembly
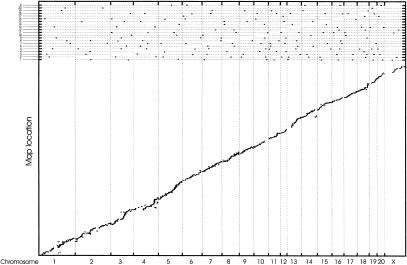
Hybridization data derived from IRS-PCR markers link RPCI-32 BACs and RPCI-31 PACs to at least one YAC in each of the 605 contigs in the YAC map. On average, there were two BACs or PACs associated with each YAC, and each BAC or PAC was associated on average with nine YACs. The mean number of BACs and PACs linked to each YAC contig was seven, with a median value of three. In total, 90% of YAC contigs had between 1 and 24 associated BACs or PACs. In the alignment of the YAC map to the fingerprint map we discarded chimeric YACs, and used only those with unique YAC contig and chromosome assignments. Of the 605 YAC contigs, 598 were anchored to the fingerprint map through RPCI-31 and RPCI-32 clones in common to both maps. We used end sequence or in silico coordinates of RPCI-31 and RPCI-32 clones associated with the YAC map to anchor 14,288 YACs to the sequence assembly. Using these anchored YACs, we determined sequence locations for 577 YAC contigs (95%). The consistency between clones in the YAC map and the rat sequence assembly is shown in Figure 3. Views of the integration between the fingerprint map, YAC map, and the sequence assembly are available at http://mkweb.bcgsc.ca/rat/mapview.
Figure 3.
Representation of consistency between the order of YACs in the YAC physical map and sequence assembly. The plot shows positions of identified sequence anchors within the YAC map plotted against their derived position on the assembled rat genome sequence
DISCUSSION
The rat genome fingerprint map we have constructed comprises 376 contigs incorporating 95% of the clones in the map. Positional annotations to the sequence assembly were derived for 95% of all clones in contigs, using BAC-end sequence and in silico coordinates. Using these annotations, we identified overlapping map clones providing coverage for 99% of the sequence assembly localized to chromosomes (chr1-20, X), and for 78% of the unlocalized sequence assembly regions (chrUn). This is likely an underestimate of the sequence coverage by the map due to the conservative nature of the in silico derived coordinates and the effect of unfinished regions of the sequence assembly on the anchoring process. The high degree of integration between the fingerprint map and the sequence assembly is perhaps not unexpected given that the methodology used to generate the rat genome sequence used BAC clone as well as whole genome shotgun based reads (Rat Genome Sequencing Project Consortium 2004); however, it does indicate that essentially complete genome coverage can be achieved from a clone-based physical map. The high percentage of map clones with sequence coordinates represents a degree of integration between a fingerprint map and sequence assembly that is unique for a large genome. This relationship ties together the BAC-based and sequence-based resources that, together, will be of use to a broad community of researchers.
Given that fingerprint map assembly and sequence assembly are independent processes, the two resources can be used for cross-validation once they have been interrelated. The fingerprint map can be used to evaluate the accuracy of the assembly and aid in the assembly of repeat-rich regions and other areas known to confound assembly programs, and the sequence assembly can be used to evaluate the accuracy of fingerprint map merges and to identify contig overlaps not recognized by fingerprint comparisons. The degree to which the fingerprint map can be used to guide the construction of another genome-ordered resource, such as a sequence assembly, largely depends on the contiguity of the map. The current paradigm in fingerprint map construction begins with fingerprint generation, followed by automated fingerprint assembly and manual editing, assisted where possible by automated contig orientation and merging based on a closely related reference genome. In the case of the rat fingerprint map, automated and manual editing improved the contiguity by more than an order of magnitude (Table 3). This optimized fingerprint map was, in turn, used to guide efficient selection of BAC clones to fill sequence assembly gaps (Rat Genome Sequencing Project Consortium 2004). In addition, unlocalized portions of the rat sequence assembly can be linked to chromosomes because of their association with fingerprint contigs anchored to chromosome assemblies. However, the manual editing phase of map construction is time-consuming and requires dedicated and highly trained staff. The timely application of fingerprint map data to enhance the sequence assembly process will therefore require continued efforts to streamline the map-building process and, ideally, to convert the editing and merging processes required for map contiguity into a series of purely automated steps, particularly in the absence of a closely related genome resource.
Clone-based laboratory methods maintain importance in the study of large genomes through applications such as fluorescent in situ hybridization (FISH; du Manoir et al. 1993; Joos et al. 1994; Levsky and Singer 2003) and comparative genomic hybridization (CGH; Kallioniemi et al. 1992; Houldsworth and Chaganti 1994; Lapierre et al. 1998; Pinkel et al. 1998; Snijders et al. 2001; Fiegler et al. 2003). The generation of high-depth fingerprint maps will therefore continue to be a desired component of the generation of integrated genomic resources. For example, the detailed localizations of nearly all clones in the rat fingerprint map on the sequence assembly serve as an entry point into generating clone-based resources for genomic regions of interest. Moreover, the integrated map and assembly will facilitate the creation of a BAC-based whole-genome array for the rat, which we are planning to undertake to complement an existing whole-genome array for human (http://www.bcgsc.ca/lab/mapping/bacrearray/human; M. Krzywinski, in prep.) and a clone set for mouse (M. Krzywinski, unpubl.), which is currently undergoing validation.
METHODS
Large Insert Libraries
Clones from the following libraries were used: Rat YAC ICRF/BWH/Liege SHRSP (MPMGy916; Cai et al. 1997) and Rat YAC WI/MIT (WIBRy933; Cai et al. 1997; Haldi et al. 1997) libraries, obtained from the Resource Center Primary Database (RZPD; http://www.rzpd.de); RPCI-32 (Osoegawa et al. 2004) rat BAC library and RPCI-31 rat PAC library (Woon et al. 1998), obtained from BACPAC Resources (http://bacpac.chori.org); CHORI-230 rat BAC library (Osoegawa et al. 2004), obtained from BACPAC Resources (http://bacpac.chori.org). The CHORI-230 library was constructed from female brown Norway DNA (BN/SsNHsd/MCW) and is comprised of two segments. Segment 1 was derived from genomic DNA partially digested with EcoRI and EcoRI Methylase, and Segment 2 was derived from DNA partially digested with MboI (Osoegawa et al. 2004).
IRS-PCR
IRS-PCR was performed with a single rat ID element-derived primer (IDR; 5′-CCACTGAGCTAAATCCCCAACCCC-3′) in a 60-μL reaction volume. The PCR reaction mix contained 1 μg of Rat IDR primer, 250 μM each dNTP, 0.2 U of Taq polymerase, 50 mM KCl, 1.5 mM MgCl2, 35 mM Tris base, 15 mM Tris-HCl, 0.1% Tween 20, and 15 μM cresol red. PCR conditions were initial denaturation for 4 min at 94°C, followed by 35 cycles of 30 sec at 94°C, 60 sec at 65°C, 3 min at 72°C, and a final extension for 5 min at 72°C.
Nylon Filter Production
Nylon filters for YAC marker content mapping were spotted in duplicate in a 5 × 5 pattern as described previously (Gösele et al. 2000). IRS-PCR products of the 106 T55 RH clones were transferred onto nylon membranes by Southern blotting as described (Gösele et al. 2000).
Probe Preparation, Hybridization, and Filter Analysis
IRS-PCR products were excised from low melting point agarose gel and melted by heating in 20 μL of 1× TE buffer. Then 18 μL of melted IRS-PCR fragment was further used for labeling with 20 μCi of [α-32P] by random hexamer priming (Feinberg and Vogelstein 1984). Hybridizations against three-dimensional rat YAC pool filters and rat radiation hybrid filters were carried out overnight at 65°C in 15 mL of Church buffer (0.5 M Na2HPO4 at pH 7.2, 5% SDS, 2.5 mM EDTA at pH 8.0). Filters were washed for 30 min at 65°C in 2× SSC, 0.1% SDS, followed by a second wash for 30 min at 65°C in 0.5× SSC, 0.1% SDS. Filters were exposed to autoradiographic films for 1-3 d. To minimize errors in the radiation hybrid data vectors produced, radiation hybrid mapping data were generated in duplicate. Autoradiograms were marked, checked for errors, and subsequently entered directly into a database. The database was implemented with MySQL 3.22.32. Input tools used a PHP module in conjunction with an Apache Web server. HTML forms were used to record the textual experimental details (filter number, hybridization date, data quality assessment). A Java applet using HTTP allowed hybridization results to be entered (or edited) graphically by clicking on positions in a grid representing the spotting pattern. Positive filter coordinates were converted into clone names by the server using a deconvolution routine written in C, which assigned a weight of 3 to complete, unambiguous positive clones, 2 to complete, ambiguous addresses (i.e., where there is more than one positive clone in a block of eight plates), and 1 to incomplete addresses (e.g., row and column, but no plate). Only clones with weight 3 were used for contig building (31,757).
Radiation Hybrid Map Placement
Placement of all markers was carried out with respect to the previously published radiation hybrid framework maps (Steen et al. 1999; Watanabe et al. 1999) using RHMAPPER software (Stein 1998). Marker placement was compared with a framework map order that was calculated using the traveling-salesman problem (TSP) approach using the Lin-Kernighan heuristic from the CONCORDE package (Applegate et al. 1998). We then checked the relative ordering of these conserved segments for consistency against the Rat Genome Database (RGD) map.
Because TSP transformations are strictly valid only for haploid, error-free data, we re-evaluated the map likelihoods and intermarker distances with the radiation hybrid maxlink program (Boehnke et al. 1991).
YAC Map Construction
The co2 software package (http://www-genome.wi.mit.edu) was designed to integrate map information from multiple sources. It searches marker orders to maximize a scoring function. The scoring function awards a high score for YACs hitting a pair of adjacent markers, assesses large penalties for violating the genetic map order, and awards smaller penalties for introducing gaps or breaks in clones. The costs are optimized to approximate the log likelihood of the given order, so that the chosen marker is consistent with as much of the data as possible.
BAC-End Sequences
BAC-end sequences for CHORI-230 and RPCI-32 clones were generated at The Institute for Genomic Research (TIGR) and are publicly available from http://www.tigr.org/rat/bac_end_intro.shtml. BAC-end sequences were available for 164,768 CHORI-230 clones in the fingerprint map. Of these, 129,088 have paired end sequences, and 35,680 have single end sequences.
BAC Clone Fingerprinting
An agarose-gel-based fingerprinting methodology (Marra et al. 1997; McPherson et al. 2001; Schein et al. 2004) was used to generate HindIII fingerprints of clones from the CHORI-230 BAC library, RPCI-32 BAC library, and RPCI-31 PAC library. Briefly, bacterial clones were inoculated for culturing in 96-well format directly from the 384-well library plates. The bacterial clones were cultured overnight, and bacterial pellets were collected by centrifugation. BAC DNA was isolated by alkaline lysis purification, digested with HindIII, and the resulting restriction fragments were resolved via electrophoresis on 1.2% agarose gels. Each gel contained 121 lanes, comprised of all 96 samples from a single microtiter plate as well as 25 marker lanes containing a mixture of commercial size standards (Analytical Marker DNA Wide Range, Promega; and Marker V, Roche Applied Science). Gels were stained postelectrophoresis with SYBR Green I (Molecular Probes, Inc.), and digital images were acquired using a Molecular Dynamics Fluorimager 595. Positions of lanes on the gel images were identified (lane tracked) interactively using the program Image (Sulston et al. 1988; http://www.sanger.ac.uk/Software/Image). The lane-tracked gel images were analyzed by our automated restriction fragment identification software, BandLeader v2.3.3 (Fuhrmann et al. 2003), to identify and size the restriction digest fragments. HindIII fragments derived from sequences internal to the vector were subsequently removed from the fragment data. Fragment sizes <600 bp were also removed from the fragment data because of the variability in detection of fragments <600 bp.
Automated checks were performed to identify potential loading or plate tracking errors. These are described in detail in the Supplemental material.
Automated Fingerprint Assembly
Automated assemblies of the fingerprint data were performed using a parallelized version of FPC (Soderlund et al. 1997, 2000; Ness et al. 2002). Assemblies were performed on a weekly basis as fingerprint data accumulated and were made publicly available (http://www.bcgsc.ca/lab/mapping/data). The automated FPC assembly bins similar clones together into contigs, where similarity is assessed according to a user-defined cutoff value for the Sulston score (Sulston et al. 1988). Clones that do not have fingerprint similarity scores with other clones in the database below this defined value are not placed into contigs, and are called singletons. Additionally, the assembly algorithm attempts to identify the nonredundant set of clones within each contig, and these are called canonical clones. The last automated assembly that contained only CHORI-230 fingerprint data was performed using the following parameters: cutoff 10-17, tolerance 7, min bands 3, bury 0.100, best 10, CpM Off. A copy of this automated assembly was made and subjected to manual review and editing, and this became the official, edited map.
Manual Editing of the Automated Fingerprint Assembly
Manual review and editing of the rat fingerprint assembly was performed using the following general approach, called the pathfinder process: (1) review and correction of clone order within each contig, and identification and correction of chimeric fingerprint contigs; (2) extension of contig ends using singleton clones and identification of contig merges, using fingerprint comparisons for contig end clones with a reduced stringency from that used for the original assembly; (3) alignment of contigs to the sequence assembly of a closely related genome to assist with identification of contig merges; and (4) additional editing and analysis of contig terminal ends to ensure that all possible merges between contigs were made. All clone positions and contig compositions were edited within the FPC interface with the assistance of several scripted external tools used to evaluate the integrity of the contigs.
To assist with intercontig orientation and contig merges during the manual editing process, rat contig merges were made based on rat BAC-end sequence comparisons to the MGSC Version 3 mouse genome sequence assembly (http://www.ncbi.nih.gov/genome/guide/mouse). The 306,779 masked end sequences for CHORI-230 BAC clones were compared with the mouse assembly by WU-BLAST (W. Gish; http://blast.wustl.edu) using the following parameters: kap M = 17 N = -21 X = 140 S2 = 340 gapX = 240 gapS2 = 425 Q = 51 R = 22 e = 1e-06 topcomboN = 2 nonnegok novalidctxok gapsepsmax = 2000, requiring matches that were ≥50 bases long and keeping only the best hit for each BAC end. There were 195,989 ends from clones present in the rat FPC clone database that hit the mouse assembly, representing 134,815 unique clones. The positions on the assembly of each BAC end were treated as map positions, and contigs were assigned to mouse chromosomes based on majority rule. The orientation of each contig relative to the assembly was found, and contig orientations were reversed where necessary. The contigs were then ordered by their midpoints on the assembly. The boundaries of each contig were determined after removing spurious BAC-end hits that were far from the contig midpoint, giving an unreasonable size to the contig. The size in base pairs of a contig was restricted to 4000 times the number of clones in the contig, with a minimum size of 800 kb allowed. BAC-end “markers” were removed from the ends until the contigs were less than this maximum size. Overlapping contigs were joined into a single contig, provided at least two BAC-end sequences supported the contig positions.
Automated Insertion of RPCI-31 and RPCI-32 Singleton Clones Into Edited Fingerprint Map Contigs
Where possible, the automated fingerprint assembly containing clones from all three libraries was used to inform the placement of RPCI-31 and RPCI-32 singleton clones in the edited map. For each RPCI-31 or RPCI-32 singleton clone in a contig in the automated assembly, a neighborhood of adjacent clones was determined. The corresponding neighborhood was identified in the edited map, and the singletons were inserted into the most suitable location. Clones for which a neighborhood could not be determined were compared with all clones within the edited map to determine a location. Details on the insertion process can be found in the Supplemental material.
BAC-End Sequence Alignment to the v3.1 Rat Genomic Sequence
Genome sequence assembly coordinates for CHORI-230 end sequences were obtained from the BCM HGSC (ftp://ftp.hgsc.bcm.tmc.edu/pub/analysis/rat/bacendmap.dat). Sequence assembly coordinates for the RPCI-32 clones were determined by aligning the end sequences to the genomic sequence. The end sequences were first masked with RepeatMasker (A.F.A. Smit and P. Green, unpubl.; ftp://ftp.genome.washington.edu/RM/RepeatMasker.html) using the -rod option. The masked sequences were then searched against the genomic sequence using blastall (J. Ryan, unpubl.; http://genome.nhgri.nih.gov/blastall) with the following options: -p blastn -e 10e-20 -v 20 -b 20. The best hit for each end sequence was identified. Only those hits confirming the chromosome assignment on the YAC map were considered. Paired end coordinates spanning regions >500 kb were not used.
There were end sequence alignments for 228,871 CHORI-230 BAC-end sequences (86,483 clones with paired end alignments and 55,905 clones with single end alignments) and 2943 alignments for RPCI-32 BACs (1160 clones with paired end alignments and 623 clones with single end alignments). We found that the paired end coordinates for 1180 of the CHORI-230 clones mapped to different chromosomes, and that paired end coordinates for 1230 CHORI-230 clones and 72 RPCI-32 clones were >500 kb apart on the same chromosome. These were removed from the data set as they represented likely artifacts. We therefore were left with genomic sequence positions for 141,689 clones, including paired end positions for 85,161 (60%) clones and single end positions for 56,528 (40%) clones. Of the 141,689 clones with end sequence-based coordinates, 137,106 were also in the fingerprint map.
Derivation of Sequence Neighborhoods for Fingerprint Map Clones
The sequence neighborhood for each contig clone was determined using the five nearest fingerprint map neighbors with BAC-end coordinates on the rat assembly (see Supplemental material). The neighborhood represents the region of the genome assembly from which the BAC insert is derived. The neighborhood for each clone was calculated independently of any BAC-end sequence coordinates associated with the clone itself. This was purposefully done to allow for cross-validation between the genomic location predicted by the sequence coordinates and that predicted by the fingerprint map.
BAC Sequence Localizations Derived by In Silico Mapping
The experimental fingerprint of each clone with a sequence neighborhood was compared with the in silico digest fingerprint of the corresponding sequence. The entire neighborhood was first sampled using a sliding window of 120 consecutive fragments. Once the best matching region was found, a second round of comparisons was performed between the experimental fingerprint and a sliding subwindow within the matching region. The position of the clone within the region was determined on the basis of the number and arrangement of matching fragments within the best matching subwindow. See the Supplemental material for details.
Validation of In Silico Localizations
To validate the accuracy of the in silico anchoring approach, we identified a test clone set comprised of clones that (1) had paired BAC-end sequence coordinates and in silico anchors with the two coordinates overlapping; (2) had a fingerprint size of 121-237 kb, corresponding to 90% of all fingerprinted clones; (3) had a difference between BAC-end sequence and fingerprint size of <10 kb; and (4) had <10 kb of undetermined nucleotides in their sequence neighborhoods. The difference in the left and right ends, as well as the difference in size between the BAC-end sequence and in silico coordinates was calculated and used as a measure of validation.
Determination of Clone Coordinates on the Rat Sequence Assembly
The level of correspondence between the BAC-end sequence and in silico or neighborhood coordinates was used to determine which type of coordinate would be used for each clone in subsequent analysis. This was done to limit the number of potential inconsistencies arising from clone tracking errors and end sequence misalignments. Coordinates based on BAC-end sequence localization were used when no in silico anchor could be found or when the in silico anchor overlapped with the BAC-end sequence localization. In silico anchors were used when no BAC-end sequence localization was available or when the BAC-end sequence localization did not overlap with the in silico anchor. When the two coordinate types did not overlap, the in silico anchors were used because they were consistent with the clone fingerprints and the position of the clones in the edited fingerprint map. For clones with only one type of coordinate, those coordinates were used to localize the clone.
Anchoring Rat Fingerprint Contigs to the Rat Sequence Assembly
Each map contig was delineated into groups of clones that overlapped by sequence, using the sequence coordinates determined as defined above. These structures, called sequence regions, represented groups of clones from the fingerprint contigs that mapped to the same region of the sequence assembly. To avoid effects caused by spurious alignments, such as those from individual clones with end sequence coordinates inconsistent with their map positions, we did not consider sequence regions that contained fewer than three clones that were located on a different chromosome than another region from the same contig with more than 10 clones. Some regions of map contigs were exclusively comprised of clones without sequence coordinates, and in these cases the contig structure was used to inform the sequence placement. Multiple sequence regions within 3 Mb of one another derived from the same contig were considered to be contiguous if bridged by overlapping clones. In this case, overlap was inferred if any of the following applied to the bridging clone set: (1) adjacent clones had a Sulston score <10-10; (2) adjacent clones shared >80% of their fragments; or (3) there were >10 conserved bands across five left and five right neighbors. In addition, we constructed sequence regions from singletons with end sequence coordinates.
Contig Splits and Merges Based on Rat Sequence Assembly Anchors
Contig and singleton regions were ordered and oriented with respect to their anchored locations on each rat chromosome assembly. Each ordered list was scanned computationally to identify contigs with discrepant anchors, and for contigs or singleton regions that could be joined. Contigs containing regions mapping to more than one rat sequence region were manually examined to determine if the fingerprints were internally consistent. If the fingerprint data were internally consistent, the contig was not altered. If the fingerprint data were inconsistent (e.g., the discrepancy coincided with a subcontig boundary), the contig was split. However, contigs that had sequence regions anchored to unordered, unlocalized regions of the sequence assembly (chrUn) were not split if the contigs were otherwise anchored to a single chromosomal region, or if manual review of the contigs indicated that the contig was internally consistent. Contigs for which all regions mapped to chrUn sequences were not altered. Refer to the Supplemental material for additional details.
Contigs located adjacent to each other on the sequence were considered to be potentially overlapping, and the regions were analyzed to determine if contig overlap could be established. Two contigs were considered to overlap if (1) their anchored regions overlapped by sequence; (2) the best fingerprint match between the three edge clones from each of the contigs had a Sulston score <10-7 (60% shared fragments), and the matching clones match their respective contig neighbors better than 10-7; or (3) a subset of three edge clones can be found from each contig for which fingerprints created from shared fragments between the edge clones have >10 matching fragments (bridge), or at least half of the smaller number of fragments (>5 always). Map contigs with internally consistent, overlapping fingerprints but with inconsistent sequence alignments (i.e., alignment to two or more sequence regions) were excluded from the merging process.
Anchoring the YAC Map to the Rat Sequence Assembly
Sequence coordinates for YAC clones were determined using the hybridization-associated RPCI-32 BACs that also had sequence coordinates. To obtain contiguous YAC contig localizations, we screened the hybridization results to remove associations with YACs that hybridized to multiple chromosomes. The screened BAC-YAC relationships were used to annotate each YAC with the sequence coordinates of the associated BACs. These coordinates were used to determine the extent of the anchored part of the YAC, although the actual YAC insert may extend beyond its first/last BAC coordinate. The coordinates of the individual YACs were used to anchor the YAC map on the sequence assembly by associating the YAC coordinates with their cognate contigs.
Data Availability
The FPC database is available for download from http://www.bcgsc.ca/lab/mapping/data. The data can also be viewed via the Internet using iCE (Fjell et al. 2003), a Java-based application for viewing FPC data (http://www.bcgsc.ca/about/news/ice). The integrated maps and sequence assembly can be viewed at http://mkweb.bcgsc.ca/rat/mapview, which includes a link to the FPC clone map tracks in the UCSC Genome Browser. All YAC map data are freely available at http://www.mdc-berlin.de/ratgenome/ or http://www.molgen.mpg.de/~ratgenome/.
Acknowledgments
The authors wish to thank Catharine Gray, Reta Kutche, Candice McLeavy, Soo-Sen Lee, Sheryle Taylor, Robin Coope, and Nelson Siu for technical assistance. This study was supported by a grant-in-aid from the German Ministry of Science and Education (BMBF/DHGP) to N.H. Construction of the CHORI-230 library was supported by a grant from NIH (HG01165-06) to P.D.J. BAC clone fingerprinting, map editing, and mapping bioinformatics was supported by NIH grant U01 HG02155. M.A.M. is a scholar of the Michael Smith Foundation for Health Research.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2336604.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Adams, M.D., Celniker, S.E., Holt, R.A., Evans, C.A., Gocayne, J.D., Amanatides, P.G., Scherer, S.E., Li, P.W., Hoskins, R.A., Galle, R.F., et al. 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185-2195. [DOI] [PubMed] [Google Scholar]
- Agarwala, R., Applegate, D.L., Maglott, D., Schuler, G.D., and Schaffer, A.A. 2000. A fast and scalable radiation hybrid map construction and integration strategy. Genome Res. 10: 350-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitman, T.J., Glazier, A.M., Wallace, C.A., Cooper, L.D., Norsworthy, P.J., Wahid, F.N., Al-Majali, K.M., Trembling, P.M., Mann, C.J., Shoulders, C.C., et al. 1999. Identification of Cd36 (Fat) as an insulin-resistance gene causing defective fatty acid and glucose metabolism in hypertensive rats. Nat. Genet. 21: 76-83. [DOI] [PubMed] [Google Scholar]
- Anzai, K., Kobayashi, S., Suehiro, Y., and Goto, S. 1987. Conservation of the ID sequence and its expression as small RNA in rodent brains: Analysis with cDNA for mouse brain-specific small RNA. Brain Res. 388: 43-49. [DOI] [PubMed] [Google Scholar]
- Applegate, D., Bixby, R., Chvatal, V., and Cook, W. 1998. On the solution of traveling salesman problems. Documenta Mathematica Journal der Deutschen Mathematiker-Vereinigung International Congress of Mathematics III 645-656.
- The Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796-815. [DOI] [PubMed] [Google Scholar]
- Barry, G.F. 2001. The use of the Monsanto draft rice genome sequence in research. Plant Physiol. 125: 1164-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehnke, M., Lange, K., and Cox, D.R. 1991. Statistical methods for multipoint radiation hybrid mapping. Am. J. Hum. Genet. 49: 1174-1188. [PMC free article] [PubMed] [Google Scholar]
- Cai, L., Schalkwyk, L.C., Schoeberlein-Stehli, A., Zee, R.Y., Smith, A., Haaf, T., Georges, M., Lehrach, H., and Lindpaintner, K. 1997. Construction and characterization of a 10-genome equivalent yeast artificial chromosome library for the laboratory rat, Rattus norvegicus. Genomics 39: 385-392. [DOI] [PubMed] [Google Scholar]
- Daniels, G.R. and Deininger, P.L. 1985. Repeat sequence families derived from mammalian tRNA genes. Nature 317: 819-822. [DOI] [PubMed] [Google Scholar]
- Deininger, P.L. 1989. SINEs: Short interspersed repeated DNA elements in higher eukaryotes. In Mobile DNA (eds. D.E. Berg and M.M. Howe), pp. 619-636. American Society for Microbiology, Washington, DC.
- du Manoir, S., Speicher, M.R., Joos, S., Schrock, E., Popp, S., Dohner, H., Kovacs, G., Robert-Nicoud, M., Lichter, P., and Cremer, T. 1993. Detection of complete and partial chromosome gains and losses by comparative genomic in situ hybridization. Hum. Genet. 90: 590-610. [DOI] [PubMed] [Google Scholar]
- Feinberg, A.P. and Vogelstein, B. 1984. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Addendum. Anal. Biochem. 137: 266-267. [DOI] [PubMed] [Google Scholar]
- Fiegler, H., Carr, P., Douglas, E.J., Burford, D.C., Hunt, S., Smith, J., Vetrie, D., Gorman, P., Tomlinson, I.P., and Carter, N.P. 2003. DNA microarrays for comparative genomic hybridization based on DOP-PCR amplification of BAC and PAC clones. Genes Chromosomes Cancer 36: 361-374. [DOI] [PubMed] [Google Scholar]
- Fjell, C.D., Bosdet, I., Schein, J.E., Jones, S.J., and Marra, M.A. 2003. Internet Contig Explorer (iCE)-A tool for visualizing clone fingerprint maps. Genome Res. 13: 1244-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrmann, D.R., Krzywinski, M.I., Chiu, R., Saeedi, P., Schein, J.E., Bosdet, I.E., Chinwalla, A., Hillier, L.W., Waterston, R.H., McPherson, J.D., et al. 2003. Software for automated analysis of DNA fingerprinting gels. Genome Res. 13: 940-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gösele, C., Hong, L., Kreitler, T., Rossmann, M., Hieke, B., Gross, U., Kramer, M., Himmelbauer, H., Bihoreau, M.T., Kwitek-Black, A.E., et al. 2000. High-throughput scanning of the rat genome using interspersed repetitive sequence-PCR markers. Genomics 69: 287-294. [DOI] [PubMed] [Google Scholar]
- Green, E.D., Hieter, P., and Spencer, F.A. 1999. Yeast artificial chromosomes. In Genome analysis-A laboratory manual (eds. B. Birren et al.), pp. 479-487. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Gregory, S.G., Sekhon, M., Schein, J., Zhao, S., Osoegawa, K., Scott, C.E., Evans, R.S., Burridge, P.W., Cox, T.V., Fox, C.A., et al. 2002. A physical map of the mouse genome. Nature 418: 743-750. [DOI] [PubMed] [Google Scholar]
- Haldi, M.L., Lim, P., Kaphingst, K., Akella, U., Whang, J., and Lander, E.S. 1997. Construction of a large-insert yeast artificial chromosome library of the rat genome. Mamm. Genome 8: 460. [DOI] [PubMed] [Google Scholar]
- Hoskins, R.A., Nelson, C.R., Berman, B.P., Laverty, T.R., George, R.A., Ciesiolka, L., Naeemuddin, M., Arenson, A.D., Durbin, J., David, R.G., et al. 2000. A BAC-based physical map of the major autosomes of Drosophila melanogaster. Science 287: 2271-2274. [DOI] [PubMed] [Google Scholar]
- Houldsworth, J. and Chaganti, R.S. 1994. Comparative genomic hybridization: An overview. Am. J. Pathol. 145: 1253-1260. [PMC free article] [PubMed] [Google Scholar]
- Hudson, T.J., Stein, L.D., Gerety, S.S., Ma, J., Castle, A.B., Silva, J., Slonim, D.K., Baptista, R., Kruglyak, L., Xu, S.H., et al. 1995. An STS-based map of the human genome. Science 270: 1945-1954. [DOI] [PubMed] [Google Scholar]
- Jacob, H.J. and Kwitek, A.E. 2002. Rat genetics: Attaching physiology and pharmacology to the genome. Nat. Rev. Genet. 3: 33-42. [DOI] [PubMed] [Google Scholar]
- James, M.R. and Lindpaintner, K. 1997. Why map the rat? Trends Genet. 13: 171-173. [DOI] [PubMed] [Google Scholar]
- Joos, S., Fink, T.M., Ratsch, A., and Lichter, P. 1994. Mapping and chromosome analysis: The potential of fluorescence in situ hybridization. J. Biotechnol. 35: 135-153. [DOI] [PubMed] [Google Scholar]
- Kallioniemi, A., Kallioniemi, O.P., Sudar, D., Rutovitz, D., Gray, J.W., Waldman, F., and Pinkel, D. 1992. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science 258: 818-821. [DOI] [PubMed] [Google Scholar]
- Kass, D.H., Kim, J., and Deininger, P.L. 1996. Sporadic amplification of ID elements in rodents. J. Mol. Evol. 42: 7-14. [DOI] [PubMed] [Google Scholar]
- Kim, J. and Deininger, P.L. 1996. Recent amplification of rat ID sequences. J. Mol. Biol. 261: 322-327. [DOI] [PubMed] [Google Scholar]
- Kim, J., Martignetti, J.A., Shen, M.R., Brosius, J., and Deininger, P. 1994. Rodent BC1 RNA gene as a master gene for ID element amplification. Proc. Natl. Acad. Sci. 91: 3607-3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Lapierre, J.M., Cacheux, V., Da Silva, F., Collot, N., Hervy, N., Wiss, J., and Tachdjian, G. 1998. Comparative genomic hybridization: Technical development and cytogenetic aspects for routine use in clinical laboratories. Ann. Genet. 41: 56-62. [PubMed] [Google Scholar]
- Levsky, J.M. and Singer, R.H. 2003. Fluorescence in situ hybridization: Past, present and future. J. Cell Sci. 116: 2833-2838. [DOI] [PubMed] [Google Scholar]
- Marra, M.A., Kucaba, T.A., Dietrich, N.L., Green, E.D., Brownstein, B., Wilson, R.K., McDonald, K.M., Hillier, L.W., McPherson, J.D., and Waterston, R.H. 1997. High throughput fingerprint analysis of large-insert clones. Genome Res. 7: 1072-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra, M., Kucaba, T., Sekhon, M., Hillier, L., Martienssen, R., Chinwalla, A., Crockett, J., Fedele, J., Grover, H., Gund, C., et al. 1999. A map for sequence analysis of the Arabidopsis thaliana genome. Nat. Genet. 22: 265-270. [DOI] [PubMed] [Google Scholar]
- McPherson, J.D., Marra, M., Hillier, L., Waterston, R.H., Chinwalla, A., Wallis, J., Sekhon, M., Wylie, K., Mardis, E.R., Wilson, R.K., et al. 2001. A physical map of the human genome. Nature 409: 934-941. [DOI] [PubMed] [Google Scholar]
- Mozo, T., Dewar, K., Dunn, P., Ecker, J.R., Fischer, S., Kloska, S., Lehrach, H., Marra, M., Martienssen, R., Meier-Ewert, S., et al. 1999. A complete BAC-based physical map of the Arabidopsis thaliana genome. Nat. Genet. 22: 271-275. [DOI] [PubMed] [Google Scholar]
- Ness, S.R., Terpstra, W., Krzywinski, M., Marra, M.A., and Jones, S.J. 2002. Assembly of fingerprint contigs: Parallelized FPC. Bioinformatics 18: 484-485. [DOI] [PubMed] [Google Scholar]
- Olofsson, P., Holmberg, J., Tordsson, J., Lu, S., Akerstrom, B., and Holmdahl, R. 2003. Positional identification of Ncf1 as a gene that regulates arthritis severity in rats. Nat. Genet. 33: 25-32. [DOI] [PubMed] [Google Scholar]
- Ono, T., Kondoh, Y., Kagiyama, N., Sonta, S., and Yoshida, M. 2001. Genomic organization and chromosomal distribution of rat ID elements. Genes Genet. Syst. 76: 213-220. [DOI] [PubMed] [Google Scholar]
- Osoegawa, K., Zhu, B., Shu, C.L., Ren, T., Cao, Q., Vessere, G.M., Lutz, M.M., Jensen-Seaman, M.I., Zhao, S., and de Jong, P.J. 2004. BAC resources for the Rat Genome Project. Genome Res. (this issue).
- Pinkel, D., Segraves, R., Sudar, D., Clark, S., Poole, I., Kowbel, D., Collins, C., Kuo, W.L., Chen, C., Zhai, Y., et al. 1998. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat. Genet. 20: 207-211. [DOI] [PubMed] [Google Scholar]
- Rat Genome Sequencing Project Consortium. 2004. Genome sequence of the Brown Norway Rat yields insights into mammalian evolution. Nature (in press). [DOI] [PubMed]
- Sapienza, C. and St Jacques, B. 1986. `Brain-specific' transcription and evolution of the identifier sequence. Nature 319: 418-420. [DOI] [PubMed] [Google Scholar]
- Schalkwyk, L.C., Cusack, B., Dunkel, I., Hopp, M., Kramer, M., Palczewski, S., Piefke, J., Scheel, S., Weiher, M., Wenske, G., et al. 2001. Advanced integrated mouse YAC map including BAC framework. Genome Res. 11: 2142-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheetz, T.E., Raymond, M.R., Nishimura, D.Y., McClain, A., Roberts, C., Birkett, C., Gardiner, J., Zhang, J., Butters, N., Sun, C., et al. 2001. Generation of a high-density rat EST map. Genome Res. 11: 497-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schein, J., Kucaba, T.A., Sekhon, M., Smailus, D., Waterston, R.H., and Marra, M.A. 2004. High-throughput BAC fingerprinting. In Methods in molecular biology, Vol. 255. Bacterial artificial chromosomes: Library construction, physical mapping and sequencing (eds. S. Zhao and M. Stodolsky). Humana Press Inc., Totowa, NJ (in press). [DOI] [PubMed]
- Snijders, A.M., Nowak, N., Segraves, R., Blackwood, S., Brown, N., Conroy, J., Hamilton, G., Hindle, A.K., Huey, B., Kimura, K., et al. 2001. Assembly of microarrays for genome-wide measurement of DNA copy number. Nat. Genet. 29: 263-264. [DOI] [PubMed] [Google Scholar]
- Soderlund, C., Longden, I., and Mott, R. 1997. FPC: A system for building contigs from restriction fingerprinted clones. Comput. Appl. Biosci. 13: 523-535. [DOI] [PubMed] [Google Scholar]
- Soderlund, C., Humphray, S., Dunham, A., and French, L. 2000. Contigs built with fingerprints, markers, and FPC V4.7. Genome Res. 10: 1772-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen, R.G., Kwitek-Black, A.E., Glenn, C., Gullings-Handley, J., Van Etten, W., Atkinson, O.S., Appel, D., Twigger, S., Muir, M., Mull, T., et al. 1999. A high-density integrated genetic linkage and radiation hybrid map of the laboratory rat. Genome Res. 9: AP1-AP8, insert. [PubMed] [Google Scholar]
- Stein, L. 1998. RHMAPPER, Installation and user's guide. http://www.broad.mit.edu/ftp/distribution/software/rhmapper/doc/rhmapper.html.
- Stoll, M., Cowley Jr., A.W., Tonellato, P.J., Greene, A.S., Kaldunski, M.L., Roman, R.J., Dumas, P., Schork, N.J., Wang, Z., and Jacob, H.J. 2001. A genomic-systems biology map for cardiovascular function. Science 294: 1723-1726. [DOI] [PubMed] [Google Scholar]
- Sulston, J., Mallett, F., Staden, R., Durbin, R., Horsnell, T., and Coulson, A. 1988. Software for genome mapping by fingerprinting techniques. Comput. Appl. Biosci. 4: 125-132. [DOI] [PubMed] [Google Scholar]
- Watanabe, T.K., Bihoreau, M.T., McCarthy, L.C., Kiguwa, S.L., Hishigaki, H., Tsuji, A., Browne, J., Yamasaki, Y., Mizoguchi-Miyakita, A., Oga, K., et al. 1999. A radiation hybrid map of the rat genome containing 5,255 markers. Nat. Genet. 22: 27-36. [DOI] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- Woon, P.Y., Osoegawa, K., Kaisaki, P.J., Zhao, B., Catanese, J.J., Gauguier, D., Cox, R., Levy, E.R., Lathrop, G.M., Monaco, A.P., et al. 1998. Construction and characterization of a 10-fold genome equivalent rat P1-derived artificial chromosome library. Genomics 50: 306-316. [DOI] [PubMed] [Google Scholar]
- Yokoi, N., Komeda, K., Wang, H.Y., Yano, H., Kitada, K., Saitoh, Y., Seino, Y., Yasuda, K., Serikawa, T., and Seino, S. 2002. Cblb is a major susceptibility gene for rat type 1 diabetes mellitus. Nat. Genet. 31: 391-394. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- ftp://ftp.hgsc.bcm.tmc.edu/pub/analysis/rat/bacendmap.dat; BACFisher placements of BAC-end sequences on the rat assembly.
- http://bacpac.chori.org/rat230.htm; CHORI-230 BAC library at BACPAC Resources.
- http://blast.wustl.edu; WU-BLAST.
- http://ftp.genome.washington.edu/RM/RepeatMasker.html; RepeatMasker.
- http://genome.nhgri.nih.gov/blastall; blastall.
- http://mkweb.bcgsc.ca/rat/mapview; fingerprint map tables and UCSC tracks.
- http://www.bcgsc.ca/about/news/ice; Internet Contig Explorer.
- http://www.bcgsc.ca/lab/mapping/bacrearray/human; BAC-based whole-genome array for human.
- http://www.bcgsc.ca/lab/mapping/data; rat fingerprint map.
- http://www.broad.mit.edu/ftp/distribution/software/rhmapper/doc/rhmapper.html; RHMAPPER.
- http://www-genome.wi.mit.edu; co2 software.
- http://www.hgsc.bcm.tmc.edu/projects/rat/assembly.html; rat sequence assembly (Rnor3.1).
- http://www.mdc-berlin.de/ratgenome; YAC mapping data.
- http://www.molgen.mpg.de/~ratgenome; YAC mapping data.
- http://www.ncbi.nih.gov/genome/guide/mouse/; mouse sequence assembly (mm4).
- http://www.rzpd.de; clone resources.
- http://www.sanger.ac.uk/Software/Image; Image.
- http://www.tigr/org/rat/bac_end_intro.shtml; TIGR.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The FPC database is available for download from http://www.bcgsc.ca/lab/mapping/data. The data can also be viewed via the Internet using iCE (Fjell et al. 2003), a Java-based application for viewing FPC data (http://www.bcgsc.ca/about/news/ice). The integrated maps and sequence assembly can be viewed at http://mkweb.bcgsc.ca/rat/mapview, which includes a link to the FPC clone map tracks in the UCSC Genome Browser. All YAC map data are freely available at http://www.mdc-berlin.de/ratgenome/ or http://www.molgen.mpg.de/~ratgenome/.