Abstract
Mammalian neuroepithelial stem cells divide using a polarized form of cytokinesis, which is not well understood. The cytokinetic furrow cleaves the cell by ingressing from basal to apical, forming the midbody at the apical membrane. The midbody mediates abscission by recruiting many factors, including the Kinesin-6 family member Kif20b. In developing embryos, Kif20b mRNA is most highly expressed in neural stem/progenitor cells. A loss-of-function mutant in Kif20b, magoo, was found in a forward genetic screen. magoo has a small cerebral cortex, with reduced production of progenitors and neurons, but preserved layering. In contrast to other microcephalic mouse mutants, mitosis and cleavage furrows of cortical stem cells appear normal in magoo. However, apical midbodies show changes in number, shape and positioning relative to the apical membrane. Interestingly, the disruption of abscission does not appear to result in binucleate cells, but in apoptosis. Thus, Kif20b is required for proper midbody organization and abscission in polarized cortical stem cells and has a crucial role in the regulation of cerebral cortex growth.
Keywords: Microcephaly, Cortical development, Kinesin-6, Kif20b, Cytokinesis, Abscission, Neural stem cells, Mouse
INTRODUCTION
Most studies of cytokinesis are performed in isolated single cells. Yet in developing organisms, cells must divide within a tissue, often maintaining cell junctions and polarity. Furthermore, the control of symmetric versus asymmetric divisions of stem cells depends on the partitioning of cell fate determinants. This partitioning may be directed by spindle orientation, but it is accomplished through cytokinesis. The mechanisms of cytokinesis in developing vertebrate tissues are poorly understood.
Neural stem cells are highly polarized and undergo polarized cytokinesis. The early vertebrate brain is a pseudostratified epithelium of tall, bipolar stem cells that extend from the ventricle, where they have their apical endfeet, up to the basal lamina beneath the meninges. The nuclei undergo interkinetic nuclear migration during the cell cycle, moving basally within the cell for S phase, then to the apical surface for M phase, continuously maintaining apical and basal attachments. Cytokinetic furrowing starts at the basal side and ingresses toward the apical membrane, where the midbody is formed (Dubreuil et al., 2007). Cytokinetic abscission is mediated by the midbody at the ventricle only after the daughter nuclei have migrated away (Kosodo et al., 2008). The mechanisms of this specialized cytokinesis are not understood.
Neural stem cells of the cerebral cortex (also called apical progenitors) can divide symmetrically or asymmetrically. Early symmetric divisions expand the cortical surface area, and later asymmetric divisions increase cortical thickness by giving rise to basal progenitors, neurons and glia (for a review, see Tan and Shi, 2013). During symmetric division, it is thought that apical membrane and junctions must be partitioned equally between the two daughters to produce equal fates (Costa et al., 2008; Konno et al., 2008; Kosodo et al., 2004; Marthiens and ffrench-Constant, 2009). Since the apical membrane is tiny relative to the total cell membrane, splitting it requires exquisite control not only of the mitotic spindle angle, but also of cleavage furrow ingression and abscission. Asymmetric division also occurs with vertical cleavage (Noctor et al., 2008), but may require less precision. Basal progenitors, which are multipolar, can only divide symmetrically to produce two neurons (Kriegstein and Alvarez-Buylla, 2009).
Perhaps due to these complex demands on cell division, the developing cortex is especially vulnerable to mutations affecting cell division. Mutations known to cause human primary microcephaly disrupt genes involved in mitosis (Gilmore and Walsh, 2013). Surprisingly, although most microcephaly genes are ubiquitously expressed, non-brain tissues usually develop normally.
Cytokinesis in animal cells consists of two distinct processes: furrowing and abscission (Green et al., 2012). Furrowing is a rapid process in which the contractile membrane cleaves the cell body and compacts the microtubules of the central spindle into a structure called the midbody within a narrow intercellular bridge. Then, the midbody mediates abscission by recruiting factors to sever the bridge (Gromley et al., 2003; Hu et al., 2012). The midbody proteome contains over 150 proteins (Skop et al., 2004), and some key players in abscission have recently been defined (Agromayor and Martin-Serrano, 2013; Chen et al., 2012; Elia et al., 2011; Gromley et al., 2005; Guizetti et al., 2011; Zhao et al., 2006).
Among the midbody proteins involved in abscission are members of the Kinesin-6 family of plus-end-directed microtubule motors. Vertebrates have three Kinesin-6 family members (Miki et al., 2005). Kif23 (MKLP1) and Kif20a (MKLP2) each have orthologs in invertebrates and play independent roles in cytokinesis (Adams et al., 1998; Cesario et al., 2006; Glotzer, 2009; Neef et al., 2006; Raich et al., 1998; Zhu et al., 2005). Kif20b, by contrast, is vertebrate specific, and its role in cytokinesis is less understood. Kif20b was originally cloned as a mitotic phosphoprotein called Mphosph1, MPP1 (Matsumoto-Taniura et al., 1996; Westendorf et al., 1994) or KRMP1 (Kamimoto et al., 2001). KIF20B knockdown in dividing human cell lines did not disrupt cytokinetic furrowing, but increased abscission failures (Abaza et al., 2003; Kanehira et al., 2007).
Here, we present the first analysis of a mouse mutant for a Kinesin-6. We show that Kif20b mutation in mice disrupts cerebral cortex growth, with reduced neurogenesis but preserved lamination. Our analysis suggests a requirement for Kif20b in the specialized cell divisions of cortical neural stem cells, in particular for cytokinetic midbody organization. This work provides a novel model of microcephaly and suggests that the regulation of cytokinesis mechanisms plays an important role in building complex vertebrate tissues.
RESULTS
The magoo mutant has a small, thin cerebral cortex with preserved lamination
Previously, an ENU screen for defective cortical development identified the magoo mouse mutant as carrying a recessive, perinatal lethal mutation with fully penetrant microcephaly (Dwyer et al., 2011). Heterozygotes appear normal.
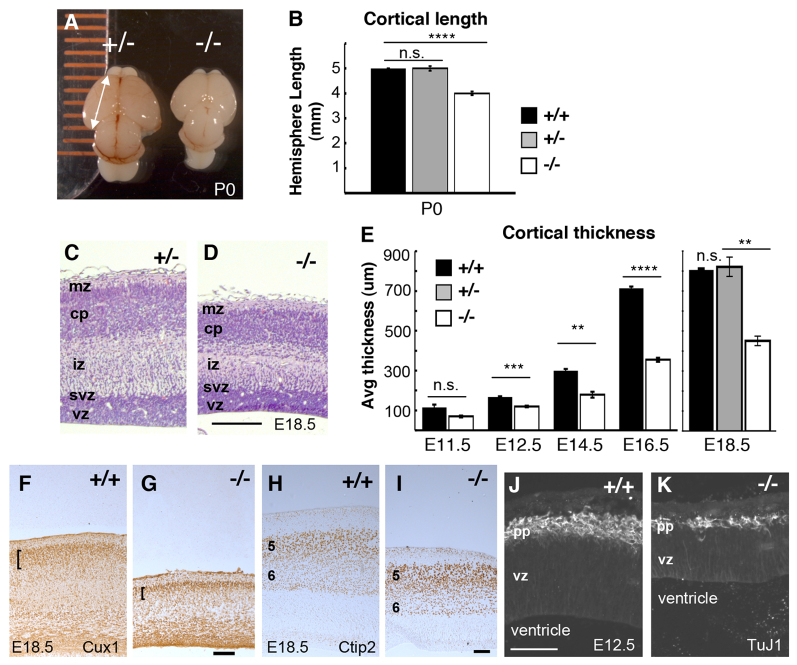
When collected at birth, the forebrains of magoo mutants are consistently smaller and rounder than those of control littermates. Cortical hemisphere lengths of magoo homozygous mutants averaged 83% of those of wild-type (+/+) or heterozygous (+/-) controls, which were indistinguishable (Fig. 1A,B). Cortical sections of E18.5 mutants show reduced thickness (Fig. 1C,D). Younger mutant cortices also have reduced thickness and area (Fig. 1E; supplementary material Fig. S1). As development proceeds, mutant cortices do increase in thickness, but remain thinner than controls. Body size is also affected, averaging 72% of control size at E16.5, but morphogenesis of the body and organs below the neck appears normal (data not shown).
Fig. 1.
magoo mutant cortex has reduced length and thickness but preserved layer structure. (A) Dorsal view of heterozygous control (+/-) and magoo mutant (-/-) newborn [postnatal day (P) 0] mouse cortices. (B) The average length (mm) ± s.e.m. of eight wild-type (+/+), 20 heterozygous and 24 mutant P0.5 cortical hemispheres. ****P<1.0×10-8; n.s., not significant. (C,D) Cortical sections from comparable levels of control and homozygous magoo mutant littermates stained with H&E. In the mutant, cortical layers are thinner but positioned normally as marginal zone (mz), cortical plate neurons (cp), intermediate zone (iz), subventricular zone (svz) and ventricular zone (vz). (E) The average cortical thickness (μm) ± s.e.m. of magoo mutants is less than that of controls at all ages tested. Number of brains of each genotype: E11.5, 2; E12.5 and E14.5, 4; E16.5, 5; E18.5, 2. **P≤0.01, ***P<0.005, ****P<0.0001. (F,G) Cux1+ cells are present in superficial layers of both control and magoo mutant caudal cortex sections (brackets). (H,I) Ctip2 immunostaining on sections of middle cortex shows that cortical layers 5 and 6 are distinguishable and in the correct order in magoo mutants. (J,K) At E12.5, the preplate (pp) and vz are both thinner in mutants, but positioned normally. TuJ1 labels neuronal tubulin. Scale bars: 100 μm.
Interestingly, the layered structure of the cortex is preserved in mutants (Fig. 1D,F-I). The cortical plate contains a superficial layer marked by Cux1 and deeper layers 5 and 6 marked by Ctip2 (Bcl11b - Mouse Genome Informatics); these are thinner than in controls. Earlier, at E12.5, the first-born neuronal layer (preplate) is thin but present and properly positioned in mutants (Fig. 1J,K). Together, these data suggest that in the magoo mutant cortex fewer neurons are generated, but they are able to migrate out of the ventricular zone to form normally ordered layers.
The magoo mutant cortex exhibits reduced production of basal progenitors
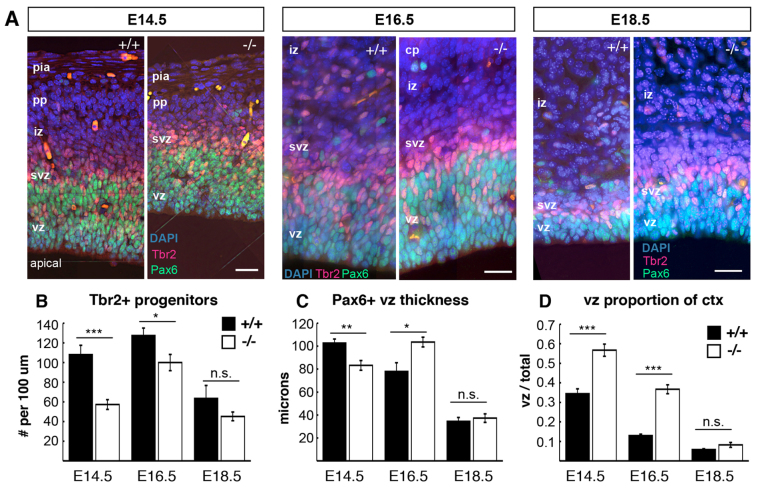
To examine the neural progenitor populations in the magoo mutant cortex, sections of control and mutant cortices at three ages were immunostained for Pax6 and Tbr2 (Eomes - Mouse Genome Informatics) to mark apical and basal progenitor nuclei, respectively (Englund et al., 2005). In both mutant and control, Tbr2+ nuclei occupy the subventricular zone (svz), basal to the Pax6+ apical progenitor nuclei in the ventricular zone (vz) (Fig. 2A). However, mutants have fewer Tbr2+ nuclei per field (Fig. 2B). The vz was reduced in thickness and length in mutants at E14.5 (Fig. 2C; supplementary material Fig. S2A). The density of apical progenitors was similar in controls and mutants at E13.5 and E15.5, although the neocortical area was smaller in mutants at E13.5 (supplementary material Fig. S2B,C), suggesting that the total number of apical progenitors is reduced at early ages. The vz thickness was slightly increased at E16.5 in mutants, which is perhaps explained by the presence of more basal progenitors in the vz at this age, suggesting a delayed peak production of Tbr2+ progenitors (supplementary material Fig. S2D). Most striking was the large proportion of cortical thickness occupied by the vz in mutants, since other layers are so thin (Fig. 2D). Together with the results shown in Fig. 1, these data suggest that in magoo mutants the output of progeny by apical progenitors is greatly reduced, but their capacity to produce daughters with ordered layer fates is intact.
Fig. 2.
magoo mutant cortex has reduced production of progenitors. (A) Pax6 (green) and Tbr2 (red) mark apical and basal progenitors, respectively, in control and magoo mutant cortical sections. Scale bars: 20 μm for each age pair. (B) The number of Tbr2+ progenitors per 100 μm length of ventricle is significantly reduced in magoo mutants at E14.5 and E16.5 compared with controls. Number of brains for +/+ and -/-: E14.5 and E16.5, 3 and 3, n=6 fields each; E18.5, 2 and 2, n=4 fields each. (C) The average thickness of the vz (Pax6+) is slightly reduced in E14.5 and increased in E16.5 mutants. Number of brains for +/+ and -/-: E14.5 and E16.5, 3 and 3, n=6 fields each; E18.5, 2 and 2, n=4 fields each. (D) The vz makes up a larger proportion of the total cortical thickness in magoo mutant cortices at E14.5 and E16.5. Number of brains for +/+ and -/-: E14.5, 3 and 3, n=5 and n=6 fields, respectively; E16.5, 3 and 3, n=4 and n=6 fields, respectively; E18.5, 2 and 2, n=4 fields each. *P<0.05, **P<0.01, ***P<0.001.
The magoo mutant carries a splice mutation in the Kif20b kinesin gene
To understand the molecular cause of the severely reduced neural stem cell productivity in the magoo phenotype, we positionally cloned the mutant gene. Previously, magoo was mapped to a 3.9 Mb interval (Dwyer et al., 2011). We further refined the interval using additional recombinant animals and a new SSLP marker (see Materials and methods; Fig. 3A). Remarkably, this 0.94 Mb interval contains only one complete gene, Kif20b, and one exon of Pank1 (UCSC Genome Browser). The exons and flanking introns of these genes were sequenced in mutants and compared with the reference C57BL/6J. A single change was identified in an intron of Kif20b: a T-to-G mutation near the splice acceptor for exon 19 (Fig. 3A, asterisk). We confirmed that this mutation is absent in parental strains, heterozygous in all carriers, and homozygous in all mutants.
Fig. 3.
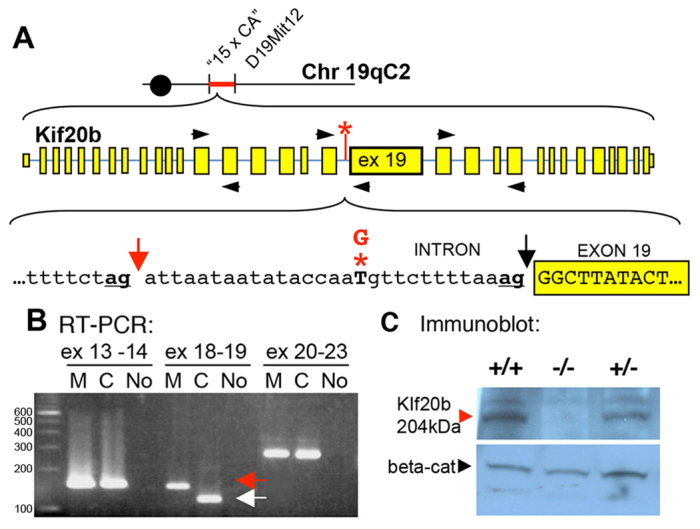
magoo is a splice mutant of Kif20b in which Kif20b protein is reduced to an undetectable level. (A) The magoo mutation was mapped between microsatellite markers ‘15 x CA’ and D19Mit12. magoo carries a T-to-G mutation (red asterisk) 13 bases upstream of exon 19 of Kif20b. This disrupts the normal splice acceptor (black arrow) and favors the ‘ag’ 29 bp upstream (red arrow). (B) RT-PCR detects aberrant splicing of Kif20b exon 19 in magoo mutants. Primers (black arrowheads in A) were designed to span splice sites between exons 13 and 14, 18 and 19, and 20 and 23. The exon 18-19 primers amplified a larger band from mutant cDNA (red arrow) than from the control (white arrow). Identical results were seen in three independent experiments. M, mutant; C, control; No, no template cDNA; ex, exon. (C) Immunoblot with antiserum recognizing the N-terminus of mouse Kif20b detects a 204 kDa band in brain lysates from +/+ and +/- E12.5 embryos, but not -/- embryos, even after film overexposure. Anti-β-catenin (beta-cat) is a loading control.
To determine whether this intronic mutation in Kif20b causes a splicing defect, we performed RT-PCR to compare Kif20b mRNA in mutant and normal tissue. Primers spanning the splice junction of exons 18 and 19 produced a longer than expected band from mutant samples (Fig. 3B). Sequencing of this band showed that 29 bases of intronic sequence are included in the mutant mRNA due to mis-splicing to an ‘ag’ 29 bases upstream of the normal splice acceptor (Fig. 3A, underlined ‘ag’ at red arrow). This addition of 29 bases to the message would cause a frameshift and multiple premature stop codons about halfway through the open reading frame. The premature stop codons appear to cause nonsense-mediated decay of the mRNA, since semi-quantitative RT-PCR showed that the mRNA was less abundant in magoo mutants than in controls (data not shown).
To test whether the splice site mutation reduces Kif20b protein levels, we compared immunoblots of control and magoo mutant brain lysates. We raised rabbit antisera against peptides of mouse Kif20b, since the published antibody to human KIF20B (Abaza et al., 2003) did not detect mouse Kif20b on denaturing blots. Using antisera to either the N-terminal or C-terminal peptides, we were able to detect a 204 kDa band in brain lysates from controls and heterozygotes, but not from magoo mutants (Fig. 3C).
To further confirm that the magoo phenotype is caused by the Kif20b mutation, we performed a genetic complementation test between magoo and another recessive allele of Kif20b. The 33c mutant in Kif20b was isolated in a separate genetic screen (García-García et al., 2005). It carries a missense mutation that changes an R residue to G in loop 11 of the motor head domain of Kif20b. By analogy to similar mutations in other kinesins, this change is likely to disrupt microtubule affinity and motor function (Hirokawa and Noda, 2008; Ebbing et al., 2008; Reid et al., 2002; Shimizu et al., 2000). We intercrossed the two mutant lines and collected 13 transheterozygous embryos. The two alleles failed to complement: all 13 transheterozygotes had small, misshapen forebrains and heads. Since this test confirmed that both mutations are in Kif20b, 33c was renamed mcbarker, after Mr Magoo’s pet dog.
Kif20b protein localizes to cytokinetic microtubule structures of normal but not magoo mutant embryonic fibroblasts
Previous work in human cell lines indicated that KIF20B protein localizes to the central spindle during cytokinetic furrowing and the midbody during abscission (Abaza et al., 2003; Kanehira et al., 2007). To examine mouse Kif20b protein localization in control and mutant cells, we performed immunocytochemistry on mouse embryonic fibroblasts (MEFs). Aurora B kinase was used as a marker for the central spindle in furrowing cells and the midbody during abscission. In control MEFs, Kif20b staining was evident in the nucleus in interphase, dispersed in prophase and metaphase, associated with the central spindle during cytokinetic furrowing and with the midbody during abscission (Fig. 4, +/+ cells). These findings match those from human cells, suggesting a role for Kif20b in late cytokinesis (Abaza et al., 2003; Kanehira et al., 2007). Interestingly, Kif20b appeared concentrated at the outer flanks of the aurora B midbody signal (Fig. 4, late cytokinesis, +/+ cell). By contrast, in magoo mutant MEFs, Kif20b signal was undetectable, confirming that Kif20b protein is absent or severely reduced in magoo mutants (Fig. 4, -/- cells).
Fig. 4.
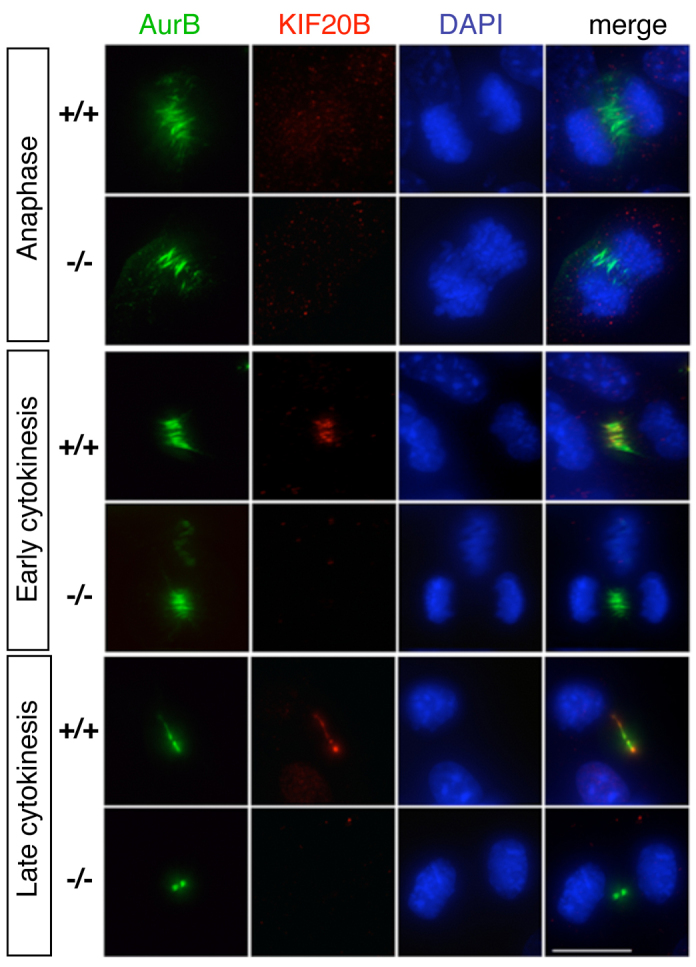
Immunostaining detects Kif20b protein in the central spindle and midbody of normal but not magoo mutant MEFs. Fibroblasts cultured from E12.5 magoo mutant and control littermate mouse embryos were immunostained for Kif20b (red) and aurora B kinase (AurB, green). In control anaphase cells, aurora B localizes to microtubules of the central spindle, and Kif20b is barely seen as a few small puncta. In early cytokinesis, Kif20b protein is detected on the compacting central spindle of control but not mutant cells. In late cytokinesis, Kif20b appears concentrated at the inner and outer flanks of the midbody in control cells but is not detectable in mutant midbodies. Scale bar: 15 μm.
Kif20b is expressed in germinal zones of the brain and localizes to midbodies of dividing progenitors
To examine whether Kif20b could play a role in the division of neural stem/progenitor cells, we examined its expression in the developing brain. Indeed, Kif20b mRNA is detected in the germinal zones lining the brain ventricles, being highest at E14.5 (Fig. 5A-D). Interestingly, the signal appears stronger at the apical side of the vz, suggesting that cortical apical progenitors upregulate Kif20b mRNA during G2/M, as HeLa cells do (Abaza et al., 2003). Kif20b message was very low or undetectable in postmitotic layers.
Fig. 5.
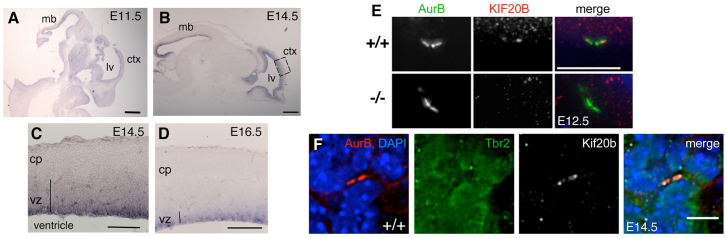
Kif20b is expressed in cortical apical and basal progenitors. (A,B) Kif20b mRNA is detected in the neuroepithelial progenitor layers of control brains. ctx, cortex; lv, lateral ventricle; mb, midbrain. (C,D) Kif20b mRNA is expressed most strongly in the apical vz of the cortex. (E) Kif20b protein co-localizes with AurB in midbodies of normal apical progenitors but is undetectable in mutants. (F) Kif20b protein is detected in midbodies of presumed basal progenitors in the svz. n=6 of 6 midbodies. Scale bars: 1 mm in A,B; 250 μm in C,D; 7.5 μm in E; 5 μm in F.
Kif20b protein localization in cortical stem and progenitor cells is similar to that in MEFs. It is detected in cytokinetic midbodies of control apical progenitors at the ventricular surface, but is absent in magoo mutants (Fig. 5E). In the svz, midbodies of dividing basal progenitors also contain Kif20b (Fig. 5F).
Indices of mitosis and S phase appear normal in Kif20bmagoo mutant cortex
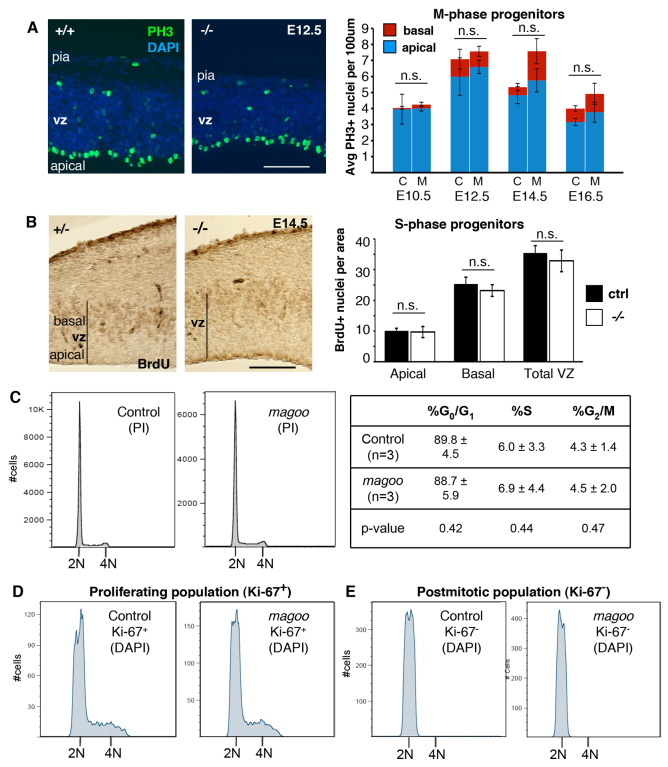
Since known human and mouse microcephaly genes disrupt mitosis in cortical progenitors (Feng and Walsh, 2004; Lizarraga et al., 2010), we analyzed mitosis in magoo mutant cortices. Cortical sections were immunolabeled with anti-phospho-histone H3 (PH3) to detect M-phase nuclei and determine whether mitosis occurred at the proper location and frequency. In both control and mutant brains, PH3+ nuclei were primarily detected at the ventricular surface, with a smaller number in the svz, indicating the M-phase apical and basal progenitors, respectively (Fig. 6A). No significant differences were observed in the positions or densities of these mitotic nuclei in mutant cortices. These data suggest that in magoo mutant brains mitosis occurs at the proper locations and rates.
Fig. 6.
Mitotic parameters are similar in control and magoo mutant developing cortices. (A) (Left) Sections through control and mutant cortex stained with DAPI and immunostained for anti-phospho-histone H3 (PH3, green), a marker of cells in prophase through anaphase. Many PH3+ nuclei line the ventricle, indicating M-phase apical progenitors; the few PH3+ nuclei positioned basally represent basal progenitors. (Right) The average (± s.e.m.) numbers of apical and basal PH3+ nuclei per 100 μm length of ventricle were similar in control (C) and mutant (M) cortical sections. Number of brains for control (+/+, unless otherwise stated) and -/-: E10.5, 4 each; E12.5, 3 and 6; E14.5, 3 (2 +/+, 1 +/-) and 5; E16.5, 4 (2 +/+, 2 +/-) and 3. Two fields of several hundred microns were counted per brain. (B) (Left) Uptake of BrdU labels S-phase progenitors at the basal side of the ventricular zone (vz) in both control and mutant. (Right) The average (± s.e.m.) number of BrdU+ nuclei in the apical vz, basal vz, or total in a defined field (10,000 μm2). n=6 control (3 +/+, 3 +/-) and n=5 mutant fields. (C-E) No significant difference in DNA content was detected by flow cytometry in magoo mutant cortical cells. (C) DNA content of dissociated cells was analyzed by propidium iodide (PI) staining. The average percentage of cells in each cell cycle phase (± s.d.) is shown from three experiments. (D,E) The DNA content of neither proliferative (Ki-67+) nor postmitotic (Ki-67-, presumed neurons) cells differs in mutant cortices, with no evidence of tetraploidy. Histograms are representative of three independent experiments with pairs of E14.5 mutant and control cortices. Scale bars: 100 μm.
To label cortical progenitors in S phase, bromo-deoxyuridine (BrdU) was injected into pregnant dams for a 1-hour uptake by embryos, and detected in cortical sections with anti-BrdU antibody. In both control and mutant brains, BrdU+ nuclei were primarily detected in the basal vz as expected (Fig. 6B). No significant differences were detected in the density or distribution of S-phase (BrdU+) nuclei.
To complement these histological analyses, flow cytometry was used to assay the DNA content of dissociated cortical cells (Fig. 6C,D). Neither the total population nor the subpopulations of progenitors and neurons showed an increase in the proportion of cells in mitosis, in S phase or with polyploidy in the mutant brains. Together with the mitotic index (PH3) and S-phase index (BrdU) analyses, these data indicate that Kif20b mutant neural progenitors are not blocked in mitosis, and that loss of Kif20b does not cause any significant increase in polyploid neurons.
Cytokinetic midbodies of apical progenitors are misaligned and wider in Kif20bmagoo mutant cortex
KIF20B knockdown in human cells increases cytokinesis failures (Abaza et al., 2003; Kanehira et al., 2007), so we hypothesized that magoo mutant cortical progenitors have defects in cytokinesis. Cytokinesis of apical cortical progenitors occurs with predominantly vertical cleavages (perpendicular to the ventricular surface) (Noctor et al., 2008), in which furrows ingress asymmetrically from basal to apical and form the midbody at the ventricle. Abscission occurs after daughter nuclei migrate basally in G1 (Dubreuil et al., 2007).
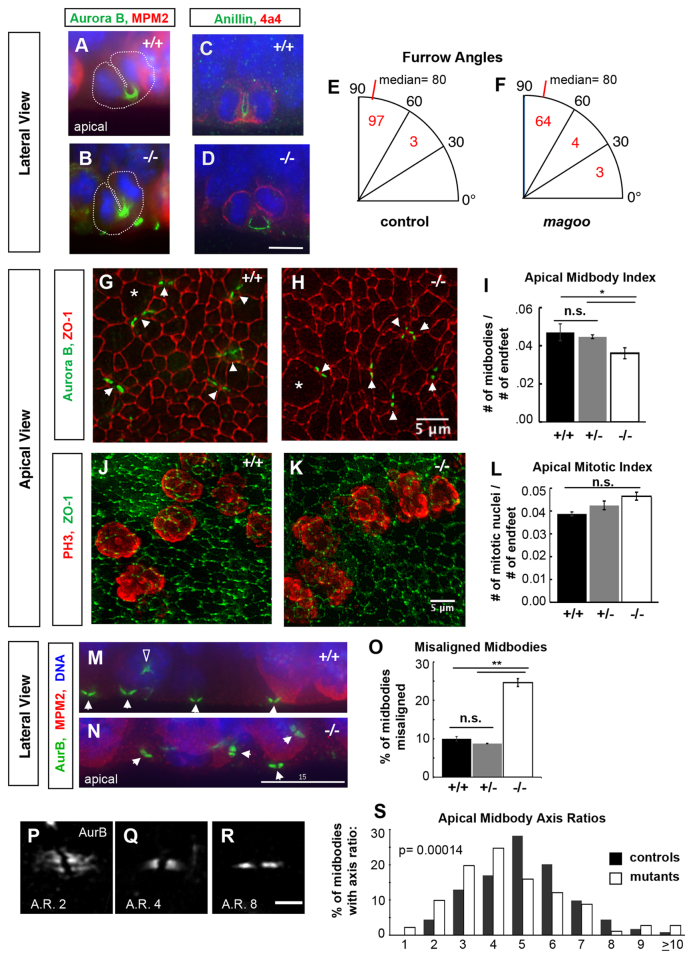
To examine whether cytokinetic furrows were normal in magoo mutant apical cortical progenitors, we immunostained for furrow markers on fixed cortical sections. In both control and mutant progenitors in late anaphase, aurora B kinase labels a U-shaped bundle of central spindle microtubules that is being compacted within the cleavage furrow (Fig. 7A,B). Anillin, a cytokinetic scaffolding protein, marks the contractile furrow membrane (Piekny and Maddox, 2010). In both control and mutant cortex, anillin staining labels the asymmetrically ingressing furrow membrane (Fig. 7C,D). To test whether furrow plane specification was normal in mutant brains, we compared cleavage furrow angles in control and mutant apical progenitors in anaphase or telophase. The angle distributions were not significantly different between control and mutant cells, and both had a median of 80° (Fig. 7E,F). These analyses suggest that Kif20b is not required for cytokinetic furrow specification or ingression in cortical apical progenitors.
Fig. 7.
Cytokinetic cleavage furrows appear normal but midbodies are abnormal in apical progenitors of magoo mutant cortex. (A,B) Aurora B (green) localizes to microtubules within cytokinetic furrows in both control and mutant apical progenitors in E12.5 cortex. Cytoplasm is outlined with a dotted white line. MPM2 (red) marks the cytoplasm of prophase and metaphase cells strongly, anaphase weakly. (C,D) Anillin (green) labels the cleavage furrow membrane in both control (C) and mutant (D) apical progenitors. Phospho-vimentin (4a4 epitope, red) marks cells in prophase through anaphase. (E,F) Cleavage furrow angles are mostly vertical in both control and mutant apical progenitors in E12.5 cortex. Numbers of anaphase or telophase cells with a furrow angle (relative to the apical surface) in each bin are indicated. The angles had similar skewed distributions in controls and mutants (P=0.17, Kolmogorov-Smirnov test), each with a median of 80° (P=0.07, Mann-Whitney U-test), and most between 60° and 90° (P=0.19, chi-squared test). n=100 control cells (+/+ and +/-), n=71 mutant cells. (G,H) Apical views of the ventricular surfaces of cortical slab whole-mounts from E13.5 embryos, immunostained for ZO-1 (Tjp1; red) to outline apical endfoot junctions and aurora B (green, arrowheads) to mark cytokinetic midbodies. Apical endfeet enlarge for M phase (asterisks indicate examples) and shrink in G1/S. Images are maximum intensity projections of deconvolved 8-20 μm deep z-stacks. (I) The average (± s.e.m.) apical midbody index (number of midbodies per apical progenitor) is decreased in magoo mutant cortices compared with +/+ or +/- controls at E13.5. n=5 +/+, n=4 +/-, n=8 -/- hemispheres. *P<0.05 (when +/+ and +/- controls are pooled, P<0.01). (J,K) Apical surfaces of cortical slabs immunostained for phospho-histone H3 (PH3, red) to mark apical progenitors in prophase through anaphase, and for ZO-1 (green). (L) The average mitotic index (± s.e.m.) of apical progenitors is not significantly different in mutant cortex versus wild-type or heterozygous controls at E13.5 (P=0.08 and P=0.15). n=2 +/+, n=12 +/-, n=9 -/- hemispheres. (M,N) In control E12.5 cortical sections, most midbodies (white arrowheads) are aligned parallel to the ventricular surface, but in mutants many midbodies appear misaligned. The open arrowhead in M indicates aurora B in a prometaphase cell, not a midbody. Aurora B, green; MPM2, red; DAPI, blue. (O) The average percentage (± s.e.m.) of misaligned midbodies is significantly increased in mutant cortices compared with wild-type or heterozygous controls at E13.5. n=3 +/+, n=3 +/-, n=4 -/- cortices (∼150 midbodies examined per cortex). **P<0.01 (if +/+ and +/- controls pooled, P<0.0001). (P-R) Examples of aligned apical midbodies with different axis ratios (AR, length/width). Apical views of the ventricular surface labeled by aurora B; maximum intensity projections of 2 μm deep z-stacks. (S) Midbodies in magoo mutant cortices tend to have smaller axis ratios. Shown is the percentage of midbodies with a given axis ratio in control and magoo mutant E13.5 cortices. Bin 1 includes ratios of 1.0 to 1.9, bin 2 includes 2.0-2.9, etc. Only midbodies aligned with the apical surface were measured. Mutants and controls had remarkably different axis ratio medians (4.7 versus 5.5; P=1.4×10-4, Mann-Whitney U-test) and distributions (P=2.5×10-5, Komogorov-Smirnov test). +/+ and +/- controls had similar distributions (P=0.71, Kolmogorov-Smirnov test) and medians (5.5 and 5.6; P=0.53, Mann-Whitney U-test), so they were combined in this histogram for visual clarity. n=117 +/+ and n=106 +/- control midbodies from 2 cortices each; n=182 midbodies from 4 -/- cortices. Scale bars: 7.5 μm in D; 5 μm in H,K,R; 15 μm in N.
Next, we examined whether cytokinetic midbodies, which mediate abscission, were normal in magoo mutant progenitors. In human cells, knockdown of KIF20B does not disrupt furrowing, but increases abscission failures (Abaza et al., 2003). No straightforward method had been established to analyze midbodies or abscission in mouse cortex. Therefore, we devised a cortical slab whole-mount preparation to view midbodies at the apical surface en face (Fig. 7G,H). This enabled the analysis of midbodies in a large population of apical progenitors. We coined the term ‘midbody index’, analogous to mitotic index, meaning the percentage of cells with midbodies.
Interestingly, in magoo mutant cortices, the apical midbody index was decreased compared with normal littermates (Fig. 7I). To control for the possibility that the reduced midbody index was due to fewer apical progenitors entering mitosis, we remeasured the apical mitotic index in the cortical whole-mount preparation (Fig. 7J,K). However, as found previously with cortical sections, the apical mitotic index was not significantly different in magoo mutants (Fig. 7L). Therefore, the decreased apical midbody index in magoo mutants suggests a defect in late cytokinesis at the midbody stage.
Aside from the altered midbody index, significant differences in midbody organization and shape were observed in magoo mutant cortices. In sections of control cortex, most apical midbodies are aligned at the ventricular plane and have a stereotypical shape (Fig. 7M). But in magoo mutant cortex, more midbodies appear misaligned (Fig. 7N). Again we used the cortical slab whole-mount preparations to quantify this finding in a large population of apical midbodies. By stepping through z-stacks of apical plane images, midbodies could be scored as aligned or misaligned to the apical surface. Indeed, homozygous mutant cortices had a significantly higher percentage of misaligned midbodies than wild-type or heterozygous controls (Fig. 7O). In addition, even the midbodies that were aligned parallel to the ventricle in mutant cortices appeared abnormal in shape. We measured the lengths and widths of properly aligned midbodies to calculate the axis ratios (Fig. 7P-R). Strikingly, the mutant midbodies displayed a shift toward smaller axis ratios (Fig. 7S) - that is, mutant midbodies tended to be shorter and wider. This phenotype has not been reported previously, but in HeLa cells midbodies are wider at early stages of their formation and then narrow as the microtubules are condensed (Hu et al., 2012). Thus, magoo mutant apical cortical progenitors may have defects in midbody formation, maturation or maintenance.
Apoptosis is increased during early cerebral cortex development of Kif20bmagoo mutants
The consequences of abscission defects in intact polarized epithelia have not been defined. When abscission is blocked in dissociated human cells, unseparated sister cells may re-merge to try division again, but ultimately arrest as syncytia connected by thin cytoplasmic bridges, or undergo apoptosis (Gromley et al., 2003; Zhao et al., 2006). Similarly, when abscission is perturbed by KIF20B knockdown in dissociated human cells, binucleates and apoptosis increase (Abaza et al., 2003; Kanehira et al., 2007). In neuroepithelium, however, loss of Kif20b does not apparently increase the occurrence of binucleate cells (Fig. 6C-E). Therefore, we tested for the predicted increase in apoptosis.
In normal developing cortex, very little apoptosis is detectable with anti-cleaved caspase 3 (CC3), whereas in magoo mutants apoptotic cells are frequently observed (Fig. 8A,B). At E10.5, when apical progenitors predominate and neurogenesis has not yet begun, mutant cortices had ∼6-fold more apoptotic cells than controls. Apoptosis was also robustly increased in mutants during early neurogenesis at E12.5 and E14.5, but was less significant at E16.5 (Fig. 8C). The apoptotic cells were present in both the progenitor and neuron layers, in percentages directly proportional to the thickness of each layer (Fig. 8D). The striking increases in apoptosis at E10 and E12, when there are very few basal progenitors and neurons, coupled with the finding of abnormal midbodies in apical progenitors (Fig. 7I-S), strongly suggest that a large portion of the apoptosis that occurs in magoo cortex is due to stochastic failures of abscission in apical progenitors.
Fig. 8.
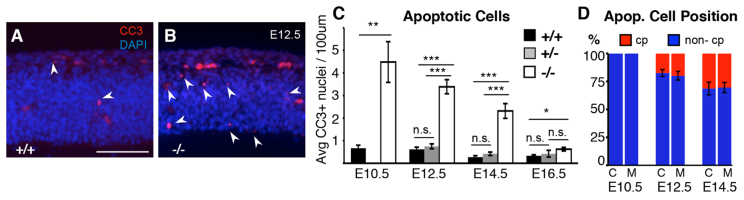
magoo mutant cortex shows increased apoptosis during corticogenesis. (A,B) Cryosections through wild-type and mutant cortices labeled with anti-cleaved caspase 3 (CC3; red, arrowheads) for apoptotic cells and stained with DAPI (blue) for all nuclei. Blood vessels have distinguishable autofluorescence and were not counted. Scale bar: 100 μm. (C) The mean number (± s.e.m.) of apoptotic cells per 100 μm length of ventricle is significantly higher in mutant cortices than in controls. Number of brains: E10.5, 2 +/+, 2 -/-; E12.5, 4 +/+, 3 +/-, 6 -/-; E14.5, 3 +/+, 2 +/-, 5 -/-; E16.5, 5 +/+, 2 +/-, 6 -/-. Average 5 fields per brain. *P≤0.01, **P≤0.001, ***P≤10-5. (D) The percentages of apoptotic cells in proliferative layers (blue) versus neuronal layer (red, cp) were proportional to layer thicknesses at E10.5, E12.5 and E14.5, and similar in controls (C) and mutants (M). Number of brains: E10.5, 2 +/+, 2 -/-; E12.5, 4 +/+, 3 +/-, 6 -/-; E14.5, 3 +/+, 2 +/-, 5 -/-.
DISCUSSION
We have identified mutations in mouse Kif20b that demonstrate a requirement for this Kinesin-6 in the proper abscission of cortical stem cells and growth of the cerebral cortex. Specifically, our analyses in Kif20bmagoo mutant cortex show that mitotic parameters and cytokinetic furrows appear normal, but the number, positioning and shape of cytokinetic midbodies are disrupted. These findings indicate that Kif20b regulates apical midbody formation or maintenance until abscission. Furthermore, we show that loss of Kif20b causes increased apoptosis during early corticogenesis and reductions in the cortical progenitor pool. Despite reduced neurogenesis, the potential for producing sequential layer fates remains intact.
This work underscores the power of forward genetics to discover subtle or specialized gene functions. Surprisingly, Kif20b was not pulled out of recent comprehensive biochemical and siRNA screens for cytokinetic proteins in mammalian cells (Kittler et al., 2004; Neumann et al., 2010; Skop et al., 2004). This could be because Kif20b is both rare and sensitive to proteolysis (Westendorf et al., 1994). In addition, the effects of Kif20b knockdown in cultured cells might be relatively subtle; a developing embryo provides a more sensitive test of its requirement.
The findings in Kif20b mutants highlight the importance of understanding the different phenotypes that may arise after disruption of cytokinesis in vivo versus in vitro, or in furrowing versus abscission. In dissociated cells, disrupting furrowing or abscission can increase binucleate cells. However, careful analysis of live imaging showed that disrupting abscission causes only transient binucleate intermediates but longer-lasting syncitia of mononucleate cells connected by cytoplasmic bridges, often followed by apoptosis (Gromley et al., 2003). In the developing brain, disruption of furrowing can produce binucleate cells, as seen in the citron mouse mutant (Di Cunto et al., 2000). But in magoo brains, in which abscission is disrupted, the nuclear migration away from the apical midbody before abscission may prevent ill-separated sister cells from re-merging, resulting instead in apoptosis. The signaling pathway leading from abscission defects to apoptosis is not understood, but might be more sensitive in vivo than in cultured cells.
Kif20b does not appear to be absolutely required for cytokinesis, since invertebrates and many mouse tissues develop normally without it; even the forebrain, although severely affected, does grow in the magoo mutant. By contrast, mutations in the cytokinetic furrowing protein mgcRacGAP (Racgap1 - Mouse Genome Informatics) cause preimplantation lethality in mice (Van de Putte et al., 2001). It is not yet clear whether most tissues do not express Kif20b mRNA or simply do not need it. It is expressed most highly in germinal zones of the nervous system, but might be present in other tissues at very low levels. Kif23 and Kif20a mRNAs appear more abundant (data not shown) and might suffice for most tissues, whereas cortical stem cells, with their specialized cytokinesis and demanding schedule for producing neuron layers, may need all three Kinesin-6 proteins. Kif20b might have specialized functions crucial for neural stem cell division.
KIF20B knockdown reduces abscission success in human cell lines (Abaza et al., 2003), but the precise role of Kif20b in abscission is not known. Our midbody phenotypes provide clues to its specific roles. In magoo mutant cortex, the reduced number and altered morphology of midbodies suggest that Kif20b may be involved in the formation or maturation of midbodies or in their structural maintenance until abscission. It could do this by localizing or linking abscission factors within the midbody. In addition, the increase in misaligned midbodies in mutant cortices suggests that Kif20b might also regulate midbody positioning in relation to apical structures. A molecular understanding of these phenotypes awaits future work.
Downstream effectors of Kif20b for cytokinesis remain to be discovered. Kif20b is highly similar to its sister Kinesin-6 proteins Kif23 and Kif20a in the motor head domain, but divergent in the stalk and tail, predicting different cargo binding (Miki et al., 2005). The only proteins shown to interact with KIF20B during cytokinesis, PIN1 and PRC1, are upstream regulators (Kamimoto et al., 2001; Kanehira et al., 2007; Lee et al., 2012; Lu and Zhou, 2007). Other candidate cargos of Kif20b (Maliga et al., 2013; Sapir et al., 2013) do not have known roles in cytokinesis.
The spatial and temporal regulation of abscission influence the partitioning of cytoplasm and membrane between daughters, but also determine inheritance of the midbody. In some cells, abscission occurs on one side of the midbody, so one daughter inherits it (Gromley et al., 2003; Guizetti and Gerlich, 2010; Guizetti et al., 2011). Others have observed abscission on both sides of the midbody (Elia et al., 2011; Ettinger et al., 2011). Intriguingly, midbody inheritance was recently linked to cell fate (Chen et al., 2013; Schink and Stenmark, 2011). One group showed that embryonic stem cells and cancer cells tend to accumulate midbodies (Kuo et al., 2011). Another group reported that neural stem cell divisions release midbodies with two abscissions, and hypothesized that this is important to maintain symmetric daughter fates (Dubreuil et al., 2007; Ettinger et al., 2011). Loss of Kif20b could disrupt double abscissions or the control of midbody inheritance in neural stem cells. Much more work is needed to understand the roles of abscission and midbody inheritance in cell fate.
In conclusion, future insights into development and disease will require a better understanding of cytokinesis mechanisms in different cell types, complex tissues and symmetric versus asymmetric divisions. The Kif20bmagoo mutant offers a new model of microcephaly, different from previous mouse models, that shows abnormalities specific to the mitosis of cortical progenitors (Feng and Walsh, 2004; Lizarraga et al., 2010). This and other work suggest that the regulation of cytokinetic abscission, in addition to the regulation of mitosis and cleavage, could play a previously unappreciated role in stem cell renewal and the control of cerebral cortex size.
MATERIALS AND METHODS
Animals and breeding
Mouse colonies were maintained in accordance with NIH guidelines and approved by the IACUC. Embryos were harvested by c-section, and the morning of the vaginal plug was considered embryonic day (E) 0.5. Littermate embryos served as controls for all experiments. The magoo mutation was induced on the C57BL/6J (BL6) background and backcrossed to FVB/N for mapping. It is maintained on both BL6 and FVB/N backgrounds, and mixed background embryos are used for experiments. Genotyping tests for a RFLP due to a StyI site created by the magoo mutation.
After initial mapping (Dwyer et al., 2011), the magoo interval was further refined to 1 Mb of chromosome 19, with D19Mit12 on the right and a new SSLP marker ‘15 x CA’ on the left at 34.95 Mb near the first exon of Pank1. Genomic exons with flanks of Pank1 and Kif20b were sequenced as described (Dwyer et al., 2011).
The 33c mutant was a gift from Kathryn V. Anderson (Sloan-Kettering Institute, New York, NY, USA). It was induced on BL6 and maintained on C3H. The screen that isolated 33c is described by García-García et al. (García-García et al., 2005). Subsequently, 33c/mcbarker was mapped between D19Mit46 and D19Mit63. Candidates in the interval were sequenced and the missense mutation was found in Kif20b. Genotyping utilized PCR for a RFLP due to a DdeI site created by the point mutation.
Immunoblotting
Brain lysates were prepared with RIPA lysis buffer (150 mM NaCl, 1% NP40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris-HCl pH 8) with protease and phosphatase inhibitors. Protein concentration in lysates was determined by bicinchoninic acid (BCA) assay, and 50-100 μg total protein was loaded per lane on 7.5% polyacrylamide gels. Proteins were transferred by electroblotting onto a 0.2 μm PVDF membrane overnight at 30 mA. Membranes were blocked in 150 mM NaCl, 100 mM Tris-HCl pH 7.5 and 0.5% Tween 20 (TBST) with 5% dried milk (blocking buffer) for 1 hour. Rabbit anti-mouse Kif20b antiserum (1:2000) or 5 μg/ml affinity-purified primary antibody in blocking buffer was incubated with the membrane overnight at 4°C. Rabbit anti-β-catenin (1:1000) served as loading control. After three washes, goat anti-rabbit IgG-HRP secondary antibody (1:3000) in blocking buffer was applied for 1 hour at room temperature. After washing, immune complexes were visualized using the Supersignal Western Blotting Kit (Thermo Fisher Scientific).
Immunostaining and histology
For fibroblast culture and immunostaining, MEFs were collected as described (Favero et al., 2013) and plated on fibronectin-coated coverslips, grown overnight, and fixed with 4% paraformaldehyde (PFA) for 2 minutes and cold methanol for 10 minutes. Immunohistochemistry was performed as described (Favero et al., 2013).
Immunohistochemistry, BrdU labeling, and Hematoxylin and Eosin (H&E) staining on fixed cryosections or paraffin sections of cerebral cortex were performed as described (Favero et al., 2013). For S-phase index, pregnant mice were injected intraperitoneally with a single dose of BrdU at 100 mg/kg at E14.5, embryos harvested after 60 minutes, and brains fixed with 4% PFA and embedded for cryosectioning. BrdU+ cells were counted in rectangular fields of ∼10,000 μm2. The apical bin was within 50 μm of the ventricle and the basal bin was 50-100 μm away from the ventricle. For mitotic index assessed by PH3 staining, the apical bin was defined as within 30 μm of the ventricle, and the basal bin was greater than 30 μm away. Images were collected on either a Zeiss Axio ImagerZ1 microscope with AxioCam MRm or a Leica MZ16F microscope with DFC300FX camera. Ventricle length measurements for each section and cell counts were performed in ImageJ. Statistical analyses were performed using Excel (Microsoft) or PAST (Hammer et al., 2001). P-values are for t-test unless otherwise stated.
Rabbit polyclonal anti-mouse Kif20b antiserum was raised against two pairs of co-injected peptides from the unstructured N-terminal tail and the C-terminus of mouse Kif20b. Peptides from the unstructured N-terminal tail were (C)GVPRPSYVFSADPIARPLE and (CKK)FDGVLDLSHEFSLVAS. Peptides from the C-terminal domain were (C)VTIKIPKARKRKSGEVEE and (C)SKENVSQPKKAKRKLYRNE (Covance). Affinity purification gave qualitatively similar but reduced signal compared with antisera in immunostaining and immunoblotting at 1:300 dilution. The N- and C-terminal peptide antibodies showed similar localization to rabbit anti-human KIF20B (MPP1) antibody used at 1:500 [a gift from Fabienne Pirollet, Grenoble, France (Abaza et al., 2003)].
For all other primary and secondary antibodies used see supplementary material Table S1.
Cortical slab whole-mount preparation, staining and analysis
Brains were fixed with 2% PFA for 20 minutes and rinsed. Cortices were pinched off with fine forceps and placed ventricular side up in PBS in a 250 μm deep slide-well. A micro-scalpel was used to trim off ganglionic eminences and cortical hems, and the resulting ‘cortical slabs’ were fixed again in 2% PFA for 2 minutes.
For immunostaining, cortical slabs were incubated for 1 hour at room temperature in blocking buffer. Then cortices were incubated in primary antibody in blocking buffer for 1 hour at room temperature and 4°C overnight. The next day, cortices were rinsed and incubated in species-appropriate Alexa Fluor secondary antibody (see supplementary material Table S1) diluted in blocking buffer, in the dark at room temperature for 30 minutes. Cortical slabs were rinsed, covered with mounting medium and coverslipped.
Apical membrane fields of cortical slabs were imaged at 40× using an inverted DeltaVision with TrueLight deconvolution microscope and softWoRx Suite 5.5 image acquisition software (Applied Precision). z-stack depth was 8-20 μm and step size was 0.5 μm. 3D stacks were deconvolved and collapsed. Apical endfeet, midbodies and mitotic nuclei were counted over an area of 3500 μm2 using a counting grid in ImageJ. Midbodies were considered misaligned if the two halves of the midbody (aurora B label) were not in the same z-plane or within two adjacent z-planes.
RNA in situ hybridization
mRNA analysis was performed on cryosections (16-25 μm) as described (Favero et al., 2013). Sections were hybridized overnight at 66°C with digoxigenin-labeled riboprobes at a final concentration of 500 ng/ml. Kif20b riboprobes against the 5′ or 3′ end of the mRNA were synthesized from cDNA templates of IMAGE clones BE199511 or BC036147. The expression pattern was the same with 5′ or 3′ probes.
Flow cytometry
Cells from E13.5 or E14.5 brains were dissociated using the Papain Dissociation Kit (Worthington Biochemical Corporation). Single-cell suspensions were obtained by filtering through a 40 μm filter (BD Falcon Cell Strainer, blue nylon mesh, catalog #352340). For propidium iodide (PI) staining, cells were resuspended in 500 μl PBS and added to 4.5 ml ice-cold 70% ethanol for at least 2 hours. Samples were stored at 4°C. Fixed cells were rinsed in PBS and resuspended in 1 ml solution containing 100 μg/ml RNase A, 0.1% Triton X-100 and 50 μg/ml PI, and incubated at room temperature for 30 minutes in the dark. Four independent experiments with one pair of mutant and control littermate brains each were analyzed (two experiments at E13.5 and two at E14.5). For Ki-67 and DAPI analysis, single-cell suspensions of E14.5 brains (n=3 pairs of magoo mutants and littermate controls) were fixed in 1.5% PFA for 15 minutes on ice. Cells were then washed twice with FACS buffer (2% BSA, 1 mM EDTA, 0.01% sodium azide, PBS) and permeabilized in 0.1% Triton X-100 in PBS for 30 minutes on ice. Cells were washed twice in FACS buffer and 2×106 cells were incubated in 100 μl FACS buffer with 1 μg/ml DAPI and 2 μl anti-Ki-67 antibody (monoclonal rat anti-Ki-67 Alexa Fluor 647, clone SolA15, eBioscience). Following three washes with FACS buffer, fluorescence was measured using a FACSCanto II flow cytometer (Becton Dickinson). At least 20,000 events were collected per sample. Data were analyzed using FlowJo software (TreeStar).
Supplementary Material
Acknowledgments
We regret not being able to cite more of the original literature due to space limitations. We are very grateful to Kathryn V. Anderson for the 33c/mcbarker line, to Evan H. Feinberg for mapping help and to Fabienne Pirollet for the anti-human KIF20B antibody. We thank Christine Savoie for help with data quantification and Xiaowei Lu, Jing Yu, Sofia Lizarraga, Yuanyi Feng, Seonhee Kim, Stephen Turner, Sue Delos, Maureen Bjerke, Kevin Pfister, Bettina Winckler, Sally Parsons and Todd Stukenberg for discussions and advice.
Footnotes
Competing interests
The authors declare no competing financial interests.
Author contributions
N.D.D., V.M.V. and K.M.J. conceived and designed experiments. V.M.V., K.M.J., M.S.F., A.S., C.M.G.-M., B.A.R., S.A.N., T.D.C., J.M.K. and N.D.D. performed experiments and analyzed data. V.M.V., K.M.J., C.M.G.-M., J.M.K. and N.D.D. interpreted data. K.F.L. isolated and cloned the mcbarker allele. N.D.D. supervised the study and wrote the manuscript.
Funding
These studies were supported in part by a Basil O’Connor Starter Scholar Research Award [#5-FY09-101] from the March of Dimes Foundation (N.D.D.), a Hartwell Foundation Postdoctoral Fellowship (V.M.V. and C.M.G.-M.), the UVA Fund for Excellence in Science and Technology (N.D.D.), ACS-IRG#81-001-26 from the American Cancer Society through the UVA Cancer Center (N.D.D.) and by National Institutes of Health grants [K01MH69647 and R01NS076640 to N.D.D., and R01NS044385 to K.V.A.]. Deposited in PMC for release after 12 months.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.093286/-/DC1
References
- Abaza A., Soleilhac J. M., Westendorf J., Piel M., Crevel I., Roux A., Pirollet F. (2003). M phase phosphoprotein 1 is a human plus-end-directed kinesin-related protein required for cytokinesis. J. Biol. Chem. 278, 27844–27852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams R. R., Tavares A. A., Salzberg A., Bellen H. J., Glover D. M. (1998). pavarotti encodes a kinesin-like protein required to organize the central spindle and contractile ring for cytokinesis. Genes Dev. 12, 1483–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agromayor M., Martin-Serrano J. (2013). Knowing when to cut and run: mechanisms that control cytokinetic abscission. Trends Cell Biol. 23, 433–441 [DOI] [PubMed] [Google Scholar]
- Cesario J. M., Jang J. K., Redding B., Shah N., Rahman T., McKim K. S. (2006). Kinesin 6 family member Subito participates in mitotic spindle assembly and interacts with mitotic regulators. J. Cell Sci. 119, 4770–4780 [DOI] [PubMed] [Google Scholar]
- Chen C. T., Hehnly H., Doxsey S. J. (2012). Orchestrating vesicle transport, ESCRTs and kinase surveillance during abscission. Nat. Rev. Mol. Cell Biol. 13, 483–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. T., Ettinger A. W., Huttner W. B., Doxsey S. J. (2013). Resurrecting remnants: the lives of post-mitotic midbodies. Trends Cell Biol. 23, 118–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M. R., Wen G., Lepier A., Schroeder T., Götz M. (2008). Par-complex proteins promote proliferative progenitor divisions in the developing mouse cerebral cortex. Development 135, 11–22 [DOI] [PubMed] [Google Scholar]
- Di Cunto F., Imarisio S., Hirsch E., Broccoli V., Bulfone A., Migheli A., Atzori C., Turco E., Triolo R., Dotto G. P., et al. (2000). Defective neurogenesis in citron kinase knockout mice by altered cytokinesis and massive apoptosis. Neuron 28, 115–127 [DOI] [PubMed] [Google Scholar]
- Dubreuil V., Marzesco A. M., Corbeil D., Huttner W. B., Wilsch-Bräuninger M. (2007). Midbody and primary cilium of neural progenitors release extracellular membrane particles enriched in the stem cell marker prominin-1. J. Cell Biol. 176, 483–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer N. D., Manning D. K., Moran J. L., Mudbhary R., Fleming M. S., Favero C. B., Vock V. M., O’Leary D. D., Walsh C. A., Beier D. R. (2011). A forward genetic screen with a thalamocortical axon reporter mouse yields novel neurodevelopment mutants and a distinct Emx2 mutant phenotype. Neural Dev. 6, 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbing B., Mann K., Starosta A., Jaud J., Schöls L., Schüle R., Woehlke G. (2008). Effect of spastic paraplegia mutations in KIF5A kinesin on transport activity. Hum. Mol. Genet. 17, 1245–1252 [DOI] [PubMed] [Google Scholar]
- Elia N., Sougrat R., Spurlin T. A., Hurley J. H., Lippincott-Schwartz J. (2011). Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. Proc. Natl. Acad. Sci. USA 108, 4846–4851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund C., Fink A., Lau C., Pham D., Daza R. A., Bulfone A., Kowalczyk T., Hevner R. F. (2005). Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J. Neurosci. 25, 247–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettinger A. W., Wilsch-Bräuninger M., Marzesco A. M., Bickle M., Lohmann A., Maliga Z., Karbanová J., Corbeil D., Hyman A. A., Huttner W. B. (2011). Proliferating versus differentiating stem and cancer cells exhibit distinct midbody-release behaviour. Nat. Commun. 2, 503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favero C. B., Henshaw R. N., Grimsley-Myers C. M., Shrestha A., Beier D. R., Dwyer N. D. (2013). Mutation of the BiP/GRP78 gene causes axon outgrowth and fasciculation defects in the thalamocortical connections of the mammalian forebrain. J. Comp. Neurol. 521, 677–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Walsh C. A. (2004). Mitotic spindle regulation by Nde1 controls cerebral cortical size. Neuron 44, 279–293 [DOI] [PubMed] [Google Scholar]
- García-García M. J., Eggenschwiler J. T., Caspary T., Alcorn H. L., Wyler M. R., Huangfu D., Rakeman A. S., Lee J. D., Feinberg E. H., Timmer J. R., et al. (2005). Analysis of mouse embryonic patterning and morphogenesis by forward genetics. Proc. Natl. Acad. Sci. USA 102, 5913–5919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore E. C., Walsh C. A. (2013). Genetic causes of microcephaly and lessons for neuronal development. Wiley Interdiscip. Rev. Dev. Biol. 2, 461–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M. (2009). The 3Ms of central spindle assembly: microtubules, motors and MAPs. Nat. Rev. Mol. Cell Biol. 10, 9–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R. A., Paluch E., Oegema K. (2012). Cytokinesis in animal cells. Annu. Rev. Cell Dev. Biol. 28, 29–58 [DOI] [PubMed] [Google Scholar]
- Gromley A., Jurczyk A., Sillibourne J., Halilovic E., Mogensen M., Groisman I., Blomberg M., Doxsey S. (2003). A novel human protein of the maternal centriole is required for the final stages of cytokinesis and entry into S phase. J. Cell Biol. 161, 535–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromley A., Yeaman C., Rosa J., Redick S., Chen C. T., Mirabelle S., Guha M., Sillibourne J., Doxsey S. J. (2005). Centriolin anchoring of exocyst and SNARE complexes at the midbody is required for secretory-vesicle-mediated abscission. Cell 123, 75–87 [DOI] [PubMed] [Google Scholar]
- Guizetti J., Gerlich D. W. (2010). Cytokinetic abscission in animal cells. Semin. Cell Dev. Biol. 21, 909–916 [DOI] [PubMed] [Google Scholar]
- Guizetti J., Schermelleh L., Mäntler J., Maar S., Poser I., Leonhardt H., Müller-Reichert T., Gerlich D. W. (2011). Cortical constriction during abscission involves helices of ESCRT-III-dependent filaments. Science 331, 1616–1620 [DOI] [PubMed] [Google Scholar]
- Hammer O., Harper D. A. T., Ryan P. D. (2001). PAST: Paleontological statistics software package for education and data analysis. Paleontologia Electronica 4, http://palaeo-electronica.org/2001_1/past/past.pdf [Google Scholar]
- Hirokawa N., Noda Y. (2008). Intracellular transport and kinesin superfamily proteins, KIFs: structure, function, and dynamics. Physiol. Rev. 88, 1089–1118 [DOI] [PubMed] [Google Scholar]
- Hu C. K., Coughlin M., Mitchison T. J. (2012). Midbody assembly and its regulation during cytokinesis. Mol. Biol. Cell 23, 1024–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimoto T., Zama T., Aoki R., Muro Y., Hagiwara M. (2001). Identification of a novel kinesin-related protein, KRMP1, as a target for mitotic peptidyl-prolyl isomerase Pin1. J. Biol. Chem. 276, 37520–37528 [DOI] [PubMed] [Google Scholar]
- Kanehira M., Katagiri T., Shimo A., Takata R., Shuin T., Miki T., Fujioka T., Nakamura Y. (2007). Oncogenic role of MPHOSPH1, a cancer-testis antigen specific to human bladder cancer. Cancer Res. 67, 3276–3285 [DOI] [PubMed] [Google Scholar]
- Kittler R., Putz G., Pelletier L., Poser I., Heninger A. K., Drechsel D., Fischer S., Konstantinova I., Habermann B., Grabner H., et al. (2004). An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature 432, 1036–1040 [DOI] [PubMed] [Google Scholar]
- Konno D., Shioi G., Shitamukai A., Mori A., Kiyonari H., Miyata T., Matsuzaki F. (2008). Neuroepithelial progenitors undergo LGN-dependent planar divisions to maintain self-renewability during mammalian neurogenesis. Nat. Cell Biol. 10, 93–101 [DOI] [PubMed] [Google Scholar]
- Kosodo Y., Röper K., Haubensak W., Marzesco A. M., Corbeil D., Huttner W. B. (2004). Asymmetric distribution of the apical plasma membrane during neurogenic divisions of mammalian neuroepithelial cells. EMBO J. 23, 2314–2324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosodo Y., Toida K., Dubreuil V., Alexandre P., Schenk J., Kiyokage E., Attardo A., Mora-Bermúdez F., Arii T., Clarke J. D., et al. (2008). Cytokinesis of neuroepithelial cells can divide their basal process before anaphase. EMBO J. 27, 3151–3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A., Alvarez-Buylla A. (2009). The glial nature of embryonic and adult neural stem cells. Annu. Rev. Neurosci. 32, 149–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo T. C., Chen C. T., Baron D., Onder T. T., Loewer S., Almeida S., Weismann C. M., Xu P., Houghton J. M., Gao F. B., et al. (2011). Midbody accumulation through evasion of autophagy contributes to cellular reprogramming and tumorigenicity. Nat. Cell Biol. 13, 1214–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. Y., Davies T., Mishima M. (2012). Cytokinesis microtubule organisers at a glance. J. Cell Sci. 125, 3495–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizarraga S. B., Margossian S. P., Harris M. H., Campagna D. R., Han A. P., Blevins S., Mudbhary R., Barker J. E., Walsh C. A., Fleming M. D. (2010). Cdk5rap2 regulates centrosome function and chromosome segregation in neuronal progenitors. Development 137, 1907–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K. P., Zhou X. Z. (2007). The prolyl isomerase PIN1: a pivotal new twist in phosphorylation signalling and disease. Nat. Rev. Mol. Cell Biol. 8, 904–916 [DOI] [PubMed] [Google Scholar]
- Maliga Z., Junqueira M., Toyoda Y., Ettinger A., Mora-Bermúdez F., Klemm R. W., Vasilj A., Guhr E., Ibarlucea-Benitez I., Poser I., et al. (2013). A genomic toolkit to investigate kinesin and myosin motor function in cells. Nat. Cell Biol. 15, 325–334 [DOI] [PubMed] [Google Scholar]
- Marthiens V., ffrench-Constant C. (2009). Adherens junction domains are split by asymmetric division of embryonic neural stem cells. EMBO Rep. 10, 515–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto-Taniura N., Pirollet F., Monroe R., Gerace L., Westendorf J. M. (1996). Identification of novel M phase phosphoproteins by expression cloning. Mol. Biol. Cell 7, 1455–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H., Okada Y., Hirokawa N. (2005). Analysis of the kinesin superfamily: insights into structure and function. Trends Cell Biol. 15, 467–476 [DOI] [PubMed] [Google Scholar]
- Neef R., Klein U. R., Kopajtich R., Barr F. A. (2006). Cooperation between mitotic kinesins controls the late stages of cytokinesis. Curr. Biol. 16, 301–307 [DOI] [PubMed] [Google Scholar]
- Neumann B., Walter T., Hériché J. K., Bulkescher J., Erfle H., Conrad C., Rogers P., Poser I., Held M., Liebel U., et al. (2010). Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature 464, 721–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor S. C., Martínez-Cerdeño V., Kriegstein A. R. (2008). Distinct behaviors of neural stem and progenitor cells underlie cortical neurogenesis. J. Comp. Neurol. 508, 28–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekny A. J., Maddox A. S. (2010). The myriad roles of Anillin during cytokinesis. Semin. Cell Dev. Biol. 21, 881–891 [DOI] [PubMed] [Google Scholar]
- Raich W. B., Moran A. N., Rothman J. H., Hardin J. (1998). Cytokinesis and midzone microtubule organization in Caenorhabditis elegans require the kinesin-like protein ZEN-4. Mol. Biol. Cell 9, 2037–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid E., Kloos M., Ashley-Koch A., Hughes L., Bevan S., Svenson I. K., Graham F. L., Gaskell P. C., Dearlove A., Pericak-Vance M. A., et al. (2002). A kinesin heavy chain (KIF5A) mutation in hereditary spastic paraplegia (SPG10). Am. J. Hum. Genet. 71, 1189–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapir T., Levy T., Sakakibara A., Rabinkov A., Miyata T., Reiner O. (2013). Shootin1 acts in concert with KIF20B to promote polarization of migrating neurons. J. Neurosci. 33, 11932–11948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schink K. O., Stenmark H. (2011). Cell differentiation: midbody remnants -junk or fate factors? Curr. Biol. 21, R958–R960 [DOI] [PubMed] [Google Scholar]
- Shimizu T., Thorn K. S., Ruby A., Vale R. D. (2000). ATPase kinetic characterization and single molecule behavior of mutant human kinesin motors defective in microtubule-based motility. Biochemistry 39, 5265–5273 [DOI] [PubMed] [Google Scholar]
- Skop A. R., Liu H., Yates J., 3rd, Meyer B. J., Heald R. (2004). Dissection of the mammalian midbody proteome reveals conserved cytokinesis mechanisms. Science 305, 61–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X., Shi S. H. (2013). Neocortical neurogenesis and neuronal migration. Wiley Interdiscip. Rev. Dev. Biol. 2, 443–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Putte T., Zwijsen A., Lonnoy O., Rybin V., Cozijnsen M., Francis A., Baekelandt V., Kozak C. A., Zerial M., Huylebroeck D. (2001). Mice with a homozygous gene trap vector insertion in mgcRacGAP die during pre-implantation development. Mech. Dev. 102, 33–44 [DOI] [PubMed] [Google Scholar]
- Westendorf J. M., Rao P. N., Gerace L. (1994). Cloning of cDNAs for M-phase phosphoproteins recognized by the MPM2 monoclonal antibody and determination of the phosphorylated epitope. Proc. Natl. Acad. Sci. USA 91, 714–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W. M., Seki A., Fang G. (2006). Cep55, a microtubule-bundling protein, associates with centralspindlin to control the midbody integrity and cell abscission during cytokinesis. Mol. Biol. Cell 17, 3881–3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Zhao J., Bibikova M., Leverson J. D., Bossy-Wetzel E., Fan J. B., Abraham R. T., Jiang W. (2005). Functional analysis of human microtubule-based motor proteins, the kinesins and dyneins, in mitosis/cytokinesis using RNA interference. Mol. Biol. Cell 16, 3187–3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.