Abstract
Changes in redox status have been observed during immune responses in different organisms, but the associated signaling mechanisms are poorly understood. In plants, these redox changes regulate the conformation of NPR1, a master regulator of salicylic acid (SA)–mediated defense genes. NPR1 is sequestered in the cytoplasm as an oligomer through intermolecular disulfide bonds. We report that S-nitrosylation of NPR1 by S-nitrosoglutathione (GSNO) at cysteine-156 facilitates its oligomerization, which maintains protein homeostasis upon SA induction. Conversely, the SA-induced NPR1 oligomer-to-monomer reaction is catalyzed by thioredoxins (TRXs). Mutations in both NPR1 cysteine-156 and TRX compromised NPR1-mediated disease resistance. Thus, the regulation of NPR1 is through the opposing action of GSNO and TRX. These findings suggest a link between pathogen-triggered redox changes and gene regulation in plant immunity.
Innate immune responses are evolutionarily conserved among plants and animals (1, 2) and are often associated with changes in cellular oxidative and reductive states. In plants, these redox changes are sensed by the NPR1 protein, a master regulator of defense gene expression (3). In unchallenged plants, NPR1 resides in the cytoplasm as an oligomer maintained through redox-sensitive intermolecular disulfide bonds. Upon pathogen challenge, the plant defense signaling molecule salicylic acid (SA) increases and changes the cellular redox state, leading to reduction of the disulfide bonds in NPR1. Reduction of the NPR1 oligomer releases monomer that translocates to the nucleus where it activates the expression of a battery of pathogenesis-related (PR) genes (4). Mutations at residues Cys82 and Cys216 in NPR1 result in increased monomer accumulation, constitutive nuclear localization, and NPR1-mediated gene expression in the absence of pathogen challenge (3). On the basis of these results, it has been proposed that conformational changes in NPR1 (that is, oligomer-monomer exchange) regulate its nuclear translocation and activity (3).
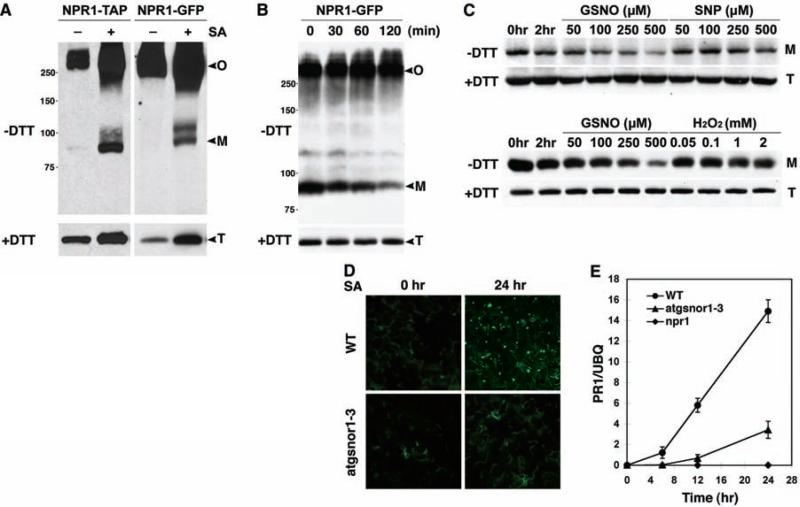
Oligomerization of proteins through intermolecular disulfide bonds is unusual under reductive cytosolic conditions (5). However, treatment with SA not only induced NPR1 monomer release but also facilitated oligomerization in wild-type plants (fig. S1A). Similar results were obtained with biologically active NPR1 fused with green fluorescent protein (NPR1-GFP) (4) or with tandem affinity purification tag (NPR1-TAP) (fig. S2), driven by the constitutive 35S promoter (Fig. 1A). On the basis of these observations, we hypothesized that a catalyst of cysteine thiol oxidation was involved in the formation of the NPR1 oligomer. To search for cellular oxidants facilitating NPR1 oligomerization, we established a cell-free assay in which total protein extract from 35S::NPR1-GFP plants was treated with the reducing agent dithiothreitol (DTT) to partially convert the NPR1-GFP oligomer to monomer. Removal of DTT by dialysis allowed reformation of the oligomer as shown by a decrease in monomer, while the total protein amount remained constant (Fig. 1B). These data suggest that, depending on the cellular environment, NPR1 switches between the oligomeric and monomeric states.
Fig. 1.
GSNO facilitates the oligomerization of NPR1. (A) SA induces NPR1 monomer release as well as oligomerization. Nonreducing (–DTT) and reducing (+DTT) immuno-blot analysis revealed oligomeric (O), monomeric (M), and total (T) NPR1. (B) Re-oligomerization of monomeric NPR1-GFP in a cell-free assay. (C) NPR1 monomer disappears with an increasing concentra tion of GSNO but not of SNP or H2O2, while total protein levels are unaffected. (D) SA induces nuclear localization of the NPR1-GFP monomer in the wild type but not in the atgsnor1-3 mutant. (E) SA-induced PR-1 gene expression is compromised in atgsnor1-3 plants. PR-1 expression was determined by real-time polymerase chain reaction (PCR) and normalized with ubiquitin (UBQ). Error bars represent SD.
Using the cell-free assay shown in Fig. 1B, we tested the effect of hydrogen peroxide (H2O2) as well as the nitric oxide (NO) donors sodium nitroprusside (SNP) and S-nitrosoglutathione (GSNO) on the NPR1 oligomer-monomer equilibrium, because these oxidants accumulate during innate immune responses (6, 7). The treatment of protein extracts with H2O2 and SNP had no effect on NPR1 conformation (Fig. 1C). In contrast, GSNO, a natural NO donor, markedly facilitated oligomerization of NPR1 as shown by the disappearance of the monomer while total NPR1 levels remained unchanged (Fig. 1C). This is consistent with the finding that unlike SNP, the treatment of plant cell cultures with GSNO caused protein S-nitrosylation (8), a process in which NO is covalently attached to a reactive cysteine thiol to form an S-nitrosothiol (SNO) (9). To further confirm this specific effect of GSNO, NPR1 activity was monitored in the GSNO reductase knockout mutant atgsnor1-3, which displays increased S-nitrosylation activity (7). We found that SA-induced monomerization of the endogenous NPR1 (fig. S1B) and nuclear translocation of monomeric NPR1-GFP were inhibited (Fig. 1D). NPR1 oligomer accumulated to higher levels in the atgsnor1-3 mutant as compared to the wild type (fig. S1B). Accordingly, SA-induced expression of the NPR1-dependent defense gene PR-1 was also suppressed in atgsnor1-3 plants (Fig. 1E). These data suggest that GSNO affects the conformation of NPR1 and consequently its activity in innate immunity.
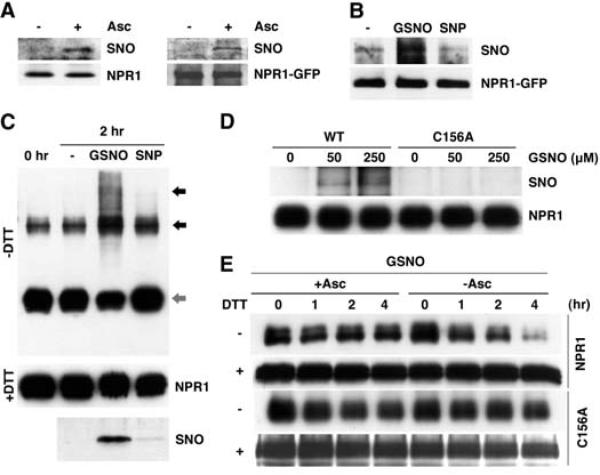
In Arabidopsis, pathogen infection induces an increase in cellular SNO levels, and elevated SNO levels in atgsnor1-3 are associated with enhanced susceptibility to disease (7). The effect of GSNO on NPR1 oligomerization, together with the fact that some of the cysteine residues in NPR1 are critical for oligomer formation (3), suggests that one or more NPR1 cysteine thiols are directly modified by GSNO. To test this, we examined whether NPR1 is S-nitrosylated in planta, using the biotin-switch method (10), which specifically detects S-nitrosylated proteins (11). Total protein was extracted from SA-treated wild-type and transgenic 35S::NPR1-GFP plants and then incubated with or without ascorbate, which specifically reduces SNO groups (10). The resulting free thiols were then covalently coupled to biotin-HPDP (biotin–N-[6(biotinamido)hex1]-3′-(2′-pyridyldithio)propionamide) and immuno-precipitated with streptavidin beads. Immunoblot analysis revealed that both endogenous NPR1 and transgenic NPR1-GFP were pulled down only in ascorbate-treated samples, indicating that these proteins were specifically S-nitrosylated in vivo (Fig. 2A). Treatment with SA enhanced the S-nitrosylation of NPR1 (fig. S3A). We then applied the biotin-switch method to our cell-free assay and found that GSNO increased S-nitrosylation of NPR1, whereas SNP was ineffective in this respect (Fig. 2B). These results suggest that GSNO may facilitate NPR1 oligomerization directly through thiol S-nitrosylation.
Fig. 2.
S-nitrosylation of Cys156 facilitates the assembly of NPR1 oligomer. (A) SA induces S-nitrosylation of endogenous NPR1 and the NPR1-GFP proteins in vivo. Sodium ascorbate (Asc) was used to specifically detect S-nitrosylated (SNO) NPR1. Equal loading was verified with antibodies against NPR1 or NPR1-GFP. (B) GSNO, but not mock (–) or SNP treatment, induces S-nitrosylation of NPR1-GFP in plant extracts. S-nitrosylated NPR1-GFP was detected with the biotin-switch assay. An antibody against NPR1-GFP was used to verify equal loading. (C) GSNO, but not mock (–) or SNP treatment, induces S-nitrosylation and multimerization (black arrows) of recombinant His6-NH (NPR1 residues 1 to 246) monomer (gray arrow). Equal loading was verified with an antibody to NPR1. (D) Cys156 is the principal site of S-nitrosylation in NPR1. Recombinant His6-NH and His6-NH-C156A proteins were incubated with different GSNO concentrations, and S-nitrosylation was detected by the biotin-swich assay. Equal loading was verified with an antibody to NPR1. (E) The C156A mutation impairs GSNO-induced oligomerization. Recombinant His6-NH and His6-NH-C156A proteins were treated with GSNO and with (+) or without (–) sodium ascorbate. Subsequently, monomers were allowed to re-oligomerize for the indicated times. Monomeric (–DTT) and total (+DTT) proteins were detected with an antibody to NPR1.
Previously, we demonstrated that Cys82, Cys150, Cys155, Cys160, and Cys216 in and adjacent to the BTB/POZ domain of NPR1 (fig. S4) are important in the oligomer-monomer exchange (3). This suggests that the N-terminal half of NPR1 is sufficient for oligomerization. We purified recombinant protein containing the hexa-histidine (6xHis)–tagged N-terminal half of NPR1 (His6-NH, residues 1 to 246) and examined its oligomerization properties in response to NO donors. As compared with the control, treatment of purified His6-NH with GSNO resulted in increased S-nitrosylation and multimerization of His6-NH (Fig. 2C). Similar results were obtained with the NO donor diethylamine-NO (DEA/NO) (fig. S3, B and C), further supporting a role of SNO in stimulating NPR1 oligomerization. In contrast, SNP treatment failed to S-nitrosylate and multi-merize His6-NH (Fig. 2C). Thus, at least one NO-sensitive cysteine lies within the 246 N-terminal residues of NPR1. Indeed, mutation of Cys156 abolished both GSNO-triggered S-nitrosylation (Fig. 2D) and oligomerization (Fig. 2E). Taken together, these findings indicate that GSNO S-nitrosylates NPR1 at Cys156. Similar to the SNO-mediated disulfide bond formation in myoglobulin (12, 13), S-nitrosylation of Cys156 may directly facilitate disulfide linkage between NPR1 monomers. Computational modeling of the NPR1 BTB domain according to previously published BTB crystal structures (14, 15) predicts that one or more disulifide bonds may form between Cys150, Cys155, Cys156, and Cys160 in the NPR1 oligomer (fig. S5). Alternatively, S-nitrosylation of Cys156 may lead to conformational changes in NPR1 that favor oligomerization as reported for SNO-facilitated dynamin oligomerization (16).
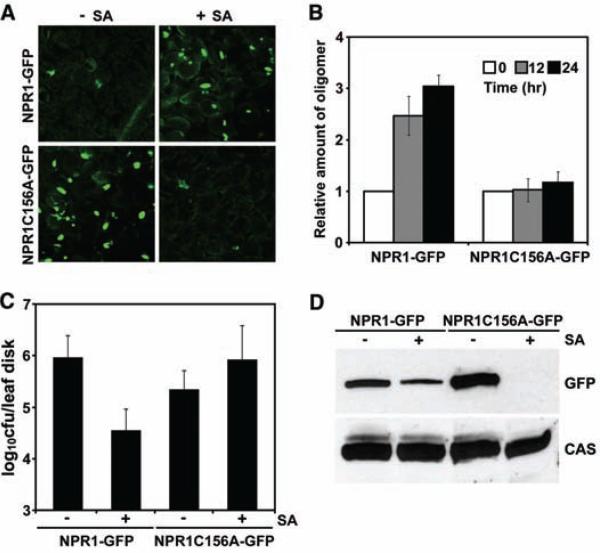
Transformation of 35S::NPR1C156A-GFP into mutant npr1 plants consistently resulted in constitutive nuclear fluorescence relative to un-induced 35S::NPR1-GFP plants (Fig. 3A), suggesting that the NPR1C156A-GFP (C156A, Cys156→Asp156) protein does not form oligomers as efficiently as wild-type protein. Although NPR1C156A-GFP protein still formed oligomers before induction, it lacked any SA-induced increase in oligomerization (Fig. 3B), indicating that Cys156 is required for SNO-facilitated oligomerization in vivo.
Fig. 3.
S-nitrosylation of Cys156 is essential for NPR1 protein homeostasis and SA-induced disease resistance. (A) SA treatment reduces the constitutive nuclear fluorescence of NPR1C156A-GFP. (B) The C156A mutation impairs NPR1 oligomer formation in response to SA. 35S::NPR1-GFP and 35S::NPR1C156A-GFP plants were treated with SA. The relative amount of NPR1 oligomer was determined by calculating densitometric ratios between induced and uninduced samples and normalized against total NPR1 protein. Error bars represent SD (n = 3 measurements). (C) SA-induced resistance is compromised in NPR1C156A plants. Error bars represent 95% confidence limits (n = 8 xxxxx). cfu, colony-forming unit. (D) SA treatment decreases NPR1C156A protein levels. 35S::NPR1-GFP and 35S::NPR1C156A-GFP plants were treated with (+) or without (–) SA for 48 hours. NPR1-GFP protein was detected with an antibody to GFP, and equal loading was verified with an antibody against constitutively expressed Ca2+-sensing receptor (CAS).
The effect of the NPR1C156A mutation on plant defense was demonstrated when plants were challenged by Pseudomonas syringae pv. maculicola ES4326 (Psm ES4326). Consistent with the nuclear accumulation of NPR1C156A-GFP (Fig. 3A) and its normal interaction with TGA transcription factors (fig. S6), untreated 35S::NPR1C156A-GFP plants showed enhanced resistance to this pathogen as compared with 35S::NPR1-GFP plants (Fig. 3C). However, unlike 35S::NPR1-GFP, treatment with SA for 48 hours did not enhance resistance in 35S:: NPR1C156A-GFP plants. These findings indicate that SNO-Cys156-mediated oligomerization is necessary to maintain NPR1 homeostasis upon SA activation. Immunoblot analysis (Fig. 3D) and GFP fluorescence (Fig. 3A) showed that the NPR1C156A protein was depleted 48 hours after SA treatment, explaining the compromised pathogen resistance.
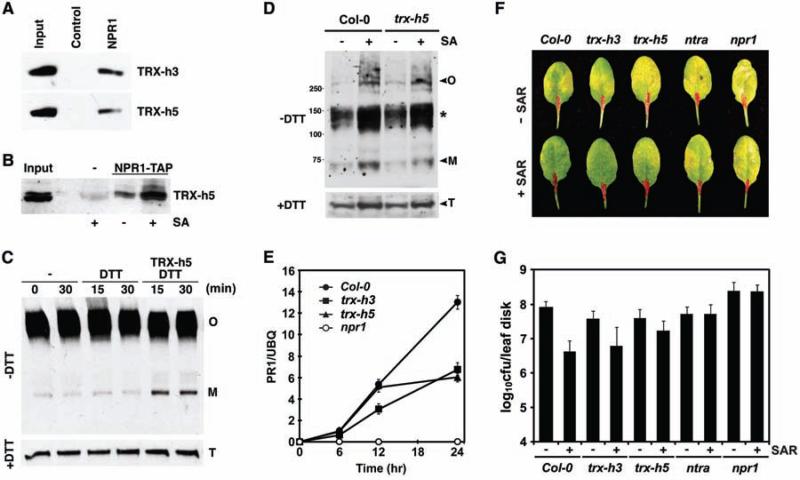
To counter the effect of SNO-facilitated NPR1 oligomerization, reducing agents must be engaged upon SA induction to catalyze the NPR1 oligomer-to-monomer switch. To identify such agents, we performed pull-down assays with recombinant His6-NH protein and identified two thioredoxins (TRXs), TRX-h3 and TRX-h5 (Fig. 4A). Among the eight cytosolic TRX-h genes in Arabidopsis, TRX-h5 is substantially up-regulated upon infection with P. syringae (17), whereas TRX-h3 is the most highly constitutively expressed TRX-h (18) (fig. S7). In a reverse experiment, we covalently trapped NPR1 using mutant TRXs (TRX-h3M and TRX-h5M), in which the second catalytic cysteine was changed to serine to prevent the completion of substrate reduction (19) (fig. S8A). Pull-down experiments showed that the NPR1-binding affinity of TRX-h was inversely correlated with its enzymatic activity (fig. S8, B and C), suggesting that TRX-h is the enzyme catalyzing NPR1 oligomer reduction. The transient nature of this interaction made it difficult to examine the NPR1/TRX-h interaction in vivo. Therefore, we fixed the enzyme-substrate intermediate [see supporting online material (SOM) text] and coimmunoprecipitated TRX-5h with NPR1-TAP (Fig. 4B). Treatment with SA further increased the interaction. In vivo interaction of TRX-h5 with NPR1 suggests that it may be involved in catalysis of the NPR1 oligomer-tomonomer reaction during plant defense. We added recombinant TRX-h5 to cell lysates containing NPR1-GFP oligomer and showed that, compared to the control, the amount of NPR1-GFP monomer increased within 15 min of incubation (Fig. 4C).
Fig. 4.
TRX is a redox mediator of NPR1. (A) Immobilized His6-NH pulls down TRXs in vitro. TRXs were detected with protein-specific antibodies. (B) 35S::NPR1-TAP transformed and untransformed plants were treated with or without SA. SA treatment enhanced coimmunoprecipitation of TRX-h5 with NPR1-TAP as detected by an antibody to TRX-h5. (C) NPR1-GFP oligomer (O) is reduced to monomer (M) by TRX-h5 in vitro. DTT (0.33 mM) was added to recycle TRX activity. Detection of total (T) NPR1-GFP verified equal loading. (D) In vivo reduction of NPR1 oligomer to monomer in Col-0 and trx-h5 plants 12 hours after SA treatment. Non-reducing (–DTT) and reducing (+DTT) SDS-polyacrylamide gel electrophoresis were performed in parallel, and the endogenous NPR1 was detected with an antibody to NPR1. The asterisk indicates possible NPR1 dimer complexes. (E) SA-induced PR-1 gene expression is compromised in trx mutants. PR-1 expression was determined by real-time PCR and normalized with ubiquitin (UBQ). Error bars represent SD. (F and G) Induction of SAR significantly decreased disease symptoms and Psm ES4326 growth in Col-0 plants but not in the trx, ntra, and npr1-1 mutants. Error bars represent 95% confidence limits (n = 8 samples).
TRX-h5 was required in vivo for SA-induced monomer release (Fig. 4D). Both TRX-h3 and TRX-h5 were required for full induction of PR genes (Fig. 4E). Additionally, in the trx mutants, NPR1-dependent systemic acquired resistance (SAR) against Psm ES4326, triggered after local inoculation of avirulent Psm ES4326/avrRpt2, was partially impaired (Fig. 4, F and G). Consistently, the TRX reductase knockout mutant ntra (20), which blocks the regeneration of cytosolic TRXs, showed a complete loss of SAR.
Our study provides a molecular mechanism to explain how cellular redox changes during pathogen challenge lead to transcriptional reprogramming and disease resistance (fig. S9). We propose that redox signals are conveyed through SNO and cytosolic TRXs, which directly catalyze the NPR1 oligomer-monomer exchange. Upon pathogen challenge, SA induces TRX-5h to catalyze the release of NPR1 monomer and possibly prevent oligomerization of some of the monomer. Induction also leads to S-nitrosylation of NPR1, which facilitates oligomerization to prevent protein depletion. SA-induced NPR1 oxidation and reduction may occur sequentially as the application of inducers of SAR results in transient oxidative and reductive fluctuations (3). To test this hypothesis, we treated plants with a combination of the translation inhibitor cycloheximide, the proteasome inhibitor MG115, and SA. In the absence of protein synthesis and degradation, SA-induced monomer accumulation was highest 12 hours after treatment. However, after 16 hours, NPR1 monomer re-oligomerized (fig. S10). The biological importance of controlling NPR1 homeostasis is demonstrated by the impaired immune responses of the NPR1C156A and trx mutants.
NO has long been proposed to be involved in responses to plant hormones, salt stress, ultra-violet light, and pathogens (21, 22). However, our knowledge of NO direct targets and its molecular effects on gene expression is limited. Recently, S-nitrosylation of Arabidopsis peroxiredoxin II E was shown to cause the accumulation of peroxynitrite (ONOO–) (23). Increased ONOO– levels induced tyrosine nitration of proteins, which might activate the plant defense mechanism known as the hypersensitive response. Even though the in vivo concentration and subcellular localization of GSNO have yet to be determined, genetic studies with Arabidopsis atgsnor mutants indicated that GSNO functions as an endogenous signal in plant defense responses (7). The identification of NPR1 as a direct target of S-nitrosylation may explain the phenotype of the atgsnor mutants. In mammals, NO functions as an anti-inflammatory signal by S-nitrosylating IκB kinase β (IKKβ) (24), the catalytic subunit of IKK, required for activation of the transcriptional immune regulator nuclear factor kB (NF-κB). S-nitrosylation of IKKβ inactivates IKK and retains NF-κB in the cytoplasm. This response is reminiscent of NPR1 oligomerization by S-nitrosylation, which prevents NPR1 from entering the nucleus (fig. S9). This suggests that redox-mediated transcription regulatory mechanisms are a common feature of immune responses in both plants and animals.
Supplementary Material
Acknowledgments
We thank M. Delledonne, J. Stamler, K. Murase, A. Matsumoto, K. Kojima, and K. Ozawa for discussion; G. Loake and N. Spivey for the atgsnor1-3 mutant and microarray data on TRXs, respectively; J. Yoo for technical assistance; Y. Meyer and Z. Pei for TRX-h3/TRX-h5 and CAS antibodies, respectively; and J. Stamler, J. Siedow, Z. Pei, and N. Spivey for critiquing the manuscript. Supported by a grant from NIH (1R01-GM69594) to X.D.
Footnotes
Supporting Online Material
References and Notes
- 1.Jones JD, Dangl JL. Nature. 2006;444:323. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 2.Nurnberger T, Brunner F, Kemmerling B, Piater L. Immunol. Rev. 2004;198:249. doi: 10.1111/j.0105-2896.2004.0119.x. [DOI] [PubMed] [Google Scholar]
- 3.Mou Z, Fan W, Dong X. Cell. 2003;113:935. doi: 10.1016/s0092-8674(03)00429-x. [DOI] [PubMed] [Google Scholar]
- 4.Kinkema M, Fan W, Dong X. Plant Cell. 2000;12:2339. doi: 10.1105/tpc.12.12.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dietz KJ. Int. Rev. Cytol. 2003;228:141. doi: 10.1016/s0074-7696(03)28004-9. [DOI] [PubMed] [Google Scholar]
- 6.Delledonne M, Xia Y, Dixon R, Lamb C. Nature. 1998;394:585. doi: 10.1038/29087. [DOI] [PubMed] [Google Scholar]
- 7.Feechan A, et al. Proc. Natl. Acad. Sci. U.S.A. 2005;102:8054. doi: 10.1073/pnas.0501456102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindermayr C, Saalbach G, Durner J. Plant Physiol. 2005;137:921. doi: 10.1104/pp.104.058719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stamler JS, et al. Proc. Natl. Acad. Sci. U.S.A. 1992;89:444. [Google Scholar]
- 10.Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Nat. Cell Biol. 2001;3:193. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- 11.Forrester MT, Foster MW, Stamler JS. J. Biol. Chem. 2007;282:13977. doi: 10.1074/jbc.M609684200. [DOI] [PubMed] [Google Scholar]
- 12.Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Nat. Rev. Mol. Cell Biol. 2005;6:150. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 13.Arnelle DR, Stamler JS. Arch. Biochem. Biophys. 1995;318:279. doi: 10.1006/abbi.1995.1231. [DOI] [PubMed] [Google Scholar]
- 14.Ahmad KF, et al. Mol. Cell. 2003;12:1551. doi: 10.1016/s1097-2765(03)00454-4. [DOI] [PubMed] [Google Scholar]
- 15.Li X, et al. Cancer Res. 1999;59:5275. [PubMed] [Google Scholar]
- 16.Wang G, Moniri NH, Ozawa K, Stamler JS, Daaka Y. Proc. Natl. Acad. Sci. U.S.A. 2006;103:1295. doi: 10.1073/pnas.0508354103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laloi C, Mestres-Ortega D, Marco Y, Meyer Y, Reichheld JP. Plant Physiol. 2004;134:1006. doi: 10.1104/pp.103.035782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reichheld J-P, Mestres-Ortega D, Laloi C, Meyer Y. Plant Physiol. Biochem. 2002;40:685. doi: 10.1104/pp.103.035782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Motohashi K, Kondoh A, Stumpp MT, Hisabori T. Proc. Natl. Acad. Sci. U.S.A. 2001;98:11224. doi: 10.1073/pnas.191282098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reichheld JP, et al. Plant Cell. 2007;19:1851. doi: 10.1105/tpc.107.050849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Besson-Bard A, Pugin A, Wendehenne D. Annu. Rev. Plant Biol. 2008;59:21. doi: 10.1146/annurev.arplant.59.032607.092830. [DOI] [PubMed] [Google Scholar]
- 22.Grun S, Lindermayr C, Sell S, Durner J. J. Exp. Bot. 2006;57:507. doi: 10.1093/jxb/erj053. [DOI] [PubMed] [Google Scholar]
- 23.Romero-Puertas MC, et al. Plant Cell. 2007;19:4120. doi: 10.1105/tpc.107.055061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reynaert NL, et al. Proc. Natl. Acad. Sci. U.S.A. 2004;101:8945. doi: 10.1073/pnas.0400588101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.