Abstract
Systems using immobilized enzymes are attractive for a wide range of industrial and medical applications because they allow for the fabrication of stable, reusable substrates with highly specific functionality. The performance of these systems is greatly dependent upon the orientation and conformation of the adsorbed enzymes. To investigate these relationships, we have developed and applied methods to quantitatively assess the secondary structure of adsorbed enzyme layers on planar surfaces using circular dichroism (CD) spectroscopy and evaluate their bioactivity using colorimetric assays. These combined measurements provide molecular-level insights regarding whether observed changes in adsorbed enzyme bioactivity are due to the adsorbed orientation of an enzyme or adsorption-induced changes in its conformation. Using this approach, we investigated the adsorption behavior of lysozyme (HEWL), xylanase (XYL), and glucose oxidase (GOx) on OH-, CH3-, NH2-, and COOH-terminated alkanethiol self-assembled monolayer (SAM) surfaces. The bioactivities of the small enzymes, HEWL and XYL, had pronounced variations between the different SAM surfaces despite their structural stability, highlighting the role of adsorbed orientation on bioactivity. In contrast, GOx, which is a much larger enzyme, exhibited wide variations in both its structure and bioactivity after adsorption, with adsorption-induced conformational changes actually enhancing its bioactivity. These results provide new insights into protein-surface interactions at the molecular level and demonstrate that adsorption can either promote or inhibit bioactivity depending on how the surface chemistry influences the orientation and conformational state of the enzyme on the surface.
Introduction
The adsorption of bioactive proteins (e.g., enzymes) to surfaces is critically important in many technological and biomedical processes. Systems using surface-immobilized biomolecules to present molecularly recognized surfaces that elicit highly specific, predictable responses have been used for many applications, such as biomimetic materials, bioelectronics, biosensors, immunosensors, genetics, drug development, and cell screening.1–9 These systems are desirable because of the ease in constructing miniaturized arrays and the potential for running numerous analyses in parallel.2, 10 A major concern with the implementation of these systems is the reproducibility of their fabrication. For this reason, alkanethiol self-assembled monolayers (SAMs) have been researched extensively for these applications because they form well-ordered, dense, thin films of a precise thickness that are stable even after the immobilization of biomolecules.2, 11
The structural and functional stability of the biomolecules that are immobilized on the surface, as well as any biomolecules that subsequently bind, is vital to the performance of these systems. The development of techniques that are effective in measuring the molecular structure of adsorbed biomolecules and assessing their bioactivity would aid in the development of highly efficient biomimetic surfaces. Circular dichroism (CD) has been shown to be effective in quantitatively measuring the secondary structure of protein layers adsorbed onto bare quartz slides, gold-coated quartz slides, and polymer coated quartz slides.12–15 While these studies provide very useful information regarding the effect of adsorption on the structure of proteins, they provide little insight regarding how these structural changes translate to changes in the bioactive state of the enzymes.
The objective of our research study was therefore to develop a technique that can be used to not only determine how surface chemistry influences the structure of adsorbed proteins, but how these structural changes translate into changes in the bioactive state of the enzymes on the surface. For these studies, we have selected a series of enzymes with well documented molecular structures and bioactive sites as our model bioactive proteins and alkanethiol self-assembled monolayer (SAM) surfaces on gold-coated quartz slides as our functionalized surfaces. By using thin gold coatings (i.e., 100 Å) for the preparation of these surfaces, these substrates provide sufficient optical transparency in the ultraviolet and visible light wavelength ranges to enable both CD and colorimetric bioactivity assays to be performed on the same enzyme-coated surfaces. This combination of methods enables assessment to be made at the molecular level regarding how the adsorbed orientation and adsorption-induced changes in conformation of the enzyme influence its bioactivity, thus providing new insights into how surface chemistry can be used to control the bioactive state of an adsorbed enzyme layer.
Experimental Methods
Alkanethiol SAMs
Bare quartz slides purchased from Chemglass (Vineland, NJ) were coated with a 30 Å chromium adhesion layer followed by a 100 Å gold layer using a thermal vapor deposition (TVD) evaporator (Model E 12 E, Edwards High Vacuum Ltd.). Prior to TVD, the slides were thoroughly cleaned by incubating them at 50°C for 30 minutes in each of the following solutions, in order, with this cycle being then repeated a second time: “piranha” wash (7:3 H3SO4/H2O2), a basic solution (1:1:5 NH4OH/H2O2/H2O), and an acidic solution (1:1:5 HCl/H2O2/H2O). After completion of the cleaning process, the slides were dried with flowing nitrogen gas before being placed in the evaporator for deposition. The gold-coated slides were then sonicated at room temperature for 1 min in a sulfuric acid solution (8:2 H3SO4/H2O2) and rinsed with nanopure water. The cleaned/coated slides were rinsed with 100% ethanol and placed into the appropriate 1.0 mM alkanethiol solution (purchased from Aldrich, Asemblon, and Prochimia) in ethanol for a minimum of 16 hours. All of the alkanethiols (HS-(CH2)n-R) used in these experiments have an 11-carbon alkyl chain and one of the following terminal groups: R= CH3, OH, NH2, or COOH.
After surface modification, the slides were sonicated for 15 seconds in ethanol, followed by an ethanol rinse and then stored in ethanol. Prior to surface characterization and protein adsorption the SAMs substrates were sonicated in a 10 mM phosphate buffer containing 0.005% (v/v) Triton® X-100 for 5 minutes then thoroughly rinsed with pure buffer. The buffer was adjusted to a pH of 7.4 by mixing the appropriate amounts of 10 mM K2HPO4 and KH2PO4 solutions. COOH-, OH-, and NH2-SAMs were then sonicated in ethanol followed by acetone for 5 minutes each and thoroughly rinsed with pure buffer, whereas the nonpolar CH3-SAMs were sonicated for 5 minutes each in ethanol, hexane, and ethanol again to remove all traces of the detergent (i.e., Triton® X-100), and then rinsed thoroughly with buffer.
Ellipsometry
Ellipsometry measurements were taken on a Sopra GES 5 variable-angle spectroscopic ellipsometer prior to surface modification and after surface treatment at six different spots to establish the bare-substrate optical constants and to calculate the thickness of the monolayer on each surface, respectively. The spectra were collected at an incidence of 70° in the wavelength range of 250–800 nm at 10 nm intervals. The layer thickness was calculated using the regression method in Sopra’s Winelli (ver. 4.07) software.
Contact Angle Goniometry
The surface energy of the gold and the SAM surfaces were characterized by contact angle goniometry using a CAM 200 optical contact-angle goniometer from KSV Instruments, Ltd. The advancing contact angles, using nanopure water (pH 7), from six separate drops were measured on each of the sensor chips before and after surface modification.
X-Ray Photoelectron Spectroscopy
The alkanethiol SAMs were characterized by X-ray photoelectron spectroscopy (XPS) to obtain the chemical composition of the films to ensure purity. After surface treatment and SAM formation, the slides were dried and packaged in a nitrogen environment and sent to NECSAC/BIO at the University of Washington for all XPS analysis. Samples were analyzed using a Surface Science Instrument (SSI) X-Probe spectrometer (Mountain View, CA) or a Kratos-Axis Ultra DLD spectrometer, equipped with a monochromatic Al Kα source (KE = 1486.6 eV), a hemispherical analyzer and a multichannel detector. Spectra were collected at a photoelectron takeoff angle of 55° and at 80 eV for survey spectra and 20 eV for high resolution C1s and S2p spectra. Elemental compositions were determined from the peak areas in the spectra, using the SSI data analysis software or Kratos Vision 2 software program.
Model Enzymes
The following enzymes, as well as their buffers and substrates, were all purchased from Sigma unless stated otherwise: lysozyme (HEWL) derived from hen egg whites (14.4 kDa, pI = 11.0, PDB# 1GXV), endo-1,4-β-xylanase (XYL) derived from thermomyces lanuginosus (21 kDa, pI = 4.1, PDB# 1YNA), and glucose oxidase (GOx) derived from aspergillus niger (160 kDa, pI = 4.2, PDB# 1CF3). Stock solutions of each enzyme (5.0 mg/mL) were prepared in a 10 mM potassium phosphate buffer.
Circular Dichroism
CD spectra were collected using a Jasco J-810 spectropolarimeter over the wavelength range of 190–300 nm. All CD spectra were obtained using a water-cooled sample holder (Jasco) attached to a circulating water bath operating at 15°C to reduce buffer evaporation. The structural contents of the enzymes in a 10 mM phosphate buffer (pH ~ 7.4) solution were determined using 1.0 mg/mL enzyme solutions in a demountable quartz cuvette with a 0.10 mm path length (Starna). The demountable cuvettes have two quartz slides, one of which has a 0.10 mm deep groove ground into it that serves as a buffer well.
The structure of each of the enzymes was first determined in solution by filling the well with the 1.0 mg/mL enzyme solutions and scanning over the 190–300 nm range to generate the CD spectrum. Previous studies conducted by our group have shown that the unavoidable presence of an adsorbed layer of protein on the surfaces of the quartz cuvette has negligible influence on the measured solution structure of the protein.16
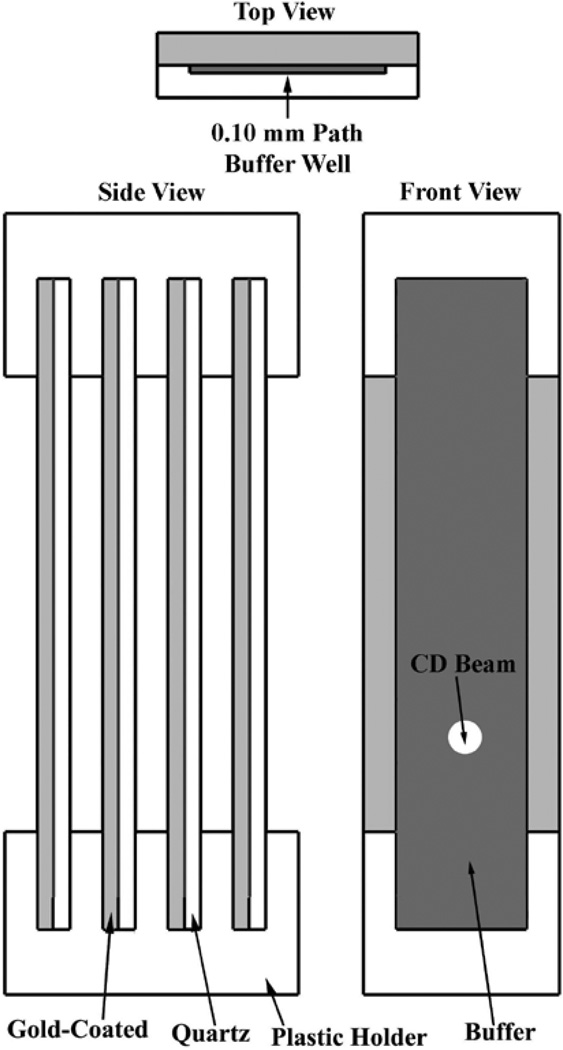
For the adsorption experiments, a SAM-coated quartz slide replaced the flat quartz window of the demountable cuvette. A custom cuvette holder (Figure 1) capable of supporting four individual slides was designed for these studies to increase the signal-to-noise ratio for a more accurate measurement of the structure of the adsorbed protein layers. Background spectra were first collected of four sets of four SAM substrates immersed in the buffer solution for each SAM composition prior to enzyme adsorption. Afterwards, the slides were immersed in a Petri dish filled with pure buffer and the appropriate amount of the enzyme stock solution was then added to yield an enzyme concentration of 1.0 mg/mL. The slides were incubated for 24 hours to allow all the surfaces to fully saturate. The solutions were infinitely diluted with buffer to remove reversibly bound protein and the slides were only then removed from the remaining pure buffer solution. This process enables the enzymes to adsorb to the SAM surfaces from solution without passing the surfaces through the air/water interface, which can be expected to contain a relatively thick layer of denatured protein. The slides were transferred to a clean dish filled with buffer, then mounted with the grooved slide, immersed in pure buffer solution, and placed in the cuvette holder.
Figure 1.
Customized CD cuvette holder designed to hold four 0.10 mm demountable window quartz cuvettes for the analysis of surface-adsorbed protein layers. Top view of demountable cuvette (top), side view of cuvettes in holder (bottom left), front view of cuvettes in holder (bottom right).
The background spectra were subtracted from the spectra of samples containing protein and the resulting spectra were converted to molar ellipticity ([Θ]) according to the following equations:17
| (1) |
| (2) |
where [Θ] is the molar ellipticity (deg∙cm2/dmol), Θ is the raw ellipticity obtained from the instrument (mdeg), M0 is the mean residue molecular weight (118 g/mol), Csoln is the enzyme solution concentration (g/mL), Cads is the enzyme surface concentration (g/cm2), and L is the optical path length (cm).
The enzyme concentrations in solution (Csoln) and on the SAM surfaces (Cads) were determined using the peptide absorbance peak at 195 nm (A195). For these determinations, a calibration curve was first constructed for each enzyme by plotting A195 vs. its concentration multiplied by the path length (Csoln × L) using serial dilutions of stock solutions of each enzyme, with the concentration of the stock solution being verified by a bicinchoninic acid (BCA) assay (Pierce). Based on the Beer’s Law relationship, which is expressed as:18, 19
| (3) |
the slope of each calibration curve, which is referred to as the extinction coefficient (εsoln; with units of mL/(g∙cm)·or cm2/g), was then used to calculate the solution concentration (Csoln, in g/mL) and the surface concentration (Cads, in g/cm2) by:
| (4) |
It should be noted that in the Beer’s Law relationship for the surface adsorbed proteins, the Csoln × L term in Eqn. (3) is replaced by the surface concentration of protein (Cads). Preliminary studies were conducted using ellipsometry to confirm the validity of using CD to measure Cads via Eqn. (4).20 By plugging the relationships expressed in Eqn. (4) into their respective relationships shown in Eqns. (1) and (2), the spectra were converted to units of molar ellipticity, and were then deconvoluted using CDPro software to obtain a quantitative assessment of the secondary structural content (i.e., percent α-helix and β-sheet) of the proteins in solution and on each of the functionalized SAM-surfaces.21, 22 With respect to helicity, the algorithm used for deconvolution reports the total helical content (i.e., α-helix and 310-helix combined together to give total helicity).
Bioactivity Assays
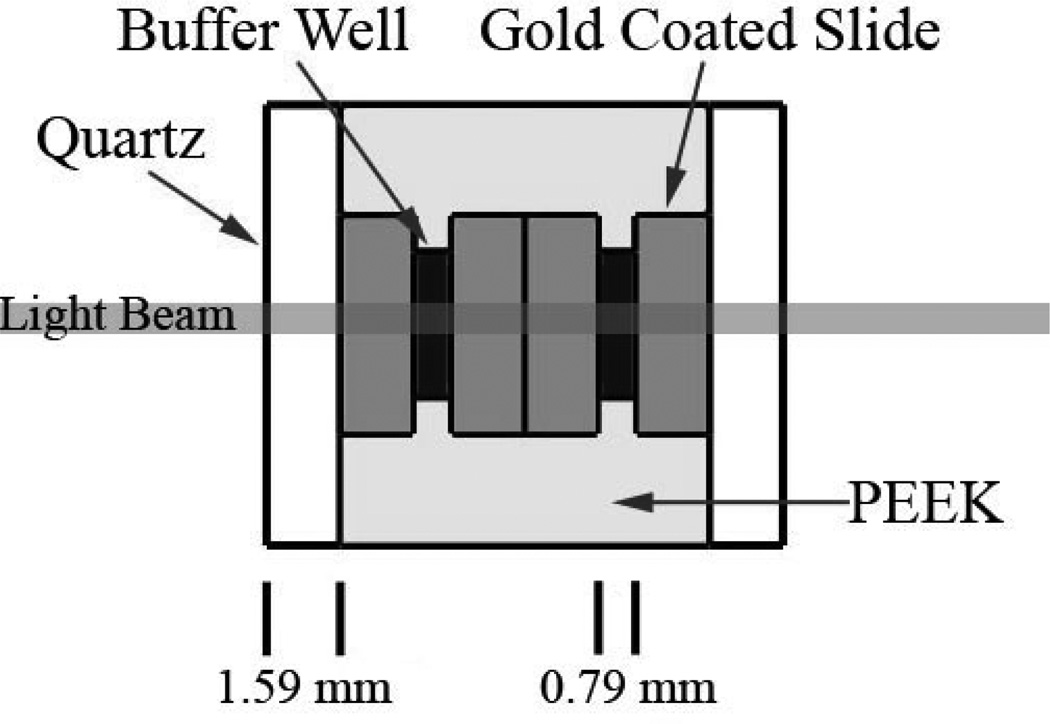
A cuvette was custom designed for the spectropolarimeter to hold 4 gold coated slides for the purpose of performing the bioactivity assays for the adsorbed proteins (Figure 2). Quartz windows were secured to a polyetheretherketone (PEEK) holder, allowing for the transmission of light in the visible wavelength range for colorimetric activity assays as well as the far UV range for the quantification of the amount of protein adsorbed on the surface (Cads). This cuvette was used due to the high surface to volume ratio, minimal amount of substrate needed to fill cuvette (400 μL), and the ability to easily pipette substrate solution in and out of the cuvette. Although CD measurements for structural determination could also be taken using this cuvette,20 the setup mentioned in the previous section (Figure 1) was used for CD because its shorter path length through the buffer provides a higher signal-to-noise ratio at a given scanning speed, which was found to be beneficial for the determination of the adsorbed structure for small proteins such as lysozyme.
Figure 2.
Top view of custom cuvette designed for the Jasco spectropolarimeter for operation in the wavelength range of 190 to 590 nm for protein quantification and spectrophotometric bioactivity assays.
In preparation for the bioactivity studies, the proteins were adsorbed onto the slides and Cads was quantified according the methods described in the previous section. The value for Cads was compared to the value obtained during CD analysis to ensure that the adsorbed layers were stable. This was necessary to make certain that the CD spectra and bioactivities of the adsorbed enzymes layers were not influenced by soluble protein in the buffer solution that may have desorbed from the surface after the slides were mounted in the holder. After measuring Cads, the buffer was replaced with 400 μL of the appropriate enzyme substrate solution to assay the bioactivity of the adsorbed protein layer. The rates of change in the absorbencies of the substrates were determined and specific activities (activity per mg of enzyme) were calculated accordingly. Control studies were also conducted to measure the specific activities of the enzymes in solution (1.0–100 μg/mL) for comparison with the adsorbed enzyme layers. The specific activity of each adsorbed enzyme layer was normalized by its specific activity in solution and multiplied by 100% to determine the relative activities of the surface adsorbed enzymes.
The following dyed peptidoglycan suspensions were used as substrates for HEWL and XYL, respectively: 3 mg/L peptidoglycan from staphylococcus aureus dyed with Remazol Brilliant Blue R (RBB-R) read at 595 nm23 and 0.1 mg/mL 4-O-methyl-D-glucuron-D-xylan dyed with RBB-R read at 590 nm.24 Dyed, low molecule weight fragments are released from each of the peptidoglycan substrates as they are enzymatically digested. After 2 minutes of incubation, 200 μL of the substrate was pipetted out of the cuvette and added to 600 μL of ethanol to terminate the reaction and precipitate the undigested, high molecule weight fragments. The mixtures were then vortexed and left to equilibrate for 10 minutes, after which they were centrifuged at 1,500 g for 10 minutes and the supernatant was then collected and the absorbance read at the appropriate wavelength. The bioactivity of GOx was assessed using a mixture of the following solutions: 2.4 mL of 0.66 mg/mL o-dianisidine, 0.5 mL of 10% (w/v) β-D-glucose, and 0.1 mL of 5 mg/mL horseradish peroxidase (HRP) read at 540 nm.25 To avoid potential complications from competitive adsorption between GOx and HRP,26 the slides were incubated in the o-dianisidine/β-D-glucose mixture for 2 minutes (i.e., without the addition of HRP), 290 μL of the solution was pipetted into a 48 well plate, and then 10 μL of the HRP solution was added, following which the absorbance was read.
Statistical Analysis
The mean and 95% confidence interval (CI) were calculated for all of the sets of the experimental data that were collected. Statistical differences were determined using a Student’s unpaired t-test with values of p ≤ 0.05 considered to be statistically significant.
Results
Surface Characterization
The alkanethiol SAMs formed on gold-coated slides were characterized using contact angle goniometry and ellipsometry to obtain the advancing contact angle and thickness for the various functionalized surfaces, respectively (Table 1). The contact angle values are in close agreement with other published values.27, 28 The ellipsometry measurements showed that the film thicknesses correspond with a single alkanethiol monolayer on each surface. The XPS results (Table 1) also verified that we were forming single alkanethiol monolayers with negligible amounts of oxidized sulfur or contaminates. The presence of oxygen on the NH2-SAM and an excess of oxygen on the OH-SAM are believed to be due to tightly bound water on the surface.29
Table 1.
Atomic composition (%) measured by XPS of all elements, with the exception of Au, present on the alkanethiol SAMs (N=3, mean ± 95% CI). Advancing contact angle (10 mM phosphate buffer solution at pH 7.4) and thickness of SAM surfaces (N=6, mean ± 95% CI).
| Alkanethiol SAM | C1s | S2p | N1s | O1s | Contact Angle (°) |
Thickness (Å) |
|---|---|---|---|---|---|---|
| HS-(CH2)11-NH2 | 85.3 ± 1.4 | 3.2 ± 0.4 | 6.3 ± 0.5 | 5.2 ± 0.5 | 47.6 ± 1.8 | 14.7 ± 2.5 |
| HS-(CH2)11-COOH | 83.8 ± 3.2 | 2.8 ± 0.2 | -- | 13.4 ± 0.6 | 17.9 ± 1.3 | 15.8 ± 1.9 |
| HS-(CH2)11-CH3 | 95.9 ± 1.0 | 4.1 ± 0.3 | -- | -- | 100.9 ± 1.9 | 11.5 ± 2.2 |
| HS-(CH2)11-OH | 84.6 ± 1.2 | 4.2 ± 0.8 | -- | 11.2 ± 0.3 | 17.6 ± 1.9 | 12.1 ± 1.4 |
In a previous study, we measured the surface pKd of the COOH- and NH2-SAMs using SPR spectroscopy.30 We determined that the pKd, based on the pH of the bulk solution, was 7.4 ± 0.2 and 6.5 ± 0.3 (N=4, mean ± 95% CI) for the COOH- and NH2-SAMs respectively. According to those results, when the bulk pH is 7.4 approximately 50% of the COOH groups are deprotonated and negatively charged and 11% of the NH2 groups are protonated and positively charged.
Solution Structures
HEWL is a small, globular enzyme (14.4 kDa)34 that has a net positive charge at the physiological pH of 7.4 (pI=11.0).35 Refaee et al. reported that the solution structure of HEWL consists of five helices that account for 38% of the protein and one β-sheet in which 10% of the residues reside (Figure 3).36 Our CD measurements determined the solution structure of HEWL to be 38 ± 1% helix and 12 ± 1% β-sheet, which is in very good agreement with the NMR solution structure in the PDB file.
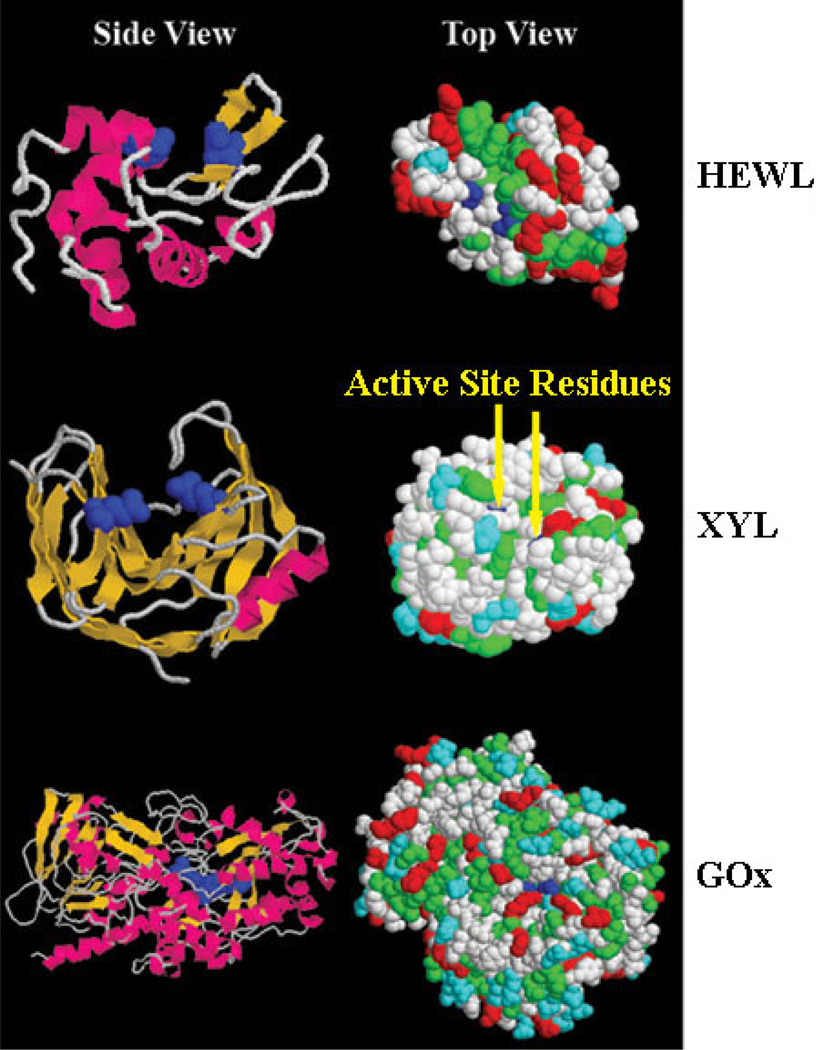
Figure 3.
Left Side: Protein data bank (PDB) images of the structures of the proteins with their active sites represented in dark blue, helices in pink, β-sheets in yellow, random segments in white. Right Side: PDB images of the proteins with their active sites represented in dark blue, nonpolar residues in green, polar residues in white, positively charged residues in red, negatively charged residues in light blue. The PDB numbers and the active site amino acid residues are as follows: HEWL (PDB #1GXV) Glu35 and Asp52,31 XYL (PDB #1YNA) Glu86 and Glu178,32 and GOx (PDB #1CF3) Glu412, His516, and His559.33
XYL from thermomyces lanuginosus is a compact (21 kDa), acidic (pI=4.1) strain that is one of the most thermally stable because it has nearly twice as many salt bridges as other strains and a disulfide bond that secures its α-helix to the β-sheet.32 The crystal structure of this strain of XYL consist of two large, twisted β-sheets that account for 63% of the protein and 5% of the residues reside in its one α-helix (Figure 3).32 The percentages of β-sheet and α-helix for XYL in solution were measured by CD to be 39 ± 2% and 7.0 ± 2%, respectively, with the β-sheet content thus not being in close agreement with the reported PDB structure. The β-sheets in XYL are highly curved, resulting in wide variations in the bond angles. It is believed that these variations may be responsible for the underestimation of β-sheet by CDPro.37 The enzyme still retained a substantial level of activity leading us to believe that low β-sheet values measured in CD is an artifact caused by the contorted β-sheets.
GOx is an acidic, dimeric glycoprotein (160 kDa) consisting of two identical 80 kDa subunits, each containing one non-covalently bound flavin adenine dinucleotide (FAD) coenzyme. The monomer subunit (Figure 3) is made up of 583 residues, 34% of them in helical structures and 23% in β-sheets, with only one disulfide bond.38 Our CD results of the solution structure were again found to be in close agreement with the PDB structure, with the secondary structure measured to be 36 ± 2% helix and 21 ± 2% β-sheet.
As shown from these results, our CD results for the solution structures of two out of our three enzymes were in very close agreement with their published PDB structures, with the β-sheet structure of XYL being substantially lower than the expected structure. While we do not fully understand the reason for the discrepancy with XYL, the excellent agreement for the other enzymes along with the α-helix content of XYL gives us reason to trust that our CD values are correct for the enzyme samples used in our studies.
Conformation of Adsorbed Enzyme Layers
Adsorption on the hydrophobic CH3-SAM surface caused the most significant degree of change in conformation for each of the proteins (Figure 4). Each enzyme tended to adsorb in a manner that led to a general increase in β-sheet structures and decrease in helices. This is consistent with results reported by others that proteins adsorb more strongly with a greater degree of conformational change with increasing surface hydrophobicity.39–41 HEWL and XYL proved to be substantially more resistant to conformational changes on the CH3-SAM surface than GOx, which was expected since the melting temperature of GOx is 47°C,42 whereas the melting temperatures of HEWL and XYL are in excess of 65°C,43, 44 thus reflecting the higher stability of HEWL and XYL.
Figure 4.
The percentage of helices and β-sheets present in 1.0 mg/mL enzyme solutions and after adsorbing on the different SAMs for 24 hours as determined by CD. The asterisk denotes adsorbed values that are statistically different than its corresponding solution value. (N=4, mean ± 95% CI)
The adsorption of GOx on the methyl surface resulted in a large amount of structural change with a 41% reduction in its α-helical structure and a 57% increase in β-sheets. It has been previously reported that an increase in β-sheet may often accompany a decrease in α-helix as a protein adsorbs, with the loss in α-helix reflecting a destabilization of the native structure of the protein and the increase in β-sheet being due to the surface acting as a planar template for the alignment of the polypeptide chain segments as the protein unfolds and spreads out on the surface over time.45 In distinct contrast to this, adsorption of each enzyme on the hydrophilic OH-SAM tended to not only result in a lesser degree of structural change, but also induced a different type of structural change, with a general decrease in both the α-helical and β-sheet structures. These results indicate that while both the CH3- and the OH-SAM surfaces tend to destabilize the native-state structure of the proteins, the hydrophobic CH3-SAM surface is able to additionally act as a template to induce the refolding of the protein with the formation of new β-sheet structure, which is believed to form as the protein spreads out on the surface.
Similarly, there was a greater degree of conformational changes for each of the enzymes after adsorption onto the mildly hydrophobic NH2-SAM surface than on the hydrophilic COOH-SAM regardless of the net charge of the enzyme. On the NH2-SAM, all three enzymes exhibited a significant loss in helices post-adsorption and a significant gain in β-sheets was seen in the adsorbed XYL and GOx layers. Conversely, for the COOH-SAM there was only a significant decrease in the amount of helical structures for the adsorbed GOx layer and no statistical difference in the β-sheet content of any of the enzymes with respect to their native structure. The fact that both the acidic and basic enzymes showed a greater degree of adsorption-induced conformational change on the NH2-SAM, which has a much higher water contact angle than the COOH-SAM, suggests that hydrophobic interactions are the dominate factor behind protein unfolding even on the charged surfaces.
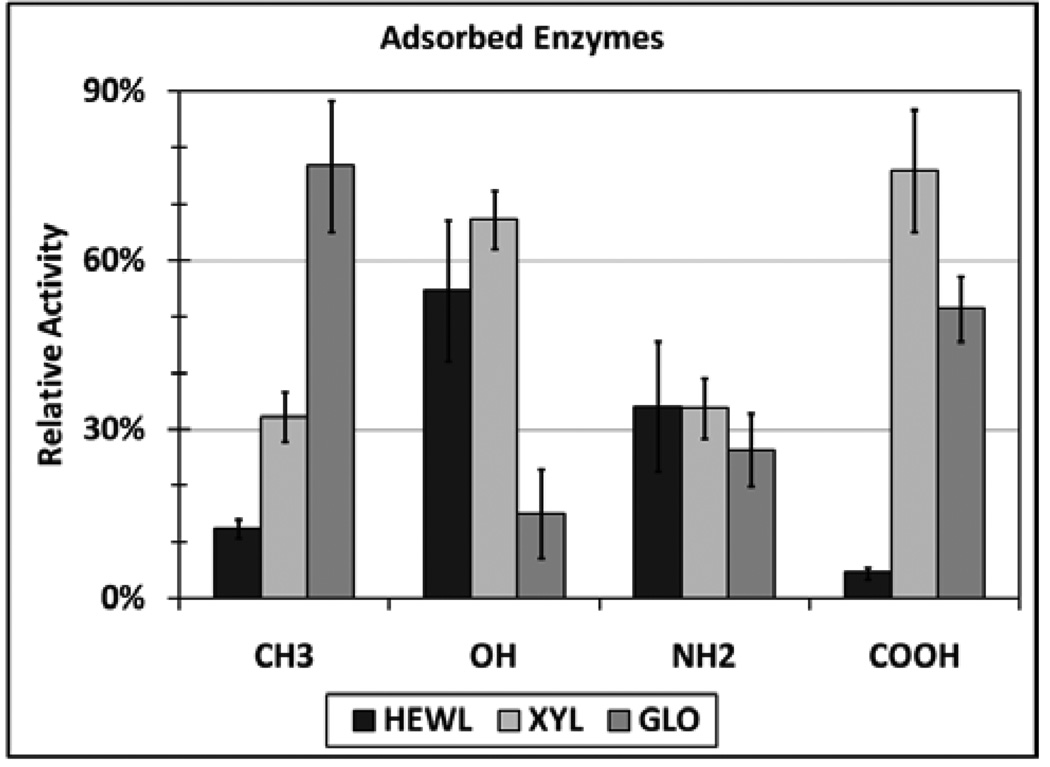
Bioactivity of Adsorbed Enzyme Layers
There were stark differences between the relative activities of the adsorbed enzyme layers on the two neutral surfaces, as well as the two charged surfaces (Figure 5). HEWL and XYL both retained a relatively high amount of activity on the OH-SAM surface while their activity was drastically reduced on the CH3-SAM surface. GOx showed the exact opposite response with its maximum activity being exhibited on the CH3-SAM surface and its minimum activity on the OH-SAM. There was no statistical difference in the relative activities of any of the adsorbed enzyme layers on the NH2-SAM surface. However, there was a substantial increase in the activities of the adsorbed XYL and GOx layers on the COOH-SAM and a drastic reduction in the activity of HEWL. It is important to note that the COOH-SAM has a much higher charge density than the NH2-SAM at physiological pH with approximately 50% of the carboxyl groups being negatively charged and only 11% of amine groups being positively charged.30 Also, as indicated by the contact angle results shown in Table 1, the NH2-SAM surface is much less hydrophilic than the COOH-SAM. Therefore hydrophobic interactions can be expected to play a role in the activities of the enzymes on the NH2-SAM in addition to the effects of surface charge.
Figure 5.
Graph of the relative activities of the adsorbed enzyme layers on the various SAMs. The surfaces were incubated in 1.0 mg/mL enzyme solutions for 24 hours and after which activity assays were carried out for 2 minutes. (N=4, mean ± 95% CI)
Discussion
The data presented in Figure 4 is the average secondary structure of the enzymes in solution and adsorbed on the different SAMs. The structure from molecule to molecule is expected to be fairly consistent when the enzymes are in solution. Conversely, the conformation and orientation of the adsorbed molecules on a given surface can be expected to vary significantly due to differences in protein-protein and/or protein-surface interactions, which are influenced by factors such as the surface concentration and unfolding kinetics, protein packing, surface charges, ionic strength, and local pH, as the adsorption process proceeds. Differences in the structure and/or orientation of molecules within the monolayer are likely to result in a greater variance between the functionality of the individual enzymes that comprise the monolayer. Additionally, it has been reported that the activity of a surface-adsorbed enzyme monolayer can be heavily dependent on the level of saturation.46–49 This is likely due to the effects of protein-protein interactions altering the diffusion of substrate in and out of the active site sterically and/or causing changes in the local electrostatics at the active sites of the adsorbed enzymes.
In this study, all adsorption experiments were carried out at the same concentration (1.0 mg/mL) with a 24 hour incubation period to allow the enzymes to fully saturate each SAM surface. The adsorption-induced changes in enzyme conformation and activity were measured for the adsorbed monolayers as a whole and compared to each other as such to investigate the effects of surface chemistry. The relative activities are reported (Figure 5) to illustrate the loss that occurs upon adsorption. While the enzyme kinetics parameters Km and Vmax can provide information about changes in substrate affinity and whether the reactions are rate- or diffusion-limited, their determination for all three enzymes on the four different SAMs would require an experimental data set that was well beyond the scope and objectives of this present study. Instead focus was placed on determining differences in the relative activities of the monolayers formed on the various SAMs with an attempt to discriminate between adsorption-induced conformational effects from adsorbed orientation. Based upon these present results, further studies may be planned to investigate how the adsorbed state of these enzymes influences their kinetic behavior.
HEWL showed no statistical change in secondary structure upon adsorption onto the oppositely charged COOH-SAM (Figure 4). Despite the preservation of secondary structure, the relative activity of the adsorbed enzyme layer was the lowest on this SAM (Figure 5), which is consistent with previous reports of the inactivity of HEWL on negatively charged surfaces at high surface concentrations.46 Although the active cleft itself contains negatively charged residues, there is a concentration of positively charge residues outside of both sides of the active cleft (Figure 3). Therefore the active face should tend to be electrostatically attracted to the COOH-SAM and repelled by the NH2-SAM. These results suggest that orientational differences contribute to the differences in relative activity on these two surfaces, with substrate access to the active site of the enzyme being sterically blocked, by the surface and/or other molecules, on the COOH-SAM but not for the NH2-SAM. While HEWL retained higher activity on the NH2-SAM surface compared to the COOH-SAM, its activity was still only about 33% of that in solution. This finding, combined with the significant loss in helicity of the protein on the NH2-SAM, suggests that adsorption-induced conformational changes may also contribute to the loss of activity on this surface.
The two acidic proteins, XYL and GOx, also showed markedly different levels of activity on the two charged surfaces. The relative activities of the adsorbed XYL and GOx layers on the COOH-SAM were at least twice the value of the respective enzymes on the NH2-SAM. When combined with the greater degree of adsorption-induced secondary structural changes for both of these enzymes on the NH2-SAM compared to the COOH-SAM, these results suggest that the changes in adsorbed activity may be due to either orientational effects induced by the differences in surface charge or conformational effects induced by the more hydrophobic NH2-SAM, or a combination of both of these effects. Electrostatically induced orientation effects can be expected if the charged residues are not uniformly distributed over the surface of the enzymes. For XYL, this distribution can be clearly seen in Figure 3 with the presence of positively charged residues being preferentially located on the right side of the bioactive cleft and negatively charged residues on the left. There are also two negatively charged glutamic acid residues in the cleft that comprise the active site of XYL, one of which retains its negative charge while the other forms an ion pair with a positively charged residue located on the right side of the cleft.32 The bioactive site is thus likely to be repelled on the COOH-SAM due to electrostatic repulsion, leading to increased substrate accessibility compared to the NH2-SAM, thus resulting in higher activity on the COOH-SAM due to adsorbed orientation effects. A previous study on XYL strains adsorbed onto a negatively charged methacrylic acid and methyl-methacrylate copolymer reported reductions in activity of around 40%, which correlates well with what we observed.50 A similar separation of charges is not readily apparent on the GOx protein and thus it is difficult to predict how differences in surface charge will affect the orientation of this enzyme. However, as shown in Figure 4, the adsorption of GOx on the NH2-SAM did result in a greater degree of structural change compared to the COOH-SAM surface, which suggests a greater possibility of the influence of structural changes on its adsorbed activity.
It can be seen in the PDB images of the enzymes (Figure 3) that both the nonpolar and the polar residues on their surfaces are distributed in a fairly uniform manner. Therefore, in the absence of electrostatic interactions between the enzymes and the SAMs, it can be expected that they will adsorb in a fairly random orientation on either of these surfaces, with the nonpolar amino acid residues tending to favorably adsorb to the CH3-SAM through hydrophobic interactions and the polar amino acid residues tending to favorably adsorb to the OH-SAM surface through hydrogen bonding. When comparing the effect of adsorption on the relative activities of HEWL and XYL, Figure 5 shows that both enzymes retained over 50% of their solution activities when adsorbed on the OH-SAM but lost substantially more activity when adsorbed on the CH3-SAM surface. As shown in Figure 4, the amount of adsorption-induced loss in secondary structure for these enzymes on these two surfaces was not significantly different from one another. Therefore the results from these studies provide no evidence to indicate that the measured difference in relative activity of HEWL and XYL is due to differences in adsorbed conformation or orientation. This suggests that some other factor(s) may be acting to reduce the ability of the substrate to complex with the active site of these enzymes in their adsorbed state. Although speculative at this point, we hypothesize that this other factor may be related to differences in the mobility and the tightness of binding of the adsorbed enzymes on these two very different types of surfaces. AFM studies1 and molecular simulations51 have indicated that a hydrophobic surface tends to adsorb proteins much more tightly than a hydrophilic surface. The increased freedom of motion of the enzymes on the very hydrophilic OH-SAM surface, combined with a very exposed cleft-type active site (see Figure 3), may thus facilitate the ability of the substrate to be able to interact with the active site of these adsorbed enzymes, thus enabling them to retain a much higher degree of activity on this type of surface. HEWL, in particular, has a highly exposed hydrophobic region in its active cleft (Figure 3) that would be susceptible to binding strongly to the hydrophobic surface. Further studies are planned to investigate these types of interactions using molecular simulation.
The most surprising result from these studies was exhibited by the effect of adsorption on GOx on the OH-SAM and CH3-SAM surfaces. The CD results show that GOx underwent a very large degree of structural change on the CH3-SAM surface with a loss of about 41% of its helical content combined with about a 57% increase in β-sheet. As previously noted, the substantial increase in β-sheet content following adsorption indicates that the protein underwent a substantial amount of spreading on the CH3-SAM surface. In comparison, adsorption of GOx on the OH-SAM surface resulted in relatively minor changes in its structure, with only about a 15% loss in both its α-helix and β-sheet structures. Conventional reasoning would predict this would result in a dramatic reduction in GOx’s bioactivity on the CH3-SAM surface with much higher bioactivity on the OH-SAM surface. However, the relative activities (Figure 5) showed the exact opposite response, with maximum relative activity (about 75%) corresponding to the enzyme layer on the CH3-SAM surface and the lowest relative activity (about 15%) corresponding to the layer adsorbed on the OH-SAM surface.
It is apparent from these results that the adsorption-induced structural changes in GOx somehow facilitated substrate access to its bioactive site. In GOx, the active site resides within a deep pocket in the center of each subunit along with the FAD coenzyme moiety. Residues 75–98 form a lid that closes the pocket, holding the FAD molecule within the protein and leaving a narrow funnel leading to the active site (see Figure 6).52 Although the CH3-SAM induces the most conformational changes within the protein, the buried active site is apparently remaining intact and accessible to the substrate. It is proposed that the large degree of spreading of the GOx on the CH3-SAM surface causes the bioactive funnel to be opened up, rendering it more accessible to its substrate than on the OH-SAM, with an increased the rate of diffusion for both the substrate and its reaction products in and out of the pocket, respectively. While the enzyme may be expected to retain greater mobility on the OH-SAM surface, the adsorbed state of the enzyme apparently impedes the ability of the substrate to access the deep-pocket binding site in GOx.
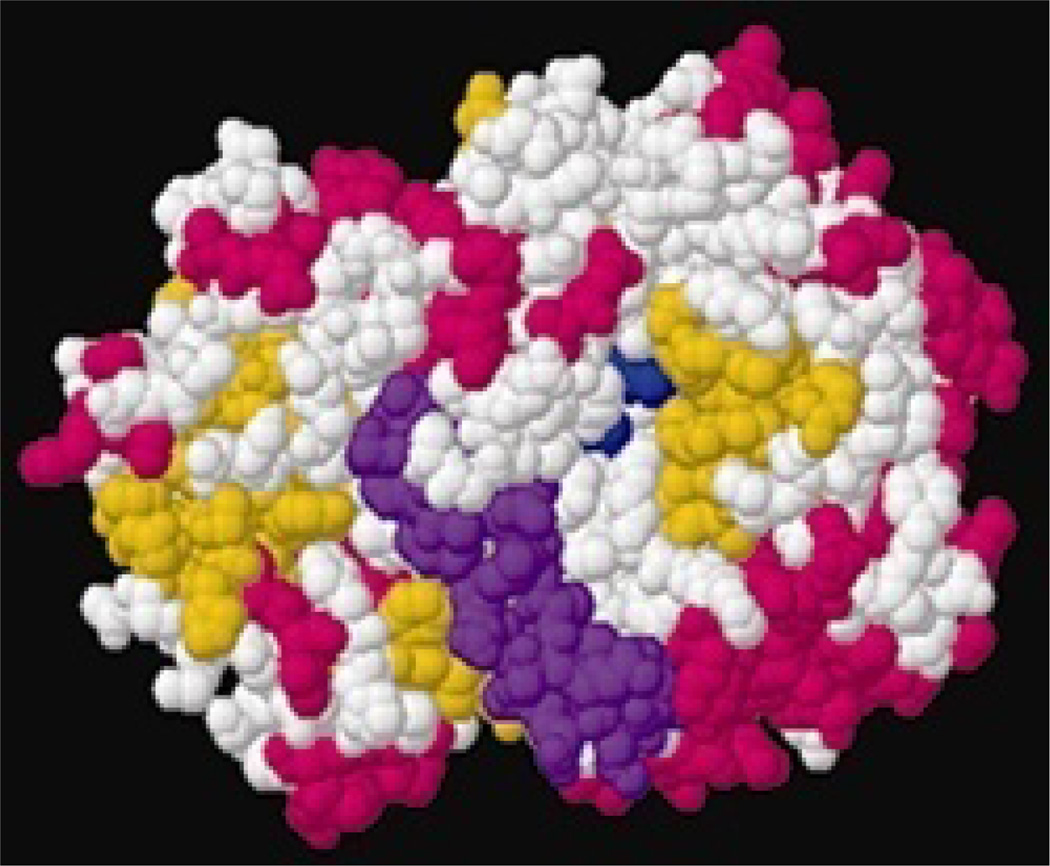
Figure 6.
View of the active face of GOx with the active site residues colored dark blue, the pocket lid colored purple, helices colored pink, and β-sheets colored yellow.
Conclusions
In this study, methods were developed to quantitatively assess the effects of adsorption on both the secondary structure and the bioactive state of a set of three enzymes that ranged from 14.4 to 160 kDa in size. When used in combination with the known native-state structures of the enzymes, these methods provide a means to gain molecular-level insights into how surface chemistry influences their adsorbed-state orientation, conformation, and bioactivity. The combined results from these studies indicate that surface chemistry has a very dramatic influence on the bioactive state of adsorbed enzymes. In our studies, enzymes with active sites within clefts (HEWL and XYL) behaved quite differently than GOx whose active site is inside a deep pocket. Surprisingly, the adsorption behavior and subsequent activity of GOx demonstrated that the loss of secondary structure does not always translate into the loss of bioactivity. Although speculative at this time, these results suggest that a possible mechanism for the enhanced bioactivity is that adsorption-induced conformational changes are leading to increased access to the binding site.
The methods developed in this study are readily applicable for a wide variety of enzymes and surfaces to quantitatively investigate how adsorption influences the orientation and conformation of enzymes at the molecular level, and how this in turn influences their adsorbed-state bioactivity. The development of a molecular-level understanding of the cause-and-effect relationships governing these types of interactions has the potential to lead to the design of adsorbed-enzyme systems with improved performance for a broad range of applications in biomedical engineering and biotechnology.
Acknowledgment
The authors would like to thank Dr. Jim Harriss (Clemson University) for performing TVD on the bare quartz slides. Also, we would like to acknowledge Dr. Lara Gamble of NESAC/BIO for the conducting XPS analysis on the SAMs. Funding was provided by NSF Award No. EEC-9731680 through the Center of Advanced Engineering Fibers and Film (CAEFF) at Clemson University.
References
- 1.Agnihotri A, Siedlecki CA. Langmuir. 2004;20(20):8846–8852. doi: 10.1021/la049239+. [DOI] [PubMed] [Google Scholar]
- 2.Chaki NK, Vijayamohanan K. Biosens. Bioelectron. 2002;17(1–2):1–12. doi: 10.1016/s0956-5663(01)00277-9. [DOI] [PubMed] [Google Scholar]
- 3.Chicurel ME, Dalma-Weiszhausz DD. Pharmacogenomics. 2002;3(5):589–601. doi: 10.1517/14622416.3.5.589. [DOI] [PubMed] [Google Scholar]
- 4.Gooding JJ, Praig VG, Hall EAH. Anal. Chem. 1998;70(11):2396–2402. doi: 10.1021/ac971035t. [DOI] [PubMed] [Google Scholar]
- 5.Kasemo B, Gold J. Adv. Dent. Res. 1999;13:8–20. doi: 10.1177/08959374990130011901. [DOI] [PubMed] [Google Scholar]
- 6.Mao HB, Yang TL, Cremer PS. Anal. Chem. 2002;74(2):379–385. doi: 10.1021/ac010822u. [DOI] [PubMed] [Google Scholar]
- 7.Puleo DA, Nanci A. Biomaterials. 1999;20(23–24):2311–2321. doi: 10.1016/s0142-9612(99)00160-x. [DOI] [PubMed] [Google Scholar]
- 8.Sarikaya M, Tamerler C, Schwartz DT, Baneyx FO. Ann. Rev. Mat. Res. 2004;34:373–408. [Google Scholar]
- 9.Wnek G, Bowlin G. The Encyclopedia of Biomaterials and Bioengineering. New York: Taylor & Francis; 2005. [Google Scholar]
- 10.Arenkov P, Kukhtin A, Gemmell A, Voloshchuk S, Chupeeva V, Mirzabekov A. Anal. Biochem. 2000;278(2):123–131. doi: 10.1006/abio.1999.4363. [DOI] [PubMed] [Google Scholar]
- 11.Mrksich M, Whitesides GM. Ann. Rev. Biophys. Biomol. Struct. 1996;25:55–78. doi: 10.1146/annurev.bb.25.060196.000415. [DOI] [PubMed] [Google Scholar]
- 12.Hylton DM, Shalaby SW, Latour RA. J. Biomed. Mat. Res. A. 2005;73A(3):349–358. doi: 10.1002/jbm.a.30295. [DOI] [PubMed] [Google Scholar]
- 13.McMillin CR, Walton AG. J. Coll. Inter. Sci. 1974;48(2):345–349. [Google Scholar]
- 14.Shimizu M, Kobayashi K, Morii H, Mitsui K, Knoll W, Nagamune T. Biochem. Biophys. Res. Comm. 2003;310(2):606–611. doi: 10.1016/j.bbrc.2003.09.054. [DOI] [PubMed] [Google Scholar]
- 15.Vermeer AWP, Norde W. J. Coll. Inter. Sci. 2000;225(2):394–397. doi: 10.1006/jcis.2000.6769. [DOI] [PubMed] [Google Scholar]
- 16.Sivaraman B, Fears KP, Latour RA. Langmuir. 2009;25:3050–3056. doi: 10.1021/la8036814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akaike T, Sakurai Y, Kosuge K, Senba Y, Kuwana K, Miyata S, Kataoka K, Tsuruta T. Kobunshi Ronbunshu. 1979;36(4):217–222. [Google Scholar]
- 18.Layne E. Meth. Enzymol. 1957;3:447–455. [Google Scholar]
- 19.Stoscheck CM. Meth. Enzymol. 1990;182:50–68. doi: 10.1016/0076-6879(90)82008-p. [DOI] [PubMed] [Google Scholar]
- 20.Sivaraman B, Fears KP, Latour RA. Langmuir. 2009 doi: 10.1021/la8036814. , in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sreerama N, Venyaminov SY, Woody RW. Anal. Biochem. 2000;287(2):243–251. doi: 10.1006/abio.2000.4879. [DOI] [PubMed] [Google Scholar]
- 22.Sreerama N, Woody RW. Anal. Biochem. 2000;287(2):252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 23.Zhou RQ, Chen SG, Recsei P. Anal. Biochem. 1988;171(1):141–144. doi: 10.1016/0003-2697(88)90134-0. [DOI] [PubMed] [Google Scholar]
- 24.Biely P, Mislovicova D, Toman R. Meth. Enzymol. 1988;160:536–541. [Google Scholar]
- 25.Bergmeyer HU, Bernt E. Methods of Enzymatic Analysis. 2nd ed. New York: Academic Press; 1974. pp. 1205–1212. [Google Scholar]
- 26.Wojciechowski P, Tenhove P, Brash JL. J. Coll. Inter. Sci. 1986;111(2):455–465. [Google Scholar]
- 27.Bain CD, Troughton EB, Tao YT, Evall J, Whitesides GM, Nuzzo RG. J. Am. Chem. Soc. 1989;111(1):321–335. [Google Scholar]
- 28.Faucheux N, Schweiss R, Lutzow K, Werner C, Groth T. Biomaterials. 2004;25(14):2721–2730. doi: 10.1016/j.biomaterials.2003.09.069. [DOI] [PubMed] [Google Scholar]
- 29.Gamble L. Assistant Director of NESAC/BIO. University of Washington, Seattle, WA.: Personal Communication; 2008. Aug, [Google Scholar]
- 30.Fears KP, Creager SE, Latour RA. Langmuir. 2008;24(3):837–843. doi: 10.1021/la701760s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsumura I, Kirsch JF. Biochem. 1996;35(6):1881–1889. doi: 10.1021/bi951671q. [DOI] [PubMed] [Google Scholar]
- 32.Gruber K, Klintschar G, Hayn M, Schlacher A, Steiner W, Kratky C. Biochem. 1998;37(39):13475–13485. doi: 10.1021/bi980864l. [DOI] [PubMed] [Google Scholar]
- 33.Wohlfahrt G, Trivic S, Zeremski J, Pericin D, Leskovac V. Mol. Cell. Biochem. 2004;260(1–2):69–83. doi: 10.1023/b:mcbi.0000026056.75937.98. [DOI] [PubMed] [Google Scholar]
- 34.Jolles P. Angewandte Chemie-International Edition. 1969;8(4):227–250. doi: 10.1002/anie.196902271. [DOI] [PubMed] [Google Scholar]
- 35.Alderton G, Lewis JC, Fevold HL. Science. 1945;101(2615):151–152. doi: 10.1126/science.101.2615.151. [DOI] [PubMed] [Google Scholar]
- 36.Refaee M, Tezuka T, Akasaka K, Williamson MP. J. Mol. Biol. 2003;327(4):857–865. doi: 10.1016/s0022-2836(03)00209-2. [DOI] [PubMed] [Google Scholar]
- 37.Oakley AJ, Heinrich T, Thompson CA, Wilce MCJ. Acta Crystallogr. Sect. D: Biol. Crystallogr. 2003;59:627–636. doi: 10.1107/s0907444903001227. [DOI] [PubMed] [Google Scholar]
- 38.Wohlfahrt G, Witt S, Hendle J, Schomburg D, Kalisz HM, Hecht HJ. Acta Crystallogr. Sect. D: Biol. Crystallogr. 1999;55:969–977. doi: 10.1107/s0907444999003431. [DOI] [PubMed] [Google Scholar]
- 39.Andrade JD, Hlady V. Adv. Poly. Sci. 1986;79:1–63. [Google Scholar]
- 40.Latour RA. The Encyclopedia of Biomaterials and Bioengineering. 2nd ed. New York, NY: Taylor & Francis; 2005. Biomaterials: Protein-Surface Interactions; pp. 270–284. [Google Scholar]
- 41.Roach P, Farrar D, Perry CC. J. Am. Chem. Soc. 2005;127(22):8168–8173. doi: 10.1021/ja042898o. [DOI] [PubMed] [Google Scholar]
- 42.Godjevargova T, Vasileva N, Letskovska Zl. J. Appl. Poly. Sci. 2003;90(5):1393–1397. [Google Scholar]
- 43.Collins T, Gerday C, Feller G. FEMS Microbiol. Rev. 2005;29(1):3–23. doi: 10.1016/j.femsre.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 44.Maeda Y, Yamada H, Ueda T, Imoto T. Protein Eng. 1996;9(5):461–465. doi: 10.1093/protein/9.5.461. [DOI] [PubMed] [Google Scholar]
- 45.Castillo EJ, Koenig JL, Anderson JM. Biomaterials. 1984;5(6):319–325. doi: 10.1016/0142-9612(84)90029-2. [DOI] [PubMed] [Google Scholar]
- 46.Ding HM, Shao L, Liu RJ, Xiao QG, Chen JF. J. Coll. Inter. Sci. 2005;290(1):102–106. doi: 10.1016/j.jcis.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 47.Lei CH, Soares TA, Shin YS, Liu J, Ackerman EJ. Nanotechnol. 2008;19(12) doi: 10.1088/0957-4484/19/12/125102. [DOI] [PubMed] [Google Scholar]
- 48.Crapisi A, Lante A, Pasini G, Spettoli P. Process Biochem. 1993;28(1):17–21. [Google Scholar]
- 49.Datta R, Armiger W, Ollis DF. Biotechnol. Bioeng. 1973;15(5):993–1006. [Google Scholar]
- 50.Gawande PV, Kamat MY. J. Biotechnol. 1998;66(2–3):165–175. doi: 10.1016/s0168-1656(98)00146-1. [DOI] [PubMed] [Google Scholar]
- 51.Agashe M, Raut V, Stuart SJ, Latour RA. Langmuir. 2005;21(3):1103–1117. doi: 10.1021/la0478346. [DOI] [PubMed] [Google Scholar]
- 52.Zhong DP, Zewail AH. Proc. Natl. Acad. Sci. U.S.A. 2001;98(21):11867–11872. doi: 10.1073/pnas.211440398. [DOI] [PMC free article] [PubMed] [Google Scholar]