Abstract
The androgen receptor (AR) is important in reproductive organ development, as well as tissue homeostasis of the pancreas, liver, and skeletal muscle in adulthood. The trinucleotide (CAG)n repeat polymorphism in exon 1 of the AR gene is thought to regulate AR activity, with longer alleles conferring reduced receptor activity. Therefore, the evaluation of the allelic distribution of the AR (CAG)n repeat in various ethnic groups is crucial in understanding the interindividual variability in AR activity. We evaluated ethnic variation of this AR polymorphism by genotyping individuals from the multiethnic Hyperglycemia and Adverse Pregnancy Outcome study cohort. We genotyped 4421 Caucasian mothers and 3365 offspring of European ancestry; 1494 Thai mothers and 1742 offspring; 1119 Afro-Caribbean mothers and 1142 offspring; and 780 Hispanic mothers and 770 offspring of Mexican ancestry from Bellflower, California. The distributions of (CAG)n alleles among all 4 ethnic groups are significantly different (P < .0001). Pairwise tests confirmed significant differences between each pair of ethnicities tested (P < 10−28). The relative AR (CAG)n repeat length in the different groups was as follows: Afro-Caribbean (shortest repeat lengths and greatest predicted AR activity) < Caucasian < Hispanic < Thai (longest repeat length and lowest predicted AR activity). Significant interethnic differences in the allele frequencies of the AR exon 1 (CAG)n polymorphism exist. Our results suggest that there may be potential ethnic differences in androgenic pathway activity and androgen sensitivity.
Keywords: Polyglutamine repeat, X-linked, polymorphism, HAPO, pregnancy
The human androgen receptor (AR) gene is located on chromosome Xq11.2–q12 and codes for a protein with 3 major functional domains: the N-terminal transactivation domain, the DNA-binding domain, and the androgen-binding domain. The protein functions as a steroid hormone–activated transcription factor, which signals through classical and nonclassical signaling pathways (Katzenellenbogen and Katzenellenbogen, 1996; Rahman and Christian, 2007). AR is expressed in many tissues during development and adulthood, influencing an enormous range of physiologic processes including organ and tissue growth and differentiation, cognition, muscle hypertrophy, bone density, and insulin levels (Gelmann, 2002).
The AR gene contains a highly polymorphic (CAG)n repeat in exon 1 encoding a glutamine tract in the N-terminal transactivation domain of the protein, which becomes active only after AR binds to its ligand (Palazzolo et al, 2008). The polyglutamine tract length is inversely correlated to the transcriptional competence of the receptor, with longer tracts being associated with lower levels of AR-mediated transcription in both normal and disease states (Beilin et al, 2000; Buchanan et al, 2004; Harada et al, 2010).
Expansion of the AR (CAG)n beyond the normal range (>38 repeats) causes spinal and bulbar muscular atrophy (SBMA), also known as Kennedy disease, which is an X-linked recessive form of spinal muscular atrophy. The expansion of AR (CAG) repeat has been implicated in SBMA either through loss of normal AR function, which induces neuronal degeneration, or through the pathogenic AR, which acquires a toxic property, thereby damaging motor neurons (Katsuno et al, 2006). Interestingly, some populations of affected individuals display a variety of endocrinologic symptoms including gynecomastia, infertility, androgen insensitivity, and increased incidence of type 2 diabetes (Mariotti et al, 2000), which would indicate a global disruption of AR function.
AR (CAG)n repeats within the normal range (10–36 repeats) are associated with numerous endocrine cancers, male and female infertility, and many other neurological and endocrine conditions (Palazzolo et al, 2008; Wu et al, 2008; Chatterjee et al, 2009; Ludwig et al, 2009; Shaik et al, 2009). There is a continuous relationship between AR (CAG)n repeat length and metabolic traits in Caucasian men, where repeat number correlates negatively with AR sensitivity and positively with body fat, insulin levels, and leptin in healthy and diabetic men (Zitzmann et al, 2003). Additionally, testosterone and luteinizing hormone levels are higher in diabetic men with >24 repeats, reflecting reduced negative feedback through a less sensitive receptor (Stanworth et al, 2008). In women with polycystic ovary syndrome, a disorder diagnosed by hyperandrogenism, association between testosterone and insulin resistance may be modified by the (CAG)n repeat polymorphism (Mohlig et al, 2006). Therefore, ethnic differences in allele distributions of this marker could have far-reaching impact on diverse conditions in men and women.
Here we report the allelic distribution of the (CAG)n polymorphism in Caucasian, Afro-Caribbean, Hispanic, and Thai populations from the Hyperglycemia and Adverse Pregnancy Outcome (HAPO) cohort. To examine this, we genotyped 14 833 mothers and neonates of these 4 ethnicities and tested for differences in allele distributions of the AR (CAG)n repeat.
Materials and Methods
Participants
The HAPO study is a multiethnic, multicenter observational epidemiologic study assembled to investigate the associations of maternal glycemia with adverse pregnancy outcomes. HAPO study protocols were in compliance with the Helsinki Declaration and were approved by the institutional review boards at all centers. Written informed consent was obtained from all participants (HAPO Study Cooperative Research Group et al, 2008). The present study included Caucasian individuals of European ancestry from centers in the United Kingdom (Manchester and Belfast), Australia (Brisbane and Newcastle), and North America (Toronto); Thai individuals from Bangkok; Afro-Caribbean individuals from Barbados; and Hispanic individuals of Mexican ancestry. All Hispanic participants were enrolled at 1 field center in Bellflower, California, and self-identified as white Hispanic. The Afro-Caribbean population represents Caribbeans of African origin, and all Afro-Caribbean participants were collected at 1 center in Barbados and self-identified as black. Other studies have shown that Barbadians of African descent have 89.8% West African ancestry and 10.2% European ancestry (Benn-Torres et al, 2008).
Genotyping
DNA was extracted from blood at the Northwestern Center for Genetic Medicine Genomics Core (Chicago, Illinois). DNA samples and 3 Centre d’Etude du Polymorphisme Humain (CEPH) control DNA samples were organized randomly in each quadrant of 384-well microtiter plates. The AR (CAG)n repeat was amplified as previously described (Ackerman et al, 2010) with primers (forward [5′-NED-GTGCGCGAAGTGATCCAGAA-3′] and reverse [5′-TAGCCTGTGGGGCCTCTACG-3′]). Polymerase chain reaction products were separated on the Applied Biosystems 3130XL capillary DNA sequence analysis system and genotypes were assigned using GENEMAPPER software v.4.0 (Applied Biosystems, Foster City, California). Visual examination of each chromatogram confirmed the genotype assignment.
Statistical Analysis
Pearson’s χ2 and Monte Carlo tests were performed on maternal genotypes to test for differences in allele distribution between ethnicities, with P < .05 considered statistically significant. χ2 tests were performed on alleles with an expected count of greater than = in all populations. Monte Carlo tests were performed using all alleles in each population, with 108 iterations and 99.9% accuracy. The above statistical tests were performed using SPSS (version 17.0; SPSS Inc, Chicago, Illinois). Hardy-Weinberg equilibrium (HWE) tests were performed from each ethnicity on maternal genotypes with frequencies >.05 using the Genepop software package (Rousset and Raymond, 1995; Rousset, 2008).
Results
Characteristics of the HAPO Study participants and outcomes have been previously reported (HAPO Study Cooperative Research Group et al, 2008; HAPO Study Cooperative Research Group, 2009). The Caucasian population (4421 mothers, 3365 neonates) represented the largest group in this study, and the Hispanics (780 mothers, 770 neonates) were the smallest group. The numbers of samples and chromosomes genotyped for each center and ethnicity are given in Table 1. The ethnic distribution was 52.5% Caucasian, 21.8% Asian, 15.2% Afro-Caribbean, and 10.5% Hispanic.
Table 1.
Maternal and neonate DNA samples by ethnicity successfully genotyped at the AR (CAG)n locus
| Mothers | Neonates | Total | |||||
|---|---|---|---|---|---|---|---|
| Center | Ethnicity | No. Samples |
No. Chromosomes |
No. Samples |
No. Chromosomesa |
No. Samples |
No. Chromosomes |
| Belfast | Caucasian (United Kingdom) | 1331 | 2662 | 1045 | 1537 | 2376 | 4199 |
| Manchester | Caucasian (United Kingdom) | 982 | 1964 | 548 | 805 | 1530 | 2769 |
| Brisbane | Caucasian (Australia) | 963 | 1926 | 703 | 1043 | 1666 | 2969 |
| Newcastle | Caucasian (Australia) | 491 | 982 | 418 | 618 | 909 | 1600 |
| Toronto | Caucasian (North America) | 654 | 1308 | 651 | 977 | 1305 | 2285 |
| Total Caucasian | 4421 | 8842 | 3365 | 4980 | 7786 | 13 822 | |
| Bangkok | Thai | 1494 | 2988 | 1742 | 2606 | 3236 | 5594 |
| Barbados | Afro-Caribbean | 1119 | 2238 | 1142 | 1695 | 2261 | 3933 |
| Bellflower | Hispanic (North America) | 780 | 1560 | 770 | 1156 | 1550 | 2716 |
| Total | 7814 | 15 628 | 7019 | 10 437 | 14833 | 26 065 | |
Neonate chromosome numbers are not 2n because this marker is on the X chromosome and approximately 50% of the neonates were male.
Genotyping
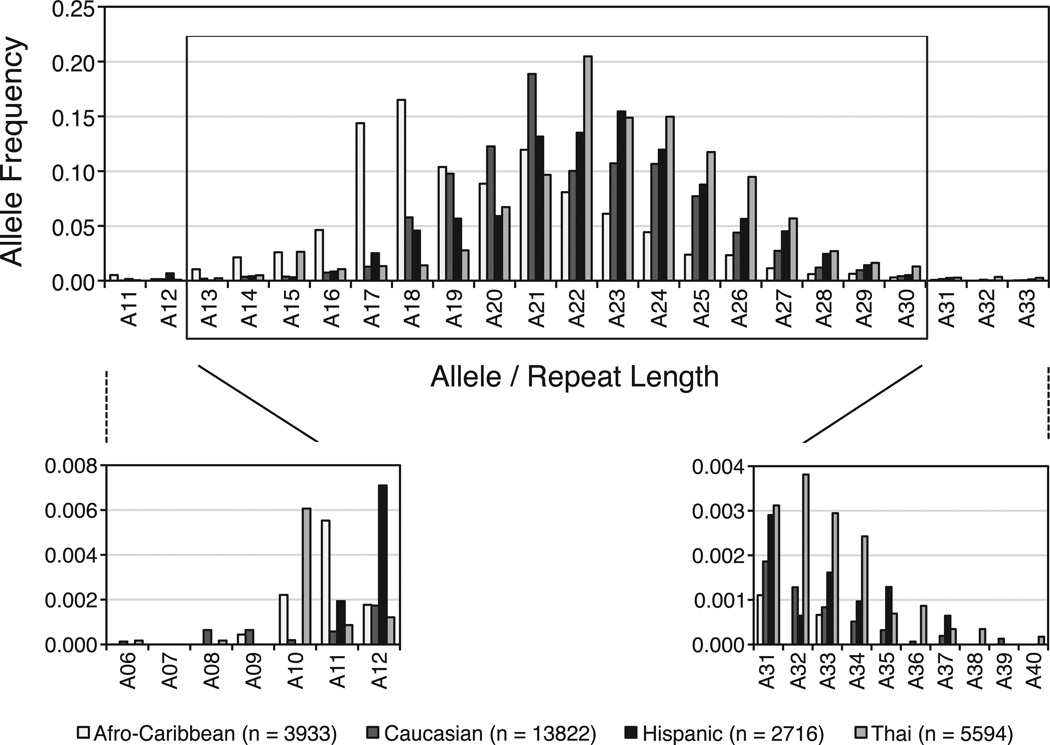
The CEPH control samples genotyped with an 89.5% success rate and a plate-to-plate concordance of 99.5%. Two percent of the HAPO participant samples were selected randomly and regenotyped without knowledge of previously assigned genotypes. Among these, 99.9% concordance was observed. We obtained AR (CAG)n genotypes for 90.6% of the samples. AR allele designations represent the number of repeats; for example, allele 6 (A06) has 6 CAG repeats. We identified 34 alleles at the AR (CAG)n locus: A06 and A08–A40. The frequency of each allele varied widely across populations, and distribution of the alleles by ethnic group for mothers and neonates combined is presented in the Figure. The range of alleles differed among ethnicities, with the Thais and Caucasians having the largest range (6–40 repeats, 32 alleles total) and the Afro-Caribbeans having the smallest range (9–37 repeats, 24 alleles total). The Caucasians, our largest group (7786 individuals, 13 822 chromosomes), had the highest number of rare alleles (frequency <.01, n = 18), and the Afro- Caribbeans, our second smallest group (2261 individuals, 3933 chromosomes), had the fewest rare alleles (n = 6). A20 through A23 were common to all ethnicities (frequency >.05). The most common CAG repeats were A18 in Afro-Caribbeans (.17), A21 in Caucasians (.19), A23 in Hispanics (.15), and A22 in Thais (.21). The mean CAG repeat lengths were Afro-Caribbean 19.6 ± 3.2, Caucasian 21.9 ± 2.9, Hispanic 22.6 ± 3.1, and Thai 23.1 ± 3.3. These results are slightly different than those published by Edwards et al (1992), and are likely due to differences in sample size, stringency of ethnic classification, and location of participant recruitment.
Figure.
Distribution of AR (CAG)n alleles by ethnic group in the Hyperglycemia and Adverse Pregnancy Outcome study. Top panel: only alleles with a frequency >.001 are shown. The alleles within the inner box are the alleles used for the χ2 analysis for all ethnicities. Lower left panel: individuals in each ethnicity with the lowest number of CAG repeats. Lower right panel: individuals in each ethnicity with the highest number of CAG repeats. White bars indicate Afro-Caribbean participants; dark gray, Caucasians; black, Hispanics; light gray, Thais. n = number of chromosomes.
Statistical Analysis
Because the maternal and fetal genotypes are not independent (eg, neonates inherit 1 of the mother’s alleles), all statistical tests were performed on maternal genotype only. The numbers of chromosomes and their corresponding frequencies for each allele used in the maternal χ2 tests are presented in Table 2. All 4 ethnic populations were in HWE. Because of the large number of rare alleles in each ethnicity and the presence of population-specific alleles, we performed a Pearson’s χ2 test on all ethnicities together using only alleles A13–30, which revealed significant differences in allele distributions between the 4 ethnic groups (P < .0001; Table 3). These differences were confirmed using the Monte Carlo test on all alleles present in each population (P < .001). Separate χ2 analyses were also performed for pairwise comparisons of ethnicities, for example, Caucasian vs Hispanic, for all 6 pairwise comparisons. For these tests we maximized the alleles used for each pair, because a slightly different subset of alleles qualified for the pairwise χ2 analyses. These analyses confirmed that there are highly significant differences between each pair of ethnicities tested (P < 10−28), even after adjusting for multiple testing (Table 3).
Table 2.
Number of chromosomes and their corresponding frequencies for each allele used in the maternal χ2 analysesa
| Caucasian | Afro-Caribbean | Thai | Hispanic | |||||
|---|---|---|---|---|---|---|---|---|
| Allele | No. Chromosomes |
Frequency | No. Chromosomes |
Frequency | No. Chromosomes |
Frequency | No. Chromosomes |
Frequency |
| A13 | 23 | .003 | 23 | .010 | 4 | .001 | 1 | .001 |
| A14 | 37 | .004 | 49 | .022 | 15 | .005 | 7 | .004 |
| A15 | 36 | .004 | 61 | .027 | 65 | .022 | 6 | .004 |
| A16 | 72 | .008 | 103 | .046 | 29 | .010 | 13 | .008 |
| A17 | 114 | .013 | 319 | .143 | 34 | .011 | 38 | .024 |
| A18 | 520 | .059 | 376 | .168 | 33 | .011 | 73 | .047 |
| A19 | 882 | .100 | 236 | .105 | 80 | .027 | 87 | .056 |
| A20 | 1115 | .126 | 193 | .086 | 168 | .056 | 90 | .058 |
| A21 | 1631 | .184 | 290 | .130 | 271 | .091 | 210 | .135 |
| A22 | 904 | .102 | 173 | .077 | 544 | .182 | 205 | .131 |
| A23 | 965 | .109 | 133 | .059 | 402 | .134 | 251 | .161 |
| A24 | 950 | .107 | 93 | .042 | 396 | .132 | 190 | .122 |
| A25 | 663 | .075 | 55 | .025 | 335 | .112 | 135 | .087 |
| A26 | 390 | .044 | 52 | .023 | 260 | .087 | 89 | .057 |
| A27 | 226 | .026 | 27 | .012 | 139 | .046 | 74 | .047 |
| A28 | 110 | .012 | 16 | .007 | 72 | .024 | 37 | .024 |
| A29 | 88 | .010 | 9 | .004 | 43 | .014 | 23 | .015 |
| A30 | 40 | .005 | 4 | .002 | 36 | .012 | 7 | .004 |
| A31 | 15 | .002 | 2 | .001 | 12 | .004 | 5 | .003 |
| A32 | 11 | .001 | 0 | .000 | 10 | .003 | 1 | .001 |
Frequency 5 No. chromosomes for each allele for a particular ethnic group/total No. chromosomes for that ethnic group.
Table 3.
Results of the Pearson χ2 analyses testing for differences in maternal allele distribution between ethnicitiesa
| Populations | Alleles Tested | No. Chromosomes Tested |
df | χ2 Value | P Value |
|---|---|---|---|---|---|
| All four ethnicities | A13–30 | 15 496 | 51 | 3215.9 | <.0001b |
| Afro-Caribbean vs Caucasian | A13–30 | 11 006 | 17 | 1716.7 | <.0001b |
| Afro-Caribbean vs Hispanic | A14–29 | 3756 | 15 | 750.0 | <.0001b |
| Afro-Caribbean vs Thai | A13–31 | 5162 | 18 | 1621.3 | <.0001b |
| Caucasian vs Hispanic | A14–30 | 10 334 | 16 | 203.1 | 1.86 × 10−34 |
| Caucasian vs Thai | A13–32 | 11 740 | 19 | 904.5 | 1.26 × 10−179 |
| Hispanic vs Thai | A14–29 | 4490 | 15 | 170.0 | 2.39 × 10−28 |
Analyses were performed on maternal genotypes only.
Indicates P values beyond those generated by SPSS. The actual results returned are P = .000.
Discussion
We genotyped over 14 800 individuals (26 065 chromosomes) at the AR (CAG)n locus in 4 ethnic groups from the HAPO cohort. This is the first study to document AR (CAG)n allele frequencies in 4 populations of this magnitude.
Interethnic differences in the distribution of genetic polymorphisms and how these affect human susceptibility to various disorders, including the development of type 2 diabetes, have been documented (Chambers et al, 2009; Tan et al, 2010). In multiethnic studies on SBMA, the number of AR (CAG)n repeats seen in affected individuals typically ranges from 35 to 57 repeats, but differs by ethnicity (Lund et al, 2001). Differences in the AR (CAG)n allelic distribution, which are expected to result in population-based differences in androgen sensitivity, may be due to either natural selection of alleles by environmental factors or fixation of allele frequency through founder effects.
In this study, Caucasians had the highest number of rare alleles and Afro-Caribbeans had the fewest. This is likely because of different sample sizes for each population as well as the fact that the Afro-Caribbean group is an island population and may thus have a smaller effective population size. Interestingly, within our Caucasian, Hispanic, and Thai populations, there are a very small number (<0.25%) of individuals with repeat lengths that fall within the disease range for SBMA (≥35 repeats). After sequencing individuals to confirm actual number of repeats, we had 19 mothers (9 Thai, 3 Hispanic, 7 Caucasian), 9 female neonates (3 Thai, 3 Hispanic, 3 Caucasian), and 1 male Caucasian neonate with 35–40 repeats. In addition, there are a number (<2%) of individuals in each ethnicity with very short (≤12) repeats (n = 28–61). These very short repeats have been observed to contribute to phenotypes indicative of AR malfunction, indicating that repeat lengths on either side of the normal range are associated with impaired receptor function (McPhaul et al, 1991). This has been further corroborated by cell culture studies that have revealed a critical range (16–29 CAG repeats) for maintenance of the protein’s stability and function (Buchanan et al, 2004).
Age of onset of puberty for both males and females varies by ethnicity (NHANES, 1997). These ethnic differences may be explained by differences in hormone levels or in hormone action in target tissues or potentially by the ethnic variation in the AR described here. Furthermore, these ethnic differences in AR (CAG)n may also contribute to interindividual variation in terms of disease risk, disease severity, and drug response. Given the association between AR activity and (CAG)n repeat length, our study suggests that an individual’s response to androgen and androgenic therapies might vary based on AR (CAG)n genotypes.
In conclusion, we have shown that there are significant ethnic differences in allele distribution of the AR (CAG)n polymorphism. This finding provides insight into ethnic differences in normal reproductive processes, as well as predicting risk and severity for a number of diseases. Our study provides an important starting point for better understanding the molecular basis underlying ethnic differences in androgen sensitivity and underscores the importance of using large multiethnic cohorts to examine genetic variation at known disease loci.
Acknowledgment
We would like to gratefully thank Dr Jean D. Wilson (UT Southwestern Medical Center) for his interest in our research, and for insights and suggestions for the manuscript.
Supported by the National Institutes of Health (P50 HD44405 to M.U.) and by the Natural Sciences and Engineering Research Council of Canada Postgraduate Scholarships Program (PGS D2-374259-2009 to C.M.A.). This work was also supported by the National Institute of Diabetes, Digestive, and Kidney Diseases, and by the National Human Genome Research Institute (R01 DK067459, and U01 HG004415 to W.L.L.). The HAPO study was funded by the Eunice Kennedy Shriver National Institute of Child Health and Human Development; the National Institute of Diabetes and Digestive and Kidney Diseases (R01-HD34242 and R01-HD34243); and the American Diabetes Association (to B.E.M. and L.P.L.).
Footnotes
Group Information: This work was done and manuscript prepared for the HAPO Study Cooperative Research Group. A list of the members of the HAPO Study Cooperative Research Group was published in New England Journal of Medicine (2008;358:1991–2002).
References
- Ackerman CM, Lowe LP, Lee H, Chen F, Hughes E, Cholod P, Dyer AR, Hayes MG, Metzger BE, Lowe WL, Urbanek M. The role of the polycystic ovary syndrome susceptibility locus D19S884 allele 8 in maternal glycemia and fetal size. J Clin Endocrinol Metab. 2010;95:3242–3250. doi: 10.1210/jc.2009-2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilin J, Ball EM, Favaloro JM, Zajac JD. Effect of the androgen receptor CAG repeat polymorphism on transcriptional activity: specificity in prostate and non-prostate cell lines. J Mol Endocrinol. 2000;25:85–96. doi: 10.1677/jme.0.0250085. [DOI] [PubMed] [Google Scholar]
- Benn-Torres J, Bonilla C, Robbins CM, Waterman L, Moses TY, Hernandez W, Santos ER, Bennett F, Aiken W, Tullock T, Coard K, Hennis A, Wu S, Nemesure B, Leske MC, Freeman V, Carpten J, Kittles RA. Admixture and population stratification in African Caribbean populations. Ann Hum Genet. 2008;72:90–98. doi: 10.1111/j.1469-1809.2007.00398.x. [DOI] [PubMed] [Google Scholar]
- Buchanan G, Yang M, Cheong A, Harris JM, Irvine RA, Lambert PF, Moore NL, Raynor M, Neufing PJ, Coetzee GA, Tilley WD. Structural and functional consequences of glutamine tract variation in the androgen receptor. Hum Mol Genet. 2004;13:1677–1692. doi: 10.1093/hmg/ddh181. [DOI] [PubMed] [Google Scholar]
- Chambers JC, Zhang W, Zabaneh D, Sehmi J, Jain P, McCarthy MI, Froguel P, Ruokonen A, Balding D, Jarvelin MR, Scott J, Elliott P, Kooner JS. Common genetic variation near melatonin receptor MTNR1B contributes to raised plasma glucose and increased risk of type 2 diabetes among Indian Asians and European Caucasians. Diabetes. 2009;58:2703–2708. doi: 10.2337/db08-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Singh R, Kadam S, Maitra A, Thangaraj K, Meherji P, Modi D. Longer CAG repeat length in the androgen receptor gene is associated with premature ovarian failure. Hum Reprod. 2009;24:3230–3235. doi: 10.1093/humrep/dep296. [DOI] [PubMed] [Google Scholar]
- Edwards A, Hammond HA, Jin L, Caskey CT, Chakraborty R. Genetic variation at five trimeric and tetrameric tandem repeat loci in four human population groups. Genomics. 1992;12:241–253. doi: 10.1016/0888-7543(92)90371-x. [DOI] [PubMed] [Google Scholar]
- Gelmann EP. Molecular biology of the androgen receptor. J Clin Oncol. 2002;20:3001–3015. doi: 10.1200/JCO.2002.10.018. [DOI] [PubMed] [Google Scholar]
- HAPO Study Cooperative Research Group. Hyperglycemia and Adverse Pregnancy Outcome (HAPO) Study: associations with neonatal anthropometrics. Diabetes. 2009;58:453–459. doi: 10.2337/db08-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAPO Study Cooperative Research Group. Metzger BE, Lowe LP, Dyer AR, Trimble ER, Chaovarindr U, Coustan DR, Hadden DR, McCance DR, Hod M, McIntyre HD, Oats JJ, Persson B, Rogers MS, Sacks DA. Hyperglycemia and adverse pregnancy outcomes. N Engl J Med. 2008;358:1991–2002. doi: 10.1056/NEJMoa0707943. [DOI] [PubMed] [Google Scholar]
- Harada N, Mitani T, Higashimura Y, Yamaji R, Okamoto K, Nakano Y, Inui H. Involvement of three glutamine tracts in human androgen receptor transactivation. J Steroid Biochem Mol Biol. 2010;118:77–84. doi: 10.1016/j.jsbmb.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Katsuno M, Adachi H, Waza M, Banno H, Suzuki K, Tanaka F, Doyu M, Sobue G. Pathogenesis, animal models and therapeutics in spinal and bulbar muscular atrophy (SBMA) Exp Neurol. 2006;200:8–18. doi: 10.1016/j.expneurol.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Katzenellenbogen JA, Katzenellenbogen BS. Nuclear hormone receptors: ligand-activated regulators of transcription and diverse cell responses. Chem Biol. 1996;3:529–536. doi: 10.1016/s1074-5521(96)90143-x. [DOI] [PubMed] [Google Scholar]
- Ludwig AH, Murawska M, Panek G, Timorek A, Kupryjanczyk J. Androgen, progesterone, and FSH receptor polymorphisms in ovarian cancer risk and outcome. Endocr Relat Cancer. 2009;16:1005–1016. doi: 10.1677/ERC-08-0135. [DOI] [PubMed] [Google Scholar]
- Lund A, Udd B, Juvonen V, Andersen PM, Cederquist K, Davis M, Gellera C, Kolmel C, Ronnevi LO, Sperfeld AD, Sorensen SA, Tranebjaerg L, Van Maldergem L, Watanabe M, Weber M, Yeung L, Savontaus ML. Multiple founder effects in spinal and bulbar muscular atrophy (SBMA, Kennedy disease) around the world. Eur J Hum Genet. 2001;9:431–436. doi: 10.1038/sj.ejhg.5200656. [DOI] [PubMed] [Google Scholar]
- Mariotti C, Castellotti B, Pareyson D, Testa D, Eoli M, Antozzi C, Silani V, Marconi R, Tezzon F, Siciliano G, Marchini C, Gellera C, Donato SD. Phenotypic manifestations associated with CAG-repeat expansion in the androgen receptor gene in male patients and heterozygous females: a clinical and molecular study of 30 families. Neuromuscul Disord. 2000;10:391–397. doi: 10.1016/s0960-8966(99)00132-7. [DOI] [PubMed] [Google Scholar]
- McPhaul MJ, Marcelli M, Tilley WD, Griffin JE, Isidro-Gutierrez RF, Wilson JD. Molecular basis of androgen resistance in a family with a qualitative abnormality of the androgen receptor and responsive to high-dose androgen therapy. J Clin Invest. 1991;87:1413–1421. doi: 10.1172/JCI115147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohlig M, Jurgens A, Spranger J, Hoffmann K, Weickert MO, Schlosser HW, Schill T, Brabant G, Schuring A, Pfeiffer AF, Gromoll J, Schofl C. The androgen receptor CAG repeat modifies the impact of testosterone on insulin resistance in women with polycystic ovary syndrome. Eur J Endocrinol. 2006;155:127–130. doi: 10.1530/eje.1.02195. [DOI] [PubMed] [Google Scholar]
- NHANES. NHANES III Reference Manuals and Reports. Analytic and Reporting Guidelines: The Third National Health and Nutrition Examination Survey (1988–94) Hyattsville, MD: National Center for Health Statistics, Centers for Disease Control and Prevention; 1997. [CD-ROM]. [Google Scholar]
- Palazzolo I, Gliozzi A, Rusmini P, Sau D, Crippa V, Simonini F, Onesto E, Bolzoni E, Poletti A. The role of the polyglutamine tract in androgen receptor. J Steroid Biochem Mol Biol. 2008;108:245–253. doi: 10.1016/j.jsbmb.2007.09.016. [DOI] [PubMed] [Google Scholar]
- Rahman F, Christian HC. Non-classical actions of testosterone: an update. Trends Endocrinol Metab. 2007;18:371–378. doi: 10.1016/j.tem.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Rousset F. Genepop’007: a complete reimplementation of the Genepop software for Windows and Linux. Mol Ecol Resour. 2008:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- Rousset F, Raymond M. Testing heterozygote excess and deficiency. Genetics. 1995;140:1413–1419. doi: 10.1093/genetics/140.4.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaik NA, Govindan S, Kodati V, Rao KP, Hasan Q. Polymorphic (CAG)n repeats in the androgen receptor gene: a risk marker for endometriosis and uterine leiomyomas. Hematol Oncol Stem Cell Ther. 2009;2:289–293. doi: 10.1016/s1658-3876(09)50040-4. [DOI] [PubMed] [Google Scholar]
- Stanworth RD, Kapoor D, Channer KS, Jones TH. Androgen receptor CAG repeat polymorphism is associated with serum testosterone levels, obesity and serum leptin in men with type 2 diabetes. Eur J Endocrinol. 2008;159:739–746. doi: 10.1530/EJE-08-0266. [DOI] [PubMed] [Google Scholar]
- Tan JT, Ng DP, Nurbaya S, Ye S, Lim XL, Leong H, Seet LT, Siew WF, Kon W, Wong TY, Saw SM, Aung T, Chia KS, Lee J, Chew SK, Seielstad M, Tai ES. Polymorphisms identified through genome-wide association studies and their associations with type 2 diabetes in Chinese, Malays, and Asian-Indians in Singapore. J Clin Endocrinol Metab. 2010;95:390–397. doi: 10.1210/jc.2009-0688. [DOI] [PubMed] [Google Scholar]
- Wu MH, Chou YC, Yu CP, Yang T, You SL, Chen CJ, Sun CA. Androgen receptor gene CAG repeats, estrogen exposure status, and breast cancer susceptibility. Eur J Cancer Prev. 2008;17:317–322. doi: 10.1097/CEJ.0b013e3282f75e7f. [DOI] [PubMed] [Google Scholar]
- Zitzmann M, Gromoll J, von Eckardstein A, Nieschlag E. The CAG repeat polymorphism in the androgen receptor gene modulates body fat mass and serum concentrations of leptin and insulin in men. Diabetologia. 2003;46:31–39. doi: 10.1007/s00125-002-0980-9. [DOI] [PubMed] [Google Scholar]