Abstract
In an effort to test whether a transition state analog is an inhibitor of the metallo-β-lactamases, a phospholactam analog of carbapenem has been synthesized and characterized. The phospholactam 1 proved to be a weak, time-dependent inhibitor of IMP-1 (70%), CcrA (70%), L1 (70%), NDM-1 (53%), and Bla2 (94%) at an inhibitor concentration of 100 µM. The phospholactam 1 activated ImiS and BcII at the same concentration. Docking studies were used to explain binding and to offer suggestions for modifications to the phospholactam scaffold to improve binding affinities.
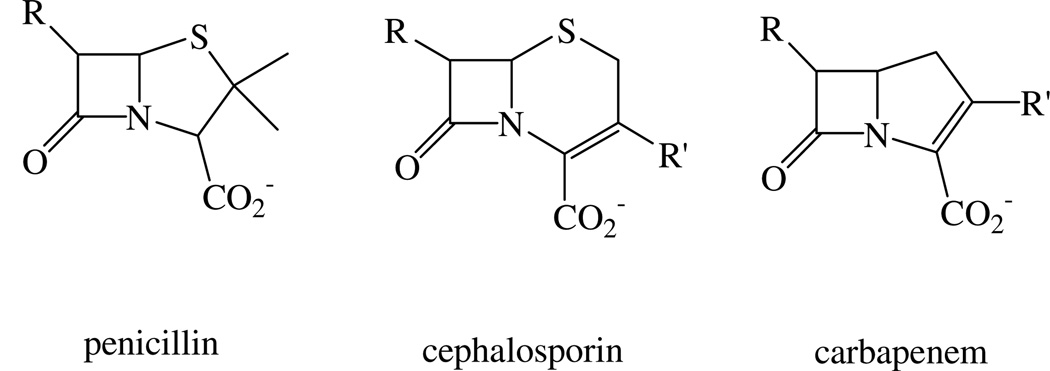
Since the antibiotic properties of penicillin were first discovered in the beginning of the last century, antibiotics have been well developed as miracle drugs in the treatment of bacterial infections in clinics.1, 2 However, overuse of antibiotics has resulted in a large number of bacteria that produce β-lactamases, and the resulting bacteria are resistant to many commonly-used β-lactam antibiotics.3–5 β-Lactamases catalyze the hydrolysis of the β-lactam ring of these antibiotics. They are divided into classes A, B, C, and D.6 The B class enzymes, called metallo-β-lactamases (MβLs), are Zn(II)-dependent and hydrolyze nearly all known β-lactam-containing antibiotics, including penicillins, cephalosporins and carbapenems (Figure 1). There are no known clinical inhibitors of the MβLs.4 The MβLs are further divided into B1, B2, and B3 subclasses.6 Both B1 and B3 subclasses have a broad-spectrum substrate profile including penicillins, cephalosphorins and carbapenems. In contrast, the B2 subclass enzymes primarily hydrolyze carbapenems.
Figure 1.
Structures of β-lactam-containing antibiotics.
Given the enormous biomedical importance of MβLs, there has been a large amount of effort in identifying novel inhibitors of these enzymes.4, 7 Succinic acid derivatives were shown to be inhibitors of IMP-1 with IC50 values in the nanomolar range.8 Lienard et al. reported that D-captopril inhibits L1 and CcrA,9 and thiomandelic acid was tested as an inhibitor for nine different MβLs.10 Picolinic acid derivatives were shown to inhibit CphA with Ki values in the micromolar range,11 while hydroxamic acid derivatives inhibit Fez-1 but not L1, IMP-1, BcII, or CphA.12 A mechanism-based inhibitor of IMP-1 was reported that exhibited irreversible inhibition.13 Penicillin-based inhibitors have been shown to be micromolar inhibitors of L1, BcII, and Bla2,14 and some N-arylsulfonyl hydrazones are inhibitors of IMP-1.15 A series of pyrrole-based inhibitors of IMP-1 have recently been reported.16 However, most of the inhibition reports have involved studies on one or two of the MβLs, and to the best of our knowledge, only three classes of inhibitors, thiomandelic acid,10 thiols,9, 17–19 and mercaptophosphonate compounds,20 have been reported to be broad-spectrum inhibitors of the MβLs. Our goal is to develop broad-spectrum, transition state analog inhibitors of MβLs and to use these inhibitors as drug/inhibitor combinations to combat bacterial infections in which the bacteria produce a MβL.
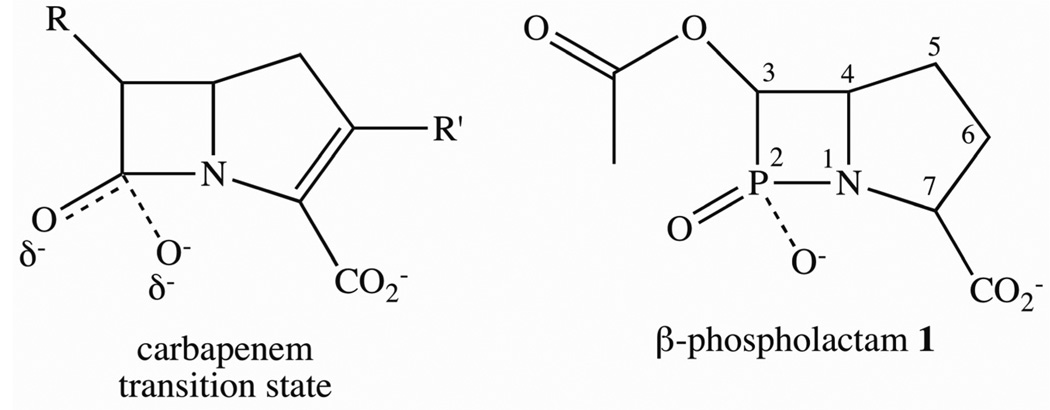
Previous mechanistic studies have suggested a ring-opened intermediate, whose breakdown is rate-limiting, for L1, CcrA, and NDM-1 when using nitrocefin or chromacef as substrate.21–23 When using other MβLs or substrates, nucleophilic attack or β-lactam ring cleavage appears to be rate-limiting.24–26 Previous studies on peptidases, many of which exhibit rate-limiting bond cleavage, suggested that a tetrahedral transition state forms during the reaction.27–29 The fact that several phosphinate, phosphonate, and phosphoramidate peptide analogs proved to be very tight binding inhibitors (one with a reported Ki of 10−15 M) strongly supported the existence of the tetrahedral transition state in these peptidases.30–37 Since β-lactam-containing antibiotics are peptide mimics and since β-lactamases catalyze peptide bond cleavage, we hypothesize that a tetrahedral transition state may form during the reaction (Figure 2) and that a chemically-stable β-phospholactam may be a very tight binding inhibitor of the MβLs.
Figure 2.
Structures of a hypothetical carbapenem transition state and β-phospholactam 1 synthesized in this study.
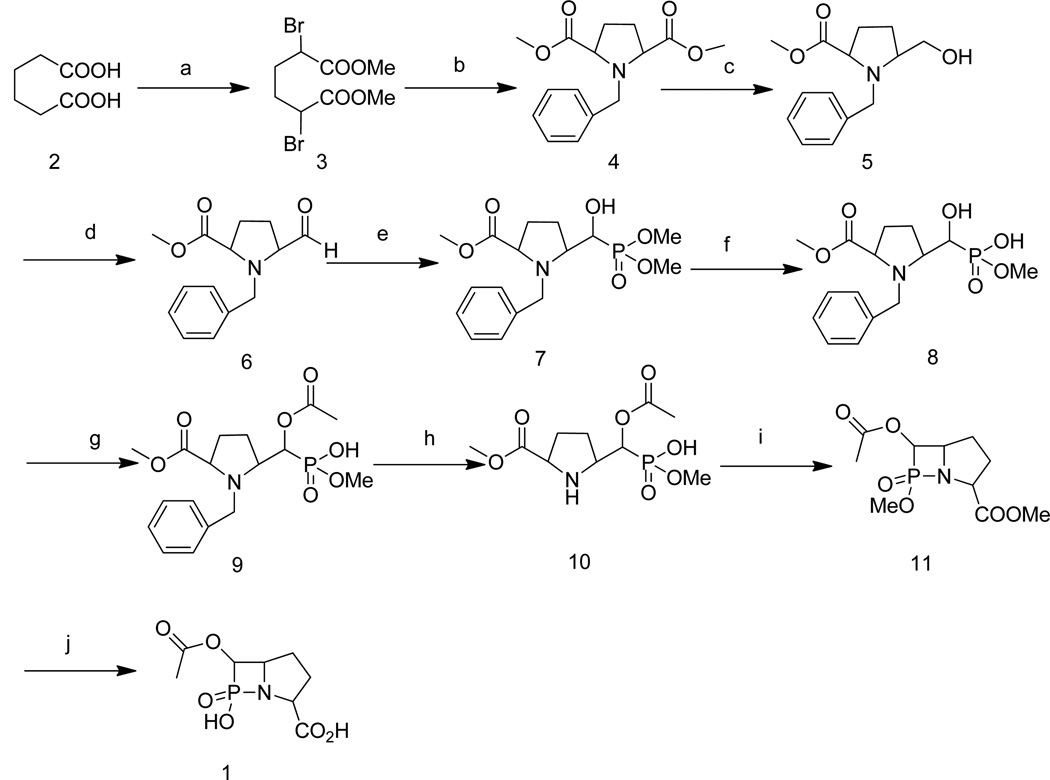
Toward this goal, a β-phospholactam analog of a carbapenem transition state (Figure 2) was synthesized by using a 12-step protocol, and the resulting compound 1 was characterized by NMR and MS. The inhibitory activities of the β-phospholactam 1 were evaluated using MβLs from the three subclasses B1 (IMP-1, CcrA, Bla2, NDM-1), B2 (ImiS), and B3 (L1). In the absence of experimental structures of MβL-ligand complexes, docking studies can provide insights into possible binding modes of inhibitors38 and β-lactam substrates.39, 40 These studies have shown that substituents with high electron density, such as, thiols, carboxylates, and carbonyl groups interact electrostatically with the zinc ions and the positively-charged Lys224 conserved in many B1 and B2 MβLs.41, 42 Here we assessed the binding mode of β-phospholactam 1 to the different MβLs using docking. Previously, non-cyclic phosphinates43 and monocyclic β-phospholactams44 were tested as inhibitors of MβLs; however, none of these compounds inhibited the tested enzymes, most likely due to the fact that these compounds did not have the correct structure to be recognized by the MβLs. Page and coworkers reported a number of studies using β-sultams and β-phospholactams as inhibitors of a serine β-lactamase, and these studies, along with a theoretical study,45 explored the stability of these compounds at different pH’s.44, 46–48 None of these compounds tested were bicyclic. Rees and coworkers reported the synthesis of a 1,2-azaphosphetidine, which is a bicyclic β-phospholactam;49 however, the compound was not tested as an inhibitor of any of the MβLs. Our efforts to synthesize this compound were unsuccessful using the published procedure; therefore, we developed a novel synthetic approach to obtain a bicyclic β-phospholactam 1 (Scheme 1). Dimethyl 2,5-dibromoadipate 3 as white solid was prepared by acylation, α-bromination, and esterification of adipic acid, using previously reported methods.50 Adipate 3 was reacted with benzylamine in alkaline medium, and the resulting product was treated by acidification and alkalization to afford pure pyrrole dicarboxylate 4. Compound 4 has a highly symmetric structure, which required harsh reduction conditions to obtain the pyrrole monocarboxylate 5. After attempting various reaction conditions, sodium borohydride was identified as the optimum reductant, and ethanol was identified as the optimum solvent. Alcohol 5 was converted to aldehyde 6 by Swern oxidation, and this product was used in the next step without further purification. Intermediate 6 was reacted with dimethyl phosphate in the presence of catalyst 1,8-diazabicyclo[5.4.0]undec-7-ene (DBU) at 0 °C to afford dimethyl phosphonate 7. Dimethyl phosphonate was converted to monomethyl phosphonate 8 by treatment of 7 with NaI in acetone. The α-hydroxy was protected by treatment of 8 in pyridine with acetic anhydride in the presence of 4-dimethylaminopyridine (DMAP) to afford 9. The removal of the benzyl protecting group was very challenging. A series of bases, including triethylamine, diisopropylethylamine, potassium acetate, sodium hydroxide, and NaH, and solvents were tested in an effort to remove the benzyl group. Fortunately, intermediate 9 was hydrogenated with 10% Pd/C in the presence of HCO2NH4 to afford 10. The ring-closure step to yield 11 was carried out using NaH. Finally, the target compound β-phospholactam 1 was obtained by hydrolysis of 11 with LiOH to remove the methyl protecting groups. The overall yield was 0.68%. The product and intermediates were characterized and confirmed by NMR and MS.
Scheme 1.
Synthetic route of β-phospholactam 1: a: (1) acetyl chloride, (2) Br2, (3) MeOH; b: benzylamine, K2CO3, CH3CN; c: NaBH4, EtOH; d: oxalyl chloride, DMSO, dichloromethane, −78 °C; e: dimethyl phosphonate, 1,8-diazabicyclo[5.4.0]undec-7-ene (DBU), anhydrous THF; f: NaI, acetone; g: acetic anhydride, pyridine; h: Pd-C, H2; i: NaH, 15-crown-5, dichloromethane; j: LiOH, H2O, MeOH.
The inhibitory activity of β-phospholactam 1 on MβLs was evaluated, and percent inhibition values are listed in Table 1. Without pre-incubation with enzymes, the β-phospholactam 1 activated CcrA and NDM-1 and showed 48, 22, and 37% inhibition against IMP-1, Bla2, and L1, respectively. After incubation with enzymes for 30 min, β-phospholactam 1 exhibited a 70% inhibition against IMP-1, CcrA, and L1, a 53% inhibition for NDM-1, and a 94% inhibition of Bla2, suggesting that β-phospholactam 1 is a time-dependent inhibitor. At this point, it is unclear whether the time-dependent inhibition is caused by intact β-phospholactam 1, a covalently-modified enzyme-inhibitor complex, or by a hydrolyzed product. Previous studies with a β-phospholactam and class C β-lactamase P99 revealed time-dependent inhibition was caused by a phosphonylated enzyme.46 Nonetheless, stability studies revealed a relatively fast hydrolysis of monocyclic β-phospholactam in water.44, 46 We believe that the different trend exhibited by the enzymes with and without pre-incubation is due to the enzymes behaving differently in DMSO, which is used as a solvent for the inhibitor. Unexpectedly, the inhibitor activates ImiS and BcII. The mechanism of this activation is unknown, but it could be due to an allosteric effect as a result of phosphonylation of solvent-exposed hydroxyl groups by β-phospholactam 1.
Table 1.
Percent inhibition of MβLs by β-phospholactam 1
| Enzyme | % inhibition |
|---|---|
| L1 | 70 ± 7 |
| IMP-1 | 70 ± 6 |
| CcrA | 69 ± 8 |
| NDM-1 | 53 ± 6 |
| Bla2 | 94 ± 4 |
The concentration of inhibitor was 100 µM.
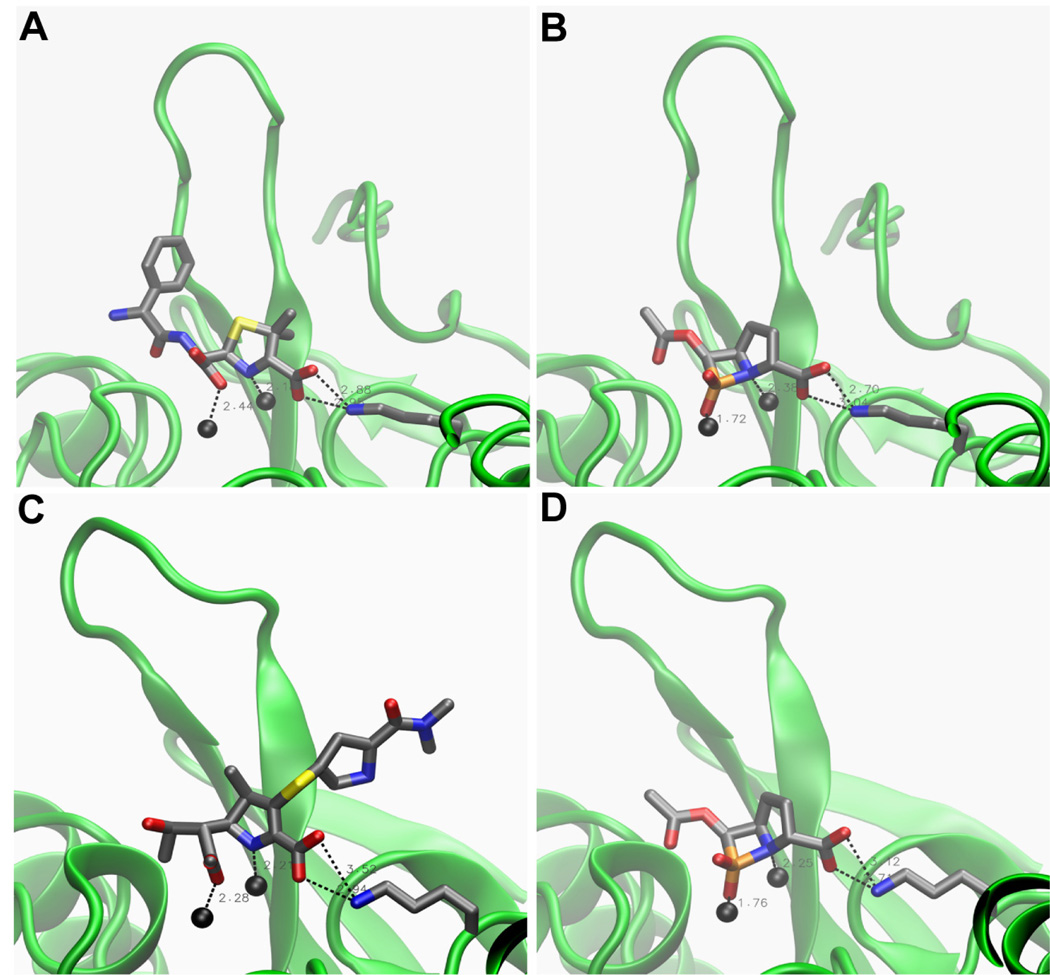
In docking calculations (computational details can be found in the Supporting Information) the majority of conformations (between 42 and 50 out of 50 per complex) were found in clusters that correspond to the expected binding mode, which is the way that the hypothetical carbapenem transition state would bind. Since no crystal structure of any MβL-carbapenem transition state complex is available, the lowest-energy conformations docked to NDM-1 and L1 are compared to enzyme-hydrolyzed β-lactam complexes in Figure 3 and Table 2. In all complexes, one phosphinate oxygen, which corresponds to the oxygen derived from the β-lactam carbonyl, coordinates Zn1, while the β-lactam nitrogen coordinates Zn2. In some complexes (IMP-1 and CcrA), the phosphinate oxygen additionally coordinates Zn2 (Figure S1), which may be due to the short Zn1-Zn2 distance of 3.6 Å and 3.5 Å, respectively, in these crystal structures. The carboxylate attached to C7 of the five-membered ring forms a salt bridge with the Lys224 side chain, as is seen in the enzyme-hydrolyzed β-lactam complexes, except in L1, which does not contain Lys224. Interestingly, in the L1-β-phospholactam 1 complex, which is compared to an L1-hydrolyzed moxalactam complex51 in Table 2 and Figure S2, one phosphinate oxygen occupies the site occupied by the carboxylate oxygen coordinating Zn1, while the other oxygen occupies the site of a water molecule that was localized between the two zinc ions, which are also very close (3.7 Å) in the crystal structure. As seen in Figure 3, the conformations of the β-phospholactam 1 docked to NDM-1 structures correspond very well to the co-crystallized hydrolyzed ampicillin and meropenem,52 consistent with the similarity of the inhibitor with both the penicillin and carbapenem core. The more substrate-like binding mode in NDM-1 (Figure 3), where the Zn1-Zn2 distance is increased to 4.6 (PDB entry 4HL2) and 4.1 Å (PDB entry 4EYL), consistent with previous findings that the Zn1-Zn2 distance varies during the catalytic cycle53, 54 as opposed to the binding mode with phosphinate oxygens coordinating both zinc ions when they are closer (Figures S1 and S2) could account for the slightly lower inhibition of NDM-1 versus that of IMP-1, CcrA, and L1 (Table 1). Based on the docking calculations, we cannot exclude the possibility that the inhibiting species is hydrolyzed β-phospholactam. When docking this product to NDM-1 (PDB entry 4EYL), the resulting phosphonate coordinated both zinc ions, but the carboxylate at C7 did not form a salt bridge with Lys244 (data not shown). The most favorable binding energy was −12.4 kcal/mol, not significantly lower than −11.2 kcal/mol observed for the intact β-phospholactam 1.
Figure 3.
Complexes of NDM-1 co-crystalized with hydrolyzed β-lactams (A and C) and with the lowest-energy docked β-phospholactam 1 conformation (B and D). The coordinates of NDM-1 in panels A and B as well as hydrolyzed ampicillin in panel A are taken from PDB entry 4HL2. The coordinates of NDM-1 in panels C and D as well as hydrolyzed meropenem in panel C are taken from PDB entry 4EYL. The images were generated with VMD.55 The protein backbone is shown as a green cartoon and zinc ions as black spheres (Zn1 on the left and Zn2 on the right). The β hairpin loop in the back is loop 3; loop 10 was removed for clarity. The hydrolyzed substrates as well as the β-phospholactam 1 inhibitor and the Lys224 side chain are shown as sticks colored by atom (C, gray; O, red, N, blue; S, yellow; P, orange). Key distances summarized in Table 2 are indicated by dashed lines.
Table 2.
Summary of geometries of lowest-energy docked MβL-β-phospholactam 1 complexes (bold) in comparison to available MβL-hydrolyzed β-lactam complexes (italic).
| Subclass | Complex | Binding Energy (kcal/mol) |
O(P2)-Zn1 O(C2)-Zn1 (Å)e |
O(P2)-Zn2 O(C2)-Zn2 (Å)e |
N1-Zn2 (Å) |
Carboxylate(C7) -Lys244 Nζ (Å)f |
|---|---|---|---|---|---|---|
| 1 | IMP-1(1DD6)-βPLa | −10.8 | 1.9 | 1.8 | 3.1 | 2.9 |
| 1 | CcrA(1A7T)-βPL | −11.9 | 1.8 | 1.9 | 3.1 | 2.7 |
| 1 | NDM-1(4HL2)-βPL | −12.6 | 1.7/2.6 | - | 2.4 | 2.9 |
| 1 | NDM-(4HL2)-hAMPb | 2.4 | - | 2.2 | 2.9 | |
| 1 | NDM-1(4EYL)-βPL | −11.2 | 1.8/2.6 | 2.7 | 2.3 | 2.9 |
| 1 | NDM-1(4EYL)-hMERc | 2.3/2.7 | 2.5 | 2.2 | 3.2 | |
| 3 | L1(2AIO)_βPL | −11.8 | 2.0/2.1 | 1.9 | 2.8 | - |
| 3 | L1(2AIO)-HOH600 | 2.0 | 2.2 | - | - | |
| 3 | L1(2AIO)-hMOXd | 2.4 | - | 2.4 | - | |
βPL = β-phospholactam 1
hAMP = hydrolyzed ampicillin
hMER = hydrolyzed meropenem
hMOX = hydrolyzed moxalactam
O(P2) and O(C2) designate the oxygen atoms bound to P2 in the β-phospholactam 1 and C2 in the β-lactams. Only atoms that are within a distance of 3.2 Å are reported. If both are within that distance, both are reported starting with the closer one.
Average distance between the two carboxylate oxygens and Lys244 Nζ.
These structures also suggest how the binding affinity of this inhibitor scaffold could be improved. Currently, it only occupies the area close to the zinc ions. However, the active site of MβLs is rather a cleft and the inhibitor could be designed to better fill out that cleft by extending the substituent at C3 to something similar to R in penicillins (Figure 3A) and adding a substituent to C7 that resembles R’ of carbapenems (Figure 3C).
We have developed a prototype β-phospholactam analog of a carbapenem transition state, which exhibits broad-spectrum and time-dependent inhibitory activity against MβLs, with an inhibition percentage of 70% for IMP-1, CcrA, and L1, 53% for NDM-1, and 94% for Bla2. This study opened a way to develop β-phospholactam compounds as broad-spectrum inhibitors of MβLs.
Supplementary Material
Acknowledgements
This work was supported by grants (21272186 to K.W.Y) from the National Natural Science Fund of China and from the National Institutes of Health (GM093987 to M.W.C.) and National Science Foundation (CHE1151658 to M.W.C.). The Center for Macromolecular Modeling and Materials Design (CM3D) at California State Polytechnic University, Pomona, where docking calculations were conducted, is supported by the W. M. Keck Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mitcher LA, Lemke TL, Gentry EJ. In: Foye's Principles of Medicinal Chemistry. 6th ed. Lemke TL, Williams DA, Roche VF, Zito SW, editors. Wolters Kluwer Lippincott Williams & Wilkins; 2007. [Google Scholar]
- 2.Bush K. Curr. Opin. Pharmacol. 2012;12:527. doi: 10.1016/j.coph.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 3.Bush K, Macielag MJ. Expert Opin. Thera. Patents. 2010;20:1277. doi: 10.1517/13543776.2010.515588. [DOI] [PubMed] [Google Scholar]
- 4.Drawz SM, Bonomo RA. Clin. Microbiol. Rev. 2010;23:160. doi: 10.1128/CMR.00037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Papp-Wallace KM, Endimiani A, Taracila MA, Bonomo RA. Antimicro. Agents Chemo. 2011;55:4943. doi: 10.1128/AAC.00296-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bush K, Jacoby GA. Antimicro. Agents Chemo. 2010;54:969. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fast W, Sutton LD. Biochim. Biophys. Acta. 2013;1834:1648. doi: 10.1016/j.bbapap.2013.04.024. [DOI] [PubMed] [Google Scholar]
- 8.Toney JH, Hammond GG, Fitzgerald PM, Sharma N, Balkovec JM, Rouen GP, Olson SH, Hammond ML, Greenlee ML, Gao YD. J. Biol. Chem. 2001;276:31913. doi: 10.1074/jbc.M104742200. [DOI] [PubMed] [Google Scholar]
- 9.Lienard BMR, Garau G, Horsfall L, Karsisiotis AI, Damblon C, Lassaux P, Papamicael C, Roberts GCK, Galleni M, Dideberg O, Frere JM, Schofield CJ. Org. Biomol. Chem. 2008;6:2282. doi: 10.1039/b802311e. [DOI] [PubMed] [Google Scholar]
- 10.Mollard C, Moali C, Papamicael C, Damblon C, Vessilier S, Amicosante G, Schofield CJ, Galleni M, Frere JM, Roberts GCK. J. Biol. Chem. 2001;276:45015. doi: 10.1074/jbc.M107054200. [DOI] [PubMed] [Google Scholar]
- 11.Horsfall LE, Garau G, Lienard BM, Dideberg O, Schofield CJ, Frere JM, Galleni M. Antimicro. Agents Chemo. 2007;51:2136. doi: 10.1128/AAC.00866-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lienard BM, Horsfall LE, Galleni M, Frere JM, Schofield CJ. Bioorg. Med. Chem. Lett. 2007;17:964. doi: 10.1016/j.bmcl.2006.11.053. [DOI] [PubMed] [Google Scholar]
- 13.Hiromasa Kurosaki YY, Toshihiro Higashi, Kimitaka Soga, Satoshi Matsueda, Haruka Yumoto, Shogo Misumi, Yuriko Yamagata, Yoshichika Arakawa, Masafumi Goto. Angew.e Chem. Intern. Ed. 2005;44:3861. [Google Scholar]
- 14.Buynak JD, Chen H, Vogeti L, Gadhachanda VR, Buchanan CA, Palzkill T, Shaw RW, Spencer J, Walsh TR. Bioorg. Med. Chem. Lett. 2004;14:1299. doi: 10.1016/j.bmcl.2003.12.037. [DOI] [PubMed] [Google Scholar]
- 15.Toney JH, Moloughney JG. Curr. Opin. Invest. Drugs. 2004;5:823. [PubMed] [Google Scholar]
- 16.Mohamed MS, Hussein WM, McGeary RP, Vella P, Schenk G, Abd El-Hameed RH. Eur. J. Med. Chem. 2011;46:6075. doi: 10.1016/j.ejmech.2011.10.030. [DOI] [PubMed] [Google Scholar]
- 17.Bounaga S, Galleni M, Laws AP, Page MI. Bioorg. Med. Chem. 2001;9:503. doi: 10.1016/s0968-0896(00)00257-1. [DOI] [PubMed] [Google Scholar]
- 18.Bounaga S, Laws AP, Galleni M, Page MI. Biochem. J. 1998;331:703. doi: 10.1042/bj3310703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang KW. Arch. Biochem. Biophys. 1999;368:1. doi: 10.1006/abbi.1999.1293. [DOI] [PubMed] [Google Scholar]
- 20.Lassaux P, Hamel M, Gulea M, Delbruck H, Mercuri PS, Horsfall L, Dehareng D, Kupper M, Frere JM, Hoffmann K, Galleni M, Bebrone C. J. Med. Chem. 2010;53:4862. doi: 10.1021/jm100213c. [DOI] [PubMed] [Google Scholar]
- 21.McManus-Munoz S, Crowder MW. Biochemistry. 1999;38:1547. doi: 10.1021/bi9826512. [DOI] [PubMed] [Google Scholar]
- 22.Wang Z, Benkovic SJ. J. Biol. Chem. 1998;273:22402. doi: 10.1074/jbc.273.35.22402. [DOI] [PubMed] [Google Scholar]
- 23.Yang H, Aitha M, Hetrick AM, Richmond TK, Tierney DL, Crowder MW. Biochemistry. 2012;51:3839. doi: 10.1021/bi300056y. [DOI] [PubMed] [Google Scholar]
- 24.Griffin DH, Richmond TK, Sanchez C, Moller AJ, Breece RM, Tierney DL, Bennett B, Crowder MW. Biochemistry. 2011;50:9125. doi: 10.1021/bi200839h. [DOI] [PubMed] [Google Scholar]
- 25.Sharma NP, Hajdin C, Chandrasekar S, Bennett B, Yang KW, Crowder MW. Biochemistry. 2006;45:10729. doi: 10.1021/bi060893t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spencer J, Clark AR, Walsh TR. J. Biol. Chem. 2001;276:33638. doi: 10.1074/jbc.M105550200. [DOI] [PubMed] [Google Scholar]
- 27.Grembecka J, Mucha A, Cierpicki T, Kafarski P. J. Med. Chem. 2003;46:2641. doi: 10.1021/jm030795v. [DOI] [PubMed] [Google Scholar]
- 28.Bradshaw RA. Encyclopedia Biol. Chemistry. 2004;1:96. [Google Scholar]
- 29.Copik AJ, Waterson S, Swierczek SI, Bennett B, Holz RC. Inorg. Chem. 2005;44:1160. doi: 10.1021/ic0487934. [DOI] [PubMed] [Google Scholar]
- 30.Wu Z, Walsh CT. Proc. Nat. Acad. Sci. 1995;92:11603. doi: 10.1073/pnas.92.25.11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu Z, Wright GD, Walsh CT. Biochemistry. 1995;34:2455. doi: 10.1021/bi00008a008. [DOI] [PubMed] [Google Scholar]
- 32.Jacobsen NE, Bartlett PA. J. Am. Chem. Soc. 1981;103:654. [Google Scholar]
- 33.Bartlett PA, Marlowe CK. Biochemistry. 1983;22:4618. doi: 10.1021/bi00289a002. [DOI] [PubMed] [Google Scholar]
- 34.Bartlett PA, Kezer WB. J. Am. Chem. Soc. 1984;106:4282. [Google Scholar]
- 35.Hanson JE, Kaplan AP, Bartlett PA. Biochemistry. 1989;28:6294. doi: 10.1021/bi00441a022. [DOI] [PubMed] [Google Scholar]
- 36.Bartlett PA, Marlowe CK. Biochemistry. 1991;22:4618. doi: 10.1021/bi00289a002. [DOI] [PubMed] [Google Scholar]
- 37.Kaplan AP, Bartlett PA. Biochemistry. 1991;30:8165. doi: 10.1021/bi00247a011. [DOI] [PubMed] [Google Scholar]
- 38.Irwin JJ, Raushel FM, Shoichet BK. Biochemistry. 2005;44:12316. doi: 10.1021/bi050801k. [DOI] [PubMed] [Google Scholar]
- 39.Yuan Q, He L, Ke H. Antimicro. Agents Chemo. 2012;57:5157. doi: 10.1128/AAC.05896-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pegg KM, Liu EM, Lacuran A, Oelschlaeger P. Antimicro. Agents Chemo. 2013 doi: 10.1128/AAC.02341-12. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garau G, Bebrone C, Anne C, Galleni M, Frere JM, Dideberg O. J. Mol. Biol. 2005;345:785. doi: 10.1016/j.jmb.2004.10.070. [DOI] [PubMed] [Google Scholar]
- 42.Widmann M, Pleiss J, Oelschlaeger P. Antimicro. Agents Chemo. 2012;56:3481. doi: 10.1128/AAC.00255-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang KW, Brandt JJ, Chatwood LL, Crowder MW. Bioorg. Med. Chem. Lett. 2000;10:1087. doi: 10.1016/s0960-894x(00)00186-4. [DOI] [PubMed] [Google Scholar]
- 44.Page MI, Laws AP. Tetrahedron. 2000;56:5631. [Google Scholar]
- 45.Yu L, Feng D, He M, Li R, Cai Z. J. Theor. Comput. Chem. 2006;5:421. Special Issue. [Google Scholar]
- 46.Page MI, Laws AP, Slater MJ, Stone JR. Pure Appl. Chem. 1995;67:711. [Google Scholar]
- 47.Slater MJ, Laws AP, Page MI. Bioorg. Chem. 2001;29:77. doi: 10.1006/bioo.2000.1192. [DOI] [PubMed] [Google Scholar]
- 48.Wood JM, Hinchliffe PS, Laws AP, Page MI. J. Chem. Soc. Perkin Trans. I. 2002;2:938. [Google Scholar]
- 49.Afarinkia K, Cadogan JIG, Rees CW. J. Chem. Soc. Chem. Comm. 1992:285. [Google Scholar]
- 50.Daly AM, Gilheany DG. Tetrahedron: Asymmetry. 2003;14:127. [Google Scholar]
- 51.Spencer J, Read J, Sessions RB, Howell S, Blackburn GM, Gamblin SJ. J. Am. Chem. Soc. 2005;127:14439. doi: 10.1021/ja0536062. [DOI] [PubMed] [Google Scholar]
- 52.Zhang H, Hao Q. FASEB J. 2011;25:2574. doi: 10.1096/fj.11-184036. [DOI] [PubMed] [Google Scholar]
- 53.Breece RM, Hu Z, Bennett B, Crowder MW, Tierney DL. J. Am. Chem. Soc. 2009;131:11642. doi: 10.1021/ja902534b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oelschlaeger P, Schmid RD, Pleiss J. Protein Eng. 2003;16:341. doi: 10.1093/protein/gzg049. [DOI] [PubMed] [Google Scholar]
- 55.Humphrey W, Dalke A, Schulten K. J. Mol. Graphics. 1996;14:33. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.