Abstract
Two 11-fold redundant bacterial artificial chromosome (BAC) libraries (RPCI-32 and CHORI-230) have been constructed to support the rat genome project. The first library was constructed using a male Brown Norway (BN/SsNHsd) rat as a DNA source long before plans for rat genome sequencing had been launched. The second library was prepared from a highly inbred female (BN/SsNHsd/MCW) rat in support of the rat genome sequencing project. The use of an inbred rat strain is essential to avoid problems with genome assembly resulting from the difficulty of distinguishing haplotype variation from variation among duplicons. We have demonstrated the suitability of the library by using a detailed quality assessment of large insert sizes, narrow size distribution, consistent redundancy for many markers, and long-range continuity of BAC contig maps. The widespread use of the two libraries as an integral part of the rat genome project has led to the database annotations for many clones, providing rat researchers with a rich resource of BAC clones that can be screened in silico for genes of interest.
Arrayed large-insert bacterial artificial chromosome (BAC) libraries play a critical role in the assembly of maps and sequence for complex genomes. Once the clones have been annotated, they become valuable as a resource for the research community. The laboratory rat (Rattus norvegicus) has been used as a model organism for a variety of biomedical studies in the areas of cardiovascular, metabolic, pharmacologic, toxicologic, and psychological research (Jacob and Kwitek 2001). A large number of quantitative trait loci (QTL) related to complex genetic diseases have been mapped to rat chromosomal regions through genetic studies (Dukhanina et al. 1997; Garrett et al. 1998). BAC clones from strain-specific libraries can be used to confirm the position of QTLs and may eventually be applied in BAC transgenic (Takahashi et al. 2000) or tissue culture experiments to recapitulate phenotypes. In support of initial rat genome mapping plans, several P1-derived artificial chromosome (PAC) and BAC libraries were created from commercially available Brown Norway (BN/ssNHsd) rats (Woon et al. 1998). The common Brown Norway strain was initially chosen because of its widespread use and availability. An additional BAC library from a highly inbred female Brown Norway rat was prepared after the rat genome sequencing project was launched in August 2000. Closely related rats from the same inbred source have been used to create the BAC library and, elsewhere, to create small insert libraries for whole-genome shotgun sequencing (WGS; Venter et al. 2001; Waterston et al. 2002). The goal was to sequence the rat genome using a hybrid strategy to generate a draft sequence of a rat genome based on the use of mapped BACs for low-coverage clone-by-clone sequencing (Lander et al. 2001), complemented by WGS and pooled-BAC sequence reads (Rat Genome Sequencing Consortium 2004). The characterization of these BAC libraries is described here, together with information on how to access the BACs and BAC-related information.
RESULTS
Modified BAC Vectors
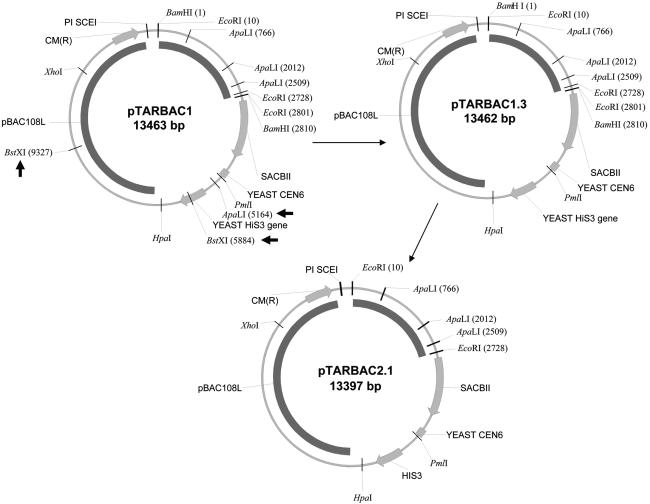
The goals were to construct a BAC library optimal for the rat genome mapping and sequencing, while also offering options for later applications. We previously reported the use of pTARBAC1 vector to clone large DNA molecules in Escherichia coli, with downstream options to reisolate related haplotypic or syngenic sequences (Zeng et al. 2001). Such sequences can be isolated from high-molecular weight genomic DNA through gap-repair of deleted linearized BAC clones. This process is implemented by cotransforming specific deleted BACs and genomic DNA into yeast spheroplasts (Zeng et al. 2001) and has been labeled Transformation-Associated Recombination, or TAR (Larionov et al. 1996). To facilitate library construction and conceivable specific recloning projects, two modified versions of the pTARBAC vector were prepared. One vector (pTARBAC1.3; Fig. 1) was modified to remove a specific ApaLI site (as described in Methods), and this vector is used for cloning DNA fragments generated by partial MboI digestion between the two BamHI sites. Cloned genomic fragments replace a 2.7-kb stuffer fragment released by BamHI digestion. The pTARBAC2.1 vector was constructed by replacing the two small BamHI-EcoRI fragments in the pTARBAC1.3 vector with synthetic adapters, resulting in elimination of the two BamHI sites. BAC clones are generated by cloning partial EcoRI-genomic fragments in the pTARBAC2.1 vector. Most of the genomic DNA in specific BACs can be deleted following BamHI digestion, resulting in relative small-insert terminal fragments attached to the linear BAC vector that are compatible with gap-repair applications.
Figure 1.
Construction of pTARBAC1.3 and pTARBAC2.1 vectors. Two BstXI (position 5884 and 9327) and one ApaLI (position 5164) sites (see arrows) were eliminated from the pTARBAC1 vector to create the pTARBAC1.3 vector. The pTARBAC2.1 vector was generated from pTARBAC1.3 by replacing the two BamHI-EcoRI fragments with synthetic adapters to regenerate the EcoRI sites while eliminating the BamHI sites.
Construction of BAC Libraries
To support the initial stage of rat genome mapping, a BAC library was constructed using EcoRI-partially digested DNA obtained from a male Brown Norway (BN/SsNHsd) rat in the pBACe3.6 vector (Frengen et al. 1999). The library was designated as RPCI-32 and is composed of approximately 230,000 clones arrayed in 624 384-well microtiter dishes. The library was divided into two segments (plates 1-288 for segment 1 and plates 289-624 for segment 2) for logistical reasons (Table 1). To support whole rat genome sequencing, a new BAC library (CHORI-230) was constructed from a highly inbred female rat (BN/SsNHsd/MCW). A female rat was selected to obtain the diploid representation of the X-chromosome in the library. To minimize conceivable cloning bias caused by nonuniform distribution of restriction sites for a particular enzyme, the new BAC library was prepared using different restriction enzymes for the two library segments. The first segment of the library resulted from EcoRI-partially digested DNA, whereas the second segment was created from MboI-partially digested DNA, and a sufficient number of clones were arrayed for at least fivefold redundancy per segment (Table 1).
Table 1.
Rattus norvegicus BAC Libraries
|
Library
|
RPCI-32
|
CHORI-230
|
||
|---|---|---|---|---|
| Segment | 1 | 2 | 1 | 2 |
| Cloning vector | pBACe3.6 | pBACe3.6 | pTARBAC2.1 | pTARBAC2.1 |
| DNA source | Male spleen | Male spleen | Female brain | Female liver |
| Restriction enzyme | EcoRI | EcoRI | EcoRI | MboI |
| Plate range | 1-288 | 289-624 | 1-240 | 241-528 |
| Total 384-well dishes | 288 | 336 | 240 | 288 |
| Empty wells (%) | 1244 (1.1%) | 1559 (1.2%) | 1742 (1.9%) | 1677 (1.5%) |
| Noninsert clones (%) | 4/159 (2.5%) | 7/156 (4.5%) | (1.1%) | 26/382 (6.8%) |
| Recombinant clones | ∼106,000 | ∼122,000 | ∼89,000 | ∼102,000 |
| Insert size (Average) | 147 kb | 153 kb | 214 kb | 168 kb |
| Standard deviation | 35 kb | 29 kb | 27 kb | 53 kb |
| Library redundancy | 5.2-fold | 6.2-fold | 6.4-fold | 5.5-fold |
The “Recombinant clones” column represents total wells after empty wells and noninsert clones were subtracted in each segment. The library redundancy was estimated using 3000 Mb as the genome size.
Average Insert Sizes and BAC End Sequences
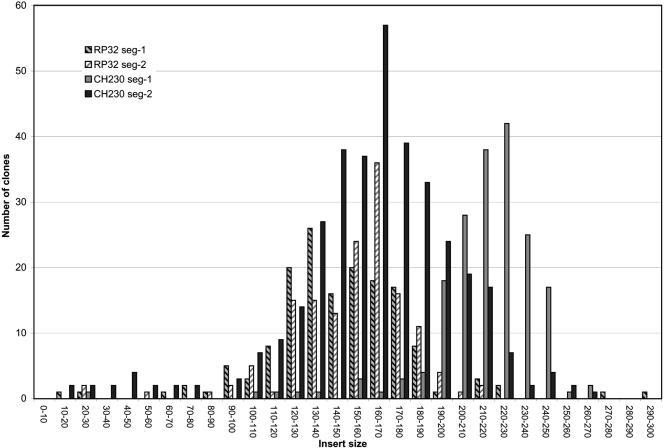
To be compatible with rat genome sequencing, the most recent BAC libraries should have large inserts (near 200 kb average), a narrow insert size distribution, and a low percentage of noninsert clones. To assess the suitability of the library, we determined average insert sizes, insert size distribution, and the percentage of noninsert clones for each library segment, as summarized in Table 1. Using pulsed-field gel electrophoresis, the average insert sizes were determined to be 147, 153, 214, and 168 kb, with a standard deviation of 35, 29, 27, and 53 kb for RPCI-32 segments 1 and 2 and CHORI-230 segments 1 and 2, respectively (Table 1). We have attempted to construct the CHORI-230 library with inserts as large as feasible to maximize fingerprint information and minimize the number of required clone-derived shotgun libraries. The percentage of noninsert clones, compared with clones containing an insert, was estimated to be 1.0%, 1.2%, 1.1%, and 6.8% for the RPCI-32 and CHORI-230 library segments, respectively (Table 1). The low percentages of noninsert clones economize the BAC fingerprinting and clone-by-clone sequencing approaches. Insert size distributions for each library segment are provided in Figure 2, as determined by pulsed-field electrophoresis. To verify the accuracy of the average insert size and the standard deviation of the CHORI-230 library, 30,110 BACs from segment 1 and 50,317 BACs from segment 2 were mapped onto the June 19, 2003, assembly of the rat genome using unique BAC end sequences (S. Zhao, unpubl.; http://genome.ucsc.edu/ and http://www.hgsc.bcm.tmc.edu/projects/rat/). The average sequence matches were 580 and 679 bp, with 98.7% and 99% identity, for segment 1 and segment 2, respectively. The average insert sizes determined from paired BAC-end mapping were 229 and 174 kb, with standard deviations of 35 and 45 kb for CHORI-230 segments 1 and 2, respectively. The average insert sizes obtained from BAC-end mapping were compatible with the sizes independently obtained from pulsed-field gel electrophoresis analysis. The insert sizes estimated through pulsed-field electrophoresis were within 7% of the more accurate size determinations derived through mapping of the paired-BAC-end sequences, revealing a unique opportunity to calibrate the accuracy of the empirically determined insert sizes. Although approximately 80,000 BACs ended up with consistent map assignments, another 3872 clones experienced unrelated chromosome locations for the paired BAC-ends. A majority of 2701 inconsistently mapped BACs were based on BAC-end sequences mapping to the same chromosome, either in the wrong orientation (2108 BACs) or at a distance incompatible with BAC cloning capacity (>400 kb; 593 BACs). The very preferential colocalization of ends to the same chromosomes indicates that the inconsistent end sequences were in most cases not derived from cloning artifacts. Chimeric clones would most likely have ends completely randomly distributed through the rat genome. One thousand one hundred seventy-one BACs have ends mapping to unrelated chromosomes, representing about 1.2% of the BACs with two uniquely mapped end sequences. This places an upper limit on possible chimeric clones of around 1.2%, whereas the real number of chimeric clones is likely much lower. The strong bias of inconsistently mapped BACs for both termini to map to a shared chromosome indicates sequence assembly errors to be the most likely reason. The low chimerism rate indicated by this analysis facilitates the use of mapped BAC-ends to evaluate possible sequence assembly errors. In addition, it is conceivable that the BAC-ends have incorrectly been mapped by BLAST as a result of the presence of nonmasked low-level repetitive sequences at the BAC-ends. On the basis of the number of clones and the average insert sizes, it is estimated that RPCI-32 segments 1 and 2 and CHORI-230 segments 1 and 2 provide 5.2-, 6.2-, 6.4-, and 5.5-fold redundancies, respectively (Table 1). These estimates are based on the assumed rat haploid genome size of 3000 Mb.
Figure 2.
Insert size distribution of the RPCI-32 and CHORI-230 libraries. The horizontal axis indicates the insert size range of BAC clones. The vertical axis refers to the number of clones in each size range. Gray background with black diagonal stripe, white background with gray diagonal stripe, gray and black bars represent RPCI-32 segment 1, RPCI-32 segment 2, CHORI-230 segment 1, and CHORI-230 segment 2, respectively.
Redundancies
To evaluate genomic representation and possible cloning bias, the RPCI-32 and CHORI-230 libraries were screened using a mixture of 22 overlapping oligonucleotide (overgo) probes designed from DNA sequences conserved between human and other organisms. All positive clones were subjected to secondary hybridization screening with individual probes to determine the probe-clone correlation. In addition, all positive clones were analyzed by DNA fingerprinting. A priori positive clones identified by a unique probe should be assembled into a single contig. It is also feasible to eliminate false-positive clones or to recover false-negative clones by combining hybridization and fingerprinting analysis. Using fingerprinting, 4.7% (27 out of 573) of hybridization positive clones were eliminated as false positive, whereas 4.5% (28 out of 619) of clones were recovered as positive. Nineteen of the 22 probes were unique, permitting the assembling of single contigs using the fingerprinting approach, whereas three probes provided multiple (two to four) contigs. The average numbers of confirmed clones per contig via hybridization and fingerprinting were estimated to be 3.5 (RPCI-32 segment 1), 5.4 (RPCI-32 segment 2), 6.6 (CHORI-230 segment 1), and 5.0 (RPCI-230 segment 2), respectively. Standard deviations from the average number of positive clones were 2.8 (RPCI-32 segment 1), 2.8 (RPCI-32 segment 2), 3.3 (CHORI-230 segment 1), and 2.6 (RPCI-230 segment 2), respectively.
Quality Assessment Using Physical Maps
Contiguous overlapping clone maps play an essential role for genome sequencing and gene identification. Contigs were built using the CHORI-230 library at two end-stage renal-failure QTLs (Rf-1: 20.7 Mb, and Rf-4: 20.6 Mb; Brown et al. 1996; Shiozawa et al. 2000), through a combination of overgo hybridizations and in silico mapping of markers as the DNA sequence of BAC clones became available. The modal number of BACs identified from hybridizing 480 probes to segment 1 was six, consistent with the expectation based on the estimated redundancy of this library. Only 44 out of 500 overgo probes failed to give a positive hit to the library. Most of these (34) were later found in the BAC sequence, indicating that experimental failure was responsible for these probes not identifying BACs in the hybridizations. The sequence of over half of the failed probes had low G+C content (∼35%), the likely cause of the failed hybridization. At most, five gaps remain in the ∼41Mb of the physical maps combined in these two QTLs, all of which are less than 125 kb, confirming the even representation of the rat genome in the CHORI-230 library.
DISCUSSION
A 10-fold redundant PAC library was constructed to assist in the early stage of the rat genome project (Woon et al. 1998). BAC and PAC clones have both been shown to have, in general, excellent clonal integrity and good stability through extensive characterization of rat, mouse, and human libraries (Woon et al. 1998; Osoegawa et al. 2000, 2001). BAC vectors became preferable for construction of genomic libraries mostly as a result of their smaller vector size (about 8 kb versus about 15 kb) because of the resulting favorable effect on the economy of clone-by-clone shotgun sequencing. A further reason for the use of the BAC libraries was that the earlier rat PAC library (RPCI-31) was prepared using DNA from multiple individuals from a less inbred stock and had relatively small insert sizes (∼140 kb; Woon et al. 1998). The first BAC library was constructed using EcoRI-partially digested DNA obtained from a single male Brown Norway (BN/SsNHsd) rat in the pBACe3.6 vector (Frengen et al. 1999). Genotyping of rats from a similar stock (BN/SsNHsd) from Harlan Sprague Dawley, using 4328 microsatellite markers, at Medical College of Wisconsin resulted in the discovery that six loci were heterozygous (Steen et al. 1999). As a consequence, when the rat genome project was started, it became desirable to construct a BAC library and small-insert sequencing libraries from a more inbred strain (Rat Genome Sequencing Consortium 2004). Improvements in the resolution of the DNA fingerprinting technology permits analysis of BACs with larger (>200 kb) insert fragments, permitting improved contig assembly (Fuhrmann et al. 2003). To facilitate the fingerprint applications and reduce the number of required BAC shotgun libraries, the newest BAC library was prepared with inserts larger than 200 kb by use of improved procedures for sizing of genomic fragments (Osoegawa et al. 1998) and more experienced cloning skills. The average insert sizes for the various library segments have been determined using pulsed-field gel electrophoresis, and the results were compatible with sizes later determined by mapping BAC-end sequences to the assembled rat genome. Narrow-insert size distribution permits better scaffolding for assembly of whole-genome shotgun sequence data because of easier distance prediction between paired end sequences. The standard deviations for the EcoRI libraries, RPCI-32 and CHORI-230 segment 1, were about 30 kb, somewhat narrower than for CHORI-230 segment 2, cloned from MboI-partially digested rat DNA (∼50 kb). The RPCI-31 PAC (Woon et al. 1998) and RPCI-32 BAC libraries have been extensively used elsewhere to construct a framework for mapping genes and genetic markers to the rat genome. As of November 19, 2003, 5803 randomly selected BAC and PAC clones derived from the RPCI-32 and RPCI-31 (Woon et al. 1998) libraries have been integrated with the rat RH map and the YAC map (Krzywinski et al. 2004; http://www.molgen.mpg.de/~ratgenome/). The CHORI-230 library has been used as the central resource for mapping and sequencing of the rat genome. The BAC library has been almost entirely (∼190,000 clones) fingerprinted and end-sequenced at the Genome Sciences Centre, Vancouver, British Columbia, and at the Institute for Genomic Research, respectively. The information on fingerprint and end-sequence data for each BAC has been deposited in publicly available databases (http://www.bcgsc.ca/lab/mapping/rat; http://www.tigr.org/tdb/bac_ends/rat/bac_end_intro.html). A subset of 5250 BAC and PAC clones anchored to YAC contigs has also been fingerprinted to allow integration with the BAC physical contig map derived from the CHORI-230 library, and 2165 BAC/PAC clones from this subset were end-sequenced to anchor the clones onto the assembled rat genomic sequence (Krzywinski et al. 2004). As of June 19, 2003, 22,267 BACs derived from CHORI-230 library have been used at Baylor College School of Medicine to prepare shotgun libraries for low-redundancy sequencing toward the rat draft genome sequence assembly (Rat Genome Sequencing Consortium 2004; http://www.hgsc.bcm.tmc.edu/projects/rat/). The CHORI-230 BAC library has been extensively screened using high-density filters with a large number of markers from two ∼21-Mb QTLs at Medical College of Wisconsin, as described above. As a result of intensive use of the RPCI-32 and CHORI-230 libraries, a highly redundant set of BACs has been annotated in databases. It has thus become feasible to identify BACs containing genes of interest through in silico screening of the library, using public databases. Many potential users are familiar with using BLAST-based screening of sequence databases to find clones with corresponding sequences; it is less well known that more redundant sets of overlapping BACs can be identified by inspecting the physical clone maps displayed by the University of California, Santa Cruz, Genome Browser (http://genome.ucsc.edu/) and the British Columbia Genome Science Center Sequence Annotated Fingerprint Map and Clone Layout (http://mkweb.bcgsc.bc.ca/rat/mapview/index.mhtml).
METHODS
Construction and Preparation of BAC Vectors
The pTARBAC1 vector has been derived from the pBACe3.6 vector by inserting yeast elements (CEN6 and His3; Zeng et al. 2001). Three restriction enzyme sites (1× ApaLI and 2× BstXI) were removed from the pTARBAC1 vector by three consecutive crossover polymerase chain reaction (PCR)/cloning procedures. For each of the three restriction sites targeted for removal, a pair of overlapping synthetic “mutant” primers was designed. These primers were used in separate PCR reactions with different distant flanking primers to create PCR products overlapping by about 20 bp. After removal of the primers, a new PCR reaction was assembled using the mixture of two overlapping fragments with the flanking primers. The resulting hybrid PCR fragment was then digested with restriction sites unique in the vector. The mutant restriction fragment was then used to replace the wild-type fragment. The following primers were used: primer 1 (P1): GGTAGTATTTGTTGGCGATC; primer 2 (P2): GAAAGTGCCT CATCCAAAG; primer 3 (P3): TTTGGATGAGGCACTTTCTA GAGCGGTGGTAGATCTT; primer 4 (P4): TTTTGCGCACGGT TATGTGG; primer 5 (P5): CGACAGCGACACACTTGC; primer 6 (P6): GGCGTTTACAAAACCTCAG; primer 7 (P7): CTGAGGTTTT GTAAACGCCGCCTTTATGGGCAGCA; primer 8 (P8): TTTA CAGCCAGTAGTGCTCG; primer 9: (P9): TCTCAGTACAATCT GCTCTG; and primer 10 (P10): CAGAGCAGATTGTACTGA GAATGCACCATAATTCCCG. For instance, to remove the BstXI site at position 9327, P6 was initially used in combination with P5 to create a 2577-bp PCR product. The mutant BstXI oligonucletide (P7) was used for PCR with the flanking primer P8 to create a 2139-bp product. The subsequent hybrid PCR product (4698 bp) was generated using the P5/P8 combination, and thus the PCR fragment was digested with XhoI (near the P8 primer) and HpaI (near P5). The original HpaI/XhoI fragment was removed from the vector by replacement with the BstXI-mutant fragment. The second BstXI site (at 5884) and the ApaLI site (at 5164) were removed using the mutant primers P3 and P10, respectively. For the BstXI removal, two initial PCR reactions were assembled with P1/P2 and P3/P4 primers to generate 1113- and 976-bp products, respectively. For the ApaLI removal, the initial PCR reactions included PCR primer pairs P1/P9 and P10/P4 to create 400- and 1691-bp products. Hybrid PCR products were created for both sites using the same flanking PCR primers (P1/P4), and the 2072-bp mutant hybrid fragments were cloned to replace the original HpaI/PmlI fragment. The final vector created by three subsequent fragment replacement steps was named pTARBAC1.3 and was designed for the cloning of MboI-partially digested fragments. This vector was then further modified to create a vector lacking BamHI sites to be used for cloning EcoRI-partially digested fragments. Double-digestion with BamHI and EcoRI resulted in two large fragments (BamHI/BamHI, 10.6 kb, and EcoRI/EcoRI, 2.8 kb) in addition to small linker fragments. Two fragments were recovered from the gel and ligated together using a synthetic adapter (AATTCCCTATGAA; GATCTTCATA GGG). After transformation of E. coli DH10B cells, a new vector (pTARBAC2.1), lacking the BamHI sites, was generated while regenerating the two EcoRI sites. The sequences of the cloned PCR fragments containing the mutations and linker insertions were confirmed by single-path sequencing and merged into the remaining vector sequences. The GenBank accession numbers for the sequences of the pTARBAC1.3 and pTARBAC2.1 vector are AY487252 and AY487253, respectively. To construct BAC libraries by cloning EcoRI fragments, pTARBAC2.1 was digested with ApaLI and EcoRI and treated with calf intestinal alkaline phosphatase (Roche). To prepare BACs by cloning MboI fragments, the pTARBAC1.3 vector was digested with ApaLI and BamHI and treated with alkaline phosphatase. The vector fragments were separated in 0.7% agarose gel in 0.5× TBE buffer and recovered by electroelution. The eluted DNA was dialyzed and concentrated using Centricon-100 (Millipore). Approximately 30 ng vector DNA was used in 50 μL ligation reaction with 50-70 ng insert DNA.
Rat Strain and DNA Isolation
Detailed procedures for preparation of high-molecular weight DNA have been described (Osoegawa et al. 1999). The RPCI-32 library was prepared from the spleen of a male Brown Norway (BN/SsNHsd) rat (Harlan Sprague Dawley). The CHORI-230 library was derived from the brain and liver of a 3-month-old female Brown Norway rat (BN/SsNHsd/MCW). Extensive inbreeding of the Brown Norway (BN/SsNHsd) rat was performed at the Medical College of Wisconsin. The rat used for construction of the CHORI-230 library belonged to the fourteenth generation after the start of local inbreeding. The BAC library was derived from the daughter of the female rat (thirteenth generation) used for WGS at Baylor College of Medicine. The animal used for WGS at Celera Genomics was a male from the tenth generation of inbreeding, but the closest common ancestor was a female of the fourth generation.
Construction of BAC Libraries
Agarose-embedded high-molecular-weight DNA was partially digested with a combination of EcoRI and EcoRI Methylase for RPCI-32 and CHORI-230 segment 1, and with MboI for CHORI-230 segment 2. The partially digested DNA was separated with a CHEF apparatus (BioRad) using the improved method (Osoegawa et al. 1998). The size-fractionated DNA was eluted and ligated to the EcoRI sites of the pBACe3.6 (Frengen et al. 1999) for RPCI-32, pTARBAC2.1 for CHORI-230 segment 1 vector, or to the BamHI sites of the pTARAC1.3 vector for CHORI-230 segment 2 (Table 1). The ligation products were transformed into E. coli DH10B cells (Invitrogen) for RPCI-32 and CHORI-230 segment 1 and E. coli DH10B T1-phage resistant cells (Invitrogen) for CHORI-230 segment 2.
Insert Size Analysis
One hundred sixty clones from each segment of the RPCI-32 library, 192 from the CHORI-230 segment 1, and 384 from the CHORI-230 segment 2 were randomly size analyzed after digestion with NotI (New England Biolabs), as described (Osoegawa et al. 1998).
Hybridization and Fingerprinting
Detailed procedures for hybridization with overgos and BAC DNA fingerprinting have been described (McPherson et al. 2001; Marra et al. 1997; Osoegawa et al. 2000). Twenty-two overgo probes of 40 bp each were designed as described (McPherson et al. 2001) for aligned conserved sequences from the HomoloGene database (Zhang et al. 2000; http://www.ncbi.nlm.nih.gov/HomoloGene/). The probes were hybridized to high-density colony filters, and the digitized hybridization images were analyzed with ArrayVision Ver6.0 (Imaging Research, Inc.). Positive clones were rearrayed into 384-well dishes and then used to prepare small colony filters for hybridization with individual probes. All candidate positive BACs were fingerprinted in parallel using HindIII, on agarose gels, stained with SYBR green, and scanned with a Fluorimager 595 (Amersham). The gel images were analyzed with Image software 1.3, and contigs were assembled using FPC software with conditions tolerance: 4, and cutoff: 1e-11. Individual probe screening results were compared with fingerprinting results. In addition, 500 overgo probes from the chromosomal regions for two renal-failure QTLs (Rf-1 and Rf-4; Brown et al., 1996; Shiozawa et al., 2000) were designed from genes, expressed sequence tags, genetic markers, and human-mouse conserved noncoding sequences as part of a large physical mapping and positional cloning project in Milwaukee. Probes were designed, labeled, and hybridized as described above, but only to the CHORI-230 segment 1 (480 probes) and segment 2 (20 probes), and using Church's hybridization solution I. Filters were washed 3× at 61°C with 1× SSC, and 0.1% SDS, and were exposed to autoradiograph film overnight. Film was scored manually using the overlay grid supplied with the filters.
Acknowledgments
We thank Joseph J. Catanese, Melanie Hierl, Aaron Mammoser, Beth A. Palka, Barbara Swiatkiewcz, and Baohui Zhao for their technical advice and assistance, and April Chan and Dr. Yuko Yoshinaga for critically reading the manuscript. The authors thank Drs. Marcelo A. Nobrega and Howard J. Jacob for their assistance in isolating rat genomic DNA for construction of CHORI-230 library. Construction of the RPCI-32 library was supported by a subcontract from the Whitehead Institute for Biomedical Research. The funding was from a National Institutes of Health grant (5R01HL55726-03) awarded to Lander (P.I.). Construction and characterization of the CHORI-230 library was supported by a grant from NIH (HG01165-06) to P.J.d.J. Hybridization of the QTL probes was supported by a grant from NIH (1R01HL69321-01) to H.J.J. K.O. and B.Z. contributed equally to this work.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2033904.
References
- Brown, D.M., Provoost, A.P., Daly, M.J., Lander, E.S., and Jacob, H.J. 1996. Renal disease susceptibility and hypertension are under independent genetic control in the fawn-hooded rat. Nature Genet. 12: 44-51. [DOI] [PubMed] [Google Scholar]
- Dukhanina, O.I., Dene, H., Deng, A.Y., Choi, C.R., Hoebee, B., and Rapp, J.P. 1997. Linkage map and congenic strains to localize blood pressure QTL on rat chromosome 10. Mamm. Genome 8: 229-235. [DOI] [PubMed] [Google Scholar]
- Frengen, E., Weichenhan, D., Zhao, B., Osoegawa, K., van Geel, M., and de Jong, P.J. 1999. A modular positive selection bacterial artificial chromosome vector with multiple cloning sites. Genomics 58: 250-253. [DOI] [PubMed] [Google Scholar]
- Fuhrmann, D.R., Krzywinski, M.I., Chiu, R., Saeedi, P., Schein, J.E., Bosdet, I.E., Chinwalla, A., Hillier, L.W., Waterston, R.H., McPherson, J.D., et al. 2003. Software for automated analysis of DNA fingerprinting gels. Genome Res. 13: 940-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett, M.R., Dene, H., Walder, R., Zhang, Q.Y., Cicila, G.T., Assadnia, S., Deng, A.Y., and Rapp, J.P. 1998. Genome scan and congenic strains for blood pressure QTL using Dahl salt-sensitive rats. Genome Res. 8: 711-723. [DOI] [PubMed] [Google Scholar]
- Jacob, H.J. and Kwitek, A.E. 2001. Rat genetics: attaching physiology and pharmacology to the genome. Nature Rev. Genet. 3: 33-42. [DOI] [PubMed] [Google Scholar]
- Krzywinski, M., Wallis, J., Gösele, C., Bosdet, I., Chiu, R., Graves, T., Hummel, O., Layman, D., Mathewson, C., Wye, N., et al. 2004. Integrated and sequence-ordered BAC and YAC-based physical maps for the rat genome. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Larionov, V., Kouprina, N., Graves, J., Chen, X.-N., Korenberg, J.R., and Resnick, M.A. 1996. Specific cloning of human DNA as YACs by transformation-associated recombination. Proc. Natl. Acad. Sci. 93: 491-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra, M.A., Kucaba, T.A., Dietrich, N.L., Green E.D., Brownstein, B., Wilson, R.K., McDonald, K.M., Hillier, L.W., McPherson, J.D., and Waterston, R.H. 1997. High throughput fingerprint analysis of large-insert clones. Genome Res. 7: 1072-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson, J.D., Marra, M., Hillier, L., Waterston, R.H., Chinwalla, A., Wallis, J., Sekhon, M., Wylie, K., Mardis, E.R., Wilson, R.K., et al. 2001. A physical map of the human genome. Nature 409: 934-941. [DOI] [PubMed] [Google Scholar]
- Osoegawa, K., Woon, P.-Y., Zhao, B., Frengen, E., Tateno, M., Catanese, J.J., and de Jong, P.J. 1998. An improved approach for construction of bacterial artificial chromosome libraries. Genomics 52: 1-8. [DOI] [PubMed] [Google Scholar]
- Osoegawa, K., de Jong, P.J., Frengen, E., and Ioannou, P.A. 1999. In Current protocols in human genetics (eds. N.C. Dracopoli, et al.), pp. 5.15.1-5.15.33. Wiley, New York. [DOI] [PubMed]
- Osoegawa, K., Tateno, M., Woon, P.-Y., Frengen, E., Mammoser, A.G., Catanese, J.J., Hayashizaki, Y., and de Jong, P.J. 2000. Bacterial artificial chromosome libraries for mouse sequencing and functional analysis. Genome Res. 10: 116-128. [PMC free article] [PubMed] [Google Scholar]
- Osoegawa, K., Mammoser, A.G., Wu, C., Frengen, E., Zeng, C., Catanese, J.J., and de Jong, P.J. 2001. A bacterial artificial chromosome library for sequencing the complete human genome. Genome Res. 11: 483-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rat Genome Sequencing Project Consortium. 2004. Genome sequence of the Brown Norway Rat yields insights into mammalian evolution. Nature (in press). [DOI] [PubMed]
- Shiozawa, M., Provoost, A.P., van Dokkum, R.P.E., Majewski, R.R., and Jacob, H.J. 2000. Evidence of gene-gene interactions in the genetic susceptibility to renal impairment after unilateral nephrectomy. J. Am. Soc. Nephrol. 11: 2068-2078. [DOI] [PubMed] [Google Scholar]
- Steen, R.G., Kwitek-Black, A.E., Glenn, C., Gullings-Handley, J., Van Etten, W., Atkinson, O.S., Appel, D., Twigger, S., Muir, M., Mull, T., et al. 1999. A high-density integrated genetic linkage and radiation hybrid map of the laboratory rat. Genome Res. 9: AP1-AP8. [PubMed] [Google Scholar]
- Takahashi, R., Ito, K., Fujiwara, Y., Kodaira, K., Kodaira, K., Hirabayashi, M., and Ueda, M. 2000. Generation of transgenic rats with YACs and BACs: preparation procedures and integrity of microinjected DNA. Exp. Anim. 49: 229-233. [DOI] [PubMed] [Google Scholar]
- Venter, J.C., Adams, M.C., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304-1351. [DOI] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- Woon, P.-Y., Osoegawa, K., Kaisaki, P.J., Zhao, B., Catanese, J.J., Gauguier, D., Cox, R., Levy, E.R., Lathrop, G.M., Monaco, A.P., et al. 1998. Construction and characterization of a 10-fold genome equivalent rat P1-derived artificial chromosomal library. Genomics 50: 306-316. [DOI] [PubMed] [Google Scholar]
- Zeng, C., Kouprina, N., Zhu, B., Cairo, A., Hoek, M., Cross, G., Osoegawa, K., Larinov, V., and de Jong, P.J. 2001. Large-insert BAC/YAC libraries for selective re-isolation of genomic regions by homologous recombination in yeast. Genomics 77: 27-34. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Schwartz S., Wagner L., and Miller W. 2000. A greedy algorithm for aligning DNA sequences. J. Comp. Biol. 7: 203-214. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://bacpac.chori.org/; BACPAC resources home page.
- http://www.bcgsc.ca/lab/mapping/rat; British Columbia Genome Science Center rat mapping page.
- http://www.tigr.org/tdb/bac_ends/rat/bac_end_intro.html; Institute for Genomic Research rat BAC ends page.
- http://www.hgsc.bcm.tmc.edu/projects/rat/; Human Genome Sequencing Center at Baylor College of Medicine Rat Genome Project page.
- http://www.molgen.mpg.de/~ratgenome/; Max Planck Institute for Molecular Genetics Rat Genomic Resources page.
- http://www.ncbi.nlm.nih.gov/HomoloGene/; National Center for Biotechnology Information HomoloGene page.
- http://genome.ucsc.edu/; University of California, Santa Cruz, Genome Bioinformatics home page.
- http://mkweb.bcgsc.bc.ca/rat/mapview/index.mhtml; British Columbia Genome Science Center Sequence Annotated Fingerprint Map and Clone Layout.