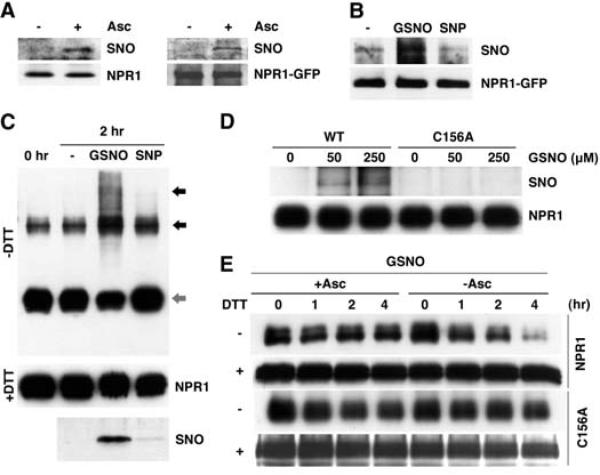
Fig. 2.
S-nitrosylation of Cys156 facilitates the assembly of NPR1 oligomer. (A) SA induces S-nitrosylation of endogenous NPR1 and the NPR1-GFP proteins in vivo. Sodium ascorbate (Asc) was used to specifically detect S-nitrosylated (SNO) NPR1. Equal loading was verified with antibodies against NPR1 or NPR1-GFP. (B) GSNO, but not mock (–) or SNP treatment, induces S-nitrosylation of NPR1-GFP in plant extracts. S-nitrosylated NPR1-GFP was detected with the biotin-switch assay. An antibody against NPR1-GFP was used to verify equal loading. (C) GSNO, but not mock (–) or SNP treatment, induces S-nitrosylation and multimerization (black arrows) of recombinant His6-NH (NPR1 residues 1 to 246) monomer (gray arrow). Equal loading was verified with an antibody to NPR1. (D) Cys156 is the principal site of S-nitrosylation in NPR1. Recombinant His6-NH and His6-NH-C156A proteins were incubated with different GSNO concentrations, and S-nitrosylation was detected by the biotin-swich assay. Equal loading was verified with an antibody to NPR1. (E) The C156A mutation impairs GSNO-induced oligomerization. Recombinant His6-NH and His6-NH-C156A proteins were treated with GSNO and with (+) or without (–) sodium ascorbate. Subsequently, monomers were allowed to re-oligomerize for the indicated times. Monomeric (–DTT) and total (+DTT) proteins were detected with an antibody to NPR1.