Abstract
Purpose
To use a simple, quantitative method to compare the HSV1sr39TK/18F-FHBG PET reporter gene/PET reporter probe (PRG/PRP) system with PRGs derived from human nucleoside kinases.
Procedures
The same adenovirus vector is used to express alternative PRGs. Equal numbers of vectors are injected intravenously into mice. After PRP imaging, quantitative hepatic PET signals are normalized for transduction by measuring hepatic viral genomes.
Results
The same adenovirus vector was used to express equivalent amounts of HSV1sr39TK, mutant human thymidine kinase 2 (TK2-DM), and mutant human deoxycytidine kinase (dCK-A100VTM) in mouse liver. HSV1sr39TK expression was measured with 18F-FHBG; TK2-DM and dCK-A100VTM with 18F-L-FMAU. TK2-DM/18F-L-FMAU and HSV1sr39TK/18F-FHBG had equivalent sensitivities; dCK-A100VTM/18F-L-FMAU was twice as sensitive as HSV1sr39TK/18F-FHBG.
Conclusions
The human PRG/PRP sensitivities are comparable and/or better than HSV1sr39TK/18F-FHBG. However, for clinical use, identification of the best PRP substrate for each enzyme, characterization of probe distribution, and consequences of over-expressing nucleoside kinases must be evaluated.
Keywords: PET reporter gene, PET reporter probe, gene therapy, cell therapy, reporter gene, positron emission tomography, adenovirus, Herpes Simplex Virus thymidine kinase, deoxycytidine kinase, thymidine kinase 2
INTRODUCTION
Both the use of exogenous platforms (e.g., liposomes, nanoparticles, viral vectors) to deliver therapeutic genes and the development of targeted cell therapies (e.g., modified T cells, embryonic stem cells, induced pluripotent stem cells) have advanced from preclinical development to clinical trials and, in some cases, to standard of care for therapy. As these new therapies are developed, monitoring the duration of their availability in vivo and their targeting selectivity, and correlating these parameters with treatment outcomes, is critical in their evaluation and adoption for standard of care.
Incorporation of “reporter genes” whose activities can be examined non-invasively by whole body imaging provides a means to monitor both pharmacokinetics and targeting of these new vector- and cell-based therapeutic agents. In murine preclinical models, bioluminescence (e.g., alternative luciferases as reporter genes and their substrates as reporter probes) has provided convenient, inexpensive reporter gene-reporter probe systems to non-invasively monitor therapeutic gene delivery and cell-based therapies [1–4]. However, reporter gene immunogenicity, tissue attenuation of the signal, and lack of adequate resolution preclude bioluminescence imaging in most clinical contexts. For clinical applications, the most common approach has been the use of “PET reporter genes” (PRGs) whose activities can be monitored non-invasively by positron emission tomography (PET) [5, 6].
Although PRGs that encode transporters, receptors and enzymes have been developed [5, 6], the most widely used PRG is the Herpes Simplex Virus type 1 thymidine kinase (HSV1-TK) gene. HSV1TK can phosphorylate a variety of nucleoside analogues, including anti-herpetic acycloguanosines. HSV1-TK mutants engineered to more effectively use anti-herpetic drugs and to be are less effective at phosphorylating endogenous thymidine have been developed as “suicide genes”, to kill cells that ectopically express these kinases. Concurrently, positron-emitting derivatives of several acycloguanosine HSV1-TK substrates have been developed as probes for detecting HSV1-TK based PRG expression. The combination of the HSV1-sr39TK PRG and 9-[4-18F-3(hydroxymethyl) butyl]guanine (18F-FHBG) as its PRP is among the most widely used PRG/PRP systems [6–10].
Despite their current utility, the immunogenicity of HSV1-TK and its derivatives limits in vivo persistence of cells expressing these PRGs, and thus their utility in clinical applications [7, 11]. Several laboratories are developing PRGs from human genes, to circumvent PRG immunogenicity. Mutated versions of two human nucleoside kinases, thymidine kinase 2 (TK2) and deoxycytidine kinase (dCK), that utilize positron-emitting nucleoside analogues as PRPs have been developed in anticipation of clinical use [12–15].
As new PRG-PRP systems are described, it becomes imperative to have reliable procedures to monitor their relative specificities (the PRP should accumulate only in cells expressing the PRG) and sensitivities (the ability to generate quantifiable signals). The most common method of evaluating PRG/PRP systems has been to express PRGs in tumor cell lines, develop mouse xenografts, and image transgene PRG activity. However differences in expression vectors (e.g., differing promoters, differing 3’ and 5’ untranslated regions), differing integration sites and copy numbers, differing rates of tumor growth, distinctions in tumor vascularization and other biological variables make it difficult to compare such reports on relative efficacies of alternative PRP/PRG technologies.
We previously described the use of a common adenovirus delivery vector and a common gene expression construct to compare in vivo efficacy of alternative luciferase reporter genes for non-invasive imaging [16]. Post-imaging measurement of hepatic viral genomes and subsequent normalization of imaging data for the hepatic reporter gene copy number eliminates any differences imposed by variations in reporter gene delivery. This assay eliminates inconsistencies due to differences in vector construction, reporter gene copy number, integration site modulation of gene expression (adenovirus genomes do not integrate into chromosomal sites), and differential vascularity of the target. Only the reporter gene and reporter probe differ in this procedure.
Our goal in establishing this procedure was to provide a common platform to evaluate reporter gene efficacy across imaging modalities. In this report we demonstrate the utility of this procedure for evaluating differences in PRG/PRP efficacies by comparing the sensitivities of a mutated human TK2 PRG (TK2-N93D/L109F) [13] and a mutated human dCK PRG (dCK-A100V/R104M/D133A) [15, 17, 18] to one another, using the same PRP, 2’-deoxy-2-18F-5-methyl-1-β-L-arabinofuranosyluracil (18F-L-FMAU) [13], and to the commonly used HSV1-sr39TK/18F-FHBG PRG/PRP reporter system.
MATERIALS AND METHODS
Adenovirus vector construction
Plasmids and adenoviral vectors are listed in Table 1; all primers are listed in Table 2. Adenovirus vectors expressing alternative PET reporter genes were constructed as described previously [16] and in the Supplemental Material for this report. Although these reporter genes have been used, in some cases, to create adenovirus vectors previously (e.g., LUC2, HSV1sr39TK) we have renamed the viruses created here as Ad.HL viruses, to facilitate the use of these viruses as proper control and comparison viruses by other laboratories in future studies.
Table 1.
Plasmids and adenoviral vectors used in this study
| Plasmid or virus | Description or relevant characteristics | Source |
|---|---|---|
| Plasmids | ||
| pMSCV-sr39TKIRES-YFP. | HSV sr39TK containing retroviral plasmid | Radu Lab |
| pMSCV-TK2 | Truncated human TK2 containing retroviral plasmid | Radu Lab |
| pMSCV-TK2-N39D/L109F | Human TK2-N93D/L109F containing plasmid | Campbell 2012 [13] |
| pMSCV hudCK WT optimized IRES-YFP | Human wild-type dCK containing retroviral plasmid | Witte Lab |
| pMSCV hudCK DM optimized IRES-YFP | Human dCK double mutant containing retroviral plasmid | Witte lab |
| pMSCV hudCK TM optimized IRES-YFP | Human dCK triple mutant containing retroviral plasmid | Witte lab |
| pENTR4D | Gateway™ shuttle vector | Invitrogen |
| pAd/CMV/V5-DEST | Gateway™ destination adenoviral vector | Invitrogen |
| pENTRHM | pENTR4D with CmR and ccdB genes replaced by a synthetic multiple cloning site. | Gil 2011 [16] |
| pENTRHM-LUC2 | pENTRHM containing Luc2 gene | Gil 2011 [16] |
| pENTRHM-TK2 | pENTRHM containing TK2 soluble mutant gene | This study |
| pENTRHM-TK2-N93D-L109F | pENTRHM containing TK2- N93D-L109F mutant gene | This study |
| pENTRJG-dCK-WT | pENTRHM containing dCK wild type gene | This study |
| pENTRJG-dCK-DM | pENTRHM containing dCK- R104M/D133A double mutant gene | This study |
| pENTRHM-dCK-TM | pENTRHM containing dCK-A100V/R104M/D133A triple mutant gene | This study |
| pAdHM-LUC2 | pAd/CMV/V5/DEST vector after recombination with pENTRHM-FLUC | Gil 2011 [16] |
| pAdHM-TK2 | pAd/CMV/V5/DEST vector after recombination with pENTRHM-TK2 | This study |
| pAdHM-TK2-N93D-L109F | pAd/CMV/V5/DEST vector after recombination with pENTRHM- TK2-N93D-L109F | This study |
| pAdJG-dCK-WT | pAd/CMV/V5/DEST vector after recombination with pENTRJG-dCK-WT | This study |
| pAdJG-dCK-DM | pAd/CMV/V5/DEST vector after recombination with pENTRJG-dCK-DM | This study |
| AdHM-dCK-TM | pAd/CMV/V5/DEST vector after recombination with pENTRHM-dCK-TRP | This study |
| Adenovirus | ||
| AdHM.LUC2 (referred to here as Ad.HL.Luc2) | Human adenovirus type 5 (Ad5) from Gateway System™ expressing Luc2 from a CMV promoter. | Gil 2011 [16] |
| Ad.HL.TK2 | Ad5 expressing TK2 cytoplasmic mutant from a CMV promoter. | This study |
| Ad.HL.TK2-DM | Ad5 expressing TK2-N93D/L109F mutant from a CMV promoter. | This study |
| Ad.HL.dCK-WT | Ad5 expressing dCK wild type from a CMV promoter. | This study |
| Ad.HL.dCK-DM | Ad5 expressing dCK-R104/D133A double mutant from a CMV promoter. | This study |
| Ad.HL.dCK-A100VTM | Ad5 expressing dCK- A100V/R104M/D133A triple mutant from a CMV promoter. | This study |
Table 2.
Primers used in this study
| Primer sequence | Description | Source |
|---|---|---|
| Fwd: TCGACCAAGCTTTCGGTACCAGCCCGGGAGAGCTCAAGGATCCAAG Rev: AATTCTTGGATCCTTGAGCTCTCCCGGGCTGGTACCGAAAGCTTGG |
Multiple Cloning site for pENTR4D: SalI - HindIII - KpnI - SmaI - SacI - BamHI - EcoRI. | Gil 2011[16] |
| Fwd: TCGACCAAGCTTGCCACCATGCCCACGCTACTGCGGGT Rev: CTAGATATCTCGAGTTATCAGTTAGCCTCCCCCATCT |
sr39TK from pMSCV-sr39TK-IRES-YFP. HindIII/XhoI directional cloning. | This study |
| Fwd: TCGACCAAGCTTGCCACCATGTCAGTGATCTGTGTCGA Rev: TCTAGATATCTCGAGTTACTATGGGCAATGCTTCCGAT |
TK2 and TK2-N93D/L109F from pMSCV-TK2 and pMSCV-TK2-N93D/L109F respectively. HindIII/XhoI directional cloning. | This study |
| Fwd: AGTCGACCAAGCTTGCCACCATGGCTACTCCTCCTAAACG Rev: TGGGTCTAGATATCTCGAGTTATCAAAGTGTTGACAGAAAT TCC |
dCK-WT, dCK-DM, and dCK-TM from pMSCV-hudCK-triple mutant-IRES-YFP. HindIII/XhoI directional cloning. | This study |
| Fwd: TAAAGCTTAGCGACGATGGCCTCCAAG Rev: TTCTCGAGCTACTGTTCGTTCTTCAGC |
RLuc8.6-535 from pRV2011. HindIII/XhoI directional cloning. | This study |
| Fwd: ACTTAAGCTTGCCACCATGGAAGATGCCAAAAACATT Rev: AACTCGAGAATTATTACACGGCGATCTTGC |
Luc2 from pGL4.13. HindIII/XhoI directional cloning. | This study |
| Fwd: ACTTAAGCTTGCCACCATGGCTTCCAAGGTGTACGAC Rev: AACTCGAGAATTATTACTGCTCGTTCTTCAGCAC |
hRLuc from phRL-TK. HindIII/XhoI directional cloning. | This study |
| Fwd: CAAGGGTTGAGTACTTGTTTAGGGTTA Rev: GGTGGGTAGAGAGAAGAAATATCTGACT Probe: TAMRA-AGGACAATGGCCTTGGCTGGACAA-BHQ2A |
TaqMan qPCR primer against mouse Oct4 promoter region | Gil 2011 [16] |
| Fwd: TTG TGGTTCTTGCAGATATGGC Rev: TCGGAATCCCGGCACC Probe: FAM-CTCACCTGCCGCCTCCGTTTCC-TAMRA |
Taqman qPCR primer against adenovirus type 5 pX gene | Gallaher 2009 [19] |
Adenovirus vector propagation and titration
Adenovirus vectors were propagated on HEK293A cells, using standard procedures [16, 19] and titrated on HeLa cells for to determine Infectious Genomes [16, 19]. Procedures have been described in detail in our comparisons of luciferase reporter genes [16], and are also described in Supplemental Material.
PET Reporter Probe Synthesis
18F-FHBG and 18F-L-FMAU syntheses were performed as previously described [9, 13], at the cyclotron facilities in the UCLA Crump Institute for Molecular Imaging and the UCLA Ahmanson Translational Imaging Division.
In vivo studies
Female hairless SKH1 mice (Charles River, San Diego, CA) were housed in accordance with the UCLA Division of Laboratory Animal Medicine guidelines. Mice between 10 to 16 weeks were used for all experiments. An optimal imaging time of 3–5 days after Ad.HL.Luc2 vector tail vein injection was determined (Supplemental Figure 1).
Titration studies to optimize numbers of viral particles per mouse were performed by injecting 1×1010, 3×1010 or 1×1011 IGU of Ad.HL.HSV1sr39TK. Three days after vector injection mice were injected via tail vein with 18F-FHBG (200 µCi). Three hours after tracer injection, mice were anesthetized (2% isoflurane), and subjected to MicroPET/CT scanning (Inveon, Siemens Medical Solutions, USA Inc.; MicroCAT, Imtek, Inc.) [13].
To compare the efficacy of the PET reporter genes, groups of three animals were each injected via the tail vein with 5×1010 IGU of the various adenovirus vectors. Four days after injection mice that received either no vector or mice that received Ad.HL.Luc2, Ad.HL.dCK-WT, Ad.HL.dCK-R104M/D133A (referred to as Ad.HL.dCK-DM), Ad.HL.dCK-A100V/R104M/D133A (referred to as Ad.HL.dCK-A100VTM), Ad.HL.TK2, or Ad.HL.TK2-N93D/L109F (referred to as Ad.HL.TK2-DM) were imaged with 18F-L-FMAU (200 µCi). Mice receiving either Ad.HL.Luc2, Ad.HL.HSV1sr39TK or no adenovirus vector were imaged with 18F-FHBG. The dCK mutant PRG expressed in Ad.HL.dCK-A100VTM (cloned and codon optimized by us; McCracken et al; Ref 18, in preparation) is distinguished from another dCK triple mutant of the dCK gene, dCK-S74E/R104M/D133A (dCK-S74ETM), described by Likar et al [12].
Mice injected with Ad.HL.Luc2 and imaged with 18F-FHBG were used to determine background values for mice injected with Ad.HL.sr39TK and imaged with 18F-FHBG; additional mice injected with Ad.HL.Luc2 and imaged with 18F-L-FMAU were used to determine background values for mice injected with all other PET reporter gene adenovirus vectors and imaged with 18F-L-FMAU.
To quantify PET data, four identical 2 mm Regions of Interest (ROI) were drawn within the liver for each mouse. The average PET signal in percent injected dose per gram (%ID/g) was determined for each mouse from these ROIs. For each mouse, a corrected %ID/g for the experimental PRG/PRP imaging pair was obtained by subtracting the average %ID/g from mice injected with Ad.HL.Luc2 and subsequently imaged with the appropriate PRP; i.e., 18F-FHBG or 18F-L-FMAU.
After imaging, the mice were euthanized, the livers were removed, and triplicate liver samples were used for adenovirus vector and mouse genomic DNA analysis. DNA was isolated from liver samples using the DNeasy protocol (Qiagen, Thousand Oaks, CA), adenoviral DNA and murine genomes were determined as described previously [16, 20] and as summarized in the Supplemental Material.
RESULTS
Creation and production of Adenovirus PRG vectors
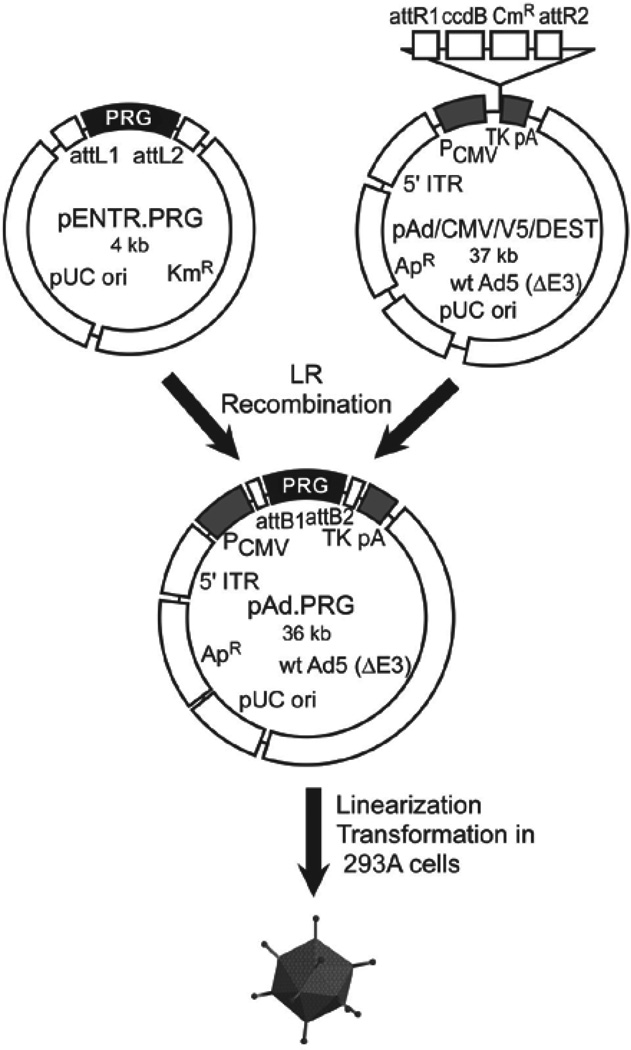
Comparisons between reporter genes are often confounded by differences in vector structure – e.g., different promoters, alternative 5’ and/or 3’ untranslated regions, and/or distinct polyadenylation signals. We mitigated these issues by using a single vector design for all PRGs. Each reporter gene was cloned into the same shuttle or “entry” vector, utilizing the commercially available Gateway System [16] and employing PCR primers with HindIII/XhoI restriction sites. Each of these entry vectors was used to insert the reporter gene, via Gateway cloning, into the viral pAd/CMV/V5-DEST vector (Fig. 1). This highly efficient and simple cloning procedure ensures that each adenovirus PRG vector is identical, with the exception of the reporter gene. Other researchers can easily create identical vectors, with the exception of their new reporter genes, and can evaluate their new reporter genes by direct comparison with our vectors as controls.
Fig. 1. Cloning and production of adenovirus vectors.
Adenovirus vectors containing coding regions for TK2, TK2-DM, dCK-WT, dCK-DM, dCK-A100VTM, HSV1sr39TK (as a reference standard) and Luc2 as a negative control were constructed using Invitrogen’s Gateway® Cloning System. The open reading frame of each reporter was inserted into an “entry” vector to create the pENTR-PRG plasmids, then transferred to the pAd/CMV/V5/DEST vector, using the LR recombination reaction, to create the seven pAd vectors. The adenoviral plasmids were linearized by PacI restriction enzyme digestion, purified, and transformed into HEK293A cells for vector rescue. After 100% cytopathic effect was observed, lysates were serially passaged on increasing numbers of HEK293A cells with each round of infection until sufficient vector was produced, following cesium chloride buoyant density ultracentrifugation and purification, for in vivo studies. Sites labeled attR1/attR2/attL1/attL2 are the initial recombination regions; attB1/attB2 are the recombination regions after the LR recombination reaction. CmR, the chloramphenicol resistance gene; KmR, the kanamycin resistance gene; ApR, the ampicillin resistance gene; ccdB, the coding region for the cytotoxic protein CcdB, used as negative-selection marker in recombined clones; PCMV, the CMV promoter; TKpA, the thymidine kinase polyadenylation signal; 5’ ITR, the viral 5’ inverted terminal repeats; wt Ad5 (DE3), Ad5 sequences that include a 3’ ITR and packaging signal.
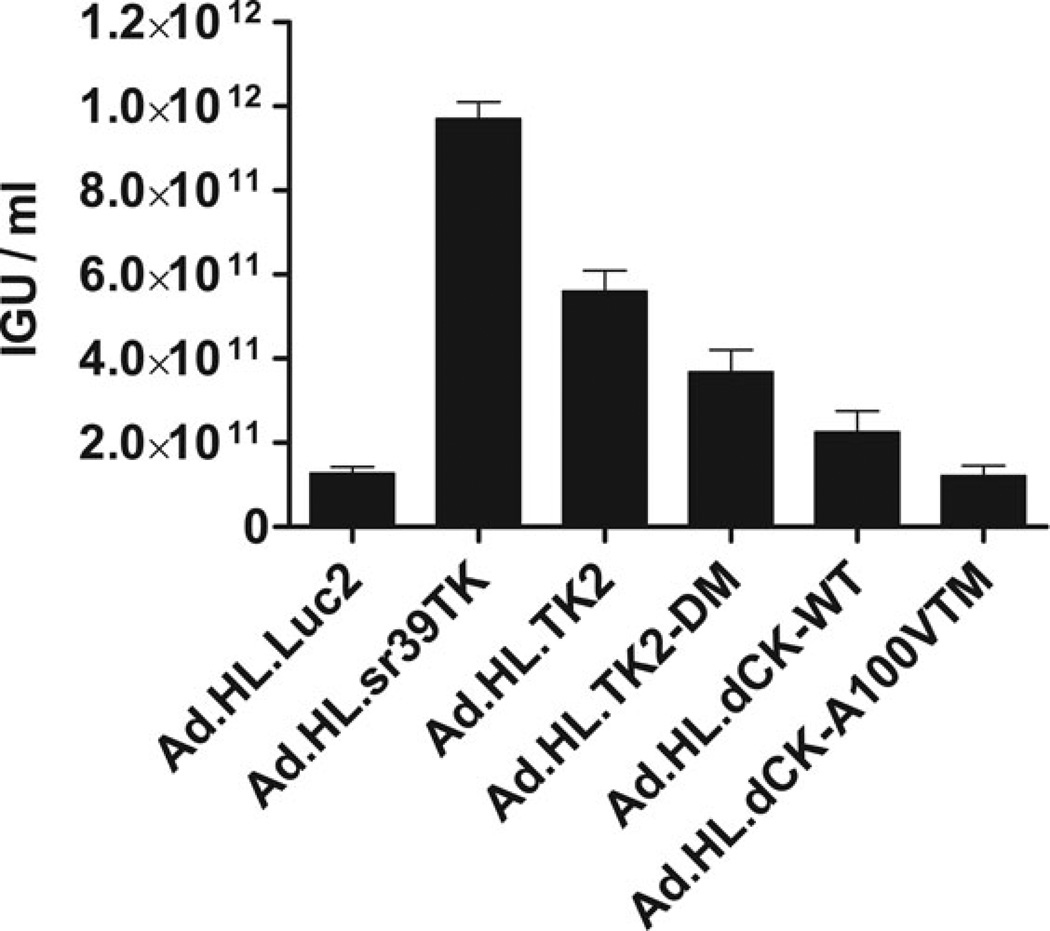
After conversion of the recombinant plasmid into a viral vector, amplification through serial rounds of infection, purification and sequencing, the adenovirus vector stocks were titrated in culture for infectious genome units (IGUs) (Fig. 2), as described in Materials and Methods and in Supplemental Material. Vector concentrations, in IGU/mL +/− S.E.M (Fig. 2), were: Ad.HL.Luc2, 1.27 ± 0.15×1011; Ad.HL.sr39TK; 9.7 ± 0.40×1011; Ad.HL.TK2, 5.61 ± 0.49× 1011; Ad.HL.TK2-DM, 3.69 ± 0.51× 1011; Ad.HL.dCK-WT, 2.25 ± 0.50×1011; and Ad.HL.dCK-A100VTM, 1.21 ± 0.24×1011.
Fig. 2. Viral titers, measured as Infectious Genome Units, for the Adenovirus vectors.
IGU values for the seven adenovirus vector preparations were determined following HeLa cell transduction. Nuclei were harvested three hours after vector addition. Vector genomes were measured by quantitative PCR as described in Materials and Methods. Data are means ± S.E.M. of duplicate qPCR assays for duplicate transductions.
Optimal time and viral titer for imaging hepatic PRG expression after intravenous adenovirus administration
To determine the optimal time between adenoviral reporter gene administration and injection of the reporter probe imaging agent, we performed a time course with the Ad.HL.Luc2/luciferin reporter gene/reporter probe system. Repeated daily luciferin imaging of SKH1 mice that received Ad.HL.Luc2 (5×1010 IGU/mouse) indicated an optimal imaging window 3–5 days after adenovirus vector injection (Supplemental Material, and Supplemental Figure 1).
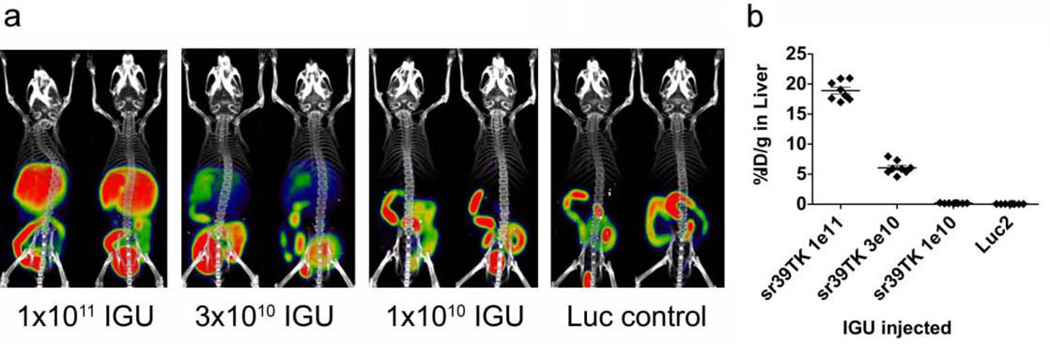
Because PET reporter probe retention is dependent on the dose of adenovirus PRG vector administration [21], we first used the Ad.HL.sr39TK/18F-FHBG PRG/PRP system to identify an Ad.HL.PRG dose for this vector that would be appropriate for comparison with other PRG/PRP combinations. Female SKH1 mice were injected intravenously with 1×1011, 3×1010 or 1×1010 IGU of Ad.HL.sr39TK or an Ad.HL.Luc2 control vector (3×1010 IGU). Three days later, mice were imaged with 18F-FHBG (Fig. 3).
Fig. 3. Titration of Ad.HL.sr39TK for MicroPET/MicroCT quantification.
(Panel a) Two mice per group were injected intravenously with 1011, 3×1010, or 1010 IGU of Ad.HL.sr39TK or with 3×1010 IGU of Ad.HL.Luc2. After three days, mice were injected with 18F-FHBG (200 µCi) and subjected to microPET/microCT imaging. (Panel b) 2 mm ROIs were drawn within the liver of each mouse, and used to determine average %ID/g per liver. Data are means ± S.E.M.
A common vector titer of 5×1010 IGU, in the midrange of the dose-response relationship, was chosen for comparison of the various PRG/PRP systems. In this way both greater and lesser efficacies relative to Ad.HL.sr39TK/18F-FHBG for the alternative human-derived PRG/PRP systems could be evaluated.
Hepatic efficacy of the alternative adenoviral PRG/PRP non-invasive imaging systems
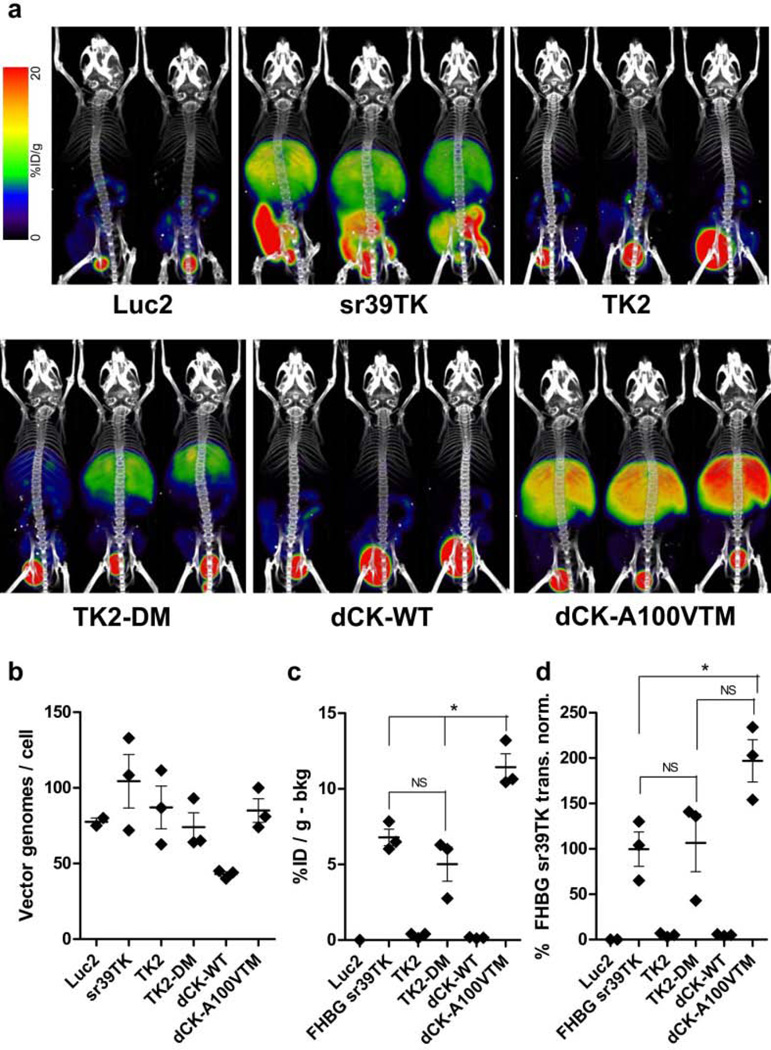
Three mice in each alternative adenoviral PRG vector experimental group and two mice in each Ad.HL.Luc2 control group were injected intravenously with the appropriate adenovirus vector (5×1010 IGU/mouse). Four days after adenovirus injection the mice received either 18F-L-FMAU (200 µCi) or 18F-FHBG (200 µCi), as appropriate. Three hours later the mice were anesthetized and subjected to PET/CT imaging (Fig. 4a). The following day, mice were euthanized and livers were removed. DNA was purified from liver samples, and DNA content was analyzed for viral and mouse genomes, to normalize the imaging data for the number of viral genomes present in the livers. The numbers of vector genomes/liver cell, ± S.E.M., were Ad.HL.Luc2, 78 ± 2.5; Ad.HL.sr39TK, 104 ± 18; Ad.HL.TK2, 87 ± 14; Ad.HL.TK2-DM, 74 ± 9.5; Ad.HL.dCK-sWT, 43 ± 1.5 and Ad.HL.dCK-A100VTM, 85 ± 7.8 (Fig. 4b).
Fig. 4. TK2 and dCK PRG efficacies, with L-FMAU as the PRP.
(Panel a) Three mice for each adenovirus vector were injected with 5×1010 IGU of Ad.HL.TK2, Ad.HL.TK2-DM, Ad.HL.dCK-WT, Ad.HL.dCK-A100VTM, Ad.HL.sr39TK (as reference standard) or Ad.HL.Luc2 (as negative control). Four days later mice were injected with 200 µCi 18F-FHBG (Ad.HL.sr39TK) or 200 µCi 18F-L-FMAU (Ad.HL.TK2, Ad.HL.TK2-DM, Ad.HL.dCK-WT, Ad.HL.dCK-A100VTM, Ad.HL.Luc2) and subjected to microPET/microCT imaging. (Panel b) One day after imaging, mice were sacrificed. Viral and liver genomes were measured in liver extracts, to determine numbers of viral vectors in the livers. Each point is the mean of three liver samples per mouse. (Panel c) Four identical ROIs were drawn within the liver of each mouse and use to determine the %ID/g for liver. Values were then corrected by subtracting ROI values determined for the mice injected with Ad.HL.Luc2 virus and imaged with the appropriate PRP (negative control, bkg). Asterisk indicates statistically significant differences (p>0.05); individual p values follow: FHBG sr39TK vs. TK2-DM, p=0.23. FHBG sr39TK vs. dCK-A100VTM, p=0.01. TK2-DM vs. dCK-A100VTM, p=0.01. (Panel d). The [%IDg – bkg] in liver for each adenovirus vector (from panel c) was normalized for the number of viral genomes per liver (from panel b). To compare the efficacies of the experimental TK2, TK2-DM, dCK-WT and dCK-A100VTM/18F-L-FMAU PRG/PRP non-invasive imaging systems with the efficacy of the HSV1sr39TK/18F-FHBG PRG/PRP system, the values for the experimental systems using 18F-L-FMAU as the PRP are expressed as a percentage of the HSV1sr39TK/18F-FHBG PRG/PRP system. Asterisk indicates statistically significant differences (p>0.05); individual p values follow: FHBG sr39TK vs. TK2-DM, p=0.86. TK2-DM vs. dCK-A100VTM, p=0.08. FHBG sr39TK vs. dCK-A100VTM, p=0.03. Data for panels b, c and d are means ± S.E.M.
PRG-dependent PRP hepatic retention was determined by choosing four identical 2 mm Regions of Interest (ROI) over the liver of each mouse, and determining the percent injected dose/gram (%ID/g). To determine the PRG-dependent PRP retention, the background hepatic signal (determined from mice injected with Ad.HL.Luc2 and the appropriate PRP) was subtracted. %ID/g values, corrected for PRP background, but not corrected for differences in the numbers of PRGs delivered to the livers of each mouse, were Ad.HL.sr39TK, 6.79 ± 0.55; Ad.HL.TK2, 0.31 ± 0.09; Ad.HL.TK2-DM, 5.02 ± 1.13; Ad.HL.dCK-WT, 0.15 ± 0.03; and Ad.HL.dCK-A100VTM, 11.43 ± 0.89 (Fig. 4c, values are means ±S.E.M.s).
To eliminate the variability caused by differences in numbers of PRGs present in livers of each mouse, the [%ID/g-bkg] was normalized to vector transduction. Results for the alternative PRG vectors were then compared to the Ad.HL.sr39TK/18F-FHBG standard. Normalized activities (Fig. 4d) were Ad.HL.sr39TK, 100 ± 19%; Ad.HL.TK2, 5.0 ± 1.2%; Ad.HL.TK2-DM, 107 ± 32%; Ad.HL.dCK-WT, 5.0 ± 0.6%; and Ad.HL.dCK-A100VTM, 197 ± 23%. Normalized PRG/PRP signal showed no significant difference between the commonly used HSV1sr39TK/18F-FHBG PRG/PRP imaging system and the TK2-DM/18F-L-FMAU imaging system by Student’s t-test (p=0.86) In contrast, a significant (p=0.03) difference between the HSV1sr39TK/18F-FHBG and dCK-A100VTM/18F-L-FMAU PRG/PRP combinations was observed.
We also compared the efficacy of the dCK-DM reporter gene described by Likar et al [13] with the dCK-A100VTM reporter gene, which has an additional amino acid substitution that was speculated to improve its utility as a PRG [17], using Ad.HL.dCK-DM and Ad.HL.dCKA100VTM adenovirus vectors. However, in this assay we could not observe a statistically significant difference in the efficacy of dCK-DM and dCK-A100VTM, using 18F-L-FMAU as a common PRP (described in Supplemental Materials and Supplemental Figure 2).
DISCUSSION
As the use of therapeutic gene delivery vectors and targeted cell therapies expands into clinical applications the need to repeatedly, non-invasively and quantitatively monitor the duration of their bioavailability, the specificity of their targeting, and their longevity at target sites become of increasingly greater importance in evaluating their relative therapeutic efficacies. As new reporter gene/reporter probe imaging systems are developed a robust, quantifiable means for their evaluation becomes essential. The procedure we developed eliminates all variables except the reporter gene and the reporter probe, restricting comparisons to a single, similarly vascularized organ and permitting post-imaging normalization for the number of reporter genomes [16].
HSV1-TK and its mutated derivatives have become the de facto “gold standard” PRGs for PRG/PRP analysis in “translational” pre-clinical models and in clinical trials. However, because of the immunogenicity of HSV1-TK and its derivatives, several groups have developed mutated human nucleoside kinases as PRGs. The “ideal” mutated nucleoside kinase PRG would (i) not be immunogenic in patients, (ii) use as its PRP substrate a positron-emitting nucleoside analogue unable to be phosphorylated by the endogenous enzyme and (iii) be unable to phosphorylate the endogenous substrate.
To date, several mutant human nucleoside kinase PRG/PRP non-invasive imaging systems have been described. Two PRG studies have been published utilizing 2’-deoxy-2-18F-5-methyl-1-β-L-arabinofuranosyluracil (18F-FEAU) as the PRP; the first used a truncated human TK2 gene as a PRG [22], while the second used a human dCK double mutant (dCK-R104M/D133A) [12]. More recently, two additional studies have used 18F-FMAU as a PRP; one of these studies used a TK2 double mutant as the PRP [13], the second used a dCK triple mutant (dCK-A100VTM) [18].
To begin the comparison of alternative PRG/PRP imaging combinations, using the rigorous system developed previously [16], we compared the efficacies of the TK2-DM and dCK-A100VTM PRGs, using the common PRP 18F-L-FMAU, to the efficacy of the HSV1sr39TK/18F-FHBG PRG/PRP imaging combination. Our data suggest that, in the adenovirus/hepatic mouse model system we developed, the TK2-DM/18F-L-FMAU PRG/PRP combination is as effective as the HSV1sr39TK/18F-FHBG combination, while the dCK-A100VTM/18F-L-FMAU PRG/PRP combination has a significant advantage over the HSV1sr39TK/18F-FHBG combination (Fig. 4).
As we developed this comparative analytical system for PRG/PRP imaging combinations, it became apparent that there are a number of additional constraints, considerations and pitfalls to take into account in comparing alternative PRG/PRP systems. We compared two reporter genes (TK2-DM and dCK-A100VTM) both to one another, using a common PRP (18F-L-FMAU), and to a distinct PRG/PRP imaging system (HSV1sr39TK/18F-FHBG). While all three PRGs are expressed from the same expression vector, and the transgene PRG copy numbers can be determined after imaging, thus assuring that the imaging results can be normalized for levels of PRG expression, we do not know if the two 18F-PRPs are present in adequate levels in the liver for comparable use as substrates. While the efficacy comparisons of TK2-DM/18F-L-FMAU and dCK-A100VTM/18F-L-FMAU in liver are controlled for essentially all variables, it is quite possible the relative availabilities of the 18F-FHBG and 18F-L-FMAU PRP substrates may be quite different in liver for these alternative PRGs. Thus substrate availability, and not PRG expression, may be limiting – perhaps in a tissue specific fashion – for PRP/PRG reporter systems. In preclinical reporter gene/reporter probe systems this is perhaps most graphically illustrated by the firefly luciferase/luciferin system; no matter what the level of luciferase expression, no image can be detected in brain – luciferin cannot penetrate the blood brain barrier.
A second pitfall for translation from preclinical to clinical applicability for PRG/PRP imaging systems in patients lies in differences between mice (and other species used in preclinical analyses) and humans for PRP biodistribution. 18F-L-FMAU biodistribution in mice, using a xenograft tumor model, showed no significant probe retention in tissues other than tumor, with variable signal in intestine. Indeed, in mice the non-specific probe retention for 18F-L-FMAU was less than that observed for 18F-FHBG [13, 23]. However, in distribution studies in humans, extensive hepatic 18F-L-FMAU retention was observed [13], suggesting that the clinical use of either the TK2-DM/18F-L-FMAU PRG/PRP reporter system or the dCK-A100VTM/18F-L-FMAU PRG/PRP monitoring system in patients will be restricted to extra-hepatic applications.
In developing the dCK-DM PRG, Likar et al [12] used 18F-FEAU as the PRP, and compared the efficacy of the dCK-DM/18F-FEAU imaging combination to the HSV1R176Qsr39TK/18F-FHBG combination. Their study used transduced xenograft tumors for comparison. The difficulties in trying to compare and evaluate alternative PRG/PRP reporter systems from various laboratories are illustrated by trying to evaluate the prior studies by Likar et al [12] of their dCK-based PRG with those of the TK2 based PRG by Campbell et al [13]; these reports used different vectors, different transduced tumor cells, different xenograft conditions likely to lead to distinct target sizes and variable vascularization, and distinct PRPs. Using a rigorous protocol that equalizes many of these conditions in preclinical studies will help to make the pursuit of clinically useful systems more effective. For example, by comparing alternative substrates (e.g., 18F-FEAU and 18F-L-FMAU) in this adenovirus based model, and evaluating the biodistribution of the two PRPs, a definitive comparison of the two presumably non-immunogenic PRGs, TK2-DM and dCK-A100VTM and the two alternative PRPs (18F-FEAU and 18F-L-FMAU) could be performed.
Differences in biodistribution for potential PRPs between mouse and human – or between human and any other species – presents an often unanticipated, but significant barrier in PRP/PRG imaging system development. 18F-L-FMAU is a case in point; its pristine lack of retention in mice is, unfortunately, not reflected in human studies [13]. As a result we are adopting a new “reverse” approach to the development of PRG/PRP imaging systems. Alternative positron-emitting nucleoside analogues are synthesized, and biodistribution studies are carried out in volunteers, to identify potential PRPs with optimal biodistribution characteristics [24]. After identification of potential PRPs with appropriate biodistribution properties, modifications of nucleoside kinases are made and in vitro analyses of their ability to phosphorylate both the modified and the endogenous substrates are evaluated, to identify mutant PRGs that can phosphorylate the potential PRP and have reduced kinase activity on the endogenous substrate.
Reducing the ability of the mutated nucleotide kinase PRG to utilize the endogenous substrate may appear to be simply a bonus, because it will effectively increase the specific activity in vivo of the PRG for the PRP; the “cold” endogenously produced compound will no longer compete with the positron-emitting PRP for the mutated PRG nucleotide kinase. However, there is another important potential value in reducing/eliminating PRG kinase activity for the endogenous substrate. While the PRPs are present in tracer, non-physiological amounts and will have no biological effect, high PRG levels – if active on an endogenous substrate – might drastically modify nucleoside/nucleotide pools in target cells and, as a result, have profound biological consequences. In principle the “ideal” PRG kinase would be orthogonal to the native enzyme; able to phosphorylate the positron-emitting PRP and unable to phosphorylate the endogenous substrate (or any other cellular component).
It is clear that reporter probe biodistribution, duration of availability, and clearance from tissues will present problems that must be overcome in optimizing non-invasive reporter gene/reporter probe combinations. Similarly, optimizing reporter gene delivery, bioavailability, and expression are among the many factors that must also be considered in developing optimal clinical reporter gene/reporter probe combinations for patient applications, whether the analysis be PET, SPECT, MRI, ultrasound, etc. We anticipate that using rigorously controlled experimental methodologies, like the one used here, will be required to bring the best methods for reporter gene/reporter probe imaging to the clinic as rapidly and cost-effectively as possible.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Arthur Catapang for technical help, David Stout, Liu Wei, and Waldemar Ladno for advice and assistance with imaging experiments, and members of the Herschman, Radu and Witte laboratories for helpful discussions. This study was funded by the National Cancer Institute In Vivo Cellular and Molecular Imaging Center (ICMIC) award P50 CA086306 (HRH), National Cancer Institute grant 5U54 CA119347 (CGR) and National Institute of Biomedical Imaging and Bioengineering grant 1R01CA16077001 (CGR). JG was supported by a Scholars in Oncologic Medical Imaging (SOMI) fellowship from the National Cancer Institute (Award R25T CA098010)
Footnotes
Conflict of interest
The authors declare they have no conflict of interest.
REFERENCES
- 1.Bhaumik S, Gambhir SS. Optical imaging of Renilla luciferase, synthetic Renilla luciferase, and firefly luciferase reporter gene expression in living mice. Journal of Biomedical Optics. 2004;9:578–586. doi: 10.1117/1.1647546. [DOI] [PubMed] [Google Scholar]
- 2.Gheysens O, Gambhir SS. Studying molecular and cellular processes in the intact organism. Progress in Drug Research. 2005;62:117–150. doi: 10.1007/3-7643-7426-8_4. [DOI] [PubMed] [Google Scholar]
- 3.Lewis Js, Achilefu S, Garbow JR, Laforest R, Welch MJ. Small animal imaging. Current technology and perspectives for oncological imaging. European Journal of Cancer. 2002;38:2173–2188. doi: 10.1016/s0959-8049(02)00394-5. [DOI] [PubMed] [Google Scholar]
- 4.Jenkins DE, Oei Y, Hornig YS, Yu S-F, Dusich J, Purchio T, Contag PR. Bioluminescent imaging (BLI) to improve and refine traditional murine models of tumor growth and metastasis. Clinical & Experimental Metastasis. 2003;20:733–744. doi: 10.1023/b:clin.0000006815.49932.98. [DOI] [PubMed] [Google Scholar]
- 5.Iyer M, Sato M, Johnson M, Gambhir SS, Wu L. Applications of molecular imaging in cancer gene therapy. Current Gene Therapy. 2005;5:607–618. doi: 10.2174/156652305774964695. [DOI] [PubMed] [Google Scholar]
- 6.Pysz MA, Gambhir SS, Willmann JK. Molecular imaging: Current status and emerging strategies. Clinical Radiology. 2010;65:500–516. doi: 10.1016/j.crad.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger C, Flowers ME, Warren EH, Riddell SR. Analysis of transgene-specific immune responses that limit the in vivo persistence of adoptively transferred HSV-TK-modified donor T cells after allogeneic hematopoietic cell transplantation. Blood. 2006;107:2294–2302. doi: 10.1182/blood-2005-08-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gambhir SS, Bauer E, Black ME, Liang Q, Kokoris MS, Barrio JR, yer M, Namavari M, Phelps ME, Herschman HR. A mutant herpes simplex virus type 1 thymidine kinase reporter gene shows improved sensitivity for imaging reporter gene expression with positron emission tomography. Procedings of the National Academy of Sciences. 2000;97:2785–2790. doi: 10.1073/pnas.97.6.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yaghoubi S, Barrio JR, Dahlbom M, Iyer M, Namavari M, Satyamurthy N, Goldman R, Herschman HR, Phelps ME, Gambhir SS. Human pharmacokinetic and dosimetry studies of [(18)F]FHBG: a reporter probe for imaging herpes simplex virus type-1 thymidine kinase reporter gene expression. Journal of Nuclear Medicine. 2001;42:1225–1234. [PubMed] [Google Scholar]
- 10.Yaghoubi SS, Jensen MC, Satymurthy N, Budhiraja S, Paik D, Czernin J, Gambhir SS. Noninvasive detection of therapeutic cytolytic T cells with 18F-FHBG PET in a patient with glioma. Nature Clinical Practice Oncology. 2009;6:53–58. doi: 10.1038/ncponc1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Traversari C, Marktel s, Magnani Z, Mangia P, Russo V, Ciceri F, Bonini C, Borgdignon C. The potential immunogenicity of the TK suicide gene does not prevent full clinical benefit associated with the use of TK-transduced donor lymphocytes in HSCT for hematologic malignancies. Blood. 2007;109:4708–4715. doi: 10.1182/blood-2006-04-015230. [DOI] [PubMed] [Google Scholar]
- 12.Likar Y, Zurita J, Dobrenkov K, Shenker L, Cai S, neschadim A, Medin JA, Sadelain M, Hricak H, Ponomarev V. A new pyrimidine-specific reporter gene: a mutated human deoxycytidine kinase suitable for PET during treatment with acycloguanosine-based cytotoxic drugs. Journal of Nuclear Medicine. 2010;51:1395–1403. doi: 10.2967/jnumed.109.074344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campbell DO, Yaghoubi SS, Su Y, Lee JT, Auerbach MS, Herschman H, Satyamurthy N, Czernin J, Lavi A, Radu CG. Structure-guided engineering of human thymidine kinase 2 as a positron emission tomography reporter gene for enhanced phosphorylation of non-natural thymidine analog reporter probe. Journal of Biological Chemistry. 2012;2887:446–454. doi: 10.1074/jbc.M111.314666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hazra S, Sabini E, Ort S, Konrad M, Lavie A. Extending thymidine kinase activity to the catalytic repertoire of human deoxycytidine kinase. Biochemistry. 2009;48:1256–1263. doi: 10.1021/bi802062w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iyidogan P, Lutz S. Systematic exploration of active site mutations on human deoxycytidine kinase substrate specificity. Biochemistry. 2008;16:4711–4720. doi: 10.1021/bi800157e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gil JS, Machado HB, Herschman HR. A Method to Rapidly and Accurately Compare the Relative Efficacies of Non-invasive Imaging Reporter Genes in a Mouse Model and its Application to Luciferase Reporters. Molecular Imaging and BIology. 2011 doi: 10.1007/s11307-011-0515-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sabini E, Ort S, Monnerjahn C, Konrad M, Lavie A. Structure of human dCK suggests strategies to improve anticancer and antiviral therapy. 2003;10:513–519. doi: 10.1038/nsb942. 2003. [DOI] [PubMed] [Google Scholar]
- 18.McCracken MN, Gschweng E, Nair-Gill E, McLaughlin J, Cooper A, Riedinger M, Chen D, Nosala C, Kohn D, Witte ON. doi: 10.1073/pnas.1221840110. (in preparation). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallaher SD, Gil Jose S, Dorigo, Oliver, Berk Arnold J. Robust In Vivo Transduction of a Genetically Stable EBV Episome to Hepatocytes in Mouse by a Hybrid Viral Vector. Journal of Virology. 2009 doi: 10.1128/JVI.01721-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gil JS, Gallaher SD, Berk AJ. Delivery of an EBV episome by a self-circularizing helper-dependent adenovirus: Long-term transgene expression in immunocompetent mice. Gene Therapy. 2010;17:1288–1293. doi: 10.1038/gt.2010.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gambhir SS, Barrio JR, Phelps ME, YIyer M, Namavari M, Satyamurthy N, W L, Green LA, Bauer E, MacLaren DC, Nguyen K, Berk AJ, Cherry SR, Herschman HR. Imaging adenoviral-directed reporter gene expression in living animals with positron emission tomography. Procedings of the National Academy of Sciences. 1999;96:2333–2338. doi: 10.1073/pnas.96.5.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ponomarev V, Doubrovin M, Shavrin A, serganova I, Beresten T, Ageyeva L, Cai C, Balatoni J, Alauddin M, Gelovani J. A human-derived reporter gene for noninvasive imaging in humans: Mitochondrial thymidine kinase type 2. Journal of Nuclear Medicine. 2007;48:819–826. doi: 10.2967/jnumed.106.036962. [DOI] [PubMed] [Google Scholar]
- 23.Alauddin MM, Shahinian A, Gordon EM, Conti PS. Direct comparison of radiolabeled probes FMAU, FHBG, and FHPG as PET imaging agents for HSV1-tk expression in a human breast cancer model. Molecular Imaging. 2004;3:76–84. doi: 10.1162/15353500200403160. [DOI] [PubMed] [Google Scholar]
- 24.Schwarzenberg J, Radu CG, Benz M, Fueger B, Tran AQ, Phelps ME, Witte ON, Satyamurthy N, Czernin J, Schiepers C. Human biodistribution and radiation dosimetry of novel PET probes targeting the deoxyribonucleoside salvage pathway. European Journal of Nuclear Medicine and Molecular Imaging. 2011;38:711–721. doi: 10.1007/s00259-010-1666-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.