Abstract
Steroid hormones trigger the onset of sexual maturation in animals by initiating genetic response programs that are determined by steroid pulse frequency, amplitude and duration. Although steroid pulses coordinate growth and timing of maturation during development, the mechanisms generating these pulses are not known. Here we show that the ecdysone steroid pulse that drives the juvenile-adult transition in Drosophila is determined by feedback circuits in the prothoracic gland (PG), the major steroid-producing tissue of insect larvae. These circuits coordinate the activation and repression of hormone synthesis, the two key parameters determining pulse shape (amplitude and duration). We show that ecdysone has a positive-feedback effect on the PG, rapidly amplifying its own synthesis to trigger pupariation as the onset of maturation. During the prepupal stage, a negative-feedback signal ensures the decline in ecdysone levels required to produce a temporal steroid pulse that drives developmental progression to adulthood. The feedback circuits rely on a developmental switch in the expression of Broad isoforms that transcriptionally activate or silence components in the ecdysone biosynthetic pathway. Remarkably, our study shows that the same well-defined genetic program that stimulates a systemic downstream response to ecdysone is also utilized upstream to set the duration and amplitude of the ecdysone pulse. Activation of this switch-like mechanism ensures a rapid, self-limiting PG response that functions in producing steroid oscillations that can guide the decision to terminate growth and promote maturation.
Keywords: Developmental timing, Ecdysone, Prothoracicotropic hormone, Prothoracic gland, Neuropeptide
INTRODUCTION
Pulses of steroid hormones are conserved temporal signals that control developmental transitions and sexual maturation in animals (Dungan et al., 2006; Rewitz et al., 2013; Tennessen and Thummel, 2011; Yamanaka et al., 2013). In insects, pulses of the steroid hormone ecdysone coordinate the developmental transition from the juvenile larval stage to a reproductively mature adult, a process called metamorphosis (Rewitz and O’Connor, 2011; Yamanaka et al., 2013). Before committing to the production of ecdysone and initiation of the maturation process, the endocrine system makes a series of complex assessments to ensure that enough growth has been completed to produce an adult of the correct size and dimensions. These checkpoints include assessment of nutrient stores, imaginal tissue growth and damage, and photoperiod (Colombani et al., 2012; Colombani et al., 2005; Garelli et al., 2012; Hackney et al., 2012; Layalle et al., 2008; Mirth and Shingleton, 2012; Rajan and Perrimon, 2012; Rewitz et al., 2013). Passage through some, if not all, of these checkpoints is thought to result in the release of prothoracicotropic hormone (PTTH), a brain neuropeptide. PTTH triggers pupariation (onset of metamorphosis) and progression to adulthood through an upregulation of ecdysone biosynthesis in the prothoracic gland (PG), the endocrine tissue of insects (Gibbens et al., 2011; McBrayer et al., 2007; Rewitz et al., 2009b; Rewitz et al., 2013). Circulating ecdysone released from the PG binds to the ecdysone receptor (EcR) and its heterodimerization partner Ultraspiracle (Usp) (King-Jones and Thummel, 2005) to initiate tissue-specific genetic responses that drive developmental progression. Key to this process is the direct induction by the EcR-Usp complex of a small number of transcription factor genes, including those encoding Broad (Br), E74 and E75 (Eip74EF and Eip75B - FlyBase), in responsive tissues that activate a larger set of secondary response genes (King-Jones and Thummel, 2005; Thummel, 1996).
PTTH stimulates the production of ecdysone through activation of its receptor Torso in the PG (Rewitz et al., 2009b). Once active, Torso induces the mitogen-activated protein kinase (MAPK) cascade that ultimately leads to increased transcription of ecdysone biosynthetic enzymes (McBrayer et al., 2007; Rewitz et al., 2009b). The details of the transcriptional regulatory responses mediated by the MAPK cascade are still largely unknown for most of the ecdysone biosynthetic enzymes, although it is known that PTTH signaling mediates nucleocytoplasmic trafficking of the nuclear receptor DHR4 (Hr4 - FlyBase), which acts as a repressor (Ou et al., 2011; Rewitz and O’Connor, 2011). Metamorphosis is initiated by a transient high-level ecdysone pulse, i.e. a temporally defined signal. The two key parameters that determine the pulse shape are the activation and repression of hormone synthesis, which must act in concert to determine both the amplitude and duration of the pulse. Although general strategies for how ecdysone production is regulated have been described, the mechanisms coordinating activation and repression to produce a high-level maturation-inducing pulse in response to PTTH are not known.
Here, we describe a pulse-generator circuit incorporated into the PG that is based on ecdysone feedback regulation of its own synthesis. The ramp up of ecdysone synthesis leading to pupariation involves a key positive-feedback loop that functions downstream of PTTH to amplify ecdysone production. We also provide evidence that, at a later stage, ecdysone elicits negative feedback to shut down PG steroidogenic activity so as to reduce the ecdysone titer during the prepupal-pupal transition. Different isoforms of the transcription factor Br mediate the feedback by regulating expression of the ecdysone biosynthetic enzymes through direct binding to their gene promoters. Thus, our work identifies a feedback circuitry that, by coordinating the rate and duration of ecdysone synthesis in the PG, generates the maturation-inducing ecdysone pulse necessary for transition to the adult stage.
RESULTS
Inactivation of ecdysone responses in the PG reduces ecdysone synthesis and delays development
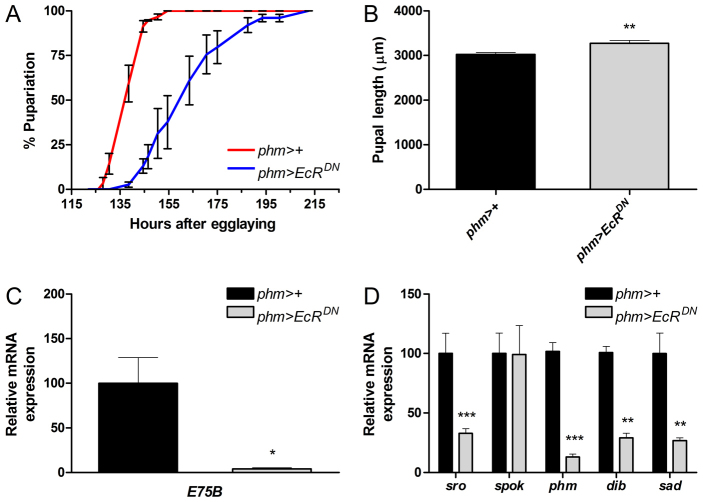
Formation of the ecdysone pulse that triggers metamorphosis requires coordination of the activation and repression of ecdysone synthesis in the PG. Earlier studies using cultured PG cells demonstrated that ecdysone can either increase or decrease its own production rate depending on its concentration and the duration of incubation (Beydon and Lafont, 1983; Sakurai and Williams, 1989). We hypothesized that an ecdysone pulse generator exists that relies on ecdysone-mediated feedback control of PG steroidogenic activity. To test this, we blocked EcR signaling by expression of a dominant-negative EcR (UAS-EcRDN) in the PG using the PG-specific phm-Gal4 (phm>) driver. To determine the requirement of EcR signaling for the normal progression of development, we analyzed the timing of pupariation in larvae expressing EcRDN in the PG (phm>EcRDN) compared with the control. Interestingly, reducing EcR activity in the PG delays pupariation by ∼1 day (Fig. 1A). Furthermore, pupariation is less synchronized in the phm>EcRDN animals compared with the phm>+ control, as indicated by the slope of the curve. These data suggest a positive feed-forward mechanism in which ecdysone produced by the PG is required to stimulate its own synthesis and trigger pupariation.
Fig. 1.
Reduced ecdysone signaling in the PG delays the ecdysone peak and pupariation. (A) Suppression of EcR activity in the prothoracic gland (PG) of phm>EcRDN Drosophila larvae is sufficient to delay timing of pupariation. (B) Effect on pupal size of EcR inactivation in the PG shows that pupal size is increased in phm>EcRDN animals (n=25). (C) Expression of E75B, as a measure of ecdysone signaling, shows reduced levels in third instar phm>EcRDN larvae 120 hours after egg laying (AEL). (D) The expression of genes involved in ecdysone biosynthesis in phm>EcRDN larvae relative to control (phm>+) larvae 120 hours AEL (n=4). Disrupting EcR activity in the PG results in reduced expression of phm, dib, sad and sro. Error bars indicate s.e.m. *P<0.05, **P<0.01, ***P<0.001, versus the phm>+ control.
Since larvae with reduced ecdysone signaling in the PG pupariate later, we asked if the prolonged larval development gives rise to an overgrowth phenotype. Analysis of the effect on size shows that phm>EcRDN larvae form slightly, but significantly, larger pupae (Fig. 1B). The increased size of phm>EcRDN indicates a prolonged growth phase, although much of the delay occurs in the non-feeding wandering stage, which indicates that these larvae fail to produce the high-level peak in the late third instar that initiates pupariation. Consistent with this, peak levels of ecdysone 120 hours after egg laying (AEL) appear to be reduced in the phm>EcRDN larvae compared with the phm>+ control as measured by expression levels of the ecdysone target gene E75B (Fig. 1C), which has been shown to serve as a proxy for the ecdysone titer (Colombani et al., 2012; Colombani et al., 2005; Layalle et al., 2008). Since EcR is a transcription factor, we also examined whether it regulates ecdysone production through transcriptional control of ecdysone biosynthetic genes (Niwa et al., 2010; Rewitz et al., 2007). Expression of the ecdysone biosynthetic genes shroud (sro), phantom (phm), disembodied (dib) and shadow (sad) were all found to be reduced in the phm>EcRDN larvae compared with the control (Fig. 1D). However, reducing EcR signaling in the PG did not affect the expression of spookier (spok), which encodes an enzyme that catalyzes an early step in the pathway (Ono et al., 2006).
We conclude from these data that ecdysone signaling through EcR acts positively in the PG to ramp up its own production, in part by either directly or indirectly controlling the transcription of several of its own biosynthetic enzyme-encoding genes.
The ecdysone-regulated factor Br binds to the promoters of genes involved in ecdysone biosynthesis
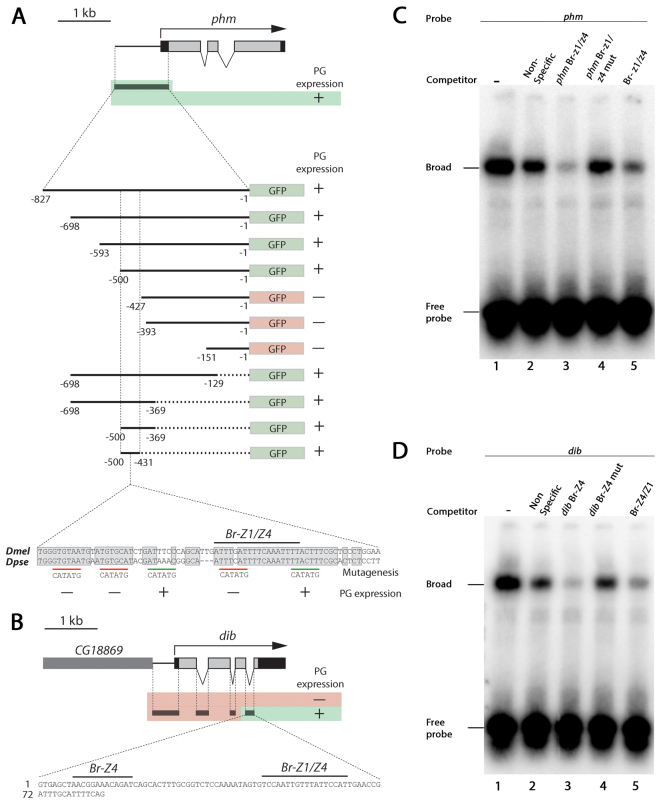
In order to identify the cis-regulatory elements responsible for the EcR-mediated control of genes involved in ecdysone biosynthesis, we conducted a promoter/enhancer characterization analysis. We identified an 827 bp genomic region upstream of the ATG start codon of phm that drives specific expression of a GFP reporter in the PG (Fig. 2A). A PG-specific dib enhancer was also identified in the small (86 bp) third intron of dib, rather than in the upstream region (Fig. 2B). To narrow down the element(s) responsible for the PG-specific expression of phm, we performed a deletion analysis that identified a conserved 69 bp element, located at -500 to -431 nucleotides relative to the ATG start codon, that was sufficient for directing PG-specific reporter expression. To identify nucleotides important for PG expression, we introduced mutations within several conserved blocks in the 69 bp sequence, as well as in one region that showed less conservation, and found three highly conserved motifs required for PG expression. Although in silico analysis of potential transcription factor binding sites did not predict any EcR sites in the 69 bp phm promoter or in the dib enhancer, both sequences contained conserved binding sites for the primary EcR target Br. The br locus encodes four isoforms of zinc-finger DNA-binding proteins called Br-Z1, Br-Z2, Br-Z3 and Br-Z4 (Riddiford et al., 2003). Although it has been shown that βFTZ-F1 (Ftz-f1 - FlyBase) is required for the expression of Phm and Dib (Parvy et al., 2005), we did not identify βFTZ-F1 consensus binding sites in their promoters, making it likely that the effects of βFTZ-F1 are indirect or are mediated through other enhancers.
Fig. 2.
The ecdysone-regulated factor Br binds to the promoters of ecdysone biosynthesis genes. Gene structure map showing the different phm (A) and dib (B) cis-regulatory elements assayed for PG-specific expression patterns. Transgenic animals expressing GFP under the control of the minimal Hsp70 promoter and different cis-regulatory elements were analyzed for PG-specific GFP expression. PG specific cis-regulatory elements were identified in the 5′ upstream region of phm and in the third intron of dib. A deletion analysis for the phm promoter identified a 69 nucleotide region (-500 to -431) in the 827 nucleotides upstream of the ATG start codon that drives PG-specific expression of GFP. Nucleotide alignment of the PG-specific cis-regulatory phm sequence shows a high degree of conservation between Drosophila melanogaster (Dmel) and Drosophila pseudoobscura (Dpse). In silico analysis of transcription factor binding sites identifies potential Br-Z1/Z4 sites in the 69 nucleotide phm promoter and in the dib enhancer located in the third intron. Underlined sequences in the alignment indicate 6 bp mutations (underlined sequence mutated to CATATG) introduced into the 69 nucleotide phm promoter conferring PG-specific expression. Mutation analysis of this element reveals sites required for PG expression in transgenic larvae, including the Br-Z1/Z4 binding site. +, PG expression; -, lack of PG expression. (C,D) Electrophoretic mobility shift assay (EMSA) was used to determine Br-Z4/Z1 binding activity of regulatory sites. Nuclear extract containing Br was incubated with [γ32]ATP-labeled probes of (C) phm or (D) dib regulatory sequences containing the Br-Z4/Z1 (phm) and Br-Z4 (dib) sites and resulted in shifted DNA-protein bands (lane 1). Competition assays were performed with unlabeled oligonucleotides of either non-specific random (lane 2), phm and dib (lane 3), phm and dib with mutated Br-Z4/Z1 sites (lane 4) or a Br-Z4/Z1 consensus motif (lane 5) sequence.
The phm and dib PG enhancers contain binding sites for both Br-Z4 and Br-Z1, which localize to a motif required for PG expression of the phm element as shown by mutagenesis (Fig. 2A,B). To examine whether Br binds to the phm Br-Z1/Z4 and dib Br-Z4 sites, we used the electrophoretic mobility shift assay (EMSA) (Fig. 2C,D). For both the phm and dib sequences, we observed a strongly shifted band upon addition of nuclear extract containing Br (supplementary material Fig. S1) (Lin et al., 2011). This complex was reduced when competed by unlabeled phm and dib oligonucleotides or a consensus Br-Z1/Z4 oligonucleotide sequence, but not by unlabeled phm and dib oligonucleotides with mutated br sites or a random oligonucleotide. Together, these data support that Br-Z4 and Br-Z1 bind directly to the phm and dib promoters/enhancers to regulate transcription.
br is induced by ecdysone and required for the rapid ramp up of ecdysone synthesis
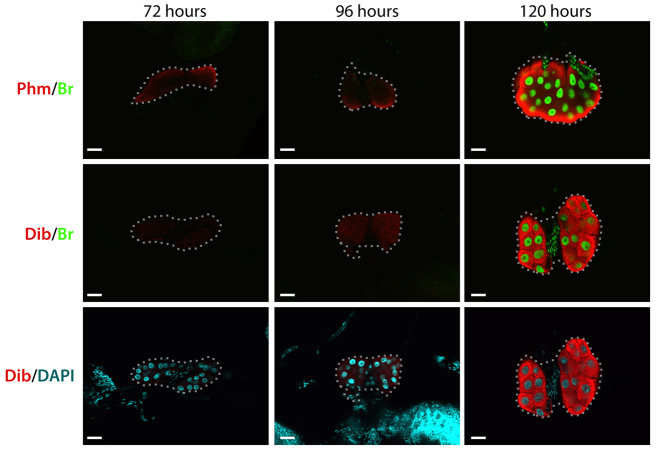
If Br is required for upregulating the genes involved in ecdysone biosynthesis in the late third instar to support high ecdysone production (Parvy et al., 2005; Rewitz et al., 2006b; Yamanaka et al., 2007) then it should be expressed in the PG during this period. In both early (72 hours AEL) and mid (96 hours AEL) third instar larvae, low Br levels were accompanied by low Phm and Dib levels in the PG (Fig. 3; supplementary material Fig. S2). During the ecdysone peak at the end of the third instar (120 hours AEL), high levels of Br coincided with increased levels of Phm and Dib. Thus, Br upregulation is temporally coordinated with the ecdysone peak and increased expression of the biosynthetic enzymes.
Fig. 3.
Br upregulation coincides with high ecdysone biosynthetic enzyme levels and the ecdysone peak. PGs from wild-type (w1118) larvae, at the indicated times AEL, were dissected and immunostained for Phm and Dib (red) and Br (green). DAPI (cyan) staining is shown for samples stained for Dib. Dotted lines encircle PG cells. All samples were processed and examined using the same microscope settings. Representative images from the PG for each experimental group (n≥5) are shown. Scale bars: 25 μm.
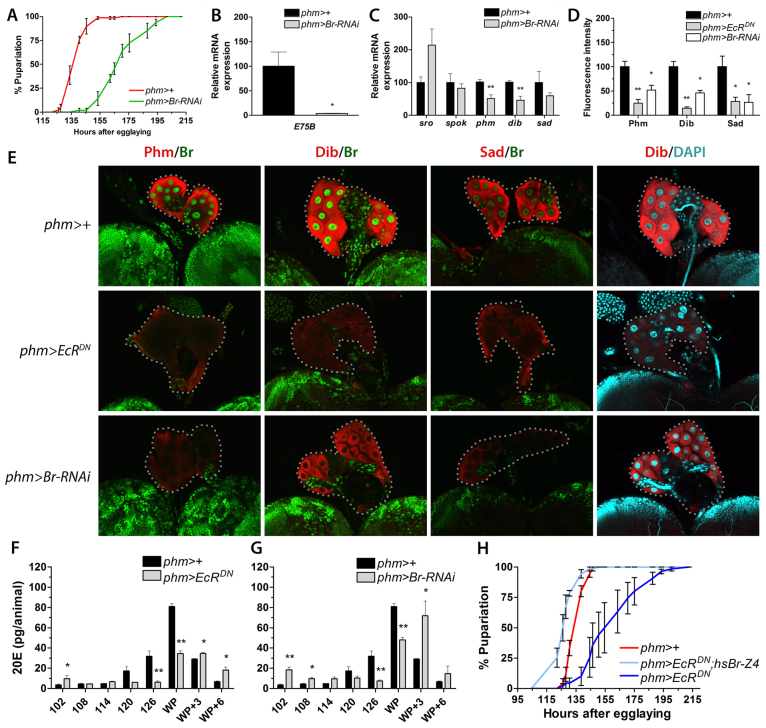
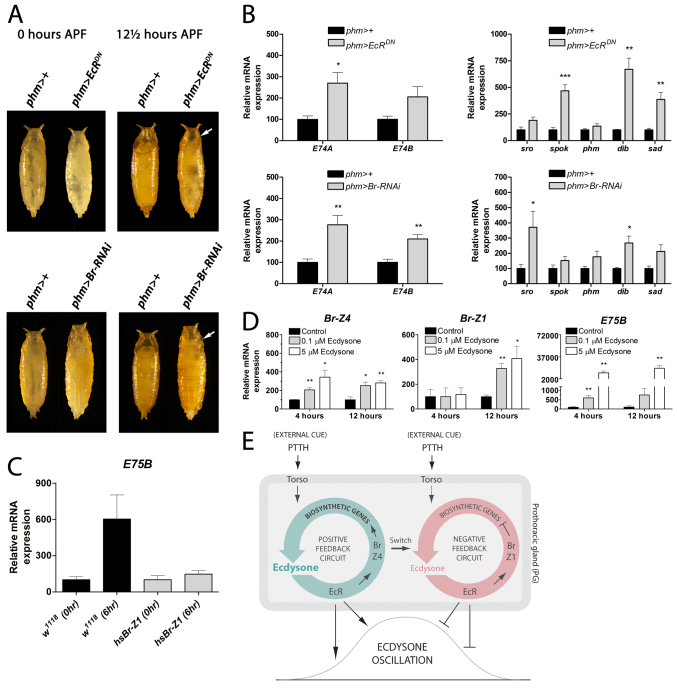
To determine whether Br has an important function in the PG, we knocked down its expression using a UAS-Br-RNAi (Br-RNAi) line driven by phm>. Knocking down br in the PG results in a developmental delay of pupariation similar to that observed in animals with reduced EcR signaling in the PG (Fig. 4A). Similar to phm>EcRDN larvae, the slope of the pupariation curve for phm>Br-RNAi larvae compared with the phm>+ controls indicates that pupariation is delayed and less synchronized in animals lacking Br. Accordingly, delayed phm>Br-RNAi larvae have reduced ecdysone peak levels, as measured by the reduced expression of E75B, compared with control larvae (Fig. 4B). This suggests that Br is required in the PG to intensify ecdysone biosynthesis at the end of the third instar, as observed for EcR. Similar to suppressing EcR signaling, knockdown of br in the PG results in reduced Phm, Dib and Sad protein levels and phm and dib transcript levels without affecting spok (Fig. 4C-E). By contrast, sro expression, which is reduced in phm>EcRDN larvae, shows no significant decrease in the phm>Br-RNAi larvae. Unlike spok, phm, dib and sad, which encode P450 enzymes, sro encodes a dehydrogenase involved in an early biosynthetic step together with Spok. Thus, ecdysone seems to stimulate sro expression through a mechanism that does not involve Br. Importantly, this also indicates that although Br is an important target, it is not the only target of EcR in the PG. Altogether, these data suggest that Br co-regulates the genes involved in late ecdysone biosynthetic steps and coordinates high expression during the late third instar in order to produce the ecdysone peak that triggers pupariation. Furthermore, induction of Br is mediated by EcR, as Br was not detected in the PG cells of phm>EcRDN larvae (Fig. 4E). Using an ELISA assay, we measured the ecdysone titer, confirming that animals with reduced EcR and Br activity in the PG fail to produce the high-level ecdysone pulse that triggers pupariation (Fig. 4F,G). Interestingly, even during the prolonged larval development, phm>EcRDN and phm>Br-RNAi animals are unable to produce the high ecdysone levels observed at the white puparium (WP) stage of the control.
Fig. 4.
Br is EcR dependent and necessary for the expression of ecdysone biosynthesis genes and the timing of pupariation. (A) Knockdown of br in the PG (phm>Br-RNAi) delays pupariation compared with the control. (B) E75B expression, as a readout for ecdysone signaling, shows that reduction of br expression in the PG (phm>Br-RNAi) results in reduced ecdysone levels in larvae 120 hours AEL (n=4). (C) Transcript levels of genes involved in ecdysone biosynthesis in phm>Br-RNAi larvae 120 hours AEL compared with the control (n=4). (D) Quantification of fluorescence intensity in PG cells (using ImageJ) shows reduced immunostaining of ecdysone biosynthetic enzymes (Phm, Dib and Sad) in the PG of phm>EcRDN and phm>Br-RNAi larvae compared with the control 120 hours AEL. (E) Immunostaining of Phm, Dib or Sad (red) and Br (green) in larval PG cells (120 hours AEL) of the indicated genotypes. DAPI (cyan) in the PG stained for Dib shows that nuclei and cells are intact. Br staining could not be detected in PG cells when EcR activity was reduced (phm>EcRDN) or when br was knocked down in these cells (phm>Br-RNAi). Dotted lines encircle PG cells. All samples were processed and examined using the same microscope settings. Representative images from the PG for each experimental group (n≥5) are shown. (F,G) Ecdysteroid levels in the indicated genotypes during the third instar and prepupal stage. (H) Resupplying Br-Z4 at 112 hours AEL (prior to the high-level ecdysone pulse) by heat-shock treatment of phm>EcRDN; hs-Br-Z4 larvae is sufficient to rescue the developmental delay resulting from PG inactivation of EcR activity. Error bars indicate s.e.m. *P<0.05, **P<0.01, versus the phm>+ control. WP, white puparium.
To determine if the lack of Br is the primary defect that results in the EcRDN-induced developmental delay and pupariation asynchrony, we attempted to rescue these phenotypes by resupplying Br through a heat shock-inducible transgene. The high-level ecdysone peak at the end of the third instar stage is accompanied by the appearance of Br-Z4, whereas Br-Z2 and Br-Z3 remain low and show little relation with the ecdysone titer (Zhou et al., 2004). Br-Z1 appears later, at the time of pupariation. Thus, Br-Z4 matches the criteria for being the isoform that induces high-level expression of phm, dib and sad during the late third instar stage. To test this notion, we expressed this isoform using a well-timed heat shock just as larvae begin to wander prior to the high-level ecdysone peak. Indeed, ectopic expression of Br-Z4 rescues the delay and asynchrony of phm>EcRDN larvae (Fig. 4H), confirming that the positive feedback through EcR induces Br-Z4 that upregulates the genes involved in ecdysone synthesis to produce the high-level pulse necessary to initiate pupariation.
Ecdysone-regulated feedback is an autonomous mechanism downstream of PTTH that amplifies ecdysone synthesis
The positive-feedback mechanism is likely to ensure a rapid and large response to neuropeptide signals such as PTTH. To examine whether the feedback system operates downstream of PTTH/Torso signaling, we reduced the expression of br in larvae expressing an activated form of Ras85D (RasV12) in the PG, which mimics constitutive PTTH signaling (Rewitz et al., 2009b). Expressing RasV12 in the PG increases ecdysone biosynthesis and results in precocious pupariation (Fig. 5A). Reducing br expression in the PG of phm>RasV12 larvae effectively attenuates the developmental acceleration. This shows that Br is necessary for the accelerated ecdysone synthesis and precocious phenotype of phm>RasV12 animals. Consistent with abolishing the precocious pupariation, phm>RasV12, Br-RNAi larvae form larger pupae than phm>RasV12 animals (Fig. 5B; supplementary material Fig. S3), indicating an extended growth period. If the EcR-mediated feed-forward mechanism is downstream of PTTH, it should not be required for expression of the PTTH receptor torso. Consistent with this, expression of torso was not affected in phm>EcRDN and phm>Br-RNAi larvae 120 hours AEL as compared with the control (Fig. 5C), confirming that the responsiveness of the PG to PTTH is not reduced. The positive feedback is therefore a PG-autonomous mechanism that helps in translating the PTTH signal into a high-level ecdysone pulse.
Fig. 5.
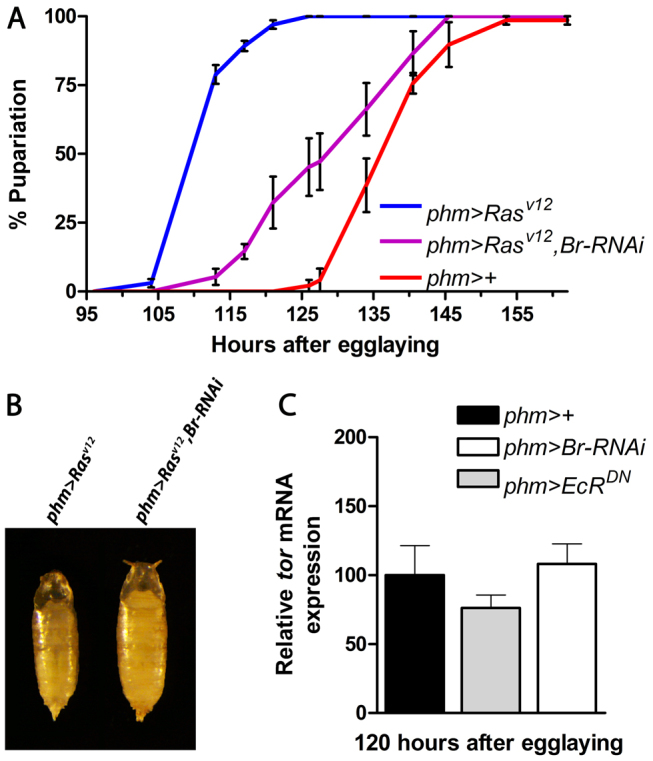
Ecdysone feedback amplifies steroid biosynthesis downstream of PTTH/Torso. (A) Knockdown of br delays precocious pupariation in larvae expressing RasV12 (a constitutively activate form) in the PG. (B) Effect on pupal size of br knockdown in animals expressing RasV12 in the PG shows that pupal size is increased. (C) Expression of torso in the PG of phm>EcRDN and phm>Br-RNAi compared with the control at 120 hours AEL. Error bars indicate s.e.m.
Negative ecdysone signaling feedback shuts down PG activity and reduces ecdysone levels
We observed that, after pupariation, phm>EcRDN and phm>Br-RNAi larvae fail to complete metamorphosis. At pupariation [0 hours after puparium formation (APF)] these animals are morphologically indistinguishable from the control, but they die in the prepupal stage as indicated by the lack of head eversion 12.5 hours APF (Fig. 6A). This phenotype is reminiscent of mutants that fail to reduce the ecdysone titer after pupariation (Rewitz et al., 2010). Recently, we showed a negative-feedback mechanism in which increasing levels of ecdysone from the PG induce a peripheral degradation/clearance mechanism that eliminates circulating ecdysone (Rewitz et al., 2010). Removal of circulating ecdysone must be accompanied by reduced synthesis to efficiently lower its concentration to create the pulse that coordinates metamorphic development. We therefore asked whether ecdysone, in addition to the positive feedback during the late third instar, forms a negative-feedback loop that dominates during the prepupal stage and represses ecdysone synthesis. If this were the case, ecdysone levels should be elevated in phm>EcRDN and phm>Br-RNAi animals compared with the control during the prepupal stage. Since the ecdysone-regulated E74 drops more rapidly in response to the declining ecdysone titer after pupariation than E75B (Andres et al., 1993), we choose to use E74A and E74B isoforms as readouts for the ecdysone titer during the prepupal stage. We observed increased expression of E74 in the phm>EcRDN and phm>Br-RNAi prepupae 6 hours APF (Fig. 6B), consistent with significantly elevated ecdysone levels at both 3 and 6 hours APF in phm>EcRDN prepupae and 3 hours APF in phm>Br-RNAi prepupae as measured by ELISA (Fig. 4F,G). Thus, despite the low ecdysone concentration in phm>EcRDN and phm>Br-RNAi animals at the WP stage (0 hours APF) compared with the control, these animals have elevated levels a few hours later, showing that the titer declines very slowly in these animals. To more directly assess PG ecdysteroidogenic activity we measured the expression of genes involved in ecdysone biosynthesis in the phm>EcRDN and phm>Br-RNAi prepupae and found increased expression of several of the genes 6 hours APF (Fig. 6B). This directly demonstrates that EcR and Br are required to suppress the expression of several biosynthetic pathway components in the PG and shut down ecdysone synthesis.
Fig. 6.
EcR and Br are required for suppressing ecdysone synthesis after pupariation. (A) Animals with reduced PG activity of EcR (phm>EcRDN) and Br (phm>Br-RNAi) form normal white puparia [0 hours after puparium formation (APF)] but fail in the prepupal-to-pupal transition as indicated by the lack of head eversion 12.5 hours APF. (B) Expression of E74A and E74B (as a readout for ecdysone signaling) and ecdysone biosynthesis genes in phm>EcRDN and phm>Br-RNAi prepupae 6 hours APF compared with the control (n=4). (C) Expression of Br-Z1 was induced 112 hours AEL by heat shocking hs-Br-Z1 larvae and expression of E75B (as a readout for ecdysone signaling) was determined at 112 hours (0 hour) and 118 hours (6 hours) AEL. Expression of Br-Z1 prevents the production of the late third instar high-level ecdysone molting peak as indicated by reduced expression of E75B. (D) Expression of Br-Z4, Br-Z1 and E75B in S2 cells incubated in the absence (control) or presence of 0.1×10-6 M or 5×10-6 M 20-hydroxyecdysone (ecdysone). (E) Model for the feedback regulation. Opposing feedback circuits that rely on a switch in the PG from Br-Z4 to Br-Z1 shape the maturation-inducing ecdysone pulse that coordinates the transition from the larval to the adult stage in response to the neuropeptide PTTH. Error bars indicate s.e.m. *P<0.05, **P<0.01, ***P<0.001, versus the phm>+ control.
Our identification of putative Br-Z1 binding sites in the phm promoter and dib enhancer suggests that Br-Z1 plays a role in regulating the ecdysone biosynthetic genes. To test if Br-Z1 is involved in the repression of ecdysone synthesis, as its expression coincides with the time (WP) when the ecdysone titer begins to decline, we ectopically expressed Br-Z1 under the control of a heat-shock promoter 112 hours AEL, just prior to the high-level ecdysone peak. Expression of Br-Z1 at this time delayed pupariation (supplementary material Fig. S4), suggesting that Br-Z1 inhibits the production of the ecdysone peak. In control animals, the expression of E75B was increased 6 hours later (118 hours AEL), whereas in larvae that ectopically express Br-Z1 expression of E75B was not increased (Fig. 6C). Furthermore, the expression of genes involved in ecdysone biosynthesis was reduced in larvae expressing Br-Z1 (supplementary material Fig. S4), indicating that Br-Z1 transcriptionally silences these genes. Overall, these results demonstrate a negative-feedback circuit relying on a switch from Br-Z4 to Br-Z1 that represses ecdysone synthesis through the transcriptional silencing of genes involved in ecdysone biosynthesis. This raises the question of what regulates the switch from Br-Z4 to Br-Z1? We reasoned that if the switch were dependent on the concentration of ecdysone, low levels should induce Br-Z4 whereas high levels should induce Br-Z1. To test this, we measured the expression of Br-Z4 and Br-Z1 in S2 cells incubated for 4 and 12 hours with ecdysone concentrations corresponding to low (0.1×10-6 M) or high (5×10-6 M) levels compared with physiological peak conditions. We observed sequential activation, with Br-Z4 being induced after 4 hours, whereas increased expression of Br-Z1 was observed after 12 hours under both conditions (Fig. 6D). Br-Z4 therefore behaves as an early ecdysone response gene, like E75B, that is induced after 4 hours in the presence of ecdysone. By contrast, even high ecdysone concentrations failed to induce Br-Z1 expression after 4 hours. Based on these data, we can exclude that the switch depends exclusively on ecdysone concentrations, but is likely to involve a sequential induction timing mechanism.
Overall, our data demonstrate the existence of opposing feedback circuits that shape the metamorphic ecdysone pulse (Fig. 6E). The positive feed-forward loop helps amplify the initial PTTH-induced rise in the ecdysone titer during the late wandering phase to trigger pupariation. The negative-feedback circuit centered on Br-Z1 expression then dominates after pupariation, ensuring a self-limiting response of the PG that help sets pulse duration and amplitude, which is crucial for the progression of development to the adult stage.
DISCUSSION
Although extensive studies have made it clear that transition to the adult stage in insects requires a high-level pulse of ecdysone, the mechanism that shapes the pulse, by determining its duration and amplitude, has remained unclear. Our experiments show that the maturation-inducing pulse that coordinates the juvenile-adult transition in Drosophila is generated by ecdysone feedback control of PG steroidogenic activity. We demonstrate that, at the end of the third larval instar, ecdysone acts through EcR in a feed-forward circuit to produce the high-level pulse that triggers pupariation in response to PTTH. This illustrates an EcR-dependent positive feedback operating downstream of PTTH to generate a sustained output in terms of biosynthesis in response to neuropeptide signaling.
The feed-forward loop described here provides an explanation for a number of previous observations. These studies have indicated that ecdysone can modulate PG steroidogenic activity and that PG cells undergo autonomous activation under long-term culture conditions (Mizoguchi and Kataoka, 2005; Sakurai and Williams, 1989; Williams, 1952; Yamanaka et al., 2007). Interestingly, autonomous activation is prevented by juvenile hormone (JH), which inhibits br expression (Riddiford et al., 2003). During the last larval instar of holometabolous insects, a drop in JH levels eventually leads to the production of a high-level ecdysone pulse that triggers metamorphosis, although the mechanism underlying this is poorly understood. Since the decline of JH is permissive for br expression, the fact that Br promotes PG steroidogenic activity is likely to explain how the drop in JH results in the production of a high-level ecdysone pulse initiating metamorphosis. Thus, our data provide a link between JH and ecdysone that might explain how the presence of JH prevents metamorphosis.
Our observations clearly show that positive feedback is required for the transcriptional upregulation of phm, dib and sad, all of which encode enzymes that act at late steps in the ecdysone biosynthetic pathway. By contrast, EcR and Br activity are not necessary for the normal activity of spok, which is involved in an earlier step in the pathway and whose transcription is regulated by Molting defective, a factor that is not involved in the regulation of the other identified biosynthetic enzymes (Neubueser et al., 2005; Ono et al., 2006). In addition, in contrast to the other ecdysone biosynthetic enzymes, Spok is also likely to be regulated at the level of translation and phosphorylation in response to PTTH signaling (Rewitz et al., 2009a). Furthermore, expression of torso is not EcR and Br dependent, consistent with levels of torso not being synchronized with the ecdysone peaks (Rewitz et al., 2009b). Together with our results demonstrating that the feedback is required downstream of Ras in the PG, this shows that the feed-forward loop functions downstream of PTTH to amplify the signal and not for endowing the PG with competence to respond to PTTH.
Our findings raise an important issue that challenges the classical view that ecdysone released from the PG is converted to its more active metabolite 20-hydroxyecdysone (20E) in peripheral target tissues, where it interacts with EcR (Rewitz et al., 2006a). Although 20E may travel back and inform the PG, a more direct route would be that ecdysone produced by the PG acts on the gland itself or that the PG produces small amounts of 20E that control the activity of the gland. Consistent with these possibilities, we have observed that reduced expression of shade, which encodes the enzyme that converts ecdysone to 20E, in the PG leads to a developmental arrest in the larval stages and that all three Drosophila EcR isoforms can induce transcription in response to ecdysone (unpublished data). Interestingly, recent reports have demonstrated the essential function of E75, DHR3 (Hr46 - FlyBase), βFTZ-F1 and DHR4 in regulating the production of ecdysone in the PG (Cáceres et al., 2011; Ou et al., 2011; Parvy et al., 2005). Although nitric oxide and PTTH regulate the activity of some of these factors, these signals alone are unlikely to explain the regulation of their function in the PG. Based on our results, an obvious possibility is that EcR controls the expression of these classical ecdysone-inducible genes in the PG. Extensive studies on these ecdysone target genes have led to the elucidation of an early response network for steroid hormone action (Ashburner et al., 1974) and the molecular characterization of the genetic architecture underlying the cellular responses to steroids (King-Jones and Thummel, 2005; Thummel, 2001; Yamanaka et al., 2013). Surprisingly, we show here that this genetic program that guides the downstream cellular decisions in response to regulatory ecdysone pulses is utilized upstream to shape the pulse by setting its duration and amplitude. Thus, the same genetic components are used for coordinating the production and reception of the steroid signals that drive directional developmental progression.
Our previous experiments demonstrated that ecdysone, produced by the PG, induces an inactivation enzyme responsible for clearance of circulating ecdysone (Rewitz et al., 2010). We show here that termination of the pulse requires negative feedback that represses PG steroid production activity in coordination with peripheral clearance. How does ecdysone stimulate and repress biosynthesis in the PG through EcR? Our results show that EcR induces different Br isoforms, forming circuits that either increase or inhibit the activity of the biosynthetic pathway by regulating the levels of the enzymatic components. Br is required specifically for the juvenile-adult transition (Kiss et al., 1988; Riddiford et al., 2003) and is expressed during the last instar. Our study shows that the appearance of Br in the PG requires EcR and correlates with the ecdysone peak. The positive effect of EcR on ecdysone biosynthesis is mediated largely through Br-Z4, which has previously been shown to induce transcription of Npc1a, which encodes a key cellular component required in the PG for the delivery of cholesterol as a substrate for steroid synthesis (Xiang et al., 2010). Together, this suggests that ecdysone-mediated positive feedback coordinates increased substrate delivery with upregulation of the biosynthetic machinery in order to produce the maturation-inducing ecdysone pulse. Conversely, the Br-Z1 isoform inhibits ecdysone synthesis, forming a negative feedback that is important for the decline of the ecdysone titer during the prepupal stage. Thus, the temporal control of these circuits relies on a dynamic switch in the PG from Br-Z4 to Br-Z1. A similar switch has been found in the imaginal discs, where Br-Z4 rapidly accumulates in response to ecdysone and then disappears several hours later when Br-Z1 is upregulated (Bayer et al., 1996). It has been suggested that the switch from Br-Z4 to Br-Z1 is regulated at the level of alternative splicing of br transcripts (Bayer et al., 1996). Our data suggest that the switch is a hard-wired genetic timing mechanism rather than being dependent on ecdysone concentrations. This switching might also occur at the enhancer level through competition of binding to overlapping Br-Z1/Z4 regulatory sites, as found in the phm promoter. Importantly, coupling a negative with a positive feedback through a common regulatory site ensures a self-limiting response by preventing ‘run away’ synthesis that would otherwise result from positive-feedback amplification alone.
In conclusion, our study shows that the maturation-inducing ecdysone pulse is shaped by an autonomous feed-forward and feedback circuitry within the endocrine tissue that modulates the rate of hormone synthesis. The coupling of these feedback circuits ensures rapid, self-limiting hormone production that translates neuropeptide signaling into a regulatory steroid pulse which functions as a switch to drive developmental progression.
MATERIALS AND METHODS
Drosophila stocks
Flies were raised on standard cornmeal food at 25°C with a 12:12 hour light:dark cycle. The Drosophila lines used were: phm-Gal4 (Ono et al., 2006); UAS-Br-RNAi (#104648) and w1118 obtained from the Vienna Drosophila RNAi Center; da-Gal4, UAS-EcRDN and UAS-RasV12 obtained from the Bloomington Stock Center; hs-Br-Z1 and hs-Br-Z4 kindly provided by Lynn Riddiford (Janelia Farm Research Campus, Howard Hughes Medical Institute, VA, USA).
Developmental timing analysis
For staging, flies were allowed to lay eggs on apple juice agar plates containing yeast paste for 4 hours in a moist chamber. The following day, first instar larvae were collected and transferred to vials containing standard cornmeal food. Pupariation was observed every 6-8 hours in three replicates each containing 20-25 animals. Images of pupae were captured using an Olympus SZX7 camera and AxioVision software (Zeiss) was used to measure pupal size. To express Br-Z1 and Br-Z4, larvae were heat shocked at 37°C and 29°C, respectively, for 20 minutes.
DNA constructs and generation of transgenic flies
DNA fragments were PCR-amplified from genomic DNA using the primers listed in supplementary material Table S1. Products were purified and cloned into the pH-Stinger GFP reporter vector (Barolo et al., 2000). Transgenic flies were generated by injection of DNA using standard procedures and analyzed for PG-specific expression of the reporter GFP. Site-directed mutations were introduced into the promoter using the QuikChange Kit (Stratagene) using the primers listed in supplementary material Table S2.
DNA binding assay
EMSA was used to assay DNA-protein binding. DNA oligonucleotides used are listed in supplementary material Table S3. Br transcription factor binding sites were identified based on analysis using TRANSFAC Professional 12.1 and Jaspar (Bryne et al., 2008) databases. The probes were prepared by annealing the complementary oligonucleotides in 0.1 M NaCl followed by 5′-end labeling with [γ32P]ATP (Perkin Elmer) using T4 polynucleotide kinase (Fermentas) and purified through Microspin G-25 columns (GE Healthcare). For the EMSA reaction, 2-5 μg nuclear extract (Active Motif) derived from Drosophila S2 cells was used. Nuclear extract was mixed with 3 μl dialysis buffer (25 mM Hepes pH 7.6, 40 nM KCl, 0.1 mM EDTA, 10% glycerol), 10 μl gelshift buffer (25 mM Tris-HCl pH 7.5, 5 mM MgCl2, 60 mM KCl, 0.5 mM EDTA, 5% Ficoll 400, 2.5% glycerol, 1 mM DTT and protease inhibitors) and 1 μg poly(dI-dC) (Sigma). For the competition assay, a 50-fold molar excess of unlabeled non-specific, wild-type, Br site mutation or consensus Br-Z4/Z1 site oligonucleotides were added to the reaction mix and incubated on ice for 20 minutes, and 0.6 pmol of the radiolabeled probe was added followed by a further incubation for 20 minutes on ice. Gelshift loading buffer [10% glycerol, 0.2% Bromophenol Blue and 0.5× Tris-borate-EDTA buffer (2× TBE: 44.5 mM Tris-HCl pH 8.0, 44.5 mM boric acid, 1 mM EDTA)] was added to the binding reaction mixture and the protein-DNA complexes were resolved on a 5% non-denaturing polyacrylamide gel. The gel was dried on a slab gel dryer (SGD4050, Savant) and exposed on a phosphorimager screen followed by scanning on a Storm 840 scanner (Molecular Dynamics). Images were analyzed using ImageQuant software version 5.2 (GE Healthcare). In addition to the Br-Z1/Z4 sites of interest, in silico analysis predicted a Br-Z3 binding site immediately downstream of the Br-Z1/Z4 site in the phm promoter. To avoid competitive interference, nucleotides were changed in the phm probe to disrupt the downstream Br-Z3 site, allowing analysis of the Br-Z1/Z4 site alone.
Immunohistochemistry
Tissues were dissected in PBS and fixed in 4% formaldehyde for 20 minutes at room temperature followed by four washes in PBS containing 0.3% Triton X-100 (PBST). Primary antibodies used were rabbit anti-Phm (1:200), rabbit anti-Dib (1:200) and rabbit anti-Sad (1:200) (Gibbens et al., 2011; Parvy et al., 2005) and mouse anti-Br core (1:400; #25E9.D7, DSHB). Secondary antibodies were goat anti-rabbit Alexa Fluor 555 (A21429, Invitrogen) and goat anti-mouse Alexa Fluor 488 (A11001, Invitrogen). Samples were incubated with primary antibodies at 4°C overnight and with secondary antibodies for 2 hours at room temperature. Nuclei were stained with DAPI (1:500). Images were acquired using a Zeiss LSM 710 confocal laser-scanning microscope and analyzed by ImageJ (NIH).
Ecdysteroid measurements
To measure ecdysone levels, ecdysone was extracted from whole animals as described (Warren et al., 2006). Briefly, ten frozen animals were homogenized in 0.5 ml methanol, followed by re-extraction in 0.5 ml methanol and 0.5 ml ethanol by vortexing. The total volume of 1.5 ml was centrifuged and 0.5 ml of the cleared supernatant was evaporated, redissolved in ELISA buffer (1 M phosphate solution containing 1% BSA, 4 M sodium chloride, 10 mM EDTA) and subjected to a commercial ELISA kit (ACE Enzyme Immunoassay, Cayman Chemical) using standard procedures. Standard curves were generated using 20E obtained from Sigma (H5142) and the absorbance was determined at 405 nm.
Quantitative PCR
Total RNA was isolated from animals using the RNeasy Mini Kit (Qiagen) followed by RNase-free DNase treatment to avoid genomic DNA contamination. cDNA was synthesized by reverse transcription using the SuperScript III First-Strand Synthesis System (Invitrogen). Relative gene expression was measured by quantitative real-time PCR (qPCR) using the SYBR Green PCR Kit (Qiagen). Primers are given in supplementary material Table S4. S2 cells were incubated in the absence or presence of 0.1×10-6 M or 5×10-6 M 20-hydroxyecdysone for 4 or 12 hours before RNA purification.
Western blotting
Western blotting was performed using S2 cell nuclear extract (Active Motif) and tissues from third instar w1118 larvae. Samples were boiled in Laemmli Sample Buffer (Bio-Rad) containing 2-mercaptoethanol for 5 minutes and the supernatant was collected after centrifugation at 14,000 rpm (17,900 g). Samples were loaded on a 4-20% polyacrylamide gradient gel (Bio-Rad) and transferred onto a PVDF membrane (Millipore). Primary antibody was monoclonal mouse anti-Br core (1:400; DSHB, 25E9.D7-C) and the secondary antibody was HRP-conjugated polyclonal rabbit anti-mouse (1:20,000; Dako, P0260). The blot was developed using Luminata Crescendo (Millipore) substrate and images were captured with an Image Station 4000 MM (Carestream).
Statistical analysis
Error bars indicate s.e.m. and the significance of the difference between data sets was calculated using two-tailed Student’s t-tests.
Supplementary Material
Acknowledgments
We thank Pierre Leopold for comments on the manuscript, Leif Søndergaard for reagents, David Zhitomirsky for technical assistance, Lynn Riddiford for fly stocks and Jesper Troelsen for helpful discussion on the optimization of EMSAs.
Footnotes
Competing interests
The authors declare no competing financial interests.
Author contributions
K.F.R. and M.B.O. conceived the study and designed the experiments; M.E.M., E.T.D. and R.H. contributed to the experimental design; M.E.M., E.T.D., R.H. and K.F.R. performed and analyzed experiments; all authors contributed to interpretation of the results and writing of the manuscript.
Funding
This work was supported by the Danish Council for Independent Research, Natural Sciences [grant 11-105446 to K.F.R.] and the National Institutes of Health [grant R01 GM093301 to M.B.O.]. Deposited in PMC for release after 12 months.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.099739/-/DC1
References
- Andres A. J., Fletcher J. C., Karim F. D., Thummel C. S. (1993). Molecular analysis of the initiation of insect metamorphosis: a comparative study of Drosophila ecdysteroid-regulated transcription. Dev. Biol. 160, 388–404 [DOI] [PubMed] [Google Scholar]
- Ashburner M., Chihara C., Meltzer P., Richards G. (1974). Temporal control of puffing activity in polytene chromosomes. Cold Spring Harb. Symp. Quant. Biol. 38, 655–662 [DOI] [PubMed] [Google Scholar]
- Barolo S., Carver L. A., Posakony J. W. (2000). GFP and beta-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. Biotechniques 29, 726, 728, 730, 732 [DOI] [PubMed] [Google Scholar]
- Bayer C. A., Holley B., Fristrom J. W. (1996). A switch in broad-complex zinc-finger isoform expression is regulated posttranscriptionally during the metamorphosis of Drosophila imaginal discs. Dev. Biol. 177, 1–14 [DOI] [PubMed] [Google Scholar]
- Beydon P., Lafont R. (1983). Feedback inhibition of ecdysone production by 20-hydroxyecdysone in Pieris brassicae pupae. J. Insect Physiol. 29, 529–533 [Google Scholar]
- Bryne J. C., Valen E., Tang M. H., Marstrand T., Winther O., da Piedade I., Krogh A., Lenhard B., Sandelin A. (2008). JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 36, D102–D106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres L., Necakov A. S., Schwartz C., Kimber S., Roberts I. J., Krause H. M. (2011). Nitric oxide coordinates metabolism, growth, and development via the nuclear receptor E75. Genes Dev. 25, 1476–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombani J., Bianchini L., Layalle S., Pondeville E., Dauphin-Villemant C., Antoniewski C., Carré C., Noselli S., Léopold P. (2005). Antagonistic actions of ecdysone and insulins determine final size in Drosophila. Science 310, 667–670 [DOI] [PubMed] [Google Scholar]
- Colombani J., Andersen D. S., Léopold P. (2012). Secreted peptide Dilp8 coordinates Drosophila tissue growth with developmental timing. Science 336, 582–585 [DOI] [PubMed] [Google Scholar]
- Dungan H. M., Clifton D. K., Steiner R. A. (2006). Minireview: kisspeptin neurons as central processors in the regulation of gonadotropin-releasing hormone secretion. Endocrinology 147, 1154–1158 [DOI] [PubMed] [Google Scholar]
- Garelli A., Gontijo A. M., Miguela V., Caparros E., Dominguez M. (2012). Imaginal discs secrete insulin-like peptide 8 to mediate plasticity of growth and maturation. Science 336, 579–582 [DOI] [PubMed] [Google Scholar]
- Gibbens Y. Y., Warren J. T., Gilbert L. I., O’Connor M. B. (2011). Neuroendocrine regulation of Drosophila metamorphosis requires TGFbeta/Activin signaling. Development 138, 2693–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney J. F., Zolali-Meybodi O., Cherbas P. (2012). Tissue damage disrupts developmental progression and ecdysteroid biosynthesis in Drosophila. PLoS ONE 7, e49105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King-Jones K., Thummel C. S. (2005). Nuclear receptors—a perspective from Drosophila. Nat. Rev. Genet. 6, 311–323 [DOI] [PubMed] [Google Scholar]
- Kiss I., Beaton A. H., Tardiff J., Fristrom D., Fristrom J. W. (1988). Interactions and developmental effects of mutations in the Broad-Complex of Drosophila melanogaster. Genetics 118, 247–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layalle S., Arquier N., Léopold P. (2008). The TOR pathway couples nutrition and developmental timing in Drosophila. Dev. Cell 15, 568–577 [DOI] [PubMed] [Google Scholar]
- Lin G. G., Kozaki T., Scott J. G. (2011). Hormone receptor-like in 96 and Broad-Complex modulate phenobarbital induced transcription of cytochrome P450 CYP6D1 in Drosophila S2 cells. Insect Mol. Biol. 20, 87–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBrayer Z., Ono H., Shimell M., Parvy J. P., Beckstead R. B., Warren J. T., Thummel C. S., Dauphin-Villemant C., Gilbert L. I., O’Connor M. B. (2007). Prothoracicotropic hormone regulates developmental timing and body size in Drosophila. Dev. Cell 13, 857–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirth C. K., Shingleton A. W. (2012). Integrating body and organ size in Drosophila: recent advances and outstanding problems. Front. Endocrinol. (Lausanne) 3, 49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi A., Kataoka H. (2005). An in vitro study on regulation of prothoracic gland activity in the early last-larval instar of the silkworm Bombyx mori. J. Insect Physiol. 51, 871–879 [DOI] [PubMed] [Google Scholar]
- Neubueser D., Warren J. T., Gilbert L. I., Cohen S. M. (2005). molting defective is required for ecdysone biosynthesis. Dev. Biol. 280, 362–372 [DOI] [PubMed] [Google Scholar]
- Niwa R., Namiki T., Ito K., Shimada-Niwa Y., Kiuchi M., Kawaoka S., Kayukawa T., Banno Y., Fujimoto Y., Shigenobu S., et al. (2010). Non-molting glossy/shroud encodes a short-chain dehydrogenase/reductase that functions in the ‘Black Box’ of the ecdysteroid biosynthesis pathway. Development 137, 1991–1999 [DOI] [PubMed] [Google Scholar]
- Ono H., Rewitz K. F., Shinoda T., Itoyama K., Petryk A., Rybczynski R., Jarcho M., Warren J. T., Marqués G., Shimell M. J., et al. (2006). Spook and Spookier code for stage-specific components of the ecdysone biosynthetic pathway in Diptera. Dev. Biol. 298, 555–570 [DOI] [PubMed] [Google Scholar]
- Ou Q., Magico A., King-Jones K. (2011). Nuclear receptor DHR4 controls the timing of steroid hormone pulses during Drosophila development. PLoS Biol. 9, e1001160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvy J. P., Blais C., Bernard F., Warren J. T., Petryk A., Gilbert L. I., O’Connor M. B., Dauphin-Villemant C. (2005). A role for betaFTZ-F1 in regulating ecdysteroid titers during post-embryonic development in Drosophila melanogaster. Dev. Biol. 282, 84–94 [DOI] [PubMed] [Google Scholar]
- Rajan A., Perrimon N. (2012). Drosophila cytokine unpaired 2 regulates physiological homeostasis by remotely controlling insulin secretion. Cell 151, 123–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rewitz K. F., O’Connor M. B. (2011). Timing is everything: PTTH mediated DHR4 nucleocytoplasmic trafficking sets the tempo of Drosophila steroid production. Front. Endocrinol. (Lausanne) 2, 108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rewitz K. F., Rybczynski R., Warren J. T., Gilbert L. I. (2006a). The Halloween genes code for cytochrome P450 enzymes mediating synthesis of the insect moulting hormone. Biochem. Soc. Trans. 34, 1256–1260 [DOI] [PubMed] [Google Scholar]
- Rewitz K. F., Rybczynski R., Warren J. T., Gilbert L. I. (2006b). Identification, characterization and developmental expression of Halloween genes encoding P450 enzymes mediating ecdysone biosynthesis in the tobacco hornworm, Manduca sexta. Insect Biochem. Mol. Biol. 36, 188–199 [DOI] [PubMed] [Google Scholar]
- Rewitz K. F., O’Connor M. B., Gilbert L. I. (2007). Molecular evolution of the insect Halloween family of cytochrome P450s: phylogeny, gene organization and functional conservation. Insect Biochem. Mol. Biol. 37, 741–753 [DOI] [PubMed] [Google Scholar]
- Rewitz K. F., Larsen M. R., Lobner-Olesen A., Rybczynski R., O’Connor M. B., Gilbert L. I. (2009a). A phosphoproteomics approach to elucidate neuropeptide signal transduction controlling insect metamorphosis. Insect Biochem. Mol. Biol. 39, 475–483 [DOI] [PubMed] [Google Scholar]
- Rewitz K. F., Yamanaka N., Gilbert L. I., O’Connor M. B. (2009b). The insect neuropeptide PTTH activates receptor tyrosine kinase torso to initiate metamorphosis. Science 326, 1403–1405 [DOI] [PubMed] [Google Scholar]
- Rewitz K. F., Yamanaka N., O’Connor M. B. (2010). Steroid hormone inactivation is required during the juvenile-adult transition in Drosophila. Dev. Cell 19, 895–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rewitz K. F., Yamanaka N., O’Connor M. B. (2013). Developmental checkpoints and feedback circuits time insect maturation. Curr. Top. Dev. Biol. 103, 1–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddiford L. M., Hiruma K., Zhou X., Nelson C. A. (2003). Insights into the molecular basis of the hormonal control of molting and metamorphosis from Manduca sexta and Drosophila melanogaster. Insect Biochem. Mol. Biol. 33, 1327–1338 [DOI] [PubMed] [Google Scholar]
- Sakurai S., Williams C. M. (1989). Short-loop negative and positive feedback on ecdysone secretion by prothoracic gland in the tobacco hornworm, Manduca sexta. Gen. Comp. Endocrinol. 75, 204–216 [DOI] [PubMed] [Google Scholar]
- Tennessen J. M., Thummel C. S. (2011). Coordinating growth and maturation - insights from Drosophila. Curr. Biol. 21, R750–R757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel C. S. (1996). Flies on steroids—Drosophila metamorphosis and the mechanisms of steroid hormone action. Trends Genet. 12, 306–310 [DOI] [PubMed] [Google Scholar]
- Thummel C. S. (2001). Molecular mechanisms of developmental timing in C. elegans and Drosophila. Dev. Cell 1, 453–465 [DOI] [PubMed] [Google Scholar]
- Wang H. B., Nita M., Iwanaga M., Kawasaki H. (2009). betaFTZ-F1 and Broad-Complex positively regulate the transcription of the wing cuticle protein gene, BMWCP5, in wing discs of Bombyx mori. Insect Biochem. Mol. Biol. 39, 624–633 [DOI] [PubMed] [Google Scholar]
- Warren J. T., Yerushalmi Y., Shimell M. J., O’Connor M. B., Restifo L. L., Gilbert L. I. (2006). Discrete pulses of molting hormone, 20-hydroxyecdysone, during late larval development of Drosophila melanogaster: correlations with changes in gene activity. Dev. Dyn. 235, 315–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C. M. (1952). Physiology of insect Diapuse. IV. The brain and prothoracic glands as en endocrine system in the cecropia silkworm. Biol. Bull. 103, 120–138 [Google Scholar]
- Xiang Y., Liu Z., Huang X. (2010). br regulates the expression of the ecdysone biosynthesis gene npc1. Dev. Biol. 344, 800–808 [DOI] [PubMed] [Google Scholar]
- Yamanaka N., Honda N., Osato N., Niwa R., Mizoguchi A., Kataoka H. (2007). Differential regulation of ecdysteroidogenic P450 gene expression in the silkworm, Bombyx mori. Biosci. Biotechnol. Biochem. 71, 2808–2814 [DOI] [PubMed] [Google Scholar]
- Yamanaka N., Rewitz K. F., O’Connor M. B. (2013). Ecdysone control of developmental transitions: lessons from Drosophila research. Annu. Rev. Entomol. 58, 497–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Zhou B., Truman J. W., Riddiford L. M. (2004). Overexpression of broad: a new insight into its role in the Drosophila prothoracic gland cells. J. Exp. Biol. 207, 1151–1161 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.