Abstract
Post-translational modifications play important roles in transcriptional regulation. Among the less understood PTMs is O-GlcNAcylation. Nevertheless, O-GlcNacylation in the nucleus is found on hundreds of transcription factors and coactivators and is often found in a mutually exclusive ying-yang relationship with phosphorylation. O-GlcNAcylation also links cellular metabolism directly to the proteome, serving as a conduit of metabolic information to the nucleus. This review serves as a brief introduction to O-GlcNAcylation, emphasizing its important thematic roles in transcriptional regulation, and highlights several recent and important additions to the literature that illustrate the connections between O-GlcNAc and transcription.
Keywords: transcription, RNA polymerase II, O-GlcNAc, nutrient sensor
The modification of proteins with N-acetylglucosamine, or O-GlcNAc, is a vastly underappreciated post-translational modification, despite its discovery nearly 30 years ago. It is nearly as common as phosphorylation and is often found as part of a ying-yang state: many serine and threonine residues are either O-GlcNAcylated or phosphorylated [1]. In contrast to the large number of protein kinases and phosphatases in the cell, there are only two enzymes directly involved in O-GlcNAcylation [1]. The O-GlcNAc transferase (OGT) adds O-GlcNAc to S/T residues using UDP-GlcNAc as the nucleotide-sugar donor. β-D-N-glucosaminidase (OGA) is responsible for removing O-GlcNAc from proteins. The regulation of OGT and OGA is not well understood but a variety of evidence indicates that O-GlcNAcylation blocks phosphorylation, ubiquitination, and directly controls target enzyme activity and specificity. It is the dynamics of these events in concert with other post-translational modifications that create the regulatory functions of OGT and OGA.
A study of O-GlcNAcylation requires that one view this post-translational modification differently than, for example, phosphorylation. Most kinases are tightly regulated at least by their substrate specificity and activation, and their relatively narrow application to a relatively small number of processes. In contrast, OGT itself is responsible for several hundred targets. Substrate specificity, as far as we know it, is not the main thematic regulatory idea here; instead, the function of O-GlcNAcylation in the cell must be considered separately for each system utilizing it and is likely mediated by protein-protein interactions to recruit OGT and OGA to the necessary sites of action [2, 3].
The substrate for OGT is UDP-GlcNAc and its synthesis occurs via a little-known pathway, the hexosamine biosynthetic pathway or HBP [4]. The HBP uses fructose-6-phosphate (F6P) as its starting material and thus splits off from the glycolytic pathway at this point. About 2–5% of glucose flux into the cell is shunted into the HBP [5]. The rate-limiting step is the next enzyme in the pathway, glutamine-fructose aminotransferase, which uses F6P and glutamine to synthesize glucosamine-6-P [4, 6]. Thereafter, acetyl CoA and uridine are brought in to synthesize UDP-GlcNAc. As a result of these influxes of metabolic intermediates, it has long been thought that O-GlcNAc is a metabolic sensor of the nutrient state of the cell [4, 7, 8].
That the flux of glucose into the cell is reflected in the [UDP-GlcNAc] suggested that insulin-resistance and type II diabetes might be causally related to perturbations in the HBP or OGT and OGA activity [9]. There is much causal data to support this hypothesis. Overexpression of OGT in muscle and adipocytes led to insulin resistance and GFAT overexpression also resulted in insulin resistance in mice. Type II diabetes patients also exhibit elevated O-GlcNAc levels and an influx of glucosamine will induce insulin-resistance [9–11]. Experiments from several groups show that insulin signaling is regulated by OGT [12–14], that O-GlcNAcylation of Foxo1, presumably by an OGT/PGC-1a recruitment mechanism, increased gluconeogenesis-specific expression in the liver [15, 16], and that the CREB coactivator, CRTC2, is O-GlcNAcylated and results in increased gluconeogenic gene expression [17]. Furthermore, the link between O-GlcNAc and insulin resistance has been extensively developed using genetics in C. elegans and Drosophila by Hanover and colleagues [18–21].
There are several hundred transcription factors that are O-GlcNAcylated [3]. Among them are the p65 subunit of NF-kB, Sp1, CREB and CRCT2, p53, and myc. OGT is also in a complex with the histone deacetylase mSin3A, and OGT/mSin3A work in concert to repress transcription [22]. Lastly, OGT is super sex combs (sxc), a member of the polycomb group genes, and polycomb response elements contain O-GlcNAcylated proteins [23–25]. Furthermore, ogt mutants lost polycomb-dependent repression. It is clear from these studies and others that O-GlcNAcylation of transcription factors can affect transcription in many ways: nuclear localization, protein stability, prevention of phosphorylation, and increased transactivation potential [3, 26].
This review will focus on advances in understanding the O-GlcNAcylation events that occur in the nucleus, how these events reflect the nutrient state of the cell and influence transcriptional output, and attempt to synthesize this material under a unifying thematic idea. The reader is referred to several more detailed and recent reviews of the literature for further reading [2, 4, 26–29].
TET family members and OGT
The Tet proteins are responsible for converting 5-methylcytosine (5mC) to 5-hydroxymethyl cytosine (5hmC). These enzymes have important functions in regulating the amount of 5mC in the cell and show that this too is a dynamic epigenetic mark. MLL family members are histone methyltransferases and often found as breakpoint fusion proteins in mixed-lineage leukemia (MLL). MLL-Tet fusions have been shown to be involved in several hematopoietic malignancies and appear to maintain the genome in a more pluripotent stem cell state [30]. The Tet proteins are not O-GlcNAcylated themselves but all three associate with OGT [31–33]. Genomically, these proteins localize to CpG islands and promoters and significantly overlap with H3 K4me3, which is a histone mark of actively transcribed genes and is predominantly localized to promoters [32]. Tet2 appears necessary for recruitment of OGT as a Tet2 shRNA abolished chromatin-associated OGT [31]. OGT and Tet2 ChIP-seq showed that they are both heavily concentrated at transcriptional start sites (TSS), as is O-GlcNAc [31–33]. Tet2 was also necessary for the H3 K4me3 events and Tet2 KO showed global decreases in O-GlcNAc [32]. Interestingly, treatment of cells with alloxan (an OGT and OGA inhibitor, [34, 35]) also showed a decrease in H3K4me3 [32]. The interaction between Tet proteins and OGT illustrates a rather complex interrelationship between DNA methylation, H3 K4me3, and O-GlcNAcylation. Further elucidation of these connections should prove to be very interesting.
O-GlcNAc and Chromatin
The Tet proteins are not the only chromatin-associated proteins associated with O-GlcNAcylation. O-GlcNAcylation of MLL5 histone methyltransferase augments retinoic-acid induced differentiation of HL60 macrophage cell line via its targeting histone H3 K4 residues for trimethylation [36]. MLL5’s methylation activity increased several-fold after O-GlcNAcylation. Additionally, OGT is part of the MLL5 complex [36]. A second set of experiments by the same group showed that histone H2B was O-GlcNAcylated at S112 and this then stimulated histone H2B K120 monubiquitinylation, which is prevalent in transcriptionally active loci. Inhibition of GFAT in the HBP blocked this ubiquitinylation [37]. A second chromatin modifier, CARM1, is O-GlcNAcylated during the cell cycle. Increased O-GlcNAcylation from OGT overexpression resulted in a decrease of H3 R27 methlylation by CARM1 suggesting that OGT plays important roles in the regulation of histone modifications [38].
Histone H3 is also O-GlcNAcylated. Using mass spectroscopy, T32 was identified as the O-GlcNAcylated residue and inhibition of OGA hindered the transition from G2 to M phase [39]. Other work showed that O-GlcNAcylation increased during G1 [38, 39]. Histone phosphorylation dynamics were also susceptible to O-GlcNAcylation. For example, phospho-H3 S10, a common mitotic mark, significantly decreased with OGT overexpression [38, 40]. Similar results were seen by Sifers and colleagues by OGA inhibition [39], but it remains to be seen whether the MSK1/2-dependent phosphorylation of H3 S10 in interphase is also dependent on the state of O-GlcNAcylation of H3. Additional work showed that all four histones in humans are O-GlcNAcylated and LC-MS/MS was used to identify those residues: H2A T101, H2B S36, and H4 S47 [41]. The O-GlcNAcylation of the histones suggests that a further expansion of the histone code is warranted.
Regulation of Circadian Clocks
O-GlcNAcylation has recently been shown to play a significant role in the regulation of circadian clocks. This system serves as an excellent illustration of the interplay between OGT and OGA and how they both work in concert with and overlay their regulatory features upon a well-understood dynamical system. The core of the circadian clock consists of a transcriptional-translational feedback loop or TTFL. The activators BMAL1 and CLOCK bind to the promoters of the Per and Cry genes and activate their expression. Per and Cry proteins in turn repress their own expression. Once Per and Cry repress their expression, they then are degraded over time and BMAL/CLOCK begin the cycle again. The periodicity of this loop is for the most part dependent on the half-life of the Per and Cry proteins. However, overlaying this core machinery are several post-translational modifications that further regulate and fine-tune the TTFL [42–44]. Recently, several papers have documented the role of O-GlcNAc in circadian clock regulation.
GSK3β is an important regulator of circadian clocks. Specifically, GSK3β inhibition shortens clock periodicity [45, 46]. Additionally, GSK3β is a substrate of OGT [47]. Using ATP analogs and mutant GSK3β OGT was identified as a GSK3 β substrate [48]. Furthermore, phosphorylation of OGT by GSK3β increased its activity. O-GlcNAc levels perturbed by OGT siRNA also shortened the clock periods while an OGA inhibitor did the opposite [48]. Similar conclusions were obtained by Kim and colleagues [49].
Several clock proteins were also identified as OGT substrates. O-GlcNAcylation of Per2 appears to block its phosphorylation, increasing Per2 stability, and thereby augmenting Per2 repression [48, 49]. Although OGT levels did not oscillate, OGA levels did oscillate in a circadian manner, suggesting that it is the regulation of OGA that is controlling periodicity. In a human cell line siRNA depletion of GFAT and OGT slightly decreased expression of BMAL1 and Cry [50]. More strikingly, the GFAT inhibitor azaserine severely affected the periodicity of expression of a BMAL-luciferase reporter. OGT overexpression increased BMAL1 and CLOCK protein half-lives. Finally, alloxan (OGT inhibitor) and OGT siRNA both decrease the amplitude of the clock oscillations and shorten the period, while PUGNAc (OGA inhibitor) while also decreasing the amplitude lengthened the period of the oscillations [48]. Increasing OGT levels also increased the amplitude of clock oscillations. These data illustrate essential functions of OGT and OGA in the regulation of circadian rhythms, controlling both period length and transcriptional output.
Circadian clocks are also regulated metabolically in order to entrain or reset the peripheral clock. This entrainment is potentially a problem as it shifts the metabolism of food intake away from the supraciasmatic clock, which is controlling the wake/sleep cycle [43]. There are several ways in which metabolism can impinge on peripheral clock regulation: AMPK and NAD+ dependent deacetylases, for example [9]. Defects in the clock regulation also have severe metabolic consequences: BMAL knockouts in the liver cause hypoglycemia and Cry overexpression results in hyperglycemia [51, 52]. Clearly the metabolic and circadian systems are interconnected and utilize some sort of feedback loop. The point made by the O-GlcNAc studies is that since UDP-GlcNAc levels are responsive to glucose flux into the cell, and thus is a nutrient sensor, then the utilization of O-GlcNAcylation is a clear metabolic imprint on circadian clock regulation.
O-GlcNAc and RNA Polymerase II Regulation
Work of Dahmus and colleagues through the 1980s and early 1990s showed that RNA polymerase II existed in two forms, pol IIA and pol IIO [53]. These two species are distinguished by post-translational modifications, specifically phosphorylation, on its C-terminal domain (CTD). Pol IIA is the unmodified species and pol IIO is modified on several serine residues (serines 2, 5, and 7) and more recently the threonine at position 4 [54–57]. The CTD in humans consists of 52 heptad repeats of the consensus sequence YSPTSPS [58]. The first 26 repeats very closely fit the consensus while the latter set of repeats, from 27–52, show more deviations from this. In general the CTD is thought to serve as a scaffold for the recruitment of the RNA processing machinery. However, it is likely to be playing a role in preinitiation events as well, given that CTD deletions are transcriptionally compromised and that the large Mediator coactivator complex binds to the CTD [59–61].
The current model of formation of a preinitation complex (PIC) on the promoter states that pol IIA is the initiation-specific species, while pol IIO represents the elongating form of pol II [62–64]. This was originally formulated by Dahmus and colleagues who elegantly showed that in crude whole cell extracts only purified pol IIA could form an initiation-competent PIC while pol IIO was specifically excluded [53]. Furthermore, pol IIO is not capable of transcription in other contexts as well [65]. Support for the functions of pol IIO in elongation events is convincingly supported by a large body of work [64, 66, 67].
Largely ignored by this model is that the calf thymus pol II is also O-GlcNAcylated (hereafter referred to as pol IIγ) [68]. This modification was found only on the pol IIA species and not pol IIO. This therefore suggests that it is not clear whether pol IIA or pol IIγ (or both) is involved in PIC formation. In addition, Comer and Hart showed that a phospho-CTD peptide could not be O-GlcNAcylated in vitro [69]. From these data, Comer and Hart suggested three models. The first was that pol IIγ was the species that entered the PIC, followed by action of OGA to remove the O-GlcNAc, thereby enabling subsequent phosphorylations during elongation. The second was that pol IIγ facilitated the formation of a pol II holoenzyme that would permit the association of pol II with promoters and quickly initiate transcription. The third hypothesis postulated that O-GlcNAcylation of the CTD would prevent the degradation of pol II.
Although tests of the latter two hypotheses are lacking thus far, there is considerable genetic and biochemical evidence supporting the basic aspects of the first hypothesis. Love et al. published ChIP-chip analysis of OGT and OGA mutations in C. elegans [20]. This analysis showed that O-GlcNAc peaks were predominantly localized to promoters. Furthermore, an OGA mutant showed increased levels of O-GlcNAc and pol II at promoters while the OGT mutant showed both decreased O-GlcNAc and pol II. These data, plus single-locus ChIP and global ChIP-seq data in mice and humans ([70] and Lewis, unpublished), showed that pol II levels at promoters are directly modulated by OGT and OGA activity. Analysis of in vitro transcription systems showed that transcription in crude HeLa nuclear extracts was O-GlcNAc-dependent and that both OGT and OGA were required [70]. Furthermore, inhibition of OGT and OGA manifested themselves as defects in PIC formation (see Figure 2). Purified human pol II was a substrate for both OGT and OGA and while pol IIA could bypass a block in OGA activity, pol IIγ could not, suggesting that pol II was a direct target of the OGA. Additionally, OGT could be detected in PICs in vitro [70] and on promoters in vivo [31] and OGT coimmunoprecipitated with pol II in nuclear extracts [70]. Perhaps most importantly, we have detected pol IIγ on promoters both in PICs formed in crude nuclear extracts and in vivo (Lewis, unpublished). These data show that there is a significant amount of O-GlcNAcylation occurring on promoters and that RNA pol II is clearly an O-GlcNAcylated species in vitro and in vivo (see additional data in [70]).
Figure 2.
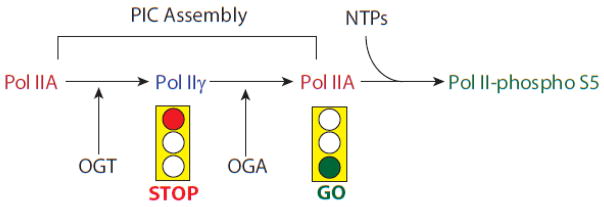
A model of the temporal activity of OGT, OGA, and pol II during PIC assembly. Pol IIA enters the PIC, and is subsequently O-GlcNAcylated on its CTD by OGT, which is a component of the PIC. The Pol IIγ exists in the PIC for a certain period while PIC assembly continues. At this stage the presence of pol IIγ serves as a “stop light” for transcription, perhaps preventing premature phosphorylation of the CTD, or as a necessary kinetic step in PIC assembly. Once PIC assembly is complete, OGA activity removes O-GlcNAc from serine 5 residues on the CTD, the “green light” is given to the PIC to indicate that transcription can now occur [70].
Genomic Localization of O-GlcNAc and OGT: Promoters as Nutrient Sensors
Several groups have also noted that the major O-GlcNAc and OGT levels across the genome are at promoters ([20, 31, 33], Lewis unpublished). These promoter-localized O-GlcNAc peaks are dependent on OGT and OGA and thus are likely dynamic features in the genome [20]. Estimates suggest as much as 40% of promoters have colocalized RNA pol II and O-GlcNAc ([20], Lewis, unpublished). These data suggest then that not only are transcription factors responsive to changing O-GlcNAc levels but conceptually it is the promoter and transcriptional output that are the true targets of OGT and OGA regulation. In other words, the promoter is acting as a nutrient sensor of the metabolic state of the cell, presumably altering its transcriptional output in response to changing levels of UDP-GlcNAc and glucose. This response makes some intuitive sense as the cell has linked its transcriptional output to the energy that is available at that point: the more glucose, the more ATP, and the more transcription.
This is not to say that this is the sole role of O-GlcNAcylation. Other functions, outside of the metabolic flux, cannot be excluded. However, this would require a pool of OGT with access to its own pool of UDP-GlcNAc and one that would be maintained independently of the glucose flux-dependent pool (or that the glucose-flux pool is at non-limiting levels). One source of this pool might be through the GlcNAc salvage pathway, turning over a steady flux of O-GlcNAcylated proteins. The glucose-independent O-GlcNAcylation would also have to be regulated by the specific recruitment of OGT to particular proteins and/or promoters. Lastly, it might be possible to regulate the OGT via regulation of any phosphatases, which would remove phosphates from residues that would subsequently be O-GlcNAcylated.
Speculations and Conclusions
The O-GlcNAc field has made tremendous strides the past several years. O-GlcNAc is finally being more widely recognized as an important post-translational modification. In many respects the most significant finding is that O-GlcNAc levels are responsive to the nutrient state of the cell. This has important ramifications in understanding metabolic diseases such as diabetes. The impact of O-GlcNAc is a genome-wide one, likely impacting at least half of the transcriptionally active genome. The global levels of O-GlcNAcylation move up or down in response to changing glucose levels, acting as a genome-wide rheostat to fine-tune gene regulation at many levels. The rheostat hypothesis has also been suggested on a larger scale as well, encompassing cellular pathways involved in responding to the nutrient state of the cell [2]. As such, the full impact of O-GlcNAc on the genome and the cell cannot be understood until systems-level approaches are implemented. This is very nontrivial and will require not only systems level data of O-GlcNAc, but also how the several subsystems, (O-GlcNAc, metabolic, circadian, and transcriptional regulation) are all interconnected. The building blocks of such understanding will require a combination of many fields, protein biochemistry, cellular and molecular biology, computational, and systems biology to create an accurate picture of nuclear O-GlcNAcylation and its regulatory impact on the genome. It is becoming more and more obvious that O-GlcNAcylation is playing an important role in transcriptional regulation and should be included alongside phosphorylation when considering post-translational regulatory problems.
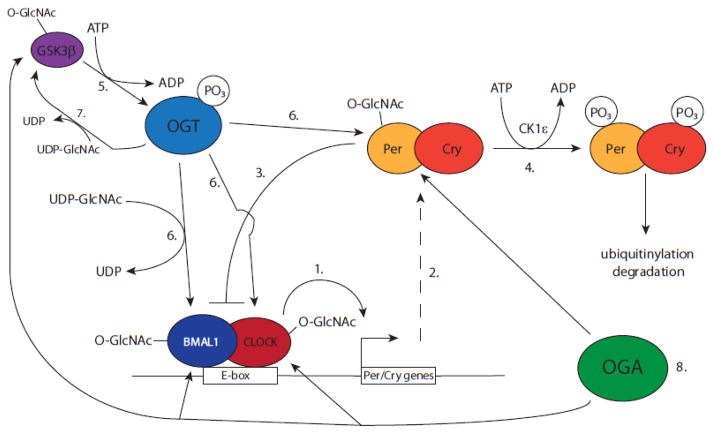
Figure 1.
Regulation of circadian clock by O-GlcNAcylation of clock components. Shown here is a schematic summary of the material in the text. 1. The clock cycle begins with the BMAL/CLOCK heterodimer HLH proteins binding to E-box sites on the Per and Cry genes. 2. Per and Cry transcription results in the synthesis of Per and Cry proteins. 3. Per and Cry proteins repress their own transcription thus completing the basic TTFL. 4. CK1ε phosphorylates Per protein which leads to its degradation via ubiquitinylation and proteasomal degradation. 5. Phosphorylation of OGT by GSK3β increases OGT activity [48]. 6. The resulting OGT activity glcnacylates Per, BMAL1, and CLOCK proteins, which increases their stability [48–50]. 7. OGT may also be regulating its phosphorylation by GSK3β, as GSK3β is a substrate of OGT [47]. 8. Finally, hypothetically, OGA may then target Per, BMAL1, and CLOCK, which would presumably result in their degradation.
Highlights.
This review article discusses recent advances in the links between O-GlcNAc and transcriptional regulation.
Discusses in detail several recent systems dependent on O-GlcNac to illustrate O-GlcNAc dynamics: Tet proteins, MLL complexes, circadian clock proteins and RNA pol II.
Suggests that promoters are nutrient sensors.
Acknowledgments
BAL would like to thank Drs. Alfonzo Fernandez and Melissa Resto for suggestions and comments. BAL acknowledges funding from the Intramural Program/CCR/NCI/NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Varki A. Essentials of glycobiology. 2. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y: 2009. [PubMed] [Google Scholar]
- 2.Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu Rev Biochem. 2011;80:825–858. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Özcan S, Andrali SS, Cantrell JEL. Modulation of transcription factor function by O-GlcNAc modification. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 1799:353–364. doi: 10.1016/j.bbagrm.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanover JA, Krause MW, Love DC. The hexosamine signaling pathway: O-GlcNAc cycling in feast or famine. Biochim Biophys Acta. 2010;1800:80–95. doi: 10.1016/j.bbagen.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marshall S, Bacote V, Traxinger RR. Discovery of a metabolic pathway mediating glucose-induced desensitization of the glucose transport system. Role of hexosamine biosynthesis in the induction of insulin resistance. J Biol Chem. 1991;266:4706–4712. [PubMed] [Google Scholar]
- 6.Broschat KO, Gorka C, Page JD, Martin-Berger CL, Davies MS, Huang HC, Gulve EA, Salsgiver WJ, Kasten TP. Kinetic characterization of human glutamine-fructose-6-phosphate amidotransferase I: potent feedback inhibition by glucosamine 6-phosphate. J Biol Chem. 2002;277:14764–14770. doi: 10.1074/jbc.M201056200. [DOI] [PubMed] [Google Scholar]
- 7.Hanover JA. Glycan-dependent signaling: O-linked N-acetylglucosamine. FASEB J. 2001;15:1865–1876. doi: 10.1096/fj.01-0094rev. [DOI] [PubMed] [Google Scholar]
- 8.Hart GW, Akimoto Y. The O-GlcNAc Modification. In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of Glycobiology. Cold Spring Harbor (NY): 2009. [PubMed] [Google Scholar]
- 9.Ruan HB, Singh JP, Li MD, Wu J, Yang X. Cracking the O-GlcNAc code in metabolism. Trends in endocrinology and metabolism: TEM. 2013;24:301–309. doi: 10.1016/j.tem.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park K, Saudek CD, Hart GW. Increased expression of beta-N-acetylglucosaminidase in erythrocytes from individuals with pre-diabetes and diabetes. Diabetes. 2010;59:1845–1850. doi: 10.2337/db09-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Virkamaki A, Daniels MC, Hamalainen S, Utriainen T, McClain D, Yki-Jarvinen H. Activation of the hexosamine pathway by glucosamine in vivo induces insulin resistance in multiple insulin sensitive tissues. Endocrinology. 1997;138:2501–2507. doi: 10.1210/endo.138.6.5172. [DOI] [PubMed] [Google Scholar]
- 12.Whelan SA, Lane MD, Hart GW. Regulation of the O-linked beta-N-acetylglucosamine transferase by insulin signaling. J Biol Chem. 2008;283:21411–21417. doi: 10.1074/jbc.M800677200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang X, Ongusaha PP, Miles PD, Havstad JC, Zhang F, So WV, Kudlow JE, Michell RH, Olefsky JM, Field SJ, Evans RM. Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature. 2008;451:964–969. doi: 10.1038/nature06668. [DOI] [PubMed] [Google Scholar]
- 14.Whelan SA, Dias WB, Thiruneelakantapillai L, Lane MD, Hart GW. Regulation of insulin receptor substrate 1 (IRS-1)/AKT kinase-mediated insulin signaling by O-Linked beta-N-acetylglucosamine in 3T3-L1 adipocytes. J Biol Chem. 2010;285:5204–5211. doi: 10.1074/jbc.M109.077818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Housley MP, Udeshi ND, Rodgers JT, Shabanowitz J, Puigserver P, Hunt DF, Hart GW. A PGC-1alpha-O-GlcNAc transferase complex regulates FoxO transcription factor activity in response to glucose. J Biol Chem. 2009;284:5148–5157. doi: 10.1074/jbc.M808890200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruan HB, Han X, Li MD, Singh JP, Qian K, Azarhoush S, Zhao L, Bennett AM, Samuel VT, Wu J, Yates JR, 3rd, Yang X. O-GlcNAc transferase/host cell factor C1 complex regulates gluconeogenesis by modulating PGC-1alpha stability. Cell metabolism. 2012;16:226–237. doi: 10.1016/j.cmet.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dentin R, Hedrick S, Xie J, Yates J, 3rd, Montminy M. Hepatic glucose sensing via the CREB coactivator CRTC2. Science. 2008;319:1402–1405. doi: 10.1126/science.1151363. [DOI] [PubMed] [Google Scholar]
- 18.Forsythe ME, Love DC, Lazarus BD, Kim EJ, Prinz WA, Ashwell G, Krause MW, Hanover JA. Caenorhabditis elegans ortholog of a diabetes susceptibility locus: oga-1 (O-GlcNAcase) knockout impacts O-GlcNAc cycling, metabolism, and dauer. Proc Natl Acad Sci U S A. 2006;103:11952–11957. doi: 10.1073/pnas.0601931103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanover JA, Forsythe ME, Hennessey PT, Brodigan TM, Love DC, Ashwell G, Krause M. A Caenorhabditis elegans model of insulin resistance: altered macronutrient storage and dauer formation in an OGT-1 knockout. Proc Natl Acad Sci U S A. 2005;102:11266–11271. doi: 10.1073/pnas.0408771102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Love DC, Ghosh S, Mondoux MA, Fukushige T, Wang P, Wilson MA, Iser WB, Wolkow CA, Krause MW, Hanover JA. Dynamic O-GlcNAc cycling at promoters of Caenorhabditis elegans genes regulating longevity, stress, and immunity. Proc Natl Acad Sci U S A. 2010;107:7413–7418. doi: 10.1073/pnas.0911857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sekine O, Love DC, Rubenstein DS, Hanover JA. Blocking O-Linked GlcNAc Cycling in Drosophila Insulin-producing Cells Perturbs Glucose-Insulin Homeostasis. J Biol Chem. 2010;285:38684–38691. doi: 10.1074/jbc.M110.155192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang X, Zhang F, Kudlow JE. Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: coupling protein O-GlcNAcylation to transcriptional repression. Cell. 2002;110:69–80. doi: 10.1016/s0092-8674(02)00810-3. [DOI] [PubMed] [Google Scholar]
- 23.Gambetta MC, Oktaba K, Müller J. Essential Role of the Glycosyltransferase Sxc/Ogt in Polycomb Repression. Science. 2009;325:93–96. doi: 10.1126/science.1169727. [DOI] [PubMed] [Google Scholar]
- 24.Myers SA, Panning B, Burlingame AL. Polycomb repressive complex 2 is necessary for the normal site-specific O-GlcNAc distribution in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2011;108:9490–9495. doi: 10.1073/pnas.1019289108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sinclair DA, Syrzycka M, Macauley MS, Rastgardani T, Komljenovic I, Vocadlo DJ, Brock HW, Honda BM. Drosophila O-GlcNAc transferase (OGT) is encoded by the Polycomb group (PcG) gene, super sex combs (sxc) Proc Natl Acad Sci U S A. 2009;106:13427–13432. doi: 10.1073/pnas.0904638106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bond MR, Hanover JA. O-GlcNAc Cycling: A Link Between Metabolism and Chronic Disease. Annual review of nutrition. 2013;33:205–229. doi: 10.1146/annurev-nutr-071812-161240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Love DC, Krause MW, Hanover JA. O-GlcNAc cycling: emerging roles in development and epigenetics. Semin Cell Dev Biol. 2010;21:646–654. doi: 10.1016/j.semcdb.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slawson C, Copeland RJ, Hart GW. O-GlcNAc signaling: a metabolic link between diabetes and cancer? Trends Biochem Sci. 2010;35:547–555. doi: 10.1016/j.tibs.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slawson C, Hart GW. O-GlcNAc signalling: implications for cancer cell biology. Nature reviews. Cancer. 2011;11:678–684. doi: 10.1038/nrc3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011;25:2436–2452. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Q, Chen Y, Bian C, Fujiki R, Yu X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2013;493:561–564. doi: 10.1038/nature11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deplus R, Delatte B, Schwinn MK, Defrance M, Mendez J, Murphy N, Dawson MA, Volkmar M, Putmans P, Calonne E, Shih AH, Levine RL, Bernard O, Mercher T, Solary E, Urh M, Daniels DL, Fuks F. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 2013;32:645–655. doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vella P, Scelfo A, Jammula S, Chiacchiera F, Williams K, Cuomo A, Roberto A, Christensen J, Bonaldi T, Helin K, Pasini D. Tet proteins connect the O-linked N-acetylglucosamine transferase Ogt to chromatin in embryonic stem cells. Mol Cell. 2013;49:645–656. doi: 10.1016/j.molcel.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 34.Konrad RJ, Zhang F, Hale JE, Knierman MD, Becker GW, Kudlow JE. Alloxan is an inhibitor of the enzyme O-linked N-acetylglucosamine transferase. Biochem Biophys Res Commun. 2002;293:207–212. doi: 10.1016/S0006-291X(02)00200-0. [DOI] [PubMed] [Google Scholar]
- 35.Lee TN, Alborn WE, Knierman MD, Konrad RJ. Alloxan is an inhibitor of O-GlcNAc-selective N-acetyl-beta-D-glucosaminidase. Biochem Biophys Res Commun. 2006;350:1038–1043. doi: 10.1016/j.bbrc.2006.09.155. [DOI] [PubMed] [Google Scholar]
- 36.Fujiki R, Chikanishi T, Hashiba W, Ito H, Takada I, Roeder RG, Kitagawa H, Kato S. GlcNAcylation of a histone methyltransferase in retinoic-acid-induced granulopoiesis. Nature. 2009;459:455–459. doi: 10.1038/nature07954. [DOI] [PubMed] [Google Scholar]
- 37.Fujiki R, Hashiba W, Sekine H, Yokoyama A, Chikanishi T, Ito S, Imai Y, Kim J, He HH, Igarashi K, Kanno J, Ohtake F, Kitagawa H, Roeder RG, Brown M, Kato S. GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature. 2011;480:557–560. doi: 10.1038/nature10656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakabe K, Hart GW. O-GlcNAc transferase regulates mitotic chromatin dynamics. J Biol Chem. 2010;285:34460–34468. doi: 10.1074/jbc.M110.158170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fong JJ, Nguyen BL, Bridger R, Medrano EE, Wells L, Pan S, Sifers RN. beta-N-Acetylglucosamine (O-GlcNAc) is a novel regulator of mitosis-specific phosphorylations on histone H3. J Biol Chem. 2012;287:12195–12203. doi: 10.1074/jbc.M111.315804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang S, Roche K, Nasheuer HP, Lowndes NF. Modification of histones by sugar beta-N-acetylglucosamine (GlcNAc) occurs on multiple residues, including histone H3 serine 10, and is cell cycle-regulated. J Biol Chem. 2011;286:37483–37495. doi: 10.1074/jbc.M111.284885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sakabe K, Wang Z, Hart GW. Beta-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proc Natl Acad Sci U S A. 2010;107:19915–19920. doi: 10.1073/pnas.1009023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang EE, Kay SA. Clocks not winding down: unravelling circadian networks. Nat Rev Mol Cell Biol. 2010;11:764–776. doi: 10.1038/nrm2995. [DOI] [PubMed] [Google Scholar]
- 43.Marcheva B, Ramsey KM, Peek CB, Affinati A, Maury E, Bass J. Circadian clocks and metabolism. Handbook of experimental pharmacology. 2013:127–155. doi: 10.1007/978-3-642-25950-0_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brown SA, Kowalska E, Dallmann R. (Re)inventing the circadian feedback loop. Dev Cell. 2012;22:477–487. doi: 10.1016/j.devcel.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 45.Iitaka C, Miyazaki K, Akaike T, Ishida N. A role for glycogen synthase kinase-3beta in the mammalian circadian clock. J Biol Chem. 2005;280:29397–29402. doi: 10.1074/jbc.M503526200. [DOI] [PubMed] [Google Scholar]
- 46.Martinek S, Inonog S, Manoukian AS, Young MW. A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell. 2001;105:769–779. doi: 10.1016/s0092-8674(01)00383-x. [DOI] [PubMed] [Google Scholar]
- 47.Lubas WA, Hanover JA. Functional expression of O-linked GlcNAc transferase. Domain structure and substrate specificity. J Biol Chem. 2000;275:10983–10988. doi: 10.1074/jbc.275.15.10983. [DOI] [PubMed] [Google Scholar]
- 48.Kaasik K, Kivimae S, Allen JJ, Chalkley RJ, Huang Y, Baer K, Kissel H, Burlingame AL, Shokat KM, Ptacek LJ, Fu YH. Glucose sensor O-GlcNAcylation coordinates with phosphorylation to regulate circadian clock. Cell metabolism. 2013;17:291–302. doi: 10.1016/j.cmet.2012.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim EY, Jeong EH, Park S, Jeong HJ, Edery I, Cho JW. A role for O-GlcNAcylation in setting circadian clock speed. Genes Dev. 2012;26:490–502. doi: 10.1101/gad.182378.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li MD, Ruan HB, Hughes ME, Lee JS, Singh JP, Jones SP, Nitabach MN, Yang X. O-GlcNAc signaling entrains the circadian clock by inhibiting BMAL1/CLOCK ubiquitination. Cell metabolism. 2013;17:303–310. doi: 10.1016/j.cmet.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okano S, Akashi M, Hayasaka K, Nakajima O. Unusual circadian locomotor activity and pathophysiology in mutant CRY1 transgenic mice. Neuroscience letters. 2009;451:246–251. doi: 10.1016/j.neulet.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 52.Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, Fitzgerald GA. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chesnut JD, Stephens JH, Dahmus ME. The interaction of RNA polymerase II with the adenovirus-2 major late promoter is precluded by phosphorylation of the C-terminal domain of subunit IIa. J Biol Chem. 1992;267:10500–10506. [PubMed] [Google Scholar]
- 54.Chapman RD, Heidemann M, Albert TK, Mailhammer R, Flatley A, Meisterernst M, Kremmer E, Eick D. Transcribing RNA Polymerase II Is Phosphorylated at CTD Residue Serine-7. Science. 2007;318:1780–1782. doi: 10.1126/science.1145977. [DOI] [PubMed] [Google Scholar]
- 55.Egloff S, Dienstbier M, Murphy S. Updating the RNA polymerase CTD code: adding gene-specific layers. Trends in genetics : TIG. 2012;28:333–341. doi: 10.1016/j.tig.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 56.Hintermair C, Heidemann M, Koch F, Descostes N, Gut M, Gut I, Fenouil R, Ferrier P, Flatley A, Kremmer E, Chapman RD, Andrau JC, Eick D. Threonine-4 of mammalian RNA polymerase II CTD is targeted by Polo-like kinase 3 and required for transcriptional elongation. EMBO J. 2012;31:2784–2797. doi: 10.1038/emboj.2012.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hsin JP, Sheth A, Manley JL. RNAP II CTD phosphorylated on threonine-4 is required for histone mRNA 3′ end processing. Science. 2011;334:683–686. doi: 10.1126/science.1206034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chapman RD, Heidemann M, Hintermair C, Eick D. Molecular evolution of the RNA polymerase II CTD. Trends in genetics : TIG. 2008;24:289–296. doi: 10.1016/j.tig.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 59.Lux C, Albiez H, Chapman RD, Heidinger M, Meininghaus M, Brack-Werner R, Lang A, Ziegler M, Cremer T, Eick D. Transition from initiation to promoter proximal pausing requires the CTD of RNA polymerase II. Nucleic Acids Research. 2005;33:5139–5144. doi: 10.1093/nar/gki802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Naar AM, Taatjes DJ, Zhai W, Nogales E, Tjian R. Human CRSP interacts with RNA polymerase II CTD and adopts a specific CTD-bound conformation. Genes Dev. 2002;16:1339–1344. doi: 10.1101/gad.987602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taatjes DJ, Naar AM, Andel F, 3rd, Nogales E, Tjian R. Structure, function, and activator-induced conformations of the CRSP coactivator. Science. 2002;295:1058–1062. doi: 10.1126/science.1065249. [DOI] [PubMed] [Google Scholar]
- 62.Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 64.Zhou Q, Li T, Price DH. RNA polymerase II elongation control. Annu Rev Biochem. 2012;81:119–143. doi: 10.1146/annurev-biochem-052610-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gebara MM, Sayre MH, Corden JL. Phosphorylation of the carboxy-terminal repeat domain in RNA polymerase II by cyclin-dependent kinases is sufficient to inhibit transcription. J Cell Biochem. 1997;64:390–402. [PubMed] [Google Scholar]
- 66.Bartkowiak B, Mackellar AL, Greenleaf AL. Updating the CTD Story: From Tail to Epic. Genetics research international. 2011;2011:623718. doi: 10.4061/2011/623718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang DW, Rodriguez-Molina JB, Tietjen JR, Nemec CM, Ansari AZ. Emerging Views on the CTD Code. Genetics research international. 2012;2012:347214. doi: 10.1155/2012/347214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kelly WG, Dahmus ME, Hart GW. RNA polymerase II is a glycoprotein. Modification of the COOH-terminal domain by O-GlcNAc. J Biol Chem. 1993;268:10416–10424. [PubMed] [Google Scholar]
- 69.Comer FI, Hart GW. Reciprocity between O-GlcNAc and O-phosphate on the carboxyl terminal domain of RNA polymerase II. Biochemistry. 2001;40:7845–7852. doi: 10.1021/bi0027480. [DOI] [PubMed] [Google Scholar]
- 70.Ranuncolo SM, Ghosh S, Hanover JA, Hart GW, Lewis BA. Evidence of the involvement of O-GlcNAc-modified human RNA polymerase II CTD in transcription in vitro and in vivo. J Biol Chem. 2012;287:23549–23561. doi: 10.1074/jbc.M111.330910. [DOI] [PMC free article] [PubMed] [Google Scholar]