Abstract
The additive genetic effects of traits can be used to predict evolutionary trajectories, such as responses to selection. Non-additive genetic and maternal environmental effects can also change evolutionary trajectories and influence phenotypes, but these effects have received less attention by researchers. We partitioned the phenotypic variance of survival and fitness-related traits into additive genetic, non-additive genetic and maternal environmental effects using a full-factorial breeding design within two allopatric populations of Atlantic salmon (Salmo salar). Maternal environmental effects were large at early life stages, but decreased during development, with non-additive genetic effects being most significant at later juvenile stages (alevin and fry). Non-additive genetic effects were also, on average, larger than additive genetic effects. The populations, generally, did not differ in the trait values or inferred genetic architecture of the traits. Any differences between the populations for trait values could be explained by maternal environmental effects. We discuss whether the similarities in architectures of these populations is the result of natural selection across a common juvenile environment.
Keywords: maternal environmental effects, additive genetic effects, non-additive genetic effects, life-history, morphology, conservation
Introduction
The genetic architecture underlying phenotypic traits can be used to predict evolutionary trajectories. In particular, responses to selection are directly related to the amount of additive genetic variance (Falconer and Mackay, 1996). Non-additive genetic effects, on the other hand, have not been considered as important in part because they cannot be used to predict the response to selection (Lynch, 1994). However, there is an increasing evidence that non-additive genetic effects are key components of phenotypes (Crnokrak and Roff, 1995; Roff and Emerson, 2006). Furthermore, non-additive genetic effects are a cause of inbreeding depression (Crnokrak and Roff, 1999; Keller and Waller, 2002) and can be converted to additive genetic effects, for example, during a bottleneck, which can then provide genetic variation for natural selection to act on (Carson, 1990; also see Neff and Pitcher, 2008).
Phenotypic variance can also be explained by maternal environmental effects (Falconer and Mackay, 1996) and these effects can also affect evolutionary trajectories (Räsänen and Kruuk, 2007). For example, maternal environmental effects can have an impact on the rate and direction of change in response to natural selection and can generate rapid phenotypic changes in offspring traits as a result of the phenotypic plasticity of female traits (Mousseau and Fox, 1998; Räsänen and Kruuk, 2007). Consequently, understanding the contributions of all of maternal environmental effects, additive genetic effects and non-additive genetic effects is needed to fully understand evolutionary trajectories and the diversity of phenotypes.
Studies examining the architecture of traits have shown that the relative contributions of genetic and maternal effects can change during development and may be influenced by the correlation between the trait and fitness. Traits expressed during the early life stage tend to be influenced mainly by maternal effects, whereas traits expressed during later life stages are influenced increasingly by genetic effects (Kruuk et al., 2008). Initial egg investments are often fully utilized during early development, leaving later life stage traits that are influenced by genetic effects (for example, Lindholm et al., 2006; Evans et al., 2010). For example, in mammals, maternal effects typically decline, whereas additive genetic effects remain constant (for example, Wilson and Réale, 2006) or increase during development (for example, Cheverud et al., 1983). In addition, life-history traits, such as survival, that have strong correlations with fitness typically have larger non-additive than additive genetic effects, whereas morphological traits, such as body size, that have weaker correlations with fitness typically have larger additive than non-additive genetic effects (Crnokrak and Roff, 1995; Roff and Emerson, 2006). Independent of trait type, directional selection, or to some extent stabilizing selection, on traits can erode additive genetic effects, fixing alleles across loci and leaving only non-additive genetic effects (Willis and Orr, 1993). For example, morphological traits that are under strong directional selection in domestic species often have larger non-additive than additive genetic variances (Roff and Emerson, 2006).
In this study, we examine the phenotypic variance of survival and fitness-related traits at three early life-history developmental stages (egg, alevin and fry) in Atlantic salmon (Salmo salar). Atlantic salmon have declined sharply throughout their North American range over the past 2 centuries (Dunfield, 1985). Lake Ontario once supported the largest freshwater population of Atlantic salmon in the world, but was extirpated by 1900 (COSEWIC, 2010). Several candidate populations are now being considered for reintroduction into Lake Ontario based on ecological evaluations, including Atlantic salmon from the LaHave River, Nova Scotia and Sebago Lake, Maine (Dimond and Smitka, 2005). We used a full-factorial quantitative genetic breeding design to partition phenotypic variance in survival and fitness-related traits to maternal environmental, additive and non-additive genetic effects. The resultant data were used to examine the relative contributions of additive and non-additive genetic effects to morphological and life-history traits, as well as any shift in contributions during early life stages.
Materials and methods
Crosses
Adult broodstock fish from each source population were provided by the Ontario Ministry of Natural Resources (OMNR). For this study, LaHave (n=25) and Sebago families (n=25) were produced on 4 November 2010 at the OMNR Harwood Fish Culture Station, Harwood, Ontario, Canada following the methods of Pitcher and Neff (2006). Five females and five males from each population were mated in all possible combinations to produce full-factorial breeding design (Lynch and Walsh, 1998, p. 598). Subsamples of eggs (n=7) from each female were measured for diameter (nearest 0.01 mm) using digital callipers and mass (nearest 0.0001 g) using a digital scale. Those eggs were then frozen at −20 °C, transported to Western University and kept frozen for subsequent energy content analysis. Remaining eggs were randomly placed into sections of Heath-style incubators and then tanks after hatching at the OMNR Codrington Research Facility, Codrington, Ontario, using two to three sections for each full-sibling family based on offspring numbers (that is, to keep densities in sections equal). Details of the broodstock and rearing of the families are provided in Supplementary Appendix A.
Survival and fitness-related traits
We collected six measures of survival as direct measures of early life fitness: egg survival (fertilized egg to hatch, days 0–83, also examined as a rate over time); alevin survival (post-hatch until yolk sac absorption, days 84–138, also examined as a rate over time); fry survival (yolk sac absorption until released into the wild, days 139–192); and overall survival (fertilized egg until released into the wild). We also measured 12 traits that are known to be related to fitness in salmonids (Metcalfe and Thorpe, 1992; Berg et al., 2001; Pakkasmaa et al., 2001; Koskinen et al., 2002): egg diameter and mass; egg contents at fertilization (relative fat, protein and energy); development time to hatch (also examined as a rate over time); body length at hatch; yolk sac volume at hatch; body length at yolk sac absorption; specific growth rate; and yolk sac conversion efficiency. Details on the methodology to estimate these parameters are provided in Supplementary Appendix A.
Statistical analysis of parental and population effects
All six survival and 12 fitness-related traits were examined for a population effect in addition to individual parental effects (dam and/or sire effects), position effects (tray and tank effects) and density effects using Akaike Information Criteria forward step-wise model selection in R 2.10.1 (available at http://www.r-project.org/). Statistical significance was set at α=0.05, and all non-proportional data were checked visually for approximate normality using histograms before the analysis with parametric statistics (Crawley, 2005). Linear models were used for normally distributed data and binomial models were used for binary data (that is, one for alive and zero for dead and one for hatched and zero for non-hatched). Effects that did not cause a change in Akaike Information Criteria of greater than 10 were considered to be poorly supported and were removed from the model (Burnham and Anderson, 2002). Remaining effects were tested for significance using an analysis of variance of a linear model or an analysis of deviance of a binomial model. Non-significant effects, starting with non-significant interactions, were removed one at a time.
If individual parental effects were retained by the model selection process, the data were analyzed using mixed-effects models that treated individual parental effects as random intercepts and examined population as a fixed effect (in addition to the fixed effects of density if retained by the selection process). Any significant position effect if retained by the selection process was treated as a random intercept. Restricted Maximum Likelihood linear mixed-effects models were used for normally distributed data, and Laplace approximation binomial generalized linear mixed-effects models were used for binary data in the lme4 package of R. The mixed-effects model output in the lme4 package does not produce significance values for fixed effects; therefore, significance for the population effect was determined using a likelihood ratio test between the full model and a reduced model without population.
Statistical analysis of architecture
In addition to parental and population effects, we examined nine out of the 18 survival and fitness-related traits for architecture. The nine traits that were not examined were the overall survival measure, because we could not control for position effects (see Supplementary Appendix A), the five egg traits (that is, diameter, mass, relative fat, protein and energy), because data were collected from only one family for each female, and the three traits examined as a rate over time (that is, egg survival, alevin survival and development time to hatch), because standard analyses cannot incorporate the inclusion of a time variable. First, the phenotypic variance was partitioned into random effects for dam ID (VD, maternal environmental and maternal additive genetic variance), sire ID (VS, paternal additive genetic variance) and dam ID × sire ID (VD × S, non-additive genetic variance) components using a mixed-effects model. We used individual estimates of traits (for example, individual survival and length) to account for within-family variation, because means of family replicates overestimates genetic effects (see Puurtinen et al., 2009; Neff et al. 2011). Means of family replicates were used for specific growth rate and yolk sac conversion efficiency because individual estimates were not available. Regardless of the Akaike Information Criteria criterion noted above, position effects were always included as a random effect to ensure that we did not overestimate non-additive genetic effects. Significances of the variance components were determined by likelihood ratio tests as above. The genetic and environmental variance components were calculated on the basis of (Lynch and Walsh, 1998, p. 509): VD=¼ VA+VM; VS=¼ VA; and VD × S=¼ VN. Negative variance components were set to a value of zero.
Using a similar method outlined in Neff and Fraser (2010), bootstrap 95% confidence intervals (CIs) were produced by first resampling with replacement the individuals within each replicate for each family until the original size was reproduced for the trait assessments. We resampled individuals to account for within-family variation and ensure that the genetic effects were not overestimated (see Puurtinen et al., 2009). We resampled means per replicate for specific growth rate and yolk sac conversion efficiency because individual estimates were not available. Using the resampled data set, additive, non-additive and maternal environmental variance components were calculated as a percentage of the phenotypic variance. The resampling and calculations were repeated 1000 times and the 95% CI was determined for each parameter. In addition, pair-wise population comparisons for each metric were done by calculating for one population the proportion of comparisons that were either larger or smaller than the other population. The proportions served as one-tailed P-values testing for differences between populations.
Results
Summary statistics of survival and fitness-related traits are presented in Table 1 and Supplementary Appendix B. Of the ‘dead' eggs that were removed from the Heath trays, 99.7% (43 397 out of 43 536 eggs) were fertilized. There was nearly 100% offspring mortality for one Sebago female (n=5 families). Thus, the offspring from those families were not used in any of the analyses. Individual parental effects and position effects (in the Heath trays and tanks) had significant influences on survival and fitness-related traits for model selection (Table 2). These effects were subsequently treated as random effects in the mixed-effects models. The examination of architecture revealed that maternal and non-additive genetic effects explained most of the phenotypic variance in survival and fitness-related traits (Figure 1; Supplementary Appendix C).
Table 1. Summary of survival and fitness-related traits from two populations of Atlantic salmon (Salmo salar).
|
Trait
|
n |
LaHave
|
n |
Sebago
|
| Egg traits | ||||
| Diameter (mm) | 35 | 5.72±0.34 | 28 | 5.33±0.40 |
| Mass (g) | 35 | 0.1051±0.0133 | 28 | 0.0864±0.0168 |
| Relative fat (g g−1 of egg) | 35 | 0.0031±0.0077 | 28 | 0.0089±0.0141 |
| Relative protein (g g−1 of egg) | 35 | 0.3702±0.0321 | 28 | 0.3780±0.0387 |
| Relative energy (kJ g−1 of egg) | 35 | 9.00±0.76 | 28 | 9.42±0.88 |
| Egg survival (%) | ||||
| Over time | 75 | −3.29 × 10−3±2 × 10−5 | 40 | −4.14 × 10−3±3 × 10−5 |
| Days 0–83 | 75 | 69.1±19.0 | 40 | 53.8±19.9 |
| Alevin survival (%) | ||||
| Over time | 75 | −3.30 × 10−2±6 × 10−3 | 40 | -2.30 × 10−2±5 × 10−3 |
| Days 84–138 | 75 | 84.0±8.2 | 40 | 79.9±8.8 |
| Fry survival (%) | ||||
| Days 139–192 | 75 | 61.3±19.5 | 40 | 58.0±19.0 |
| Overall survival (%) | 25 | 35.7±10.2 | 20 | 23.6±14.1 |
| Development time | ||||
| Over time | 75 | 2.42 × 10−1±2 × 10−3 | 40 | 1.11 × 10−1±1 × 10−3 |
| To hatch (degree-days) | 75 | 479.8±6.4 | 40 | 472.3±12.1 |
| Size traits | ||||
| Body length at hatch (mm) | 750 | 16.3±0.8 | 400 | 15.6±0.8 |
| Yolk sac volume (mm3) | 750 | 72±17 | 400 | 64±15 |
| Body length at yolk sac absorption (mm) | 750 | 25.8±1.0 | 400 | 25.7±1.2 |
| Energy conversion | ||||
| Specific growth rate (100 × ln(mm)/degree-days) | 75 | 0.146±0.007 | 40 | 0.146±0.009 |
| Yolk sac conversion efficiency (mm mm−3) | 75 | 0.136±0.016 | 40 | 0.158±0.018 |
Presented are means±1 s.d., except for over time traits that are logit estimate±1 s.e. There were 25 LaHave families (5 females × 5 males) and 20 Sebago families (4 females × 5 males). Egg traits were based on seven eggs per female. Survival, development time to hatch and energy conversion numbers (n) represent the total number of replicates: three per LaHave family and two per Sebago family. Size traits were represented by 10 individuals per replicate. For example, n of 35 for LaHave egg traits is based on seven eggs from each of the five females and n of 750 for LaHave size traits is based on 10 individuals from each of the three replicates from each of the 25 families.
Table 2. Model selection and mixed-effects model results for survival and fitness-related traits in two populations of Atlantic salmon (Salmo salar).
|
Trait
|
Model selection
|
Mixed-effects model population effect, P-value
|
| Egg traits | ||
| Diameter | dam ID | 0.022 |
| Mass | dam ID | 0.021 |
| Relative fat | No effects | |
| Relative protein | No effects | |
| Relative energy | dam ID | 0.140 |
| Egg survival | ||
| Over time | Degree-days+dam ID+tray ID+sire ID+degree-days × dam ID+degree-days × sire ID+degree-days × tray ID | <0.001 |
| Days 0–83 | dam ID+tray ID+sire ID | 0.126 |
| Alevin survival | ||
| Over time | Degree-days+dam ID+sire ID+tank ID+degree-days × dam ID+degree-days × tank ID+degree-days × sire ID | <0.001 |
| Days 84–138 | dam ID+tank ID+sire ID | 0.196 |
| Fry survival | ||
| Days 139–192 | dam ID+tank ID+sire ID | 0.451 |
| Overall survival | dam ID+sire ID | 0.104 |
| Development time | ||
| Over time | Degree-days+dam ID+tray ID+sire ID+degree-days × dam ID+degree-days × tray ID+degree-days × sire ID | <0.001 |
| To hatch | dam ID+tray ID+sire ID | <0.001 |
| Size traits | ||
| Body length at hatch | dam ID+sire ID | 0.022 |
| Yolk sac volume | dam ID+sire ID | 0.226 |
| Body length at yolk sac absorption | dam ID+tank ID+sire ID | 0.117 |
| Energy conversion | ||
| Specific growth rate | dam ID+tank ID+sire ID | 0.372 |
| Yolk sac conversion efficiency | dam ID sire ID+tank ID | <0.001 |
All mixed-effects models contained a fixed effect for population. Mixed-effects models also contained fixed effects for density and degree-days, and random effects for dam ID, sire ID, tray ID and tank ID, if these effects were identified during model selection.
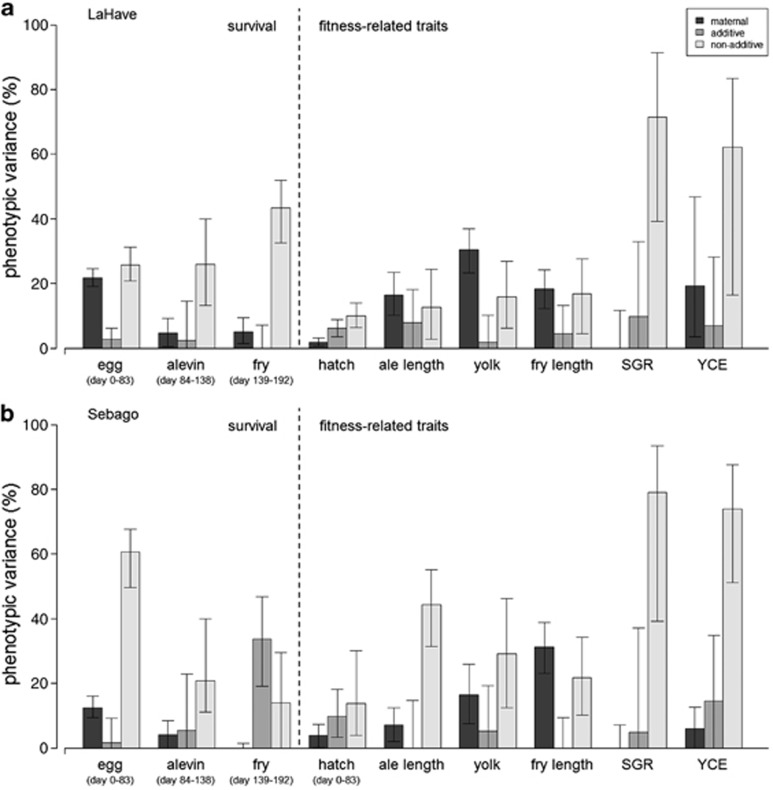
Figure 1.
The architecture underlying the phenotypic variance of survival and fitness-related traits in Atlantic salmon (Salmo salar). Shown are data from two populations: (a) LaHave and (b) Sebago. Displayed are the median and 95% confidence intervals (CI) for maternal environmental, additive genetic and non-additive genetic effects. Hatch is development time to hatch; ale length is body length at hatch; yolk is yolk sac volume; fry length is body length at yolk sac absorption; SGR is specific growth rate; and YCE is yolk sac conversion efficiency (see Supplementary Appendix A for details).
Survival
In both populations, dam effects were significant for egg survival, alevin survival (LaHave only), but not for fry survival (Supplementary Appendix C). Sire effects were not significant for either population, whereas dam × sire effects were significant for egg survival, but not for alevin survival and fry survival (Sebago only). For the LaHave population, maternal environmental and non-additive genetic effects were similar in their contribution to egg survival, but maternal environmental effects decreased, whereas non-additive genetic effects increased during the alevin and fry stages (Figure 1). On the other hand, for the Sebago population, non-additive genetic effects were larger than maternal environmental effects in their contribution to egg survival and both non-additive genetic and maternal environmental effects decreased during the alevin and fry stages. These patterns resulted in differences between populations in the genetic architecture (additive and non-additive genetic effects) of offspring survival. Sebago had significantly higher non-additive genetic effects for egg survival but lower non-additive genetic effects for fry survival than LaHave (randomization routine one-tailed P=0.001). Differences were also observed between the populations for maternal environmental effects. LaHave had significantly higher maternal environmental effects for egg and fry survival (randomization routine one-tailed P=0.001). There was no significant correlations between any of the egg content measures (relative fat, protein and energy) and offspring survival (Pearson correlations, P>0.05 for all; data not shown).
Fitness-related traits
Dam effects were significant for the LaHave population for development time to hatch, body length at hatch, yolk sac volume, body length at yolk sac absorption, specific growth rate and yolk sac conversion efficiency (Supplementary Appendix C). Dam effects were significant for fewer fitness-related traits for the Sebago population: development time to hatch, yolk sac volume and body length at yolk sac absorption. Sire effects on the fitness-related traits were not significant in either population, whereas dam × sire effects were significant for the traits; the exceptions being for LaHave development time to hatch and body length at hatch (Supplementary Appendix C). In both populations, non-additive genetic effects explained more of the phenotypic variance than maternal environmental effects for development time to hatch, body length at hatch (Sebago only), yolk sac volume (Sebago only), specific growth rate and yolk sac conversion efficiency (Figure 1). On the other hand, maternal environmental effects explained more of the phenotypic variance than non-additive genetic effects for body length at hatch (LaHave only), yolk sac volume (LaHave only) and body length at yolk absorption. There were no significant differences between populations in the majority of the genetic architecture values for the fitness-related traits (randomization routine one-tailed P>0.05), with exception that Sebago had significantly higher non-additive genetic effects for body length at hatch than LaHave (randomization routine one-tailed P=0.012; Figure 1). In contrast, there were significant differences between the populations in maternal environmental effects. LaHave had significantly higher maternal environmental effects for body length at hatch, yolk sac volume and yolk sac conversion efficiency, but lower maternal environmental effects for body length at yolk sac absorption when compared with Sebago (randomization routine one-tailed P<0.05). There was no significant correlations between any of the egg content measures (relative fat, protein and energy) and the fitness-related traits (Pearson correlations, P>0.05 for all; data not shown).
Population differences in performance
The populations differed at only 8 of the 18 traits that we measured (Table 2). These traits included egg survival (rate only), alevin survival (rate only), egg diameter and mass, body length at hatch, development time to hatch (rate and degree-days) and yolk sac conversion efficiency. For example, egg survival for the Sebago population declined at a faster rate than the LaHave population (Table 1). The opposite pattern was detected for alevin survival. However, the differences were generally small between the populations for egg diameter (0.4 mm, 7% of the mean) and mass (0.02 g, 21%), body length at hatch (0.7 mm, 4%) and development time to hatch (7 degree-days, 2%). LaHave hatched at a faster rate than Sebago. The populations were not significantly different in body length at yolk sac absorption despite Sebago possessing a higher yolk sac conversion efficiency than LaHave (Table 1 and 2). Although survival rates differed between the populations, there were no significant differences between the populations in the final egg and alevin survival values (that is, days 83 and 138) (Table 2).
Discussion
Our results detected maternal environmental and genetic effects that explained more than half (mean±1s.e.: 52±6%) of the phenotypic variance in survival and various fitness-related traits. Maternal environmental effects were prominent at early life stages, decreased during development, and non-additive effects becoming most prominent at later life stages. Across the nine traits that we could examine the genetic architecture of, we found that non-additive genetic effects were more prominent than additive effects. Our results also revealed that the LaHave and Sebago populations did not differ, or had small differences, for the majority of traits and, similarly, did not differ in the genetic architecture of those traits. The few differences between the populations that did exist were mostly attributed to differences in maternal environmental effects.
Maternal environmental and genetic effects may be important in explaining the phenotypic variance of survival and fitness-related traits (Qvarnström and Price, 2001). We found significant maternal environmental effects in the nine traits examined for architecture, and those effects explained a mean of 14±3% of the phenotypic variance across the traits. We also found sire effects in the traits, with additive genetic effects explaining a mean of 8±2% of the phenotypic variance. Similarly, 16 other studies, examining some 60 different survival and fitness-related traits in natural populations, found that maternal environmental effects explained a mean of 26±3% of the phenotypic variance in the traits and that additive genetic effects explained a bit less at a mean of 18±3% (see references in Table 1 in Puurtinen et al., 2009; also see Evans et al., 2010). Collectively, these data suggest that maternal environmental effects may be the primary factor contributing to survival and fitness-related traits during early development, although additive genetic effects also contribute to phenotypic variance during this life stage.
The amount of phenotypic variance explained by maternal environmental and genetic effects may shift during development (Fox et al., 2003; Evans et al., 2010). Early life stages that rely on maternal investments such as egg nutrients often have phenotypic variances explained more by maternal environmental effects (reviewed by Wilson and Réale, 2006). Later life stages instead have phenotypic variances largely explained by genetic effects, because maternal investments have been fully utilized (Wilson and Réale, 2006). We found that maternal environmental effects explained a mean of 21±3% of the phenotypic variance across the four traits related to egg investments (egg survival, body length at hatch, yolk sac volume and body length at yolk sac absorption), but that genetic effects also explained a similar amount of the variance in these traits (27±6%). We also found that genetic effects, largely influenced by non-additive effects, explained a mean of 47±9% of the phenotypic variance across the remaining five traits that were collected after hatch. Maternal environmental effects, on the other hand, captured only 8±4% of the variance in those traits. Similarly, other studies have found that maternal environmental and genetic effects explained about equal amounts of the phenotypic variance for early life stage traits (see references in Table 1 in Puurtinen et al., 2009; also see Evans et al., 2010). Furthermore, those studies also found that genetic effects explained 50±9% and maternal environmental effects explained only 10±4%, on average, of the phenotypic variance for traits expressed during later life stages. Thus, the data support a shift from maternal environmental effects to genetic effects during development, but also suggest that non-additive genetic effects have an important role in survival and fitness-related traits.
Life-history and morphological traits may differ in the amount of genetic variance explained by additive and non-additive genetic effects. Life-history traits, which have strong correlations with fitness, typically have large non-additive genetic effects, whereas morphological traits, which have weak correlations with fitness, tend to have large additive genetic effects (Crnokrak and Roff, 1995; Roff and Emerson, 2006). However, a review recently suggested that additive and non-additive effects contribute about equally to both life-history and morphological traits (Puurtinen et al., 2009). We found that non-additive genetic effects were larger than additive genetic effects for all nine traits that we examined for architecture, except for fry survival and development time to hatch in the Sebago population and body length at hatch in the LaHave population. Non-additive and additive genetic effects explained means of 30±6% and only 8±2%, respectively, of the phenotypic variance across the traits. In our case, the morphological traits—body length at hatch, yolk sac volume and body length at yolk sac absorption—may have possessed larger non-additive genetic effects because these traits typically have strong correlations with fitness in salmonids (see Koskinen et al., 2002); morphological traits in other wild mammal populations typically have weak correlations with fitness (see Crnokrak and Roff, 1995; Roff and Emerson, 2006). Our data support the idea that non-additive genetic effects are larger than additive genetic effects for traits that have strong correlations with fitness and that this pattern may be independent of whether the traits are life-history or morphological in nature. Some caution is warranted when making these comparisons in our data set because our analysis is based on 5 × 5 crosses (albeit both populations revealed analogous patterns).
Components that explain the phenotypic variance of fitness-related traits may be preserved across populations either by shared ancestry or by parallel evolution (Schluter et al., 2004). If populations possess similar traits, perhaps not surprisingly, the genetic architecture of those traits may also be similar because natural selection may have acted on the same genes that underlie the traits (Campbell and Bernatchez, 2004; Turner et al., 2010; Schumer et al., 2011). We found that the Atlantic salmon had similar genetic architectures (amounts of additive and non-additive effects) for seven out of the nine traits despite having a relatively large FST value. In contrast, the populations differed in the maternal environmental effects for six out of the nine traits. Similarities in the genetic architectures of these populations could be the result of parallel evolution mediated by natural selection across a common juvenile environment (that is, stream environment) (Schluter et al., 2004). For example, the LaHave River and Sebago Lake stream environments have similar temperatures (J Gibson, Department of Fisheries and Oceans and M Simpson, Maine Department of Marine Resources, unpublished data), which is a key component of natural selection on fitness-related traits in the Atlantic salmon (Garcia de Leaniz et al., 2007). Differences in maternal environmental effects of populations may instead reflect the differences in the adult environments (ocean vs lake) (see Räsänen and Kruuk, 2007), but more research is needed to better understand the source of those differences. Interestingly, both sources of broodstock (parents) used in our study came from a common captive hatchery environment, suggesting that the maternal environmental effects are not directly affected by the rearing environment of the dams. Regardless, the similar genetic architectures underlying the traits examined here may have resulted from parallel evolution.
In conclusion, our results detail the genetic and maternal environmental effects on phenotypic variance of survival and fitness-related traits in early life stages of two populations of Atlantic salmon. We found that the populations had similar trait values and underlying genetic architecture for the majority of traits. Our results support a shift from the maternal environmental to genetic effects during early development. They also indicate that non-additive genetic variance is as or more important for survival and fitness-related traits than additive genetic variance.
Data archiving
Survival and fitness-related trait data have been submitted to Dryad: doi:10.5061/dryad.75030.
Acknowledgments
The research was supported by the Natural Sciences and Engineering Research Council of Canada through a postgraduate student research award to ASH and a Strategic Project Grant to BDN. The research was also supported by the Ontario Ministry of Natural Resources, Ontario Federation of Anglers and Hunters, and Metro East Anglers. W Sloan, S Ferguson and B Lewis provided invaluable support and assistance at the OMNR Codrington Research Facility. We are grateful to the three anonymous referees for their constructive comments on an earlier manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Berg OK, Hendry AP, Svendsen B, Bech C, Arnekleiv JV, Lohrmann A. Maternal provisioning of offspring and the use of those resources during ontogeny: variation within and between Atlantic salmon families. Funct Ecol. 2001;15:13–23. [Google Scholar]
- Burnham KP, Anderson DR.2002Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach2nd ednSpringer-Verlag: New York, NY, USA [Google Scholar]
- Campbell D, Bernatchez L. Generic scan using AFLP markers as a means to assess the role of directional selection in the divergence of sympatric whitefish ecotypes. Mol Biol Evol. 2004;21:945–956. doi: 10.1093/molbev/msh101. [DOI] [PubMed] [Google Scholar]
- Carson HL. Increased genetic variance after a population bottleneck. Trends Ecol Evol. 1990;5:228–230. doi: 10.1016/0169-5347(90)90137-3. [DOI] [PubMed] [Google Scholar]
- Cheverud JM, Rutledge JJ, Atchley WR. Quantitative genetics of development: genetic correlations among age-specific trait values. Evolution. 1983;37:895–905. doi: 10.1111/j.1558-5646.1983.tb05619.x. [DOI] [PubMed] [Google Scholar]
- Committee on the Status of Endangered Wildlife in Canada [COSEWIC] 2010COSEWIC assessment and status report on the Atlantic salmon Salmo salar (anadromous form) in Canada Committee in the Status of Endangered Wildlife in Canada, Ottawa, CanadaAvailable at www.sararegistry.gc.ca/status/status_e.cfm .
- Crawley MJ. Statistics: An Introduction Using R. John Wiley and Sons: Chichester, West Sussex, UK; 2005. [Google Scholar]
- Crnokrak P, Roff DA. Dominance variance: associations with selection and fitness. Heredity. 1995;75:530–540. [Google Scholar]
- Crnokrak P, Roff DA. Inbreeding depression in the wild. Heredity. 1999;83:260–270. doi: 10.1038/sj.hdy.6885530. [DOI] [PubMed] [Google Scholar]
- Dimond P, Smitka J.2005Evaluation of selected strains of Atlantic salmon as potential candidates for the restoration of Lake Ontario Trout Unlimited Can Tech Rep ON-012Available at www.tucanada.org/reports/ON-012_AtlanticSalmonStrains.pdf .
- Dunfield RW. The Atlantic salmon in the history of North America. Can Spec Publ Fish Aquat Sci. 1985;80:181. [Google Scholar]
- Evans ML, Neff BD, Heath DD. Quantitative genetic and translocation experiments reveal genotype-by-environment effects on juvenile life-history traits in two populations of Chinook salmon (Oncorhynchus tshawytscha) J Evolution Biol. 2010;23:687–698. doi: 10.1111/j.1420-9101.2010.01934.x. [DOI] [PubMed] [Google Scholar]
- Falconer DS, Mackay TFC. Introduction to Quantitative Genetics. Longman: Harlow, Essex, UK; 1996. [Google Scholar]
- Fox CW, Bush ML, Wallin WG. Maternal age affects offspring lifespan of the seed beetle, Callosobruchus maculatus. Funct Ecol. 2003;17:811–820. [Google Scholar]
- Garcia de Leaniz CG, Fleming IA, Einum S, Verspoor E, Jordan WC, Consuegra S, et al. A critical review of adaptive genetic variation in Atlantic salmon: implications for conservation. Biol Rev. 2007;82:173–211. doi: 10.1111/j.1469-185X.2006.00004.x. [DOI] [PubMed] [Google Scholar]
- Keller LF, Waller DM. Inbreeding effects in wild populations. Trends Ecol Evol. 2002;17:230–241. [Google Scholar]
- Koskinen MT, Haugen TO, Primmer CR. Contemporary fisherian life history evolution in small salmonid populations. Nature. 2002;419:826–830. doi: 10.1038/nature01029. [DOI] [PubMed] [Google Scholar]
- Kruuk LEB, Slate J, Wilson AJ. New answers for old questions: the evolutionary quantitative genetics of wild animal populations. Annu Rev Ecol Evol Syst. 2008;39:525–548. [Google Scholar]
- Lindholm AK, Hunt J, Brooks R. Where do all the maternal effects go? Variation in offspring body size through ontogeny in the live-bearing fish Poecilia parae. Biol Lett. 2006;2:586–589. doi: 10.1098/rsbl.2006.0546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch CB.1994Evolutionary inferences from genetic analysis of cold adaptation in laboratory and wild populations of the house mouse, Mus domesticusIn: Boake CRB (ed)Quantitative Genetic Studies of Behavioral Evolution University of Chicago Press: Chicago, IL, USA [Google Scholar]
- Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. Sinauer Associates: Sunderland, MA, USA; 1998. [Google Scholar]
- Metcalfe NB, Thorpe JE. Early predictors of life history events: the link between first feeding date, dominance and seaward migration in Atlantic salmon, Salmo salar. J Fish Biol. 1992;41:93–99. [Google Scholar]
- Mousseau TA, Fox CW. The adaptive significance of maternal effects. Trends Ecol Evol. 1998;13:403–407. doi: 10.1016/s0169-5347(98)01472-4. [DOI] [PubMed] [Google Scholar]
- Neff BD, Fraser BA. A program to compare genetic differentiation statistics across loci using resampling of individuals and loci. Mol Ecol Resour. 2010;10:546–550. doi: 10.1111/j.1755-0998.2009.02786.x. [DOI] [PubMed] [Google Scholar]
- Neff BD, Pitcher TE. Mate choice for non-additive genetic benefits: a resolution to the lek paradox. J Theor Biol. 2008;254:147–155. doi: 10.1016/j.jtbi.2008.05.019. [DOI] [PubMed] [Google Scholar]
- Neff BD, Garner SR, Pitcher TE. Conservation and enhancement of wild fish populations: preserving genetic fitness versus genetic diversity. Can J Fish Aquat Sci. 2011;68:1149–1154. [Google Scholar]
- Pakkasmaa S, Peuhkuri N, Laurila A, Hirvonen H, Ranta E. Female and male contribution to egg size in salmonids. Evol Ecol. 2001;15:143–153. [Google Scholar]
- Pitcher TE, Neff BD. MHC class IIB alleles contribute to both additive and nonadditive genetic effects on survival in Chinook salmon. Mol Ecol. 2006;15:2357–2365. doi: 10.1111/j.1365-294X.2006.02942.x. [DOI] [PubMed] [Google Scholar]
- Puurtinen M, Ketola T, Kotiaho JS. The good-genes and compatible-genes benefits of mate choice. Am Nat. 2009;174:741–752. doi: 10.1086/606024. [DOI] [PubMed] [Google Scholar]
- Qvarnström A, Price TD. Maternal effects, paternal effects and sexual selection. Trends Ecol Evol. 2001;16:95–100. doi: 10.1016/s0169-5347(00)02063-2. [DOI] [PubMed] [Google Scholar]
- Räsänen K, Kruuk LEB. Maternal effects and evolution at ecological time-scales. Funct Ecol. 2007;21:408–421. [Google Scholar]
- Roff DA, Emerson K. Epistasis and dominance: evidence for differential effects in life-history versus morphological traits. Evolution. 2006;60:1981–1990. [PubMed] [Google Scholar]
- Schluter D, Clifford EA, Nemethy M, McKinnon JS. Parallel evolution and inheritance of quantitative traits. Am Nat. 2004;163:809–822. doi: 10.1086/383621. [DOI] [PubMed] [Google Scholar]
- Schumer M, Krishnakant K, Renn SCP. Comparative gene expression profiles for highly similar aggressive phenotypes in male and female cichlid fishes (Julidochromis) J Exp Biol. 2011;214:3269–3278. doi: 10.1242/jeb.055467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner TL, Bourne EC, Von Wettberg EJ, Hu TT, Nuzhdin SV. Population resequencing reveals local adaptation of Arabidopsis lyrata to serpentine soils. Nat Genet. 2010;42:260–263. doi: 10.1038/ng.515. [DOI] [PubMed] [Google Scholar]
- Willis JH, Orr HA. Increased heritable variation following population bottlenecks: the role of dominance. Evolution. 1993;47:949–957. doi: 10.1111/j.1558-5646.1993.tb01249.x. [DOI] [PubMed] [Google Scholar]
- Wilson AJ, Réale D. Ontogeny of additive and maternal genetic effects: lessons from domestic mammals. Am Nat. 2006;167:E23–E38. doi: 10.1086/498138. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.