Abstract
The recurrent evolution of adaptive clines within a species can be used to elucidate the selective factors and genetic responses that underlie adaptation. White clover is polymorphic for cyanogenesis (HCN release with tissue damage), and climate-associated cyanogenesis clines have evolved throughout the native and introduced species range. This polymorphism arises through two independently segregating Mendelian polymorphisms for the presence/absence of two required components: cyanogenic glucosides and their hydrolyzing enzyme linamarase. Cyanogenesis is commonly thought to function in herbivore defense; however, the individual cyanogenic components may also serve other physiological functions. To test whether cyanogenesis clines have evolved in response to the same selective pressures acting on the same genetic targets, we examined cyanogenesis cline shape and its environmental correlates in three world regions: southern New Zealand, the central United States and the US Pacific Northwest. For some regional comparisons, cline shapes are remarkably similar despite large differences in the spatial scales over which clines occur (40–1600 km). However, we also find evidence for major differences in both the agents and targets of selection among the sampled clines. Variation in cyanogenesis frequency is best predicted using a combination of minimum winter temperature and aridity variables. Together, our results provide evidence that recurrent adaptive clines do not necessarily reflect shared adaptive processes.
Keywords: adaptive cline, geographic mosaic, cyanogenesis, epistatic selection, parallel evolution, plant–animal interaction
Introduction
Clinal variation, a shift in phenotypes or genotypes along an environmental gradient, is frequently used as a model for studying the process of adaptive differentiation (for example, Haldane, 1948; Antonovics and Bradshaw, 1970; Endler 1977; Barton and Hewitt, 1985). Adaptive clines arise when there is spatial variation in selection that maintains adaptive differentiation across a geographic range. Clines can be used as systems to bridge the gap between evolutionary ecology and population genetics, as they can potentially reveal the ecological forces that lead to maintenance of adaptive variation (Dobzhansky, 1947; Haak et al., 2012). However, it can be difficult to discern the causative selective factors that underlie cline formation (hereafter, agents of selection) because multiple ecological variables often covary along a single environmental gradient. Furthermore, the complex genotype–phenotype relationships that exist for many adaptive traits can complicate efforts to identify which genes or gene combinations (hereafter, targets of selection) are evolving in response to these selective forces (for example, see Templeton et al., 1976).
Clines that have evolved recurrently in geographically disparate areas of a species range can be viewed as multiple replicates of adaptive evolution (Oakeshott et al., 1982; Huey et al., 2000; Steiner et al., 2009). Comparisons of these independently evolved adaptive clines can help to overcome the difficulties associated with identifying the agents and targets of selection in a single cline. Specifically, the confounding effects of covariation between environmental variables along a single cline can be reduced by comparing clines that vary with respect to environmental covariance matrices (for example, see Schmidt et al., 2007; Keller et al., 2009). Similarly, targets of selection can potentially be elucidated by comparing the genotype–phenotype relationship among clines, where the null expectation is that selection has acted on the same target(s) of selection in these independent replicates (for example, Steiner et al., 2009). Population genetic theory provides the framework for evaluating whether the agents and targets of selection are similar in independent clinal replicates: similarities in the cline shape, that is, the dispersion of phenotypic and genotypic variation that occurs across a given range of an environmental gradient, suggest that both agents and targets of selection are conserved between clines (Mallet et al., 1990; Szymura and Barton, 1991). Differences in cline shape may indicate that either the agents of selection or targets of selection differ between clines.
Recurrent evolution of cyanogenesis clines in white clover
Cyanogenesis, the ability to produce hydrogen cyanide after tissue damage, is generally considered to be an evolved chemical defense against herbivores. In white clover (Trifolium repens L.), cyanogenesis occurs as a discrete polymorphism, with cyanogenic and acyanogenic plants co-occurring in many populations (Armstrong et al., 1913; Daday, 1954a, 1954b). The cyanogenesis polymorphism arises through two independently segregating biochemical polymorphisms for the presence or absence of two required components, cyanogenic glucosides (lotaustralin and linamarin) and their hydrolyzing enzyme (linamarase; Corkill, 1942). These two components are spatially separated within photosynthetic tissue, and cell rupture leads to their mixture and subsequent release of hydrogen cyanide. The presence/absence of cyanogenic glucosides corresponds to the classically defined gene Ac/ac, with the dominant Ac allele conferring their presence. The presence/absence of linamarase corresponds to the unlinked gene Li/li, with the dominant Li allele conferring its presence (reviewed in Hughes, 1991). For both genes, a homozygous recessive genotype corresponds to the absence of the cyanogenic component. Thus, four cyanogenesis phenotypes (or cyanotypes) are possible in white clover: plants with at least one dominant Ac and Li allele are cyanogenic (the ‘AcLi' cyanotype), whereas plants lacking a dominant allele at either Ac/ac, Li/li or both genes are acyanogenic (the ‘acLi', ‘Acli' and ‘acli' cyanotypes, respectively). The molecular basis of these classically defined genetic polymorphisms has recently been described; the Ac/ac polymorphism corresponds to a gene deletion polymorphism at CYP79D15, which encodes the cytochrome P450 responsible for the first step in cyanogenic glucoside biosynthesis (Olsen et al., 2008, 2013). Similarly, the Li/li polymorphism corresponds to a gene deletion polymorphism at the Li locus, which encodes the linamarase protein (Olsen et al., 2007, 2013).
The white clover cyanogenesis polymorphism is manifested spatially in a series of recurrent clines, with higher frequencies of cyanogenic individuals in warmer climates. These clines were initially documented as a latitudinal cline across white clover's native range in Eurasia and an altitudinal cline in the Swiss Alps (Daday, 1954a, 1954b). Over the last 60 years, similar clines have been documented elsewhere in the native range of the species (for example, see de Araujo, 1976; Till-Bottraud et al., 1988; Majumdar et al., 2004), as well as in non-native populations worldwide, where white clover has been introduced as a temperate forage crop and lawn plant (for example, Daday, 1958; Ganders, 1990; Kooyers and Olsen, 2012). In a recent study, we have documented an adaptive latitudinal cyanogenesis cline in introduced populations across the central United States, where the AcLi cyanotype frequency increases from ∼11% in Wisconsin to ∼86% in southern Louisiana (Kooyers and Olsen, 2012).
Agents and targets of selection in clover cyanogenesis clines
The agents of selection associated with this clinal variation are well studied but not fully understood. In many studies, cyanogenesis has been found to be an effective deterrent against generalist clover herbivores, including slugs and snails (for example, see Angseesing, 1974; Dirzo and Harper, 1982a, 1982b; Kakes, 1989), insects (Dritschilo et al., 1979; Pederson and Brink, 1998) and small mammals (Saucy et al., 1999; Viette et al., 2000). These findings suggest that cyanogenesis is adaptive in the presence of herbivores. At the same time, cyanogenesis appears to be energetically costly and disadvantageous in environments with low herbivore pressure; acyanogenic cyanotypes tend to show greater flower production than cyanogenic plants in the absence of herbivores, suggesting a selective advantage for acyanogenic plants under these conditions (Kakes, 1989). These two agents of selection—herbivore pressure and energetic costs of cyanogenesis—could together account for the observed clinal variation if cooler climates are characterized by lower herbivore abundance or shorter periods of herbivory, a pattern that has been observed in other temperate ecosystems (for example, see Pennings and Silliman, 2005). If these are the primary agents of selection, a simple prediction would be that cyanogenesis frequency should be most closely correlated with temperature-related variables that correlate with herbivore abundance in this system (specifically, minimum winter temperature (MWT)), and less closely correlated with other abiotic variables that show a weaker correlation with herbivore abundance (for example, annual precipitation (AP); Daday, 1954a).
However, these two proposed agents of selection are not universally supported by available data. Not all studies have found that cyanogenesis in white clover deters herbivores (Hruska, 1988; Hughes, 1991; Kooyers et al., unpublished observations), nor do all studies report fitness advantages for acyanogenic plants in the absence of herbivores (reviewed by Hughes, 1991). These observations raise the possibility that agents of selection other than herbivore pressure could be acting to maintain the cyanogenesis polymorphism. Consistent with this hypothesis, cyanogenic glucosides and their metabolites have been proposed to serve physiological functions unrelated to defense, including as nitrogen storage and transport compounds (Møller, 2010), and as signaling regulators that facilitate stress responses (Segień and Bogatek, 2006). If these physiological functions are favored under specific conditions, such as limited soil nitrogen or limited moisture, and if the conditions where they are favored tend to occur in warmer locations, then spatial variability in these conditions could serve as contributing factors in cyanogenesis cline evolution. Observations of strong correlations between cyanogenesis frequency and environmental variables associated with abiotic stress (for example, AP) would be consistent with a role for these abiotic factors as agents of selection.
As with the proposed agents of selection, the genetic targets of selection in cyanogenesis clines are not fully resolved. Most previous population surveys have suggested that the clines arise through selection acting to favor or disfavor the cyanogenic phenotype (Daday, 1965; Ennos, 1982; Majumdar et al., 2004); at the genetic level, the corresponding target of selection would thus be the two-locus AcLi cyanotype. If correct, this would suggest that clines are evolving through a process of epistatic selection acting simultaneously on the two underlying cyanogenesis genes. Consistent with this hypothesis, Ac/ac and Li/li typically both show clinal variation, with dominant alleles of both genes predominating in warmer climates. In addition, a number of studies have reported that within polymorphic populations, cyanotype frequencies deviate from random expectations toward an excess of AcLi and acli cyanotypes, potentially reflecting selection against cyanotypes that are acyanogenic but that carry some of the energetic costs of producing cyanogenic components (Daday, 1954a; Ennos, 1982; Kakes, 1987; Till-Bottraud et al., 1988; Majumdar et al., 2004). On the other hand, the majority of polymorphic populations do not show skewed cyanotype frequencies (reviewed by Kooyers and Olsen, 2012), and clinal variation at the two genes could potentially be explained by selection acting on each gene individually for physiological functions unrelated to cyanogenesis. These targets of selection are not necessarily mutually exclusive; selection could be acting on Ac/ac, Li/li and the two-locus cyanotype at the same time.
The objective of this study was to investigate the agents of selection and targets of selection in the white clover cyanogenesis system by comparing populations in three geographically disparate world regions that span similar environmental gradients. Specifically, we compared these populations to examine whether: (1) cyanogenesis clines are detectable across all sampled climatic gradients; (2) clines can be explained by the same agents of selection, as assessed by correlations of clinal variation with a panel of environmental variables; (3) cline shapes are similar between the two cyanogenesis genes within each sampling transect, and similar in comparisons of transects that vary in length but that span similar environmental gradients and (4) clinal patterns can be explained by selection acting on the same genetic targets (that is, the Ac/ac gene, the Li/li gene and/or the two-locus cyanotype) in different clines.
Materials and methods
Population sampling
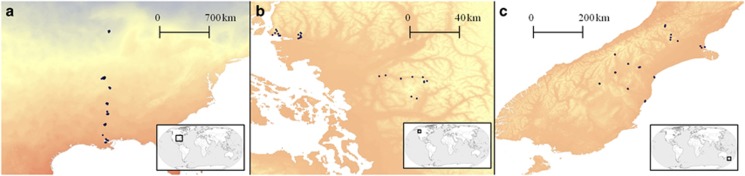
Trifolium repens is an obligately outcrossing perennial legume that also spreads vegetatively through its creeping stolons. Although native to Eurasia, it has been introduced into mesic temperate areas worldwide as a lawn plant, forage crop and nitrogen-fixing rotation crop. Here, we examined white clover from introduced portions of the species range in North America and New Zealand. Seven different population transects from three geographically disparate areas were sampled. One of these was a latitudinal transect across the central United States, consisting of 1145 individuals from 120 collection sites, which was previously documented in Kooyers and Olsen (2012; Figure 1a). We also sampled populations from three altitudinal transects in the North Cascade Mountains in the Pacific Northwest (Mt Baker, Mt Seymour and Cypress Mt; Figure 1b) and three altitudinal transects in the Southern Alps on the South Island of New Zealand (from Christchurch to Arthur's Pass, from Timaru to Mt Cook Village and from Oamaru to Lindis Pass; Figure 1c). These sampling locations were chosen because each transect spanned a similar gradient in MWT, a variable that has been found to be closely correlated with cyanotype frequencies in previous studies. Collection sites were typically public parks, roadsides, schoolyards and athletic fields.
Figure 1.
Locations of the populations sampled in (a) the central United States, (b) the North Cascades Range and (c) the Southern Alps of New Zealand. In the North Cascades, the transects sampled (from west to east) were Cypress Mt, Mt Seymour and Mt Baker. In New Zealand, the transects sampled (from northernmost to southernmost) were from Christchurch to Arthur's Pass, from Timaru to Mt Cook and from Oamaru to Lindis Pass. The blue to red gradient indicates minimum winter temperature as determined from the BIOCLIM data set (Hijmans et al., 2005). Inserts indicate locations of regions on a global map.
For the North Cascades transects, we collected stolon cuttings and seeds from 15 to 25 individuals from two to four subpopulations within three to four populations for each of the transects (523 total individuals, of which 471 survived and were included in analyses). Subpopulations and populations were defined based on spatial proximity; collection sites within 3 km of each other were considered subpopulations of a single population, whereas populations were defined as clusters of subpopulations separated by more than 3 km. Previous analyses using a similar definition of subpopulation have indicated that there is no significant genetic differentiation among white clover subpopulations within populations (Kooyers and Olsen, 2012). In New Zealand, we used a similar sampling scheme, collecting stolon cuttings from ∼15 individuals from three subpopulations within four populations sampled along each of the three altitudinal transects (540 total individuals, of which 501 survived and were included in analyses). All stolon cuttings were transplanted and grown in the Washington University greenhouse. To avoid sampling multiple ramets of the same plant, we only collected from plants that were separated by at least 3 m (Gliddon and Saleem, 1985).
Determination of cyanotypes and inferred cyanogenesis allele frequencies
Cyanotypes were determined for sampled plants using a modified Feigl-Anger HCN assay as
described elsewhere (Feigl and Anger, 1966; Olsen et al., 2007, 2008; see also Supplementary Methods).
Linkage disequilibrium between Ac/ac and Li/li was calculated
via Hill's maximum likelihood estimator (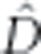 ; Hill, 1974) to test
for evidence of epistatic selection within populations.
; Hill, 1974) to test
for evidence of epistatic selection within populations.
To assess clinal variation across the sampled transects, cyanotype frequencies (AcLi, Acli, acLi and acli) and inferred Ac and Li allele frequencies were calculated for individual subpopulations and populations. Frequencies of homozygous recessive genotypes (acac or lili) can be calculated directly from cyanotype data, as they correspond to the absence of individual cyanogenic components. Allele frequencies for Ac/ac and Li/li were inferred for individual subpopulations using the observed homozygous recessive genotype frequencies under the assumption of Hardy–Weinberg equilibrium (p2+2pq+q2=1, where q2 is the frequency of homozygous recessive genotypes). Although the cyanogenesis genes are unlikely to be evolving neutrally, this violation of Hardy–Weinberg assumptions is predicted to have a relatively small effect on inferred allele frequencies at these loci where homozygotes for the dominant alleles and heterozygotes are functionally equivalent (Lachance, 2009).
Comparison of neutral markers to cyanogenesis genes
As a comparison to the cyanogenesis genes, we examined neutral markers to assess population structure within and among subpopulations, populations and transects. We used nine microsatellite markers to genotype four to eight individuals in each subpopulation in New Zealand (179 total) and in the North Cascades (144 total). We also used data from four to seven individuals per subpopulation for 38 subpopulations (349 total) from the central United States populations, as described previously (Kooyers and Olsen, 2012). DNA extraction, microsatellite amplification conditions and analysis of null alleles are described in the Supplementary Methods.
Hierarchical population structure was assessed using both Fst-based and Rst-based analysis of molecular variances implemented in Arlequin 3.5.1.2 (Excoffier et al., 2005). To investigate whether transects within New Zealand or the North Cascades could be considered samples of the same cline, neutral variation was apportioned into among-transect, among-population-within-transect and within-population levels. Significance was assessed based on a frequency distribution created via 1000 permutations of the data. We use the term ‘region' hereafter when referring to groups of transects that are not genetically differentiated from each other in the North Cascades or New Zealand. We calculated Rst and Fst for all subpopulation and population pairs using Arlequin 3.5.1.2 and ϕpt in GenAlEx 6 (Peakall and Smouse, 2006). All of these measures were similar, and we focus on ϕpt for the rest of the study because ϕpt can be calculated not only for codominant markers (for example, microsatellites), but also for dominant markers (for example, the Ac/ac and Li/li cyanogenesis genes); this measure thus allows a direct comparison of differentiation at the neutral loci and cyanogenesis genes.
Clinal variation may be adaptive, or it may arise due to neutral processes such as population history (see Kooyers and Olsen, 2012). To assess whether there was cyanogenesis clinal variation across sampled transects after accounting for neutral population structure, we used partial Mantel tests conducted in zt (Bonnet and Van de Peer, 2002). Within each region, we compared a matrix of pairwise differences between subpopulations in MWT or aridity (an index derived from precipitation and evapotranspiration; see below) with a linearized pairwise ϕpt values for the Ac allele, Li allele or the AcLi cyanotype while controlling for a matrix of linearized pairwise ϕpt values calculated from the nine microsatellite markers. Significance was determined through 1 00 000 permutations of the matrices (see Bonnet and Van de Peer (2002) for exact methods).
Identifying potential agents of selection
In each transect, we examined correlations between environmental variables for each subpopulation and frequencies of Ac alleles, Li alleles or AcLi cyanotypes. Variables included altitude, 19 bioclimatic variables and monthly precipitation (BIOCLIM; Hijmans et al., 2005), as well as annual potential evapotranspiration (Apet), monthly aridity index and monthly Apet (CGIAR: Consortium for Spatial Information; Trabucco and Zomer, 2009). From these correlations, we identified variables that were highly correlated with allele or cyanotype frequencies and that did not covary with other environmental variables. Criteria for the selection of variables and details of the correlational analyses are described in the Supplementary Methods. We selected three variables for use in multiple regression analysis: AP, MWT and Apet. Multiple regression analysis was performed using lm() in R 2.11.1 with every combination of AP, MWT and Apet, as well as interactions among these variables for each cline as well as for a pooled data set. Regressions were also performed using aridity index, a composite measure calculated as AP divided by Apet. Models were compared using akaike information criterion (AIC) values and adjusted r2 values.
Comparing cline shapes within and among clines
One way of comparing targets and agents of selection among clines is to examine whether cline shapes are similar for different cyanogenesis genes within clines and for the same locus between clines. In this study, cline shape is defined as the change in allele or phenotype frequency across a given range of an environmental gradient. Similarities in cline shapes between different genes in the same cline are consistent with shared agents of selection for the two genes; similar cline shapes in the same gene between different clines are consistent with conserved targets and agents of selection in the clines (see Supplementary Methods). We assessed the parameter ∂z/∂x for the Ac allele, the Li allele and the AcLi cyanotype in each cline, where z is the logit (allele frequency) or logit (cyanotype frequency) of a subpopulation and x is the MWT of a subpopulation. Details on these measures and data transformations are provided in the Supplementary Methods. All models were run using the lm() call in R 2.11.1; slopes (∂z/∂x) and standard error of slopes were extracted from the model and compared between cyanogenesis genes within transects and between transects for each cyanogenesis gene via Welch's t-tests to correct for unequal sampling among clines.
Identifying genetic targets of selection
To assess whether the targets of selection are the individual Ac/ac and Li/li genes or the two-gene cyanogenesis phenotype, we performed comparative analyses using the three cyanogenesis clines that our neutral marker analyses confirmed to be adaptive (see Results). We fit two general linear models for each cline; for both models, the frequency of the AcLi cyanotype was the response variable, and the Ac and Li allele frequencies were treated as independent variables. In the first analysis, we fit a model that included an interaction term for the Ac and Li factor (that is, Ac*Li); in the second, we included the Ac and Li factors without the interaction term (that is, Ac+Li). We performed likelihood ratio tests to evaluate whether the model without the interaction term provides as good of a fit as the model with the interaction term. We also performed the same analysis with the logit of frequency of AcLi cyanotype as a response variable. A significant interaction term would indicate that there is an epistatic relationship between Ac/ac and Li/li that accounts for a portion of the spatial variation in cyanotype frequency that each gene could not account for individually.
Results
As a first step in investigating clinal variation in the cyanogenesis genes, we tested whether transects within regions could be viewed as subsets of the same cline. Analysis of molecular variances indicated that no significant portion of the total genetic variance was distributed among transects in either New Zealand or the North Cascades, the two regions where multiple transects were sampled (Table 1, Supplementary Table 1). However, within the North Cascades, the Mt Baker transect differed from the other two transects with respect to geographic location (Figure 1b), transect length (∼40 km rather than 7–9 km) and climatic variation (with a greater MWT range in Mt Baker; see below). Therefore, we treated the Mt Baker transect as an independent cline from the pooled Cypress Mountain and Mt Seymour transects (referred to collectively as CYPSEY), whereas the three New Zealand transects were pooled and treated as a single cline. We report additional results for pooled data for CYPSEY and from all three North Cascade transects in the Supplementary Materials (Supplementary Figures 1–3).
Table 1. AMOVAs showing variance partitions in North Cascades and New Zealand regions for neutral microsatellite markers.
| Transect | Source of variation | d.f. | Variance components | Percentage of variation |
|---|---|---|---|---|
| North Cascades | Among transects | 2 | 0 | 0 |
| North Cascades | Between subpopulations | 20 | 0.11 | 3.64 |
| North Cascades | Within subpopulations | 259 | 2.903 | 96.36 |
| New Zealand | Among transects | 2 | 0.004 | 0.17 |
| New Zealand | Between subpopulations | 33 | 0.126 | 4.84 |
| New Zealand | Within subpopulations | 322 | 2.47 | 94.99 |
Abbreviation: AMOVA, analysis of molecular variance.
Both AMOVAs were Fst-based as implemented in Arlequin 3.5.1.2.
Statistical significance at P<0.05 is indicated by bold text.
Different patterns of clinal variation in different transects
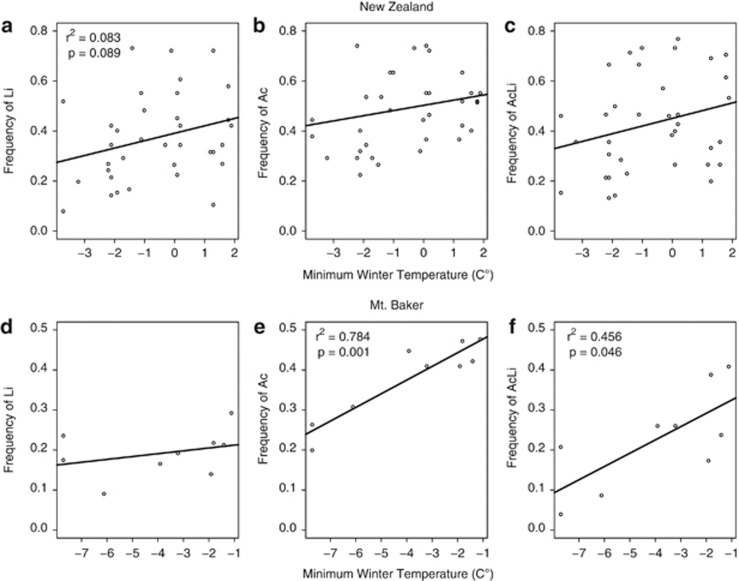
To examine whether patterns of clinal variation were present and mirrored those of previous studies, we examined correlations for the frequency of the Ac allele, the Li allele and the AcLi cyanotype with MWT. In New Zealand populations, there was a marginally significant cline in the Li/li gene with respect to MWT, with a higher frequency of the Li allele in warmer localities (r2=0.083, P=0.089; Figure 2a); there were not statistically significant correlations for the Ac allele or the AcLi cyanotype with respect to MWT, although both variables also showed a trend toward higher frequencies in warmer localities (Figures 2b and c). In the Mt Baker populations, there was significant clinal variation with respect to MWT for the Ac allele (r2=0.784, P=0.001) and the AcLi phenotype (r2=0.456, P=0.046), with higher frequencies of both in warmer areas (Figures 2e and f); there was no significant correlation for the Li/li gene, although there was again a weak trend in the same direction (Figure 2d). For the other sampled populations of the North Cascades, there was no evidence of clinal variation with respect to MWT for either of the cyanogenesis genes or cyanotype (Supplementary Figures 1 and 2). Pooling all North Cascades transects resulted in a similar pattern to Mt Baker (that is, a significant correlation between warmer temperatures and higher frequencies of Ac, with a trend toward higher frequencies of AcLi plants; Supplementary Figure 3). As has been previously reported, the central United States cline has a higher frequency of the Ac allele, the Li allele and the AcLi cyanotype in warmer climates (P<0.001 for each regression; see Figure 3 in Kooyers and Olsen, 2012). There was little evidence of linkage disequilibrium between Ac/ac and Li/li within subpopulations in any North American or New Zealand transect (13 of 179 subpopulations showed significant linkage disequilibrium; Supplementary Table 2).
Figure 2.
Regression of subpopulation frequencies of the Li allele, Ac allele or the AcLi cyanotype against average minimum winter temperature in New Zealand (a, b or c, respectively) and on Mt Baker (d, e or f, respectively).
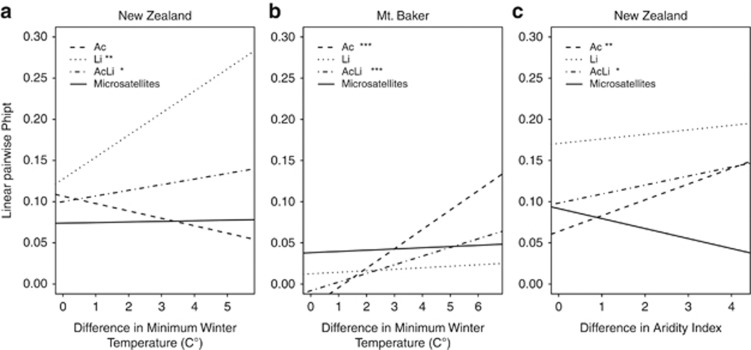
Figure 3.
Regressions of linear pairwise ϕpt values between sampling sites for microsatellite data, Ac/ac, Li/li and AcLi cyanotype against: (a) pairwise minimum winter temperature data for New Zealand, (b) pairwise minimum winter temperature data for Mt Baker and (c) pairwise aridity data for New Zealand. Statistical significance of regressions is indicated as follows: *0.01<P<0.05, **0.01<P<0.001 and ***P<0.001.
To determine whether the observed patterns of clinal variation could be artifacts of population structure, we conducted partial Mantel tests to examine correlations between cyanogenesis components and MWT while controlling for population differentiation at the microsatellite loci. Summary statistics and results for these microsatellite loci as well as pairwise genetic distances between populations for Ac/ac, Li/li and microsatellite loci can be found in (Supplementary Tables 2–4). In New Zealand, the correlation between Li allele frequencies and MWT remained after controlling for population structure (Mantel's r=0.149, P=0.005, Figure 3a), and the AcLi correlation became statistically significant (Mantel's r=0.113, P=0.018; Figure 3a). In the Mt Baker cline, controlling for population structure did not affect the observed correlations with MWT for the Ac allele frequency (Mantel's r=0.680, P<0.001) and the AcLi cyanotype frequency (Mantel's r=0.663, P<0.001; Figure 3b). As we have previously reported, correlations remain significant in the central United States cline between MWT and the Ac allele, the Li allele and the AcLi cyanotype frequencies after population structure is taken into account (Ac: P=0.057; Li: P=0.008; AcLi: P=0.04; see Table 2 in Kooyers and Olsen, 2012).
Table 2. Model-fit results for AIC analysis of the environmental correlates with variance in the population frequencies of Ac/ac, Li/li and AcLi.
| Factors in model | d.f. |
Ac/Ac
|
Li/li
|
AcLi
|
|||
|---|---|---|---|---|---|---|---|
| AIC | Adj r2 | AIC | Adj r2 | AIC | Adj r2 | ||
| MWT | 2 | −485.4 | 0.23 | −517.1 | 0.42 | −554.6 | 0.41 |
| AP | 2 | −442.3 | 0.00 | −428.9 | 0.01 | −469.4 | 0.01 |
| Apet | 2 | −463.0 | 0.12 | −444.4 | 0.09 | −499.6 | 0.18 |
| MWT+AP | 3 | −484.3 | 0.23 | −516.6 | 0.42 | −553.2 | 0.41 |
| MWT+ Apet | 3 | −489.2 | 0.25 | −515.8 | 0.42 | −561.0 | 0.44 |
| AP+Apet | 3 | −461.8 | 0.12 | −444.3 | 0.10 | −500.5 | 0.19 |
| MWT * AP | 4 | −492.5 | 0.27 | −555.4 | 0.54 | −598.4 | 0.55 |
| MWT * Apet | 4 | −493.2 | 0.27 | −549.9 | 0.53 | −595.3 | 0.54 |
| AP *Apet | 4 | −493.6 | 0.28 | −498.3 | 0.35 | −559.4 | 0.43 |
| MWT+AP+Apet | 4 | −487.6 | 0.25 | −515.1 | 0.42 | −559.3 | 0.43 |
| MWT * AP+Apet | 5 | −491.5 | 0.27 | −559.9 | 0.56 | −596.4 | 0.55 |
| MWT * Apet+AP | 5 | −492.4 | 0.27 | −553.2 | 0.54 | −595.9 | 0.55 |
| AP *Apet+MWT | 5 | −504.7 | 0.33 | −544.6 | 0.52 | −594.3 | 0.54 |
| MWT+AP+Apet+MWT AP+MWT:Apet | 6 | −490.8 | 0.27 | −561.1 | 0.56 | −599.5 | 0.56 |
| MWT+AP+Apet+MWT AP+AP:Apet | 6 | −503.0 | 0.32 | −561.1 | 0.56 | −602.6 | 0.57 |
| MWT+AP+Apet+AP Apet+MWT:Apet | 6 | −503.7 | 0.33 | −553.4 | 0.54 | −599.2 | 0.56 |
| MWT+AP+Apet+AP:Apet | 7 | −501.7 | 0.32 | −501.7 | 0.56 | −560.1 | 0.57 |
| MWT * AP * Apet | 8 | −500.7 | 0.32 | −558.4 | 0.56 | −600.1 | 0.57 |
Abbreviations: adj, adjusted; Apet, annual evapotranspiration; AP, annual precipitation; MWT, minimum winter temperature. Bold text indicates the best fitting models.
Multiple agents of selection contribute to cyanogenesis cline formation
Given that allele frequencies for the two cyanogenesis genes are not universally covarying across sampled populations, our data suggested that agents of selection other than herbivore defense could be playing a role in the distributions of allelic variation. We addressed this possibility by examining correlations between 46 environmental parameters and either Ac allele, Li allele or AcLi cyanotype frequencies. Three variables, AP, MWT and annual Apet, best accounted for variation in the frequencies of cyanogenesis alleles and cyanotype after covariance within clines was taken into account. Although several other variables were statistically significant within individual clines (Supplementary Table 5), many of these were redundant with AP, MWT or Apet. In all three clines, variation in the frequency of Ac was best explained by combinations of MWT, Apet and AP. This is not necessarily true for the frequency of Li or AcLi. The variables that best explained these frequencies differed between clines and did not involve all three variables (Supplementary Table 6). To determine whether all clines can be described collectively by the same environmental variables, we combined data from the central United States, Mt Baker, CYPSEY and New Zealand regions into a single data set. Best models for both cyanogenesis genes and the AcLi cyanotype included all three variables and various interaction terms (Table 2), indicating that all three variables are potentially correlated with agents of selection. However, there were many models that fit either the variation in Ac/ac better than Li/li or vice versa, suggesting that the agents of selection that act on the two genes are apparently not identical. Notably, all of the models that provided the best fit for Ac/ac contained all three variables and an interaction term between AP and Apet, the two variables that make up the aridity index variable (AP/Apet). This potentially suggests a close connection between aridity-related environmental variation and frequencies of plants that produce cyanogenic glucosides.
Of the three regions examined, New Zealand populations spanned a much wider aridity gradient (aridity index range: 0.5941–4.8569) than either the central United States or Mt Baker clines (0.8261–1.1208 and 1.8389–2.4037, respectively; Supplementary Figure 4). Along the aridity gradient, New Zealand populations showed significant clinal variation for the Ac allele (r2=0.213, P=0.005), the Li allele (r2=0.109, P=0.049) and the AcLi cyanotype (r2=0.184, P=0.009), with higher frequencies of cyanogenic plants in more xeric populations (Supplementary Figures 4A–C). After controlling for population structure in New Zealand, the correlations with aridity hold for Ac allele frequency (Mantel's r=0.199, P=0.002, Figure 3c) and the AcLi cyanotype frequency (Mantel's r=0.12, P=0.031; Figure 3c), although the previously significant correlation between aridity and Li allele frequency becomes nonsignificant (Mantel's r=0.04, P=0.275; Figure 3c). These results suggest that for clover populations occurring across wide aridity gradients, such as those in New Zealand, aridity or a factor closely correlated with it may be an important agent of selection that favors cyanogenic glucosides in drier areas.
Cline shape similarities and differences among regions
The observation of similar cline shapes between different genes in the same transect would be consistent with the hypothesis that there are similar agents of selection acting on each target. Similar cline shapes at the same genetic target in different regions would suggest that similar agents of selection may be acting in different regions. We used these basic predictions to examine whether the targets of selection and agents of selection were similar within and between Mt Baker, New Zealand and the central United States regions. We tested whether cline shape, expressed via the parameter ∂z/∂x, with MWT representing the independent variable x, was similar between Ac/ac and Li/li within regions and whether cline shapes were similar for Ac/ac or Li/li between regions. Cline shapes for Ac alleles and Li alleles were significantly different from each other in the Mt Baker cline (Welch's t=5.31, P=0.0001) and the central United States cline (Welch's t=20.44, P<0.0001; Table 3). This is not the case for New Zealand, where the cline shapes were not significantly different (Welch's t=5.31, P=0.38; Table 3). In comparisons between clines from different regions, the cline shape for the AcLi cyanotype was similar in the central United States and Mt Baker clines (Welch's t=0.92, P=0.19), but significantly different between each of these regions and the New Zealand cline (Mt Baker: Welch's t=3.13, P=0.004; central United States: Welch's t=5.02, P<0.001; Table 3). Cline shape of the Ac/ac gene was similar between the Mt Baker and central United States clines (Welch's t=1.347, P=0.11; Table 4). In contrast, cline shape of the Li/li gene differed between the central United States and New Zealand (Welch's t=4.305, P<0.0001, Table 3). We did not examine cline shapes for the Ac/ac gene between New Zealand and the central United States or Mt Baker, or for the Li/li gene between Mt Baker and the central United States or New Zealand, as there was not significant clinal variation in the Ac allele in New Zealand or in the Li allele in Mt Baker. Taken together, these results suggest that similar patterns of selection are occurring in Mt Baker and central United States clines, whereas a different pattern is occurring in the New Zealand cline.
Table 3. Comparison of cline shape between genes within each region and between regions for each gene.
| Region 1 | Target 1 | Slope | Standard error | Region 2 | Target 2 | Slope | Standard error | t | d.f. | P-value |
|---|---|---|---|---|---|---|---|---|---|---|
| Central United States | Ac/ac | 0.144 | 0.028 | Central United States | Li/li | 0.211 | 0.022 | 20.438 | 223.639 | <0.0001 |
| New Zealand | Ac/ac | 0.043 | 0.109 | New Zealand | Li/li | 0.152 | 0.082 | 0.315 | 80.934 | 0.377 |
| Mt Baker | Ac/ac | 0.156 | 0.025 | Mt Baker | Li/li | 0.049 | 0.055 | 5.311 | 11.205 | 0.0001 |
| Central United States | Ac/ac | 0.144 | 0.028 | Mt. Baker | Ac/ac | 0.156 | 0.025 | 1.347 | 9.601 | 0.105 |
| Central United States | Li/li | 0.211 | 0.022 | New Zealand | Li/li | 0.152 | 0.082 | 4.305 | 36.505 | <0.0001 |
| Central UnitedStates | AcLi | 0.221 | 0.027 | Mt Baker | AcLi | 0.247 | 0.086 | 0.921 | 8.121 | 0.192 |
| Central United States | AcLi | 0.221 | 0.027 | New Zealand | AcLi | 0.147 | 0.087 | 5.016 | 37.063 | <0.0001 |
| Mt Baker | AcLi | 0.247 | 0.086 | New Zealand | AcLi | 0.147 | 0.087 | 3.128 | 12.464 | 0.004 |
Statistical significance at P<0.05 is indicated by bold text.
Table 4. Log-likelihood ratio test between models including and excluding an interaction terms between Ac/ac and Li/li genes.
| Cline | Ac*Li log likelihood | Ac+Li log likelihood | Δ | X | P value (1 d.f.) |
|---|---|---|---|---|---|
| Central USA | 129.23 | 128.64 | 0.58 | 1.16 | 0.281 |
| New Zealand | 48.45 | 43.67 | 4.78 | 9.56 | 0.002 |
| Mt Baker | 18.83 | 18.83 | 0.00 | 0.00 | 0.961 |
Statistical significance at P<0.05 is indicated by bold text.
Evidence for Ac/ac and Li/li as individual targets of selection
The target of selection in this system has traditionally been assumed to be the cyanogenic phenotype, which arises through the interaction of the two cyanogenesis genes. The cline shape analysis above indicates that this may not be the case. To further test whether clinal variation could be explained most parsimoniously through selection acting on the Ac/ac and Li/li genes individually, we used likelihood ratio tests to examine two alternative models. The first modeled spatial variation in the frequency of cyanogenesis using only variation in the Ac/ac and Li/li genes (that is, Ac+Li), whereas the second model included an interaction term between the two genes (that is, Ac+Li+Ac:Li). In the two North American clines, a model with no interaction between the Ac/ac and Li/li genes provides as good of a fit as a model that includes the interaction term (central United States: Δ=1.16, P=0.281, Mt Baker: Δ<0.01, P=0.961; Table 4). However, in New Zealand the interaction between Ac/ac and Li/li does enhance the fit of the model (Δ=0.58, P=0.002; Table 4). Using the logit of the frequency of the AcLi cyanotype as a response variable does not change these results—the model fit is improved by including an interaction between the Ac/ac and Li/li genes in New Zealand (Supplementary Figure 5). This finding is consistent with cline shape analyses in suggesting evidence for epistatic selection in New Zealand, but not in the other clines.
Discussion
The recurrent evolution of cyanogenesis clines in white clover is a useful system for comparing multiple replicates of the microevolutionary process. Previous studies have reported higher frequencies of cyanogenic plants in warmer areas, and latitudinal and altitudinal cyanogenesis clines have been extensively documented in both the native and introduced species range (Daday, 1954a, 1954b, 1958; de Araujo, 1976; Till-Bottraud et al., 1988; Ganders, 1990; Majumdar et al., 2004). Here, we have investigated clinal variation in a sampling of introduced North American and New Zealand populations. We find remarkable similarities among regions in some aspects of cline shape, despite large differences in the spatial scale over which environmental gradients occur. For example, cline shape for both the Ac/ac gene and the AcLi cyanotype are not statistically significantly different between the Mt Baker cline and central US cline (Table 3), even though the former spans a 40 km altitudinal transect and the latter spans a 1600 km latitudinal transect. However, we also find evidence for regional heterogeneity in selection, where there are apparently multiple agents of selection and where the genetic targets of selection extend beyond the two-locus cyanogenesis phenotype. For example, the Mt Baker cyanogenesis cline is driven by strong clinal variation in the Ac/ac gene with no evidence for clinal variation in the Li/li gene, whereas the New Zealand cyanogenesis cline shows the opposite pattern, with clinal variation in the Li/li gene and little evidence for clinal variation in the Ac/ac gene (Figure 2, Supplementary Figure 4). Below we discuss the implications of these findings for the cyanogenesis system and more generally for the process of adaptive differentiation and cline formation.
Agents of selection in cyanogenesis clines
The potential agents of selection in cyanogenesis cline evolution have been a topic of study for over six decades (for example, see Daday, 1954a, 1954b; reviewed by Hughes 1991; Kooyers and Olsen 2012). A leading hypothesis has been that a fitness tradeoff occurs whereby cyanogenic plants have an advantage in warmer locations, where herbivore pressure is high, but are at a competitive disadvantage in cooler, low-herbivory environments because of the energetic costs of producing cyanogenic components (Kakes, 1989). However, there is also evidence that herbivore pressure may not be the sole factor driving cyanogenesis cline evolution. For example, cyanogenic plants may be less tolerant of freezing (Brighton and Horne, 1977; but see Olsen and Ungerer, 2008) or less resistant to fungal infection (Dirzo and Harper, 1982b, Lieberei et al., 1989, Ballhorn et al., 2010); both of these factors could contribute to geographical variation in cyanogenesis frequencies. In addition, there is evidence that cyanogenic glucosides and hydrogen cyanide can serve physiological functions unrelated to herbivore defense, including as mediators of stress response and as nitrogen storage or transport molecules (see reviews by Segień and Bogatek (2006) and Møller (2010)). Thus, geographical variation in nitrogen availability or other environmental factors relating to these functions might also have a role in cyanogenesis cline evolution.
Although our correlational analyses were not designed to definitively identify the specific agents of selection that are at play in this system, they do allow us to assess whether there is evidence for factors beyond those directly correlated with herbivore abundance. Our multiple regression analyses indicate that models that incorporate both MWT and aridity (but not necessarily the interaction) better describe frequencies of plants with cyanogenic glucosides (Ac_) or the cyanogenic phenotype (AcLi) than either variable alone. In addition, frequencies of the Ac allele and the AcLi cyanotype are highly correlated with aridity; in New Zealand, where transects span a marked aridity gradient, Ac_ and AcLi plants are more frequent in more arid areas (Figures 2 and 3, Supplementary Figure 4). Given that aridity has not been historically associated with herbivore richness or abundance in this system (Daday, 1954a; 1965), these observations point to adaptive functions for cyanogenic components besides herbivore defense.
One such adaptive function may involve water stress response. Usable soil nitrogen is often highly limited during the beginning stages of water stress due to the reduced activity of nitrate reductase (Nilsen and Orcutt, 1996; Larcher, 2001), and it is plausible that cyanogenic glucosides could serve as a nitrogen source during these periods of acute nitrogen limitation (Møller, 2010). Studies to date have been inconclusive regarding the relative fitness of white clover cyanotypes under water stress. One observational study speculated a fitness benefit for cyanogenic plants in areas of low water availability based on correlations between habitat aridity and cyanogenesis frequency on six New Zealand farms (Caradus et al., 1990). In contrast, Foulds and Grime (1972) reported that plants lacking cyanogenic glucosides are better able to survive extreme drought than plants producing the compounds, although in a larger scale experiment, Foulds (1977) failed to detect fitness differences among cyanotypes. Additional common garden and manipulative experiments will be useful for characterizing water stress adaptation and other aridity-associated selective factors that may be acting in this system; these experiments are currently underway.
To our knowledge, this study is the first to document white clover cyanogenesis clinal variation along an aridity gradient. One probable reason why this pattern has not been more frequently reported is that most white clover populations occur in fairly mesic conditions. For instance, in the central United States, where white clover is abundant, the range of aridity index is 10 times less than that observed in New Zealand. An additional explanation for the lack of previous documentation is that in most previously documented clines, aridity is closely correlated with MWT (for example, Daday, 1954a, 1954b; Ganders, 1990; Majumdar et al., 2004). As MWT is more closely associated with herbivore pressure than aridity (see above), the latter variable would have been more easily overlooked in studies focusing on the role of cyanogenesis as a chemical defense.
Multiple targets of selection in the cyanogenesis system
Because the white clover cyanogenesis polymorphism arises through two independently segregating biochemical polymorphisms, the genetic target(s) of selection in this system could either be the two-locus cyanotype or the individual cyanogenesis genes. In studies to date, the genetic target has often been interpreted to be the two-locus cyanotype, based on two lines of evidence: (1) both genes typically show clinal variation in cyanogenesis clines; and (2) some polymorphic populations show skewed cyanotype frequencies consistent with epistatic selection for plants that either have both required cyanogenic components (AcLi) or neither (acli). Our results indicate that although the two-locus cyanotype may indeed be the genetic target in some world regions, selection on the individual Ac/ac and Li/li genes is also likely to be occurring. Among our sampled clines, we find greatest evidence for selection on cyanotype in New Zealand. In the New Zealand populations, cline shapes are not significantly different between the Ac/ac and Li/li genes (Table 3), and variation in the frequency of cyanogenic plants is best modeled by including an interaction term between the two genes (Table 4). In contrast, for both the central United States and Mt Baker clines, cline shapes differ between the Ac/ac and Li/li genes within regions, and there is no significant support for an interaction between the two genes in cline models (Tables 3 and 4). The two North American clines also differ from each other, in that only Ac/ac shows significant clinal variation in the Mt Baker cline, whereas both genes show clinal variation in the central United States. Taken together, these patterns suggest that the two-locus cyanotype is the target of selection in the New Zealand cline, the Ac/ac gene is the sole target of selection in the Mt Baker cline, and the Ac/ac and Li/li genes may individually be targets of selection in the central United States cline. Thus, as with our inferences on the agents of selection in this system, we find evidence for multiple targets of selection in this system.
The biological significance of these regional differences in targets of selection is difficult to infer without a clearer understanding of the selective agents at play. The two-locus pattern observed in New Zealand is consistent with traditional assumptions that the cyanogenic (AcLi) phenotype is adaptive as an herbivore deterrent. However, we suggest that the most plausible scenario involves multiple agents of selection including aridity and herbivory acting on multiple different targets simultaneously.
The evolution of clines
Clines have typically been studied as a means for understanding the relative roles of natural selection and gene flow in the process of adaptive divergence and speciation. Most literature on clines assesses hybrid zones between diverging lineages. Cyanogenesis clines in white clover differ from hybrid zones, in that there is no evidence for reproductive isolation between morphs and there is an extremely low ratio of genetic drift to gene flow between populations (evidenced in the present study by low pairwise genetic distances for microsatellite markers; Supplementary Table 4), probably because of a large effective population size rather than high gene flow (Gliddon and Saleem, 1985). Unlike most hybrid zones, these clines are typically viewed under a dispersal-independent (or gradient) model because adaptive variation corresponds directly to climatic factors in the same way despite differences in transect length. On a short spatial scale (where the ratio of gene flow to selection is greater than the transect length), a dispersal-independent cline will change to a dispersal-dependent cline (Barton and Hewitt, 1985) or ecotone (Endler, 1977), where clinal variation can be maintained through a gene flow-selection balance and precise adaptation to intermediate environments is not possible. Further, transect length can be short enough that gene flow overwhelms selection and there is no resulting clinal variation. Although we see evidence that the 40 km Mt Baker transect is still a dispersal-independent cline (that is, direct correspondence to environmental variables), we interpret the lack of clinal variation in the shorter CYPSEY transects as gene flow overwhelming selection, and we predict that transects somewhere between 10 and 40 km long would be dispersal-dependent clines.
Case studies in diverse taxa have demonstrated that adaptive divergence across heterogeneous environments can be rapid when selection pressures are high and gene flow is relatively low (Antonovics and Bradshaw, 1970; Clarke et al., 1985; Umina et al., 2005). The cyanogenesis system is similar to these other study systems in that there must be intense selection acting within these populations; after all, multiple parallel clines have evolved in regions where white clover was introduced only within the last 500 years (Daday, 1958; Ganders, 1990; Kooyers and Olsen, 2012). Parallel cyanogenesis clines in white clover provide a unique perspective on adaptive divergence; we observe broadly parallel phenotypic patterns despite the fact that there are apparent differences among regions in both the agents of selection and their genetic targets. Although this conclusion is not the most parsimonious, it may be representative of organisms that adapt to similar gradients in different areas, given that many variable traits may have multiple functions that contribute to fitness.
Data archiving
Data deposited in the Dryad repository: doi:10.5061/dryad.49k4d.
Acknowledgments
This work would not have been possible without the assistance of AgResearch New Zealand, specifically Brent Barrett, Andrew Griffiths and Keith Widdup. We thank the greenhouse staff at Washington University for assistance in growing and maintaining plants. Jared Strasburg, Nick Barton and Alan Templeton provided excellent feedback that substantially improved this manuscript. This research was supported by an NSF Doctoral Dissertation Improvement Grant to NJK (DEB-1110588) and an NSF CAREER award to KMO (DEB-0845497).
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Angseesing JAP. Selective eating of the acyanogenic form of Trifolium repens. Heredity. 1974;32:73–83. [Google Scholar]
- Antonovics J, Bradshaw AD. Evolution in closely adjacent plant populations VIII. Clinal patterns at a mine boundary. Heredity. 1970;25:349–362. doi: 10.1038/sj.hdy.6800835. [DOI] [PubMed] [Google Scholar]
- Armstrong HE, Armstrong EF, Horton E. Herbage studies. II. Variation in Lotus corniculatus and Trifolium repens: (cyanophoric plants) Proc R Soc B. 1913;86:262–269. [Google Scholar]
- Ballhorn DJ, Pietrowski A, Lieberei R. Direct trade-off between cyanogenesis and resistance to a fungal pathogen in lima bean (Phaseolus lunatus L.) J Ecol. 2010;98:226–236. [Google Scholar]
- Barton NH, Hewitt GM. Analysis of hybrid zones. Annu Rev Ecol Syst. 1985;16:113–148. [Google Scholar]
- Bonnet P, Van de Peer Y. zt: a software tool for simple and partial Mantel tests. J Stat Soft. 2002;7:1–12. [Google Scholar]
- Brighton F, Horne MT. Influence of temperature on cyanogenic polymorphisms. Nature. 1977;265:437–438. [Google Scholar]
- Caradus JR, Mackay AC, Charlton JFL, Chapman DF. Genecology of white clover (Trifolium repens L) from wet and dry hill country pastures. New Zeal J Agr Res. 1990;33:377–384. [Google Scholar]
- Clarke CA, Mani GS, Wynne G. Evolution in reverse—clean-air and the Peppered Moth. Biol J Linn Soc. 1985;26:189–199. [Google Scholar]
- Corkill L. Cyanogenesis in white clover (Trifolium repens L.) V. The inheritance of cyanogenesis. NZ J Sci Technol. 1942;23:178–193. [Google Scholar]
- Daday H. Gene frequencies in wild populations of Trifolium repens L. I. Distribution by latitude. Heredity. 1954a;8:61–78. [Google Scholar]
- Daday H. Gene frequencies in wild populations of Trifolium repens II. Distribution by altitude. Heredity. 1954b;8:377–384. [Google Scholar]
- Daday H. Gene frequencies in wild populations of Trifolium repens L. III. World distribution. Heredity. 1958;12:169–184. [Google Scholar]
- Daday H. Gene frequencies in wild population of Trifolium repens IV. Mechanism of natural selection. Heredity. 1965;20:355–365. [Google Scholar]
- de Araujo AM. The relationship between altitude and cyanogenesis in white clover (Trifolium repens, L.) Heredity. 1976;37:291–293. [Google Scholar]
- Dirzo R, Harper JL. Experimental studies on slug-plant interactions III. Differences in the acceptability of individual plants of Trifolium repens to slugs and snails. J Ecol. 1982a;70:101–117. [Google Scholar]
- Dirzo R, Harper JL. Experimental studies on slug-plant interactions IV. The performance of cyanogenic and acyanogenic morphs of Trifolium repens in the field. J Ecol. 1982b;70:119–138. [Google Scholar]
- Dobzhansky T. Adaptive changes induced by natural selection in wild populations of Drosophila. Evolution. 1947;1:1–16. [Google Scholar]
- Dritschilo W, Krummel J, Nafus D, Pimentel D. Herbivorous insects colonizing cyanogenic and acyanogenic Trifolium repens. Heredity. 1979;42:49–56. [Google Scholar]
- Endler JA. Geographic Variation, Speciation, and Clines. Princeton University Press: Princeton, New Jersey; 1977. [PubMed] [Google Scholar]
- Ennos RA. Association of the cyanogenic loci in white clover. Genet Res. 1982;40:65–72. [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Feigl F, Anger V. Replacement of benzidine by copper ethylacetoacetate and tetra base as a spot-test reagent for hydrogen cyanide and cyanogen. Analyst. 1966;91:282–284. doi: 10.1039/an9669100282. [DOI] [PubMed] [Google Scholar]
- Foulds W. The physiological response to moisture supply of cyanogenic and acyanogenic phenotypes of Trifolium repens L. and Lotus corniculatus L. Heredity. 1977;39:219–234. [Google Scholar]
- Foulds W, Grime JP. The response of cyanogenic and acyanogenic phenotypes of Trifolium repens to soil moisture supply. Heredity. 1972;28:181–187. [Google Scholar]
- Ganders FR. Altitudinal clines for cyanogenesis in introduced populations of white clover near Vancouver, Canada. Heredity. 1990;64:387–390. [Google Scholar]
- Gliddon M, Saleem C.1985Gene-flow in Trifolium repens—and expanding genetic neighbourhoodIn: Jacquard P, Heim G, Antonovics J (eds)Genetic Differentiation and Dispersal in Plants Springer-Verlag: Berlin; 293–309. [Google Scholar]
- Haak DC, McGinnis LA, Levey DJ, Tewksbury JJ. Why are not all chilis hot? A trade-off limits pungency. Philos T Roy Soc B. 2012;279:2012–2017. doi: 10.1098/rspb.2011.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane JBS. The theory of a cline. J Genet. 1948;48:48–284. doi: 10.1007/BF02986626. [DOI] [PubMed] [Google Scholar]
- Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. Int J Climatol. 2005;25:1965–1978. [Google Scholar]
- Hill WG. Estimation of linkage disequilibrium in randomly mating populations. Heredity. 1974;33:229–239. doi: 10.1038/hdy.1974.89. [DOI] [PubMed] [Google Scholar]
- Hruska AJ. Cyanogenic glucosides as defense compounds—a review of the evidence. J Chem Ecol. 1988;14:2213–2217. doi: 10.1007/BF01014026. [DOI] [PubMed] [Google Scholar]
- Huey RB, Gilchrist GW, Carlson ML, Berrigan D, Serra L. Rapid evolution of a geographic cline in size in an introduced fly. Science. 2000;287:308–309. doi: 10.1126/science.287.5451.308. [DOI] [PubMed] [Google Scholar]
- Hughes MA. The cyanogenic polymorphism in Trifolium repens L (white clover) Heredity. 1991;66:105–115. [Google Scholar]
- Kakes P. On the polymorphism for cyanogenesis in natural-populations of Trifolium repens L in the Netherlands I. Distribution of the gene-Ac and gene-Li. Acta Bot Neerl. 1987;36:59–69. [Google Scholar]
- Kakes P. An analysis of the costs and benefits of the cyanogenic system in Trifolium repens L. Theor Appl Genet. 1989;77:111–118. doi: 10.1007/BF00292324. [DOI] [PubMed] [Google Scholar]
- Keller SR, Sowell DR, Neiman M, Wolfe LM, Taylor DR. Adaptation and colonization history affect the evolution of clines in two introduced species. New Phytol. 2009;183:678–690. doi: 10.1111/j.1469-8137.2009.02892.x. [DOI] [PubMed] [Google Scholar]
- Kooyers NK, Olsen KM. Rapid evolution of an adaptive cyanogenesis cline in introduced North American white clover (Trifolium repens L.) Mol Ecol. 2012;21:2455–2468. doi: 10.1111/j.1365-294X.2012.05486.x. [DOI] [PubMed] [Google Scholar]
- Lachance J. Detecting selection-induced departures from Hardy-Weinberg proportions. Genetics Selection Evolution. 2009;41:15. doi: 10.1186/1297-9686-41-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larcher W.2001Physiological Plant Ecology4th edn.Springer-Verlag: Berlin [Google Scholar]
- Lieberei R, Biehl B, Giesemann A, Junqueira NT. Cyanogenesis inhibits active defense reactions in plants. Plant Physiol. 1989;90:33–36. doi: 10.1104/pp.90.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar S, De KK, Banerjee S. Influence of two selective factors on cyanogenesis polymorphism of Trifolium repens L. in Darjeeling Himalaya. J Plant Biol. 2004;47:124–128. [Google Scholar]
- Mallet J, Barton N, Lamas G, Santisteban J, Muedas M, Eeley H. Estimates of selection and gene flow from measures of cline width and linkage disequilibrium in Heliconius hybrid zones. Genetics. 1990;124:921–936. doi: 10.1093/genetics/124.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller BL. Functional diversifications of cyanogenic glucosides. Curr Opin Plant Biol. 2010;13:338–347. doi: 10.1016/j.pbi.2010.01.009. [DOI] [PubMed] [Google Scholar]
- Nilsen ET, Orcutt DM. Physiology of Plants Under Stress. John Wiley & Sons: New York: New York; 1996. [Google Scholar]
- Oakeshott J, Gibson J, Anderson P, Knibb W, Anderson D, Chambers G. Alcohol-dehydrogenase and glycerol-3-phosphate dehydrogenase clines in Drosophila melanogaster on different continents. Evolution. 1982;36:86–96. doi: 10.1111/j.1558-5646.1982.tb05013.x. [DOI] [PubMed] [Google Scholar]
- Olsen KM, Hsu SC, Small LL. Evidence on the molecular basis of the Ac/ac adaptive cyanogenesis polymorphism in white clover (Trifolium repens L.) Genetics. 2008;179:517–526. doi: 10.1534/genetics.107.080366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen KM, Kooyers NJ, Small LL. Recurrent gene deletions and the evolution of adaptive cyanogenesis polymorphisms in white clover (Trifolium repens L.) Mol Ecol. 2013;22:724–738. doi: 10.1111/j.1365-294X.2012.05667.x. [DOI] [PubMed] [Google Scholar]
- Olsen KM, Sutherland BL, Small LL. Molecular evolution of the Li /li chemical defence polymorphism in white clover (Trifolium repens L.) Mol Ecol. 2007;16:4180–4193. doi: 10.1111/j.1365-294X.2007.03506.x. [DOI] [PubMed] [Google Scholar]
- Olsen KM, Ungerer MC. Freezing tolerance and cyanogenesis in white clover (Trifolium repens L. Fabaceae) Int J Plant Sci. 2008;169:1141–1147. [Google Scholar]
- Peakall R, Smouse PE. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson GA, Brink GE. Cyanogenesis effect on insect damage to seedling white clover in a bermudagrass sod. Agron J. 1998;90:208–210. [Google Scholar]
- Pennings SC, Silliman BR. Linking biogeography and community ecology: latitudinal variation in plant–herbivore interaction strength. Ecology. 2005;86:231–2319. [Google Scholar]
- Saucy F, Studer J, Aerni V, Schneiter B. Preference for acyanogenic white clover (Trifolium repens) in the vole Arvicola terrestris: I. Experiments with two varieties. J Chem Ecol. 1999;25:1441–1454. [Google Scholar]
- Schmidt PS, Phifer-Rixey M, Taylor GM, Christner J. Genetic heterogeneity among intertidal habitats in the flat periwinkle, Littorina obtusata. Mol Ecol. 2007;16:2393–2404. doi: 10.1111/j.1365-294X.2007.03323.x. [DOI] [PubMed] [Google Scholar]
- Segień I, Bogatek R. Cyanide action in plants—from toxic to regulatory. Acta Physiol Plant. 2006;28:483–497. [Google Scholar]
- Steiner CC, Roempler H, Boettger LM, Schoeneberg T, Hoekstra HE. The genetic basis of phenotypic convergence in beach mice: Similar pigment patterns but different genes. Mol Biol Evol. 2009;26:35–45. doi: 10.1093/molbev/msn218. [DOI] [PubMed] [Google Scholar]
- Szymura JM, Barton NH. The genetic-structure of the hybrid zone between the fire-bellied toads Bombina bombina and B. variegata—Comparisons between transects and between loci. Evolution. 1991;45:237–261. doi: 10.1111/j.1558-5646.1991.tb04400.x. [DOI] [PubMed] [Google Scholar]
- Templeton AR, Sing CF, Brokaw B. The unit of selection in Drosphila mercatorum I. The interaction of selection and meiosis in parthenogenetic strains. Genetics. 1976;82:349–376. doi: 10.1093/genetics/82.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till-Bottraud I, Kakes P, Domme B. Variable phenotypes and stable distribution of the cyanotypes of Trifolium repens L. in Southern France. Acta Oecol. 1988;9:393–404. [Google Scholar]
- Trabucco A, Zomer RJ.2009Global Aridity Index (Global-Aridity) and Global Potential Evapo-Transpiration (Global-PET) Geospatial Database. CGIAR Consortium for Spatial Information. Published online, available from the CGIAR-CSI GeoPortal at http://www.csi.cgiar.org/ .
- Umina PA, Weeks AR, Kearney MR, McKechnie SW, Hoffmann AA. A rapid shift in a classic clinal pattern in Drosophila reflecting climate change. Science. 2005;308:691–693. doi: 10.1126/science.1109523. [DOI] [PubMed] [Google Scholar]
- Viette M, Tettamanti C, Saucy F. Preference for acyanogenic white clover (Trifolium repens) in the vole Arvicola terrestris. II. Generalization and further investigations. J Chem Ecol. 2000;26:101–122. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.