Abstract
Colouration patterns have an important role in adaptation and speciation. The European crow system, in which all-black carrion crows and grey-coated hooded crows meet in a narrow hybrid zone, is a prominent example. The marked phenotypic difference is maintained by assortative mating in the absence of neutral genetic divergence, suggesting the presence of few pigmentation genes of major effect. We made use of the rich phenotypic and genetic resources in mammals and identified a comprehensive panel of 95 candidate pigmentation genes for birds. Based on functional annotation, we chose a subset of the most promising 37 candidates, for which we developed a marker system that demonstrably works across the avian phylogeny. In total, we sequenced 107 amplicons (∼3 loci per gene, totalling 60 kb) in population samples of crows (n=23 for each taxon). Tajima's D, Fu's FS, DHEW and HKA (Hudson–Kreitman–Aguade) statistics revealed several amplicons that deviated from neutrality; however, none of these showed significantly elevated differentiation between the two taxa. Hence, colour divergence in this system may be mediated by uncharacterized pigmentation genes or regulatory regions outside genes. Alternatively, the observed high population recombination rate (4Ner∼0.03), with overall linkage disequilibrium dropping rapidly within the order of few 100 bp, may compromise the power to detect causal loci with nearby markers. Our results add to the debate as to the utility of candidate gene approaches in relation to genomic features and the genetic architecture of the phenotypic trait in question.
Keywords: speciation, population genetics, bird colouration, pigmentation genes, recombination rate, linkage disequilibrium
Introduction
The diversity of animal colouration has long attracted the interest of naturalists and scientists alike. Many instances of colouration differences across and within species have been shown to be a function of habitat adaptation (Hubbard et al., 2010), while in others it may reflect the outcome of seemingly arbitrary sexual and social selection pressures (Ödeen and Björklund, 2003; Price, 2007). In either case, colouration differences within and between populations have been observed to arise rapidly and are believed to have an active role in the initial steps of divergence and speciation (Gray and McKinnon 2007; Hugall and Stuart-Fox 2012).
Birds show remarkable variation in colour and colouration patterns, including within-population polymorphisms (Galeotti et al., 2003), clinal variation (Antoniazza et al., 2010) and rapidly evolved differences between populations and species (Milá et al., 2007). The notion that colour variation can promote speciation may be particularly relevant in birds, as mate preferences are mostly learned through imprinting on parental phenotypes (Ten Cate et al., 1993). As a consequence, barriers to gene flow can quickly arise without the need for genetic coupling between mating trait and preference (Irwin and Price, 1999). One prominent example where colour differences in combination with imprinting have been invoked in the speciation process is the marked phenotypic difference between two parapatric Eurasian crow taxa, the all-black carrion crow (Corvus (corone) corone) and the grey-coated hooded crow (Corvus (corone) cornix) (Figure 1) (Brodin and Haas, 2006).
Figure 1.
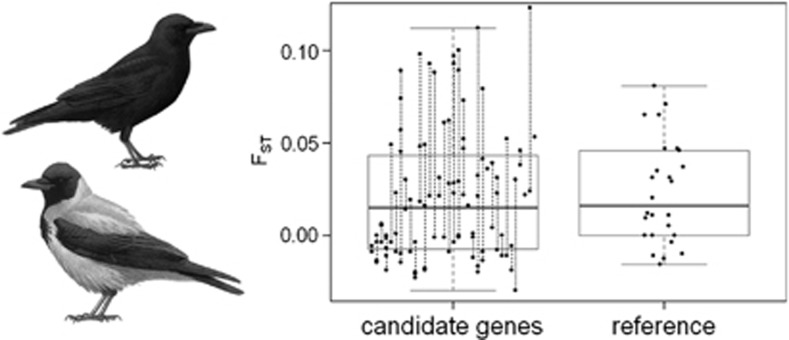
Genetic differentiation between carrion crows (all-black) and hooded crows (grey-coated). Boxplot of FST-values across 107 amplicons (dots) derived from 37 candidate genes, with vertical lines connecting amplicons from a single gene. Twenty-five random sequences from non-colour-related genes are given as a reference. Boxes include the second and third quantile; whiskers extend to the most extreme data points that lie within 1.5 times the interquartile range from the box. On the left hand side: a hooded crow (bottom) and a carrion crow (after Mullarney et al. 1999).
The two taxa, which have recently been raised to species status (Parkin et al., 2003), are geographically distributed in a leapfrog pattern, with carrion crows inhibiting western Europe and eastern Asia and hooded crows the area in between. Both in Europe and Asia, the taxa meet in hybrid zones thought to originate by secondary contact after initial isolation (Meise, 1928). These hybrid zones are narrow (50–160 km, Meise, 1928) and fairly stable, although both in Scotland and Denmark, modest hybrid zone movement to the expense of hooded crows has been described (Cook, 1975; Haas and Brodin, 2005). The marked phenotypic divergence is accompanied by assortative mating (for example, Haas et al., 2010), yet stands in contrast to limited evidence for postzygotic reproductive isolation (for example, Saino and Bolzern, 1992) and very low levels of genetic differentiation in all the genetic markers studied so far (Haas et al., 2009; Wolf et al., 2010 and references therein). Levels of between taxon separation are not distinguishable from population structure within taxa, which is largely governed by a subtle isolation-by-distance pattern (Haas et al., 2009). The prime candidate for a major reproductive barrier is the difference in colouration and is supported as such both by assortative mating and theoretical modelling (Brodin and Haas, 2009). The genetic basis of these colour differences would thus make for a prime candidate of (a) speciation gene(s) with large effect.
The genetic basis for melanin-mediated plumage colouration in birds can be assumed to follow the general pattern seen in other vertebrates (Hill and McGraw, 2006). Genetic evidence for the regulation of melanin-based plumage colouration in wild populations is, to date, mostly restricted to one gene, the melanocortin 1 receptor (Mc1r) (for example, Mundy et al., 2004). However, rich resources in vertebrate model species such as mice should allow identification of other genes in species systems for which there is no evidence of an involvement of Mc1r (such as in crows, Haas et al., 2009). Research in laboratory mice has uncovered the identity of many genes in the melanin pigmentation pathway (Bennett and Lamoreux, 2003; Hoekstra, 2006), many of which have the potential to alter pigmentation phenotypes. These include upstream signalling receptors, such as Ednrb, and less pleiotropic trans-membrane proteins like Kit (and Mc1r) and their immediate regulators (for example, agouti-signalling protein, Pomc, Kitlg). Downstream elements like Tyr, Tyrp1, Dct and Pmel17 are directly involved in eumelanin synthesis, while an important intermediate transcription factor is Mitf. Finally, genes acting, for instance, during melanoblast maturation and melanocyte development (for example, Adamts20, Muted, Mitf) or that are involved in melanosome transport and melanin deposition into keratinocytes (for example, Rab27a, Mlph) can also generate phenotypic effects (Nascimento et al., 2003).
To be able to study the genetic basis of the colour differences among carrion crows and hooded crows, we sequenced multiple regions of 37 prime pigmentation candidate genes in both taxa. We evaluate levels of genetic variation within and between species, perform outlier detection screens and quantify linkage disequilibrium (LD), which is instrumental in evaluating the utility of a candidate gene approach and devising future strategies for unravelling the genetic architecture of this hybrid zone. Our primer set was also tested for amplification in eight other species across the avian phylogeny with good success and will allow targeting of melanin pigmentation genes in most other avian species.
Materials and methods
Gene selection and sequence acquisition
We screened the Mouse Genome Informatics database (http://www.informatics.jax.org/) and extracted 142 candidate genes involved in pigmentation (pigmentation phenotype (MP:0001186)). We then identified 1:1 orthologous genes with the chicken and zebra finch genomes for a total of 95 genes from BIOMART (Ensembl 64; Supplementary Table S1). To reduce this set of genes to one of a feasible size for sequencing, we prioritized the most promising candidate genes by (1) the presumed phenotype to which the gene contributes in mouse, (2) the degree of pleiotropic effects of mutations in these genes in mouse (low degree preferred), and (3) whether genes are implicated in pigmentation changes in other vertebrate species. Details for these three selection criteria for each of the 95 genes, with extensive references for known mutations causing pigmentation changes outside of mice, can be found in Supplementary Table S1. Because carrion and hooded crows are coloured all-black and grey-pied, respectively, genes with mutations that caused tissue-specific pigmentation changes and complete melanisation were prioritized over, for example, genes with mutations that completely inhibited pigmentation or that cause very localized pigmentation aberrations, for example, in eyes or toe pads. This selection procedure resulted in a prime candidate set of 37 genes (Supplementary Table S2).
For each of these genes, we designed consensus anchored primers at multiple locations spread across the gene (Supplementary Tables S2 and S3) based on alignments of chicken and zebra finch sequences, using the Primer3 software (Rozen and Skaletsky, 1998). When the orthologue for only one bird species was known, we designed primers based on that species alone. Because of the much greater evolutionary conservation of exons, all primers were based on exonic sequences and targeted mainly the enclosed introns. We attempted to develop multiple amplicons per gene, well spaced across the gene. We also designed primers for nearby (on average 14 kb) upstream and/or downstream genes to capture nucleotide variation in potential cis-regulatory regions (17 amplicons near 14 candidate genes; see Supplementary Table S2). Part of the analyses were done on a gene-by-gene basis, and in such cases, nearby up- or downstream were included with the focal gene and are together referred to as a candidate gene unit.
PCR amplification and cross-specific amplification across the bird phylogeny
Primer pairs were initially tested on a DNA mixture of several individuals using 10 μl reactions containing 1 μl diluted DNA (at a concentration of at least 20 ng μl−1), 1 μl PCR buffer, 2.5 mM MgCl2, 0.2 μM of both the forward and reverse primer, 0.05 mM dNTP and 0.05 U μl−1 Amplitaq Gold DNA polymerase. PCR was conducted at the following basic touch-down heating scheme: (1) 10 min at 95 °C; (2) 20 cycles of (a) 30 s at 95 °C, (b) 45 s at 65 °C at cycle 1 min—0.5 °C at every step (that is, down to 55.5 °C), and (c) 60 s at 72 °C; (3) 20 cycles of (a) 30 s at 95 °C, (b) 45 s at 55 °C, and (c) 60 s at 72 °C; and (4) 5 min at 72 °C. Annealing temperatures were in several cases varied from the basic scheme to optimize amplification of the targeted amplicon. Loci that amplified properly were first sequenced on 4–8 individuals to check sequence quality. When sequences from test runs were of sub-optimal quality, we designed new species-specific primers from these crow sequences.
In total, we designed 176 primer pairs located within or near 37 different candidate colouration genes. Of the 176 primer pairs, 123 (69.9%) could be PCR amplified such that only a single clear band was consistently visible on agarose gels. Of those, 107 could be successfully sequenced with an average sequence length of 563 bp (for details, see below). Because crow and rook amplicon sequences could also be unequivocally mapped back to zebra finch using BLAST, we are confident that the amplicons represent the genes of interest. All amplicons expected to be Z-linked did not have heterozygotic sites in females, suggesting that they are also Z-linked in crows, which is consistent with a high degree of synteny found in other studies (Wolf and Bryk, 2011).
Final sequencing for each of the 107 amplicons was performed on a panel of 23 hooded crows, 23 carrion crows, and 2 rooks (Corvus frugilegus) that were used as the outgroup. All carrion crows were sampled in western Germany: 10 near Kleve (decimal degrees: 51.75N, 6.24E), 8 near Dortmund (51.57N, 7.38E), 2 near Bonn (50.66N, 6.79E), and 2 near Düsseldorf (51.26N, 6.68E). Twenty-one of the hooded crows were sampled in Poland, all near Warsaw (52.23N, 21.01E), one was sampled in Ireland, near Belfast (54.31N, 5.62W), and one in Sweden, near Uppsala (59.93N, 17.76E). On average, 44 (of the 46) crows were successfully sequenced for each amplicon.
All sequencing of PCR products was performed with traditional dye-terminator sequencing on an ABI 3730 XL instrument (Applied Biosystems, Life Technologies Ltd, Paisley, UK). Primer sequences and annealing temperatures are given in Supplementary Table S3. All sequences have been deposited in GenBank (accession numbers KF235895–KF240559).
An avian pigmentation candidate gene set
We tested PCR amplification of our primer pairs in a panel of species spread across the avian phylogeny. In cases where final sequencing primers for the crows were not the initial primers designed from zebra finch and chicken sequences, we tested the latter primers (listed in Supplementary Table S4). We used willow grouse (Lagopus lagopus, Galliformes), grey heron (Ardea cinerea, Pelecaniformes), peregrine falcon (Falco peregrinus, Falconiformes), herring gull (Larus argentatus, Charadriiformes), wood pigeon (Columba palumbus, Columbiformes) and three species of passerines (Passeriformes) from different families: pied flycatcher (Ficedula hypoleuca), blue tit (Cyanistes caeruleus), and reed bunting (Emberiza schoeniclus). Successful amplification was reported when one clear single band was visible in PCR products, with no or little non-specific amplification.
Cross-specific amplification success was high and is reported in Supplementary Table S4. On average, 76.8% of primer pairs tested amplified successfully in the eight species. Reflecting phylogenetic relationships, amplification success was better for the three passerine species (mean 87.4%) than for more divergent non-passerines (mean 70.4%). We therefore predict that this set can be used as a valuable resource for investigating the genetic basis of colour differences across most bird species.
Basic sequence analysis
Sequences were aligned using Sequencher 4.6 (Gene Codes, Ann Arbor, MI, USA) and CodonCode Aligner 3.0.1 (CodonCode Corp., Centerville, MA, USA) and exported as multifasta files. The diploid sequences were phased using the PHASE algorithm (Stephens et al., 2001) as implemented in DnaSP v5 (Librado and Rozas, 2009). Sequence diversity (π and Watterson's θ) and population differentiation estimates (FST, (Hudson et al., 1992) and exact tests of population differentiation) were computed for each amplicon in DnaSP and Arlequin 5 (Excoffier and Lischer, 2010). Recombination was tested for using the four-gamete test as implemented in DnaSP.
Outlier analysis
We initially used four FST outlier detection methods to formally evaluate the evidence for outlier loci. Genetic variation that contributes to the colour differences between hooded and carrion crows, or variation linked to such causal variation, would be expected to show elevated levels of differentiation relative to the rest of the genome and should therefore be picked up by FST outlier methods.
Firstly, we used the Bayesian method BayesFst (Beaumont and Balding, 2004), which separately estimates locus effects, population effects and the interaction between the two. Secondly, we used BayeScan (http://www-leca.ujf-grenoble.fr/logiciels.htm, Foll and Gaggiotti, 2008), a Bayesian method that directly estimates the posterior probability of each locus belonging to either a model including or a model excluding the effects of selection. Thirdly, we employed Fdist (Beaumont and Nichols, 1996) as implemented in the software Lositan (http://popgen.eu/soft/lositan, Antão et al., 2008). However, Lositan gave inconsistent results among runs and often supposedly detected tens of single-nucleotide polymorphisms (SNPs) both under balancing and positive selection, including in the latter category SNPs with an FST as low as 0.044. We also attempted to use the method of Excoffier et al. (2009) implemented in Arlequin 5 (Excoffier and Lischer, 2010), which is designed to reduce false positives by taking hierarchical population structure into account. However, this method was not able to analyse the data set, potentially because of the limited genetic differentiation present (L. Excoffier personal communication). Results are only reported for the first two approaches.
Neutrality tests
Neutrality tests were performed both on samples of hooded and carrion crows separately (to detect loci under selection in only one subspecies) as well as on samples of both taxa pooled together (to detect loci under similar selection in both subspecies). First, we used several tests that interrogate the allele frequency spectrum for each amplicon separately. We calculated Tajima's D and Fu's FS using Arlequin 5 (Excoffier and Lischer, 2010); statistical significance was assessed using 10 000 simulations. Besides identifying loci under selection, these tests are sensitive to past demographic changes, because such fluctuations perturb the allele frequency spectrum. DHEW is a recently developed compound test for neutrality. It combines Tajima's D, Fay and Wu's H and the Ewens–Watterson test, and is more robust against demography than any of these tests individually (Zeng et al., 2007). We computed the significance of the DHEW test using DH.jar, kindly provided by K. Zeng.
Second, we used two versions of the Hudson–Kreitman–Aguade (HKA) test (Hudson et al., 1987). This test compares levels of within-species polymorphism to between-species divergence to test for deviations from neutral sequence evolution. A rook (C. frugilegus) was used as the outgroup. All amplicons were analyzed simultaneously using the HKA software (http://genfaculty.rutgers.edu/hey/software#HKA), with a modification to the source code for handling more than 100 loci in one analysis. The HKA software only provides a significance value for the entire sample of loci and not for individual loci. We therefore tested amplicons with a deviation of at least 2.0 from neutrality in a HKA test using mlHKA (Wright and Charlesworth, 2004), which allows for explicit testing of candidate non-neutral loci against a sample of neutral loci.
As a ‘neutral' reference unrelated to pigmentation, we used sequence data from 25 random amplicons distributed across the genome as described in Wolf et al. (2010).
LD
LD was calculated as D' and r2 between pairs of variable sites within candidate gene units using DnaSP. Only sites with a minor allele frequency of >0.1 were used, as LD is best estimated using high-frequency polymorphisms (Reich et al., 2001). The analyses were performed for hooded and carrion crows jointly as well as separately. To avoid effects of population structure, individuals from outlying populations were excluded: two Irish and one Swedish hooded crow, and four carrion crows from Kleve and Bonn. Because we were interested in the decay of LD with physical distance also beyond our amplicon size (mostly<1 kb), we estimated LD between all sites within the same candidate gene unit (that is, all amplicons from one candidate gene as well as from potentially targeted nearby up- or downstream genes). For this, we first concatenated and then phased the sequences per candidate gene unit. We then estimated the distances between sites from different amplicons using the physical distances between the orthologous sites in the zebra finch genome according to Ensembl (based on the locations of the primer sequences used). Referring to physical distance in zebra finch is a fair assumption as divergent bird genomes have a high degree of synteny (Ellegren, 2010), and intron length is highly correlated between crow and zebra finch (r2=0.93, P<0.001, Wolf and Bryk, 2011). We calculated population recombination rates (ρ=4Ner for autosomal loci and 3Ner for Z-linked loci, with r being the per site recombination rate) for each gene separately using the software LDhat 2.0 (McVean et al., 2004) and MAXDIP (Witonsky and Di Rienzo, 2012). Predictions of r2 based on the estimate of ρ by LDhat were fitted using Equation 3 in Weir and Hill (1986).
Results
General patterns of nucleotide diversity
We obtained high-quality sequences in population samples of carrion and hooded crows (and two rooks as the outgroup) for 107 amplicons located within or near 37 candidate melanin pigmentation genes. Ninety of these amplicons were located in the candidate colouration genes themselves and the remainder in adjacent up- or downstream genes.
In total, our sequencing effort amounted to 60 213 bp for each individual, and we detected 535 polymorphic sites in the entire data set consisting of 23 birds from each taxon. Population genetic summary statistics for all amplicons can be found in Supplementary Table S5. Average nucleotide diversity π across loci (weighted by sequence length) was 0.00132, being slightly higher in carrion crows (0.00134) than in hooded crows (0.00120) (paired t-test: P=0.002). Watterson's θ per site was also higher in carrion crows (0.00174) than in hooded crows (0.00158) but not significantly so (paired t-test: P=0.239). Diversity in the three sex chromosome-linked genes (10 amplicons; average π=0.00112; average θ=0.00140) was not significantly different from the autosomes (t-test for π: P=0.540; t-test for θ: P=0.422). Finally, 41 insertion-deletion (indel) polymorphisms were detected (giving an indel to SNP ratio of ∼0.077) and were excluded from further analysis. Fixed differences between rooks and crows were numerous, with an average net sequence divergence per site (Da, Nei, 1987) of 0.006.
Population differentiation and outlier tests
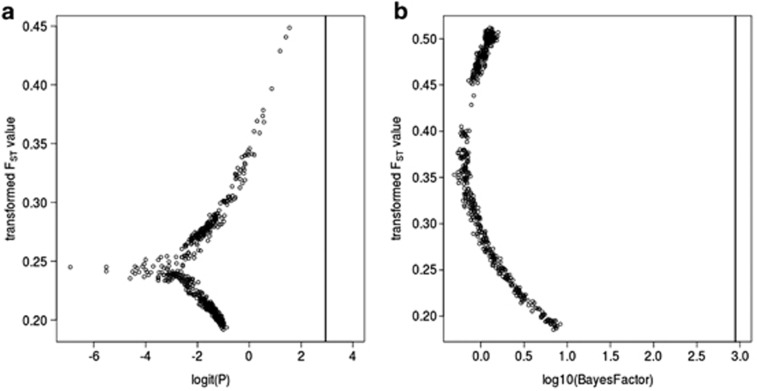
The average value of FST across the candidate pigmentation loci was generally low at 0.0209 (s.d.=0.0360, range: −0.030 to 0.119) and was not distinguishable from the average value across 25 random amplicons from Wolf et al. (2010) (0.0230, t-test: P=0.6244; Figure 1). The exact and chi-square tests of population differentiation were significant for 28 and 24 (out of 107) amplicons, respectively. Only four amplicons, all in different candidate genes, had an FST of ⩾0.1: Cno_2i (FST=0.119), Slc45a2_2 (FST=0.118), Slc24a5_1i (FST=0.111), and Kit_4i (FST=0.100). There were no fixed differences between carrion crows and hooded crows, and 290 of the 535 SNPs were shared between the two taxa. In all, 142 SNPs were unique to carrion crows, and 103 were unique to hooded crows. Average FST of sex chromosome-linked loci was 0.0281 (SD 0.0357), which is not significantly higher than that of the autosomes (Mann–Whitney U-test: P=0.288). Results from BayesFst and BayeScan both indicated a lack of significant outliers (Figure 2). In summary, we found no strong evidence for any of the candidate genes explaining differences in colouration between carrion crow and hooded crow.
Figure 2.
Results of FST outlier differentiation tests with bayesFst (a) and bayeScan (b). In both panels, the transformed FST-value for each SNP is shown, with transformed p-values on the x axis, and a vertical bar indicating significance equivalent to P=0.05. The logit function in panel (a) is logit(x)=log(x/(1−x)), see Beaumont and Balding (2004).
Neutrality tests
Tajima's D, Fu's FS and DHEW tests were carried out for all 107 amplicons and all three population configurations: (1) all crow samples together (ALL), (2) all carrion crows (CC), and (3) all hooded crows (HC). Test results are reported in Supplementary Table S5.
On average, Tajima's D was negative for all population configurations: −0.507 for ALL, −0.458 for CC, and −0.313 for HC. The difference in Tajima's D values between CC and HC was significant (paired t-test: P=0.021). Tests for seven amplicons were significantly different from zero using P<0.05 (4 All, 1 CC and 2 HC). All of these had a Tajima's D value smaller than zero, and none was significant after Bonferroni's correction.
For Fu's FS, average values were also well below zero: −2.055 for ALL, −1.451 for CC, and −0.981 for HC, and again, the difference in Fu's FS values between CC and HC was significant (paired t-test, 0.003). Fifty-two tests were significantly different from zero using P<0.05 (24 ALL, 16 CC and 12 HC), and 9 remained so after Bonferroni's correction (5 ALL, 3 CC, 1 HC). All significant tests had a negative value of Fu's FS. Two of the loci with significant Fu's FS after correction also had significant (but not after correction) Tajima's D: Asip_2b and Tpcn2_4i.
For DHEW, only five tests were significant at P<0.05: 1 for ALL, 3 for CC, and 1 for HC, and one test (for CC; Rab27a_1) remained significant after Bonferroni's correction. Rab27a_1, however, had a negative FST, and as a whole, significant DHEW tests did not overlap (same population and amplicon) with significant Tajima's D and Fu's FS tests. The HKA test, conducted with all amplicons simultaneously and separately for HC and CC, gave an overall non-significant result in both the cases. Nonetheless, as the significance of the test depends on the summed deviation from neutral expectations across many amplicons, non-neutrality of only a few amplicons might not be picked up. Using the mlHKA software to examine amplicons with a deviation of at least 2.0 in the original HKA test, Hps4_u2 (in a gene downstream of Hps4) was significantly non-neutral for both HC and CC and Pomc1_u1 (in a gene upstream of Pomc1) only for CC. However, for both amplicons, the amount of within-species polymorphism relative to between-species divergence exceeds the neutral expectation, which would suggest the potential influence of balancing rather than directional selection.
Recombination and LD
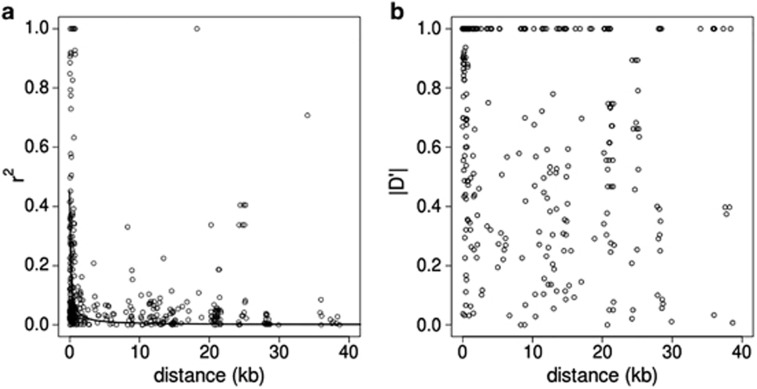
Using the four-gamete test, a total of 46 recombination events were detected within 36 amplicons (that is, in 33.6% of all amplicons). A total of 404 pairwise combinations of SNPs located within the same candidate gene unit (several amplicons but same candidate gene) had a minor allele frequency larger than 0.1 and were analyzed for LD. Of these, 144 (35.6%) were in significant LD using Fisher's test, and the average r2 was 0.118. Employing a common definition of useful LD as an r2 of at least 0.3 (Ardlie et al., 2002), useful LD was detected for 47 of all 404 (11.6%) pairwise combinations.
In Figure 3, r2 and D′ values are plotted against the expected physical distance, showing low levels of LD even at short physical distances. Nonetheless, LD also decays rapidly with physical distance. Compared with overall levels mentioned above, of 190 pairwise combinations within 1 kb of each other (the large majority of which are within the same amplicon), 96 (50.5%) were in significant LD, 37 (19.5%) in useful LD and the average r2 was 0.186. Even within this 1-kb range, decay of LD with distance is evident as shown in Supplementary Figure S1.
Figure 3.
Patterns of LD. (a) r2-Values and (b) absolute D′-values between pairs of SNPs plotted against the estimated physical distance between them. The line in panel (a) shows predictions of LD based on the value of ρ as estimated by LDhat for our data.
Analyses for hooded and carrion crows separately revealed slightly higher levels of LD in the carrion crow (average r2 0.182, 18.5% of comparisons in useful LD) than in hooded crows (average r2 0.139, 11.6% of comparisons in useful LD). LD between Z-linked sites was similar to the overall pattern: the average r2 over 24 pairwise combinations was 0.158, with 10 pairs (41.7%) in significant LD and 3 pairs (12.5%) in useful LD.
The average population recombination rate (ρ) was estimated to be 0.017/site/generation (SE across genes: 0.0058) by LDhat, whereas MAXDIP arrived at an estimate of 0.038/site/generation (SE across genes: 0.0155; see Supplementary Table S6).
Discussion
Candidate gene approaches have been successfully applied to address evolutionary questions in a variety of systems. Pigmentation genes are of prime interest as the colouration they influence is easy to observe and study, can be subject to strong natural (Linnen et al., 2009; Van't Hof et al., 2011) or sexual selection (Uy et al., 2009), and shows a clear relationship to speciation (Hubbard et al., 2010). Melanin pigmentation genes, in particular, are well suited for candidate gene approaches, as vertebrate melanogenesis is a well-characterized pathway in which many mutations with large effects have been identified.
Given the rich knowledge on this pathway, it is somewhat surprising that these approaches have generally been restricted to a very limited number of candidate genes. Here we chose a system where colouration genes are likely important in speciation and undertook an extensive candidate gene approach. We compiled a list of avian melanin pigmentation genes and sequenced, on average, three amplicons per gene in 37 of the most promising candidates of two closely related crow taxa with strikingly different colouration. We quantified levels of diversity, differentiation and LD and used this information in the context of crow speciation genetics and, more generally, as a critical test of candidate gene approaches in wild populations.
Differentiation and signatures of selection
We performed a suite of tests to identify genetic variation that may be associated with the plumage differences between all-black carrion crows and grey-coated hooded crows. Because of the fixed differences in plumage colouration and the very low background levels of genetic differentiation (average FST of 0.012), loci linked to causal variants should be easily detectable as differentiation outliers. However, no clear FST outliers were found by either of the Bayesian approaches we used. Several other tests, such as Tajima's D, DHEW and the HKA test, can detect non-neutral patterns of variation that would be expected if the plumage differences arose by selection. Although a few tests were significant, the weak signals and the lack of congruence between different tests points towards weak effects or false positives. Thus, we could not find support for any of the analyzed pigmentation genes to be involved with the phenotypic divergence between the two crow taxa.
Population genetic inferences
Deviations from neutrality are more likely to have been caused by demographic processes, as is suggested by the observation that Tajima's D and Fu's FS values were consistently negative across loci in both carrion crows and hooded crows. The signal was stronger for Fu's FS, which is known to be more sensitive to demographic change. These findings are thus consistent with a recent increase in population size, as has been inferred for many species in temperate regions (Hewitt, 2004), and likely reflects range expansion and population growth after the last glaciation. Interestingly, carrion crows showed a stronger signature of expansion than hooded crows, which hints at different recent population histories despite the low level of differentiation.
LD and the power of candidate gene approaches
The study of LD in wild organisms is only in its infancy. Results from mammals and birds (Edwards and Dillon, 2004; Backström et al., 2006; Balakrishnan and Edwards, 2009; Li and Merila, 2010) show considerable variation in levels of background LD among taxa. Our estimates of the population recombination rate (average point estimates of 0.017 and 0.037/site/generation) are only slightly lower than those from other bird species (red-winged blackbird: 0.0588 (Edwards and Dillon, 2004), zebra finch: 0.051–0.08 (Balakrishnan and Edwards, 2009)), whereas being much higher than estimates for humans (0.0004, Evans and Cardon, 2005). Similarly, levels of LD measured in D′ or r2 are close to those in for example zebra finch (Balakrishnan and Edwards, 2009), but considerably lower than in humans. This is somewhat surprising, given that levels of nucleotide variation in crows are rather similar to those in humans (π∼0.001) and much lower than in zebra finch. This may be an indication for an increased recombination rate in crows and adds to the debate concerning broad-scale recombination rate conservation across taxa (Auton et al., 2012).
The data on LD not only constitutes a valuable addition to the study of recombination in wild populations but also lays the necessary groundwork to evaluate the power of candidate gene approaches under different evolutionary scenarios. The applied tests of selection described above only detect deviation from neutrality arising by selection and/or demographic processes.
If the causative variant(s) of plumage colour segregated in the ancestral population, and drifted to fixation in one taxon, the locus would effectively look neutral. Only very closely linked markers would have the power to detect a signal of enhanced differentiation for such ancestrally segregating variants and, similarly, for old and/or weak selection events that leave but small and temporal traces of extended LD (McVean, 2007). As we mostly sequenced intronic sequence, and r2 dropped below 0.3 (that is, useful LD, Ardlie et al., 2002) within a few hundred base pairs, the power to directly observe a causal mutation is therefore small in such cases.
When sequencing genes, the problem of linkage becomes particularly acute for phenotypic changes that come about by mutations in regulatory sequences. Recent studies have shown the prevalence of especially cis-regulatory modifications, although in many cases the precise identity of the changes have remained elusive (but see for example, Chan et al., 2010). Cis-regulatory elements are often located relatively nearby their focal genes, and we therefore also sequenced parts of nearby down- and upstream genes of several, especially promising, candidates. Nonetheless, the distances between potential causal promotor or enhancer sequences and sequenced parts of genes are likely to be larger than within the genes themselves and would require even higher levels of LD.
These considerations do not preclude the utility of candidate gene approaches in general, but may raise awareness under which scenarios they are most promising. In sufficiently diverged populations, admixture provides the necessary LD for trait mapping in laboratory or natural (hybrid zone) crosses (Winkler et al., 2010). In addition, LD extends considerably further in many domesticated, and potentially in selfing, organisms (Nordborg, 2000; Andersson and Georges, 2004), as has been exploited for dog pigmentation genetics (Candille et al., 2007). Alternatively, with the increasing availability of reference genomes, sequence capture methods can be combined with next-generation sequencing to conveniently cover the entirety instead of only parts of candidate genes, alleviating reliance on LD. Eventually, in cases of very low LD, whole-genome re-sequencing will be the method of choice. Here low levels of LD come as an advantage, allowing genetic fine-mapping of a trait. Under this scenario, candidate genes can be very helpful in further narrowing of functional elements in candidate regions.
Data archiving
Sequence data have been submitted to GenBank: accession numbers KF235895–KF240559.
Acknowledgments
We thank Gunilla Kärf, Laura Vossen, and Elke Bustorf for help in the lab. Andrzej Kruszewicz and Magdalena Gajownik kindly provided the Polish samples. The project was funded by Volkswagenstiftung Grant I/83 496, the Max-Planck Society and the Swedish Research Council (to JW), the European Research Council, the Swedish Research Council and the Wallenberg Foundation (to HE).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Author contributions
JP, HE and JW designed the study, JP and JW performed the research, JP analyzed the data and all authors wrote the paper.
Supplementary Material
References
- Andersson L, Georges M. Domestic-animal genomics: deciphering the genetics of complex traits. Nat Rev Genet. 2004;5:202–212. doi: 10.1038/nrg1294. [DOI] [PubMed] [Google Scholar]
- Antoniazza S, Burri R, Fumagalli L, Goudet J, Roulin A. Local adaptation maintains clinal variation in melanin-based coloration of European barn owls (Tyto alba) Evolution. 2010;64:1944–1954. doi: 10.1111/j.1558-5646.2010.00969.x. [DOI] [PubMed] [Google Scholar]
- Antão T, Lopes A, Lopes RJ, Beja-Pereira A, Luikart G. LOSITAN: a workbench to detect molecular adaptation based on a Fst-outlier method. BMC Bionformatics. 2008;9:323. doi: 10.1186/1471-2105-9-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardlie KG, Kruglyak L, Seielstad M. Patterns of linkage disequilibrium in the human genome. Nat Rev Genet. 2002;3:299–309. doi: 10.1038/nrg777. [DOI] [PubMed] [Google Scholar]
- Auton A, Fledel-Alon A, Pfeifer S, Venn O, Ségurel L, Street T, et al. A fine-scale chimpanzee genetic map from population sequencing. Science. 2012;336:193–198. doi: 10.1126/science.1216872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backström N, Qvarnström A, Gustafsson L, Ellegren H. Levels of linkage disequilibrium in a wild bird population. Biol Lett. 2006;2:435–438. doi: 10.1098/rsbl.2006.0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan CN, Edwards SV. Nucleotide variation, linkage disequilibrium and founder-facilitated speciation in wild populations of the zebra finch (Taeniopygia guttata) Genetics. 2009;181:645–660. doi: 10.1534/genetics.108.094250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont M, Balding D. Identifying adaptive genetic divergence among populations from genome scans. Mol Ecol. 2004;13:969–980. doi: 10.1111/j.1365-294x.2004.02125.x. [DOI] [PubMed] [Google Scholar]
- Beaumont M, Nichols R. Evaluating loci for use in the genetic analysis of population structure. Proc R Soc B Biol Sci. 1996;263:1619–1626. [Google Scholar]
- Bennett D, Lamoreux M. The color loci of mice—a genetic century. Pigment Cell Res. 2003;16:333–344. doi: 10.1034/j.1600-0749.2003.00067.x. [DOI] [PubMed] [Google Scholar]
- Brodin A, Haas F. Speciation by perception. Anim Behav. 2006;72:139–146. [Google Scholar]
- Brodin A, Haas F. Hybrid zone maintenance by non-adaptive mate choice. Evol Ecol. 2009;23:17–29. [Google Scholar]
- Candille SI, Kaelin CB, Cattanach BM, Yu B, Thompson DA, Nix MA, et al. A beta-defensin mutation causes black coat color in domestic dogs. Science. 2007;318:1418–1423. doi: 10.1126/science.1147880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YF, Marks ME, Jones FC, Villarreal G, Jr., Shapiro MD, Brady SD, et al. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science. 2010;327:302–305. doi: 10.1126/science.1182213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook A. Changes in the carrion/hooded crow hybrid zone and the possible importance of climate. Bird Study. 1975;22:165–168. [Google Scholar]
- Edwards SV, Dillon M. Hitchhiking and recombination in birds: evidence from Mhc-linked and unlinked loci in red-winged blackbirds (Agelaius phoeniceus) Genet Res. 2004;84:175–192. doi: 10.1017/s0016672304007189. [DOI] [PubMed] [Google Scholar]
- Ellegren H. Evolutionary stasis: the stable chromosomes of birds. Trends Ecol Evol. 2010;25:283–291. doi: 10.1016/j.tree.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Evans DM, Cardon LR. A comparison of linkage disequilibrium patterns and estimated population recombination rates across multiple populations. Am J Hum Genet. 2005;76:681–687. doi: 10.1086/429274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Hofer T, Foll M. Detecting loci under selection in a hierarchically structured population. Heredity. 2009;103:285–298. doi: 10.1038/hdy.2009.74. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer H. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resources. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Foll M, Gaggiotti O. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics. 2008;180:977–993. doi: 10.1534/genetics.108.092221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galeotti P, Rubolini D, Dunn PO, Fasola M. Colour polymorphism in birds: causes and functions. J Evol Biol. 2003;16:635–646. doi: 10.1046/j.1420-9101.2003.00569.x. [DOI] [PubMed] [Google Scholar]
- Gray SM, McKinnon JS. Linking color polymorphism maintenance and speciation. Trends Ecol Evol. 2007;22:71–79. doi: 10.1016/j.tree.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Haas F, Brodin A. The crow Corvus corone hybrid zone in southern Denmark and northern Germany. Ibis. 2005;147:649–656. [Google Scholar]
- Haas F, Knape J, Brodin A. Habitat preferences and positive assortative mating in an avian hybrid zone. J Avian Biol. 2010;41:237–247. [Google Scholar]
- Haas F, Pointer MA, Saino N, Brodin A, Mundy NI, Hansson B. An analysis of population genetic differentiation and genotype-phenotype association across the hybrid zone of carrion and hooded crows using microsatellites and MC1R. Mol Ecol. 2009;18:294–305. doi: 10.1111/j.1365-294X.2008.04017.x. [DOI] [PubMed] [Google Scholar]
- Hewitt GM. Genetic consequences of climatic oscillations in the Quaternary. Philos Trans R Soc London Ser B Biol Sci. 2004;359:183–195. doi: 10.1098/rstb.2003.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill G, McGraw K. Bird Coloration. I. Mechanisms and Measurements. Harvard University Press: Cambridge, MA, USA; 2006. [Google Scholar]
- Hoekstra HE. Genetics, development and evolution of adaptive pigmentation in vertebrates. Heredity. 2006;97:222–234. doi: 10.1038/sj.hdy.6800861. [DOI] [PubMed] [Google Scholar]
- Hubbard J, Uy J, Hauber M, Hoekstra HE, Safran R. Vertebrate pigmentation: from underlying genes to adaptive function. Trends Genet. 2010;26:231–239. doi: 10.1016/j.tig.2010.02.002. [DOI] [PubMed] [Google Scholar]
- Hudson RR, Boos DD, Kaplan NL. A statistical test for detecting geographic subdivision. Mol Biol Evol. 1992;9:138–151. doi: 10.1093/oxfordjournals.molbev.a040703. [DOI] [PubMed] [Google Scholar]
- Hudson RR, Kreitman M, Aguade M. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugall AF, Stuart-Fox D. Accelerated speciation in colour-polymorphic birds. Nature. 2012;485:631–634. doi: 10.1038/nature11050. [DOI] [PubMed] [Google Scholar]
- Irwin DE, Price TD. Sexual imprinting, learning and speciation. Heredity. 1999;82:347–354. doi: 10.1038/sj.hdy.6885270. [DOI] [PubMed] [Google Scholar]
- Li MH, Merila J. Extensive linkage disequilibrium in a wild bird population. Heredity. 2010;104:600–610. doi: 10.1038/hdy.2009.150. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Linnen CR, Kingsley EP, Jensen JD, Hoekstra HE. On the origin and spread of an adaptive allele in deer mice. Science. 2009;325:1095–1098. doi: 10.1126/science.1175826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean G. The structure of linkage disequilibrium around a selective sweep. Genetics. 2007;175:1395–1406. doi: 10.1534/genetics.106.062828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean G, Myers SR, Hunt S, Deloukas P, Bentley DR, Donnelly P. The fine-scale structure of recombination rate variation in the human genome. Science. 2004;304:581–584. doi: 10.1126/science.1092500. [DOI] [PubMed] [Google Scholar]
- Meise W. Die verbreitung der aaskrähe (Formenkreis Corvus corone L) J Für Ornithologie. 1928;76:1–206. [Google Scholar]
- Milá B, McCormack J, Castañeda G, Wayne R, Smith T. Recent postglacial range expansion drives the rapid diversification of a songbird lineage in the genus Junco. Proc R Soc B Biol Sci. 2007;274:2653–2660. doi: 10.1098/rspb.2007.0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullarney K, Svensson L, Zetterström D, Grant P.1999. Collins Bird Guide. Harper, Collins, London.
- Mundy NI, Badcock NS, Hart T, Scribner K, Janssen K, Nadeau NJ. Conserved genetic basis of a quantitative plumage trait involved in mate choice. Science. 2004;303:1870–1873. doi: 10.1126/science.1093834. [DOI] [PubMed] [Google Scholar]
- Nascimento Aa, Roland JT, Gelfand VI. Pigment cells: a model for the study of organelle transport. Annu Rev Cell Dev Biol. 2003;19:469–491. doi: 10.1146/annurev.cellbio.19.111401.092937. [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. Columbia University Press: New York, NY, USA; 1987. [Google Scholar]
- Nordborg M. Linkage disequilibrium, gene trees and selfing: an ancestral recombination graph with partial self-fertilization. Genetics. 2000;154:923–929. doi: 10.1093/genetics/154.2.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ödeen A, Björklund M. Dynamics in the evolution of sexual traits: losses and gains, radiation and convergence in yellow wagtails (Motacilla flava) Mol Ecol. 2003;12:2113–2130. doi: 10.1046/j.1365-294x.2003.01883.x. [DOI] [PubMed] [Google Scholar]
- Parkin DT, Collison M, Helbig A, Knox A, Sangster G. The taxonomic status of carrion and hooded crows. Br Birds. 2003;96:274–290. [Google Scholar]
- Price TD. Speciation in Birds. Roberts and Company: CO, USA; 2007. [Google Scholar]
- Reich DE, Cargill M, Bolk S, Ireland J, Sabeti PC, Richter DJ, et al. Linkage disequilibrium in the human genome. Nature. 2001;411:199–204. doi: 10.1038/35075590. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H.1998. Primer3. Code available at: http://www-genome wi.mit.edu/genome_software/other/primer3.html . Accessed at: http://biotools.umassmed.edu/bioapps/primer3_www.cgi .
- Saino N, Bolzern AM. Egg volume, chick growth and survival across a carrion/hooded crow hybrid zone. Boll Zool. 1992;59:407–415. [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ten Cate C, Vos DR, Mann N. Sexual imprinting and song learning; two of one kind. Neth J Zool. 1993;43:34–45. [Google Scholar]
- Uy JAC, Moyle RG, Filardi CE. Plumage and song differences mediate species recognition between incipient flycatcher species of the Solomon Islands. Evolution. 2009;63:153–164. doi: 10.1111/j.1558-5646.2008.00530.x. [DOI] [PubMed] [Google Scholar]
- Van't Hof A, Edmonds N, Dalíková M, Marec F, Saccheri IJ. Industrial melanism in British peppered moths has a singular and recent mutational origin. Science. 2011;332:958–960. doi: 10.1126/science.1203043. [DOI] [PubMed] [Google Scholar]
- Weir B, Hill W. Nonuniform recombination within the human beta-globin gene-cluster. Am J Hum Genet. 1986;38:776–778. [PMC free article] [PubMed] [Google Scholar]
- Winkler C, Nelson GW, Smith MW. Admixture mapping comes of age. Annu Rev Genomics Hum Genet. 2010;11:65–89. doi: 10.1146/annurev-genom-082509-141523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witonsky DB, Di Rienzo A.2012. MAXDIP http://genapps.uchicago.edu/maxdip/index.html .
- Wolf JBW, Bayer T, Haubold B, Schilhabel M, Rosenstiel P, Tautz D. Nucleotide divergence vs. gene expression differentiation: comparative transcriptome sequencing in natural isolates from the carrion crow and its hybrid zone with the hooded crow. Mol Ecol. 2010;19:162–175. doi: 10.1111/j.1365-294X.2009.04471.x. [DOI] [PubMed] [Google Scholar]
- Wolf JBW, Bryk J. General lack of global dosage compensation in ZZ/ZW systems? Broadening the perspective with RNA-seq. BMC Genomics. 2011;12:91. doi: 10.1186/1471-2164-12-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SI, Charlesworth B. The HKA test revisited: a maximum-likelihood-ratio test of the standard neutral model. Genetics. 2004;168:1071–1076. doi: 10.1534/genetics.104.026500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng K, Shi S, Wu C-I. Compound tests for the detection of hitchhiking under positive selection. Mol Biol Evol. 2007;24:1898–1908. doi: 10.1093/molbev/msm119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.