Abstract
Cottus rhenanus and Cottus perifretum have formed hybrid lineages and narrow hybrid zones that can be best explained through the action of natural selection. However, the underlying selective forces as well as their genomic targets are not well understood. This study identifies genomic regions in the parental species that cause hybrid incompatibilities and tests whether these manifest in a sex-specific manner to learn about processes that affect natural hybridization in Cottus. Interspecific F2 crosses were analyzed for 255 markers for genetic mapping and to detect transmission distortion as a sign for genetic incompatibilities. The Cottus map consists of 24 linkage groups with a total length of 1575.4 cM. A male heterogametic (XY) sex determination region was found on different linkage groups in the two parental species. Genetic incompatibilities were incomplete, varied among individuals and populations and were not associated with the heterogametic sex. The variance between populations and individuals makes it unlikely that there are species-specific incompatibility loci that could affect the gene pool of natural hybrids in a simple and predictable way. Conserved synteny with sequenced fish genomes permits to genetically study the Cottus genome through the transfer of genomic information from the model fish species. Homology relationships of candidate genomic regions in Cottus indicate that sex determination is not based on the same genomic regions found in other fish species. This suggests a fast evolutionary turnover of the genetic basis of sex determination that, together with the small size of the heterogametic regions, may contribute to the absence of fitness effects related to the Haldane's rule.
Keywords: scorpaeniformes, postzygotic isolation, reference genome, hybrid speciation, hybrid zone
Introduction
Evolutionarily young and ecologically distinct lineages that diverge, although limited introgressive hybridization is still possible, are of high interest to understand how new species form. In such systems, the nature of barriers to reproduction and genetic factors that affect the fitness of hybrid offspring represent a central aspect to explain evolutionary processes. For example, an important debate relates to the relative importance of intrinsic and extrinsic barriers as species diverge (Wolf et al., 2010) or whenever hybridization occurs (Barton and Gale, 1993; Bierne et al., 2011). Besides the nature of the selective forces, little is known about numbers of incompatibility factors and how they vary between individuals for most species. Genetic factors that affect the fitness of hybrids and their offspring are also crucial to understand the evolutionary dynamics that take place when a lineage of hybrid origin emerges. An admixed gene pool initially comprises a diverse mixture of alleles with varying fitness effects in different genotype combinations. Accordingly, selection can set evolutionary trajectories that determine which combinations of parental alleles are purged from an admixed gene pool. This process has received little attention in empirical studies on the first steps of hybrid speciation to date (Mallet, 2007; Nolte and Tautz, 2010; Abbott et al., 2013), and, much like the discussion that revolves around barriers to reproduction, it is possible that loci are affected by purely intrinsic or extrinsic selective pressures.
We are studying the European freshwater sculpins Cottus rhenanus and Cottus perifretum because they can yield insights into the early evolutionary processes that lead to hybrid speciation (Nolte et al., 2005a; Nolte and Tautz, 2010; Stemshorn et al., 2011; Czypionka et al., 2012) and to learn about the genetic factors that mold narrow hybrid zones (Nolte et al., 2006, 2009). Briefly, man-made environmental changes within the past 200 years have increased the connectivity of the rivers Rhine and Scheldt and massively altered the ecological conditions in parts of these drainages. These perturbations have fostered hybridization of C. rhenanus and C. perifretum from smaller tributaries (Nolte et al. 2005a). While a phylogeographic analysis indicates that the parental taxa have been separated for up to 2 million years (Englbrecht et al. 2000, but see Volckaert et al. (2002) for an alternative view), the hybrid sculpins are possibly less than 200 generations old and were first detected at a massive scale in the Netherlands after 1980. They are referred to as ‘invasive', because they invaded perturbed habitats that are not occupied by their parental species. The ranges of invasive Cottus and C. rhenanus abut where small streams disembogue into larger rivers, an area that also represents the transition zone between different habitats along an otherwise continuous river. Our analysis of these contact zones (Nolte et al., 2006, 2009) provides evidence that selection associated with different habitats counteracts genetic admixture. With respect to the evolution of the invasive lineage itself, a small fraction of the invasive genome appears to be subject to genotypic selection (Stemshorn et al., 2011). Further, an analysis of changes in gene expression provided evidence that novel phenotypes have evolved (Czypionka et al., 2012), which highlights the need to identify the evolutionary processes at work. To date, the above results about the divergence among parental populations and hybrid lineages were discussed with a strong focus on extrinsic factors related to differentiated ecological settings. However, it can be difficult to disentangle the effects of intrinsic and extrinsic barriers to reproduction, as correlations are expected to arise between unlinked loci in structured populations and because hybrid zone clines maintained by intrinsic or extrinsic barriers are difficult to distinguish (Barton and Gale, 1993; Bierne et al., 2011; Taylor et al., 2013). Hence, testing of hypothesis related to intrinsic barriers is warranted to complement our inference that evolutionary processes in Cottus within the River Rhine are strongly determined by extrinsic, ecological factors. An intrinsic mechanism that is thought to critically affect hybrid fitness is post zygotic isolation caused by Dobzhansky–Müller incompatibilities. These result from genetic changes that, while functional in their normal genetic backgrounds, reduce the viability or fertility when recombined in F1 hybrids. The evolution of intrinsic barriers to reproduction is believed to be a slow process that is outpaced by ecologically driven adaptive evolution (Wolf et al., 2010). However, partial effects, especially those that manifest only in the F2 generation are expected to evolve more quickly (Edmands, 1999; White et al., 2011). In F2 crosses or later generation hybrids, intrinsic traits can cause selective mortality if essential regulatory interactions are disrupted when ancestral genotypes segregate (Maheshwari and Barbash, 2011). The evolution of such incompatibilities can be explained through different hypothesis related to Haldane's rule (Wolf et al., 2010). Accordingly, the first incompatibilities between young species would manifest in the heterogametic sex. Although it is not clear whether this applies to fishes in general (Russell, 2003), the genetic regions that determine the sex and the identification of the heterogametic sex are of particular interest to study genetic incompatibilities.
Genetic mapping experiments can complement or challenge the view that differentiation in Cottus is determined by ecological factors by answering whether intrinsic genetic incompatibilities affect the fitness of hybrids or not. We have previously generated low-resolution genetic maps by typing 154 random microsatellite loci in F1 crosses among invasive (hybrid lineage) Cottus and C. rhenanus populations from Germany (Stemshorn et al., 2005, 2011). These indicated that distantly related model organisms can be used to infer the gene order of genomic regions in Cottus and that major chromosomal rearrangements do not occur among different species of Cottus. In this study, we generated a more evenly covered genetic map from interspecific crosses among pure C. rhenanus and pure C. perifretum, the parental species that gave rise to invasive Cottus in nature. Given an improved genetic map, the whole genome was scanned for the presence of ancestral genetic factors that could affect the fitness of hybrids, and QTL (quantitative trait locus) analysis was used to identify the genetic basis of sex differentiation in Cottus. We evaluate whether incompatibility factors manifest in the heterogametic sex and discuss the relevance of the results for the evolutionary processes that affect hybridization and admixture of Cottus within the lower River Rhine basin. Next, we interpret our results based on patterns of evolutionary conservation in Cottus with distantly related model fish species. A combined use of Cottus EST (expressed sequence tag) sequences together with the refined map permits inferring homology relationships among genomic regions of fishes, which we employ to explore the evolutionary origins of sex differentiation in the Cottus genome.
Materials and methods
Mapping strategy and taxa used
Interspecific crosses can be used to construct linkage maps and to map regions that determine sex differentiation with the limitation that species-specific chromosomal rearrangements are difficult to resolve if they occur. Although combining mapping data is challenging, the use of several families and populations provides us with independent replicates to test the generality of patterns. The focus of this study is on four separate populations representing the ancestral species that gave rise to invasive sculpins. C. rhenanus were sampled from the small streams Broel (GIS: 50°50′N 7°22′E; Stream Broel between Bröl and Winterscheidt, Germany) and Naaf (GIS: 50°53′N 7°15′E; east of Wahlscheidt, Germany); C. perifretum were sampled from the small streams Witte Nete (WN) (GIS: 53°14′N 5°04′E; Flanders, Belgium) and Laarse Beek (LB) (GIS: 51°17′N 5°04′E; Flanders, Belgium). C. rhenanus from the Broel and Naaf populations display significant differentiation with an FST of 0.3 (Nolte et al., 2006) and the same holds for C. perifretum populations from the Witte Nete and Laarse Beek (FST=0.368, Knapen et al., 2003) (for a discussion of the divergence between C. rhenanus and C. perifretum see Englbrecht et al. (2000)). Functional differentiation in Cottus was studied using patterns of gene expression (Czypionka et al., 2012) and underlines that the same populations that were studied here have diverged within and between species.
Two independent groups of interspecific F2 crosses were obtained from pure C. perifretum and C. rhenanus, and we performed alternative cross directions with respect to the sex of the parents (Table 1). The mapping families were generated from different outbred grandparents. All fishes were kept in laboratory aquaria and fed with invertebrates. The temperature and light regime mimicked the conditions in central Europe with a winter temperature of 4 °C for at least 1 month. Spawning occurred in artificial shelters when water temperatures were raised to 8–10 °C (March 2010). A first group of 625 individuals were killed when they had reached a total length of approximately 3 cm (September 2010). The genotypes of these fish were used to estimate recombination rates, but sex was not determined. Another 273 individuals were raised until the gonads were mature for visual examination in March 2011 and used to map the genetic regions underlying sexual differentiation. Fish were killed after administering anesthesia with CO2 and preserved in RNAlater or in 70% ethanol at −20 °C. All families used for map construction and QTL analysis were shown in Table 1.
Table 1. Interspecies F2 crosses between C. rhenanus and C. perifretum were used for map construction and to identify the genetic basis of sex.
| Cross type | Mapping family | Total | Map construction | Sex mapping | Males | Females | Sex ratio | Balanced sex ratio(P-value) |
|---|---|---|---|---|---|---|---|---|
| Broel(♂) × WN(♀) | ZG11 | 104 | 104 | NA | NA | NA | NA | NA |
| ZG18 | 76 | 76 | NA | NA | NA | NA | NA | |
| ZG33 | 52 | NA | 52 | 24 | 28 | 0.462 | 0.579 | |
| ZG42 | 97 | NA | 97 | 51 | 46 | 0.526 | 0.612 | |
| WN(♂) × Broel(♀) | ZG40 | 86 | 58 | 28 | 18 | 10 | 0.643 | 0.131 |
| ZG46 | 75 | 55 | 20 | 15 | 5 | 0.750 | 0.025 | |
| LB(♂) × Naaf(♀) | ZG26 | 117 | 96 | 21 | 10 | 11 | 0.476 | 0.827 |
| ZG32 | 115 | 98 | 17 | 10 | 7 | 0.588 | 0.467 | |
| ZG35 | 38 | NA | 38 | 21 | 17 | 0.553 | 0.516 | |
| ZG38 | 70 | 70 | NA | NA | NA | NA | NA | |
| Naaf(♂) × LB(♀) | ZG39 | 68 | 68 | NA | NA | NA | NA | NA |
The cross type indicated the origin of the parental populations (Broel, Naaf, LB, WN) and the gender of the parental generation. Individual mapping families are listed together with the total number of offspring that were raised and used for map construction or to QTL map sex. The numbers of males and females are given for each mapping family. Sex ratios were found to be significantly skewed toward a male excess in family ZG46, but this pattern was not generally observed.
Cottus transcriptome sequencing
Individual transcriptome libraries were generated from four individuals for each parental population as well as six invasive Cottus from the river Sieg (GIS: 50°48′N 7°9′E; Sieg west of Siegburg, Germany). The latter samples were solely included in the assembly of EST contigs but not used to ascertain SNPs. Total RNA from whole fish was extracted using the TRIzol Reagent protocol (Invitrogen, Carlsbad, CA, USA). Enrichment for poly(A) mRNA was conducted using uMACS mRNA isolation kits (Miltenyi Biotec, Bergisch Gladbach, Germany), and full-length complementary DNA was synthesized from each poly(A) mRNA sample following the SMART PCR cDNA synthesis protocol (Clontech, Mountain View, CA, USA). All cDNA samples were PCR amplified using the Advantage 2 PCR kit (Clontech) with PCR conditions as follows: 1 min at 95 °C, followed by 16–18 cycles of 30 s at 95 °C, 30 s at 65 °C and 6 min at 68 °C. Amplification products were quantified using Agilent RNA chips (Agilent Technologies, Santa Clara, CA, USA). We followed the 454 GS FLX Titanium General Library Preparation Method Manual (April 2009), but used adapters with individual tags to assign reads back to individuals after sequencing. Equimolar amounts of all libraries were pooled and sequenced on a Roche GS-FLX DNA Sequencer following the protocol from the company.
EST Contig assembly and sequence comparison with model fish genomes
Sample-specific tags were removed from the sequences using Roche analysis software SFF Tool (Roach Genome sequencer data analysis software manual, 2008), and reads were assigned to the individuals of origin. Primer trimming and de novo assembly were performed using Roche analysis software Newbler with default criteria. The resulting EST contig sequences (>500 bp) were BLAST mapped against publicly available genomes of 5 fully sequenced teleost model fish species: zebrafish (Danio rerio), fugu (Takifugu rubripes), tetraodon (Tetraodon nigroviridis), medaka (Oryzias latipes) and stickleback (Gasterosteus aculeatus). The nucleotide, EST and protein sequences of the reference genomes were downloaded from the Ensembl Genome Browser (http://www.ensembl.org/index.html) and the International Fugu Genome Consortium (http://www.fugu-sg.org/project/info.html). BLAST searching was tuned for distant homologies using an E-value <1e−5 as thresholds to accept a hit. The genomes were searched using blastn and blastx, and the number of hits was noted at the nucleotide and at the protein sequence level.
SNP selection
We identified SNPs that have a high allele frequency difference between the parental species to analyze interspecies crosses, to generate a genetic map and to conduct QTL mapping. This ensures a high proportion of fully informative markers in a majority of the F2 families—that is, where linkage phase can be traced from the parental generation through to the F2 crosses. To identify polymorphic SNPs, individual reads were mapped against the EST contigs obtained from the de novo assembly using the Mosaik suite of programs (http://bioinformatics.bc.edu/marthlab/Main_Page). Assemblies were screened for SNPs using Gigabayes (Hillier et al., 2008). The output from Gigabayes was filtered using a custom Perl (http://www.perl.org/) script to identify candidate SNPs whether (1) the SNP distinguished all samples at the species level (2) the SNP was covered at least once for each of the four ancestral populations and (3) at least four reads covered each of the two species. The value of the resulting SNP loci to distinguish C. rhenanus and C. perifretum was validated for 220 candidate SNP loci by PCR amplification and Sanger sequencing of independent pooled DNA samples representing populations of C. perifretum (Witte Nete, Laarse Beek, Zwane Beek) and C. rhenanus (Broel, Naaf, Flaumbach) (compared with Stemshorn et al., 2011). Sequences from each parental pool were visually inspected for alternatively fixed SNPs (not shown).
Linkage map construction
SNP-carrying loci were BLAST mapped against the stickleback genome to identify a set of 384 SNPs (334 EST-based loci and 50 loci from Stemshorn et al., 2011) with inter-locus intervals of about 1.5 Mb according to the stickleback genome. A custom GoldenGate genotyping assay (Illumina) was genotyped for all of the mapping families using the Illumina BeadXpress system following the recommendations of the manufacturer. Linkage analysis was carried out using R/qtl (http://www.rqtl.org) following the latest version of the manual (Genetic map construction with R/qtl, 4-Nov-2010). Mapping families of different sizes were combined according to their population of origin and the direction of the cross as shown in Table 1. This assumes that there is no between-family variation in the genetic maps and that recombination does not differ among families, which is difficult to verify, especially when sample sizes for families are too small to gain sufficient statistical power. However, combining families is necessary in order to obtain better overall estimates of recombination fractions among loci. For map comparisons with different teleost genomes a consensus map was generated based on the combined genotyping data for all mapping families.
Individuals and markers with more than 50% missing data or with obvious genotyping problems were removed from the raw genotypic data. As the SNP markers we used were not entirely diagnostic for the ancestral species, some SNPs were not fully informative in all of the mapping families (60 loci were not fully informative in Broel × WN crosses, and 58 loci were not fully informative in Naaf × LB crosses). Genotypes for loci in non-fully informative families were marked as missing data in order to reconstruct genetic maps and to map QTL. We retained only markers that showed no strong indication for segregation distortion in the F2 offspring using χ2-test with an expected segregation ratio of 1:2:1 for all alleles (P-value<1e−10 following the manual of R/qtl) for the map construction, and the markers excluded from map construction were still used for later analysis. Independent linkage maps were constructed for the groups of crosses as defined by their population of origin. Linkage was estimated using the function est.rf to obtain pairwise recombination fractions and to calculate a LOD score for a test of rf=1/2. Two markers were placed in the same linkage groups when they have an estimated recombination fraction ⩽0.35 and LOD score ⩾4. We ordered the markers within each linkage group, then compared all possible orders and choose the best order using the ripple function.
Synteny and collinearity analysis
Conserved synteny is defined as consistent linkage between certain genes across species genomes, and this definition does not require conservation of gene order or orientation within chromosomes. This condition was visualized using oxford grids that summarize the map position of SNPs for each linkage group in the consensus Cottus map versus the BLAST hit location for the respective locus in reference genomes. Collinearity refers to the conservation of the order of orthologous loci within chromosomes between two species. Permutation of map data permits testing whether a block of n collinear markers indicates identical genome organization between two genomes. The method adopted here to assess the significance of collinear regions is based on the approaches outlined by Gaut (2001) and Lukens et al. (2003) with some modifications. We considered the strict definition, in that loci in the collinear region must be in the same order between species and not be interrupted by other markers. Accordingly, it does not matter which genome was taken as ‘reference' or as a ‘tester' to detect collinear blocks. If a collinear region with n shared markers was detected, it was scored. The score is defined as the average distance (centimorgan, cM) between the loci within the collinear region. The physical distance (Megabase, Mb) of model fish genomes was transferred into genetic distance (cM) according to the mean ratio of genetic distance to physical distance for each Cottus linkage group and the corresponding model fish chromosome. To test whether the score of a collinear region could be expected by chance or not, markers were randomly and uniformly assigned new map positions within the respective linkage group 10 000 times. In the permuted data, the largest collinear block with the reference genome was identified, and the score was calculated as above for each permutation. The probability that the observed collinear region was due to random association between loci was evaluated against the distribution of P-values (P<0.05) from the permutations. The test for collinear regions was implemented using a custom Perl script.
Mapping of regions affecting sex and survival
Sex was examined for 273 offspring after gonad maturation by raising the water temperatures after the artificial winter phase (Table 1). Male and female gonad morphology was inspected in ethanol-preserved specimens. Male fish have a pair of flat and prolonged white gonads, whereas the female gonads are rounded and have an orange or brown color, sometimes with discernible eggs inside. Available sex phenotypes were used for map-based QTL analysis in R/qtl. Sex was mapped independently for crosses combined according to the population of origin (Broel × WN and Naaf × LB) and considering the paternal direction of the cross (C. perifretum ♂ vs C. rhenanus ♂). We have carefully checked whether all loci were fully informative for individual mapping families (that is, whether grandparents were differentially fixed and where both parents from the F1 generation were heterozygous) before combining families into mapping populations. The trait considered was sex (coded as a binary character: male—0; female—1), and no other covariate was included in the model. Standard interval mapping and a binary model were used for the genome scan. Likelihood of odds (LOD) significance thresholds for each cross was determined by permutation testing (1000 permutations) using the scanone function of R/qtl. Significant QTL was above the genome-wide LOD threshold (α=0.05). The confidence interval was examined by 95% Bayes interval flanking the QTL peak using the bayesint function of R/qtl.
Evidence that genetic regions affect the survival of F2 crosses was sought by testing all loci included in the consensus map for segregation distortion. This test can reveal deviations from mendelian inheritance induced by differential survival of certain genotypic classes. χ2-test was used to detect deviations from the expected genotypic proportions of 1:2:1 for homozygotes AA, heterozygotes AB and homozygotes BB, respectively. The significant threshold to identify candidate loci for segregation distortion was set to P<0.05 and not corrected for multiple testing. We expected a corresponding number of false-positive results due to chance, and we have reported patterns of segregation distortion only when minimally two neighboring loci show a significant and congruent signal, which we take as evidence that the underlying genetic loci have a sufficiently strong effect. Further, it was assessed whether segregation distortion was observed only in single crosses or whether the pattern appeared in parallel crosses.
Importantly, the above testing approaches exclude those loci not included in the map that nonetheless constitute loci of great interest with respect to the goals of this study. Sex-linked loci may occur in a hemizygous state, and loci that affect survival will show deviations from mendelian inheritance. Alternatively, deviations from mendelian inheritance can also result from genotyping artifacts. These phenomena bias the estimation of recombination fractions. Excluding the affected loci is necessary for applying the mapping functions used here (see linkage map construction) but implicates a danger of losing particularly interesting candidates related to sex differentiation or genetic incompatibilities. Our strategy to still explore loci not included in the map construction was to (1) perform tests for individual markers and (2) to use positional genomic information from the stickleback genome (see collinearity analysis) to explore congruence of the signals with likely neighboring loci. Although the patterns derived from integrating these sources of information can contain artifacts because of genotyping error and differences in marker order between species, they were screened for loci that can add evidence for the presence of candidate regions as identified in the direct, map-based analysis. Markers that were excluded from map construction because of non-mendelian inheritance (see above) were assigned a most likely Cottus map position on the basis of BLAST hits in the stickleback genome. We then examined whether neighboring loci on the Cottus map showed a congruent signal of segregation distortion to test whether our mapping strategy excluded incompatibility loci with particularly strong effects.
To complement QTL mapping and to find possible evidence for hemizygous inheritance of sex-determining regions, raw genotypic data for each locus, including loci where the F1 generation was not fully heterozygous in some families, were screened for associations of parental genotypes with offspring's sex using the marker regression method from R/qtl. To further refine the analysis of candidate QTL intervals, additional markers were selected, based on positional information from the stickleback genome. This included three length polymorphism markers (CottE1-indel, ctg06004i-indel and Cott108-indel, Supplementary Table S1) and two microsatellite markers (CottE1 and Cott108) from Nolte et al. (2005b). These markers were typed by separating PCR products from fluorescently labeled primers on a capillary sequencer as done by Nolte et al., 2006. The inferred map position of the three length polymorphism markers was confirmed through patterns of segregation in our mapping families. The genomic fragments carrying the microsatellites CottE1 and Cott108 both contain SNP or indel markers that were already included in our SNP-based map. Hence, patterns of inheritance of chromosome-specific parental alleles could be analyzed for the actual microsatellites CottE1 and Cott108 in sexed offspring to test for the presence of a heterogametic sex. To test whether recombination was suppressed at sex-linked markers, we calculated male and female meiotic recombination frequencies for loci spanning the QTL interval for groups of F2 crosses.
Results
Sequence similarity between Cottus and model species
A total of 1 124 134 reads with an average length of 299 nucleotides were obtained from 454 sequencing of cDNA libraries. The assembly yielded 11 548 contigs larger than 500 bp that were mapped against five teleost model fish genomes and protein databases. The number of blastx hits that detect similarity at the amino-acid level was relatively similar for all reference genomes (46.2–51.2%). However, there was conspicuous variance in the number of significant BLAST hits at the level of the nucleotide sequence. The highest number of hits was found when comparing Cottus ESTs with the stickleback genome with significant hits for 94.9% of the contigs. The medaka (71.6%), tetraodon (69.3%), fugu (75.3%) and zebrafish (52.2%) genomes also permit to locate homologues of the Cottus contigs, but their genomes are less conserved.
SNP discovery and map construction
The assembly of individual reads against the EST contigs yielded 1302 SNPs that were ancestry-informative according to our criteria. These, together with SNPs from a previous study (Stemshorn et al., 2011), were used to identify a set of 384 SNPs that were evenly distributed across the 21 chromosomes contained in the most recent version of the stickleback genome assembly (http://www.ensembl.org/index.html). A custom 384-plex GoldenGate SNP genotyping assay was used to type 898 F2 individuals and parents. After excluding SNPs found to be monomorphic (9) or in which genotyping had a low success rate for most of the mapping families (77), data for 298 SNPs were available for further analysis. Genotypes for loci in non-fully informative families were marked as missing data in order to reconstruct genetic maps and to perform QTL analysis. Independent linkage maps were constructed for the groups of crosses as defined by their population of origin (Supplementary Figure S1). For the Broel × WN map, 234 SNPs mapped to 24 linkage groups totaling a length of 1437.2 cM with an average interval of 6.8 cM. For the Naaf × LB map, 238 SNPs mapped to 24 linkage groups with a length of 1319.0 cM with an average mapping interval of 6.2 cM. The two maps share a total of 201 markers.
The conservation of synteny and collinearity between the two Cottus maps was analyzed. All markers show a 1:1 representation on single linkage groups, and the maps are highly collinear (Supplementary Figure S1). Only four single markers do not show the same order between the two maps. Notably, all of them mapped to the end of linkage groups in one of the Cottus maps. The test for significance of collinearity shows that the two Cottus maps share 27 significantly collinear linkage blocks containing 2–14 markers, which covers 98.75% of the genetic map.
Given the fully conserved synteny between the two Cottus maps and under the assumption that the observed differences between maps are minor, we combined data for all mapping families to generate a consensus map for comparisons with reference genomes. A total of 252 markers (Supplementary Table S1) mapped to 24 linkage groups in the Cottus consensus map at a LOD threshold of four (Figure 1) with a total map length of 1575.4 cM and an average distance between markers of 6.8 cM. In comparison with the preliminary maps of Cottus (Stemshorn et al., 2011), the maps generated here are shorter and do not have an exceedingly large linkage group.
Figure 1.
A consensus genetic map derived from interspecies crosses between C. rhenaus and C. perifretum can be best anchored in the stickleback (Gasterosteus) genome, which permits the transfer of positional information to explore the Cottus genome. Filled blocks within linkage groups represent significantly collinear regions (P<0.05) with a perfectly conserved marker order. Grey blocks are collinear, but this was not found to be significant. Each of the parental species has a different and single heterogametic region that determines sex. These regions were identified on linkage groups 1 and 2 (see text) and are highlighted through bold marker names.
Conservation of synteny and collinearity between Cottus and model species
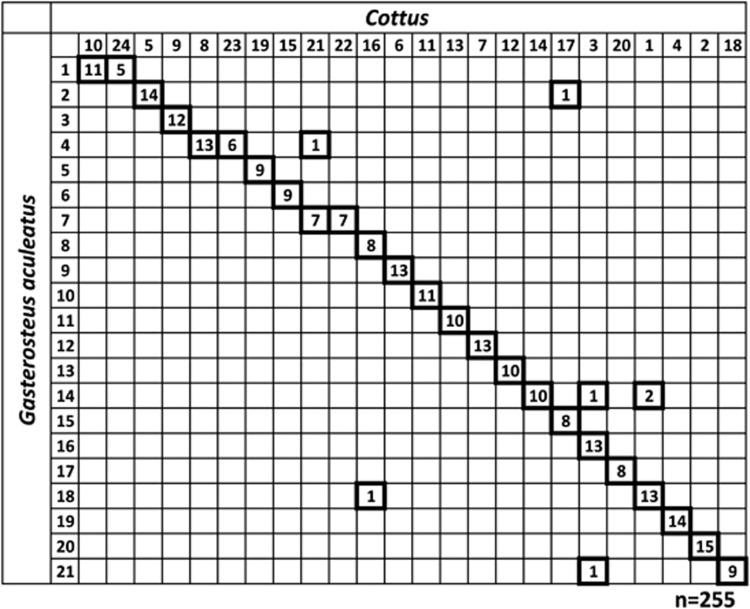
BLAST searches permitted establishing syntenic relationships of Cottus with the reference species, which was summarized in the form of Oxford grids (Figure 2 and Supplementary Figure S2). A one-to-one relationship among many known chromosomes and linkage groups can be observed. The majority of Cottus linkage groups are associated with single stickleback chromosomes with only seven markers that do not follow this rule (Figure 2). Some chromosomal rearrangements suggestive of fusion/fission events distinguish Cottus from the stickleback, which appears to have fused chromosomes 1, 4 and 7 that fall into separate linkage groups in Cottus. The correlation of genetic maps gets weaker when comparing Cottus with the medaka, tetraodon, fugu and zebrafish genomes that correspond with the decreasing pattern of nucleotide sequence conservation (Supplementary Figure S2). Testing for significance revealed that a number of collinear blocks could be expected by chance given the number of sampled markers per chromosome, and again Cottus and stickleback showed the greatest similarity (Table 2). Finally, the correlation of the physical distance in stickleback and genetic distance in Cottus within collinear blocks is shown in Supplementary Figure S3. The average physical distance in stickleback of a recombination unit in Cottus is 4.25 cM/Mb (range: 0.000006 cM/Mb to 11.67 cM/Mb).
Figure 2.
Oxford grid of synteny relationships between Cottus linkage groups (x-axis) and stickleback chromosomes (y-axis) based on BLAST mapping of Cottus EST contigs. Numbers in fat squares indicate shared markers among genetic regions. Six instances of possible interchromosomal rearrangements were noted, and chromosomal fusion events are suggested for stickleback chromosomes 1, 4 and 7.
Table 2. The number of significantly collinear regions, the markers contained in these and the proportion of the genome that is covered was determined between Cottus and sequenced fish genomes.
| Collinear blocks (n) | Markers per block | Percentage of Cottus map | |
|---|---|---|---|
| Cottus (within) | 27 | 2–14 | 98.75% |
| Stickleback | 34 | 2–11 | 60.62% |
| Medaka | 26 | 2–8 | 39.15% |
| Fugu | 21 | 3–9 | 32.44% |
| Tetraodon | 21 | 2–9 | 31.59% |
| Zebrafish | 7 | 2–3 | 7.36% |
Conservation of marker order between genomes was assessed by randomizing markers within linkage groups. Besides the almost perfectly conserved marker order between the two Cottus maps studied here, the stickleback shows the highest degree of genome collinearity with Cottus.
Genomic regions affecting sex and genetic incompatibilities
Sex could be determined for 273 individuals. The QTL analysis to detect genomic regions that determine sex differentiation revealed one significant region on LG01 (LOD=5.41) for the Broel(♂) × WN(♀) crosses. In contrast, both of the two other types of crosses revealed a single significant QTL on LG02 ((WN(♂) × Broel(♀) crosses (LOD=5.30) and LB(♂) × Naaf(♀) crosses (LOD=5.71)). Note that the first type of cross involves male C. rhenanus, while crosses that suggest an alternative QTL location involve male C. perifretum. The significance of these candidate regions was further supported when additional fragment length polymorphism markers were typed and analyzed. CottE1-indel is linked to sex QTL locus CottE1 on LG01. Likewise, ctg06004i-indel and Cott108-indel are linked to sex QTL locus ctg06004 on LG02. The LOD scores of the initial QTL candidates combined with the added markers increased for both candidate regions (Table 3). To complement this inference, raw genotypes including those for loci that were excluded from the mapping construction (46 loci) were tested for single locus association with sex. One additional marker (ctg05522, Supplementary Table S1) was found only for WN(♂) × Broel(♀) crosses. The flanking sequence of this locus had a BLAST hit on the stickleback genome that indicates that the locus is linked to the candidate region detected on LG02. Hence, it does not suggest the presence of additional genomic regions that affect sex differentiation. Sex-specific recombination rates were estimated for both candidate sex-determining regions in the respective crosses (Table 3). Recombination was observed in both sexes and was not consistently smaller in either sex. Parental microsatellite alleles permitted to trace the inheritance of alleles from the F1 into the sexed F2 offspring in six mapping families (Table 4). The segregation patterns revealed that males inherit an allele that is strongly associated with sex, which indicates a male heterogametic (XY) sex determination system. Seven families from three different types of crosses were available to compare the sex ratios in the F2 generation (Table 1). The sex ratios did not deviate significantly from balanced proportions, except for one family (ZG46) that had a significant male excess, which corresponded to an excess of the heterogametic sex.
Table 3. QTL mapping results for sex differentiation in interspecies Cottus crosses according to the origin and gender of the parental populations.
| Crosses | LG | Interval size (cM) | LOD | Linked marker | Recombination fraction (♂:♀) (cM) |
|---|---|---|---|---|---|
| Broel(♂) × WN(♀) | 1 | 6.1 | 7.12 | CottE1-indel | 7: 8.8 |
| WN(♂) × Broel(♀) | 2 | 17.1 | 5.09 | ctg06004i-indel | 24.4: 23.6 |
| LB(♂) × Naaf(♀) | 2 | 6.9 | 9.48 | ctg06004i-indel | 15.1: 12.5 |
QTL that exceeds the significance threshold (LOD) are reported with their linkage group (LG), interval size, maximum LOD score and the most closely linked marker. Recombination fractions for male and female F2 offspring were determined for the markers spanning the significant QTL intervals.
Table 4. The segregation of dam and sire microsatellite alleles could be traced for six mapping families.
| Cross type | Broel(♂) × WN(♀) | WN(♂) × Broel(♀) | LB(♂) × Naaf(♀) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Microsatellite | LG01—CottE1 | LG02—Cott108 | LG02—Cott108 | ||||||||||||
| Parents | ZG42 | Dam (♀) | Sire (♂) | ZG40 | Dam (♀) | Sire (♂) | ZG26 | Dam (♀) | Sire (♂) | ||||||
| A | B | A | C | B | A | C | A | A | A | A | B | ||||
| Offspring | ♂ | 25 | 26 | 3 | 48 | ♂ | 7 | 10 | 13 | 4 | ♂ | 7 | 0 | 0 | 7 |
| ♀ | 20 | 24 | 43 | 1 | ♀ | 4 | 6 | 1 | 9 | ♀ | 10 | 0 | 10 | 0 | |
| ZG46 | Dam (♀) | Sire (♂) | ZG32 | Dam (♀) | Sire (♂) | ||||||||||
| B | A | C | D | A | A | A | B | ||||||||
| ♂ | 10 | 0 | 10 | 0 | ♂ | 10 | 0 | 0 | 10 | ||||||
| ♀ | 0 | 3 | 0 | 3 | ♀ | 7 | 0 | 6 | 1 | ||||||
| ZG35 | Dam (♀) | Sire (♂) | |||||||||||||
| A | A | A | B | ||||||||||||
| ♂ | 21 | 0 | 0 | 21 | |||||||||||
| ♀ | 17 | 0 | 16 | 1 | |||||||||||
Each subtable summarizes results for cross type (origin and gender of the parental populations), the concerned linkage groups with the marker name and genotypic data for one of six mapping families. The segregation of alleles (labeled A, B, C, D) from the dam and sire of the F1 into the F2 generation reveals a strong correlation of the offspring sex with the presence of a single paternal allele (numbers indicate individuals carrying the respective alleles. Bold numbers highlight the sex specific allele. Note that the inheritance of identical alleles cannot be resolved). This pattern is observed in a single family (ZG42) for the paternal allele on linkage group 1 from C. rhenanus and five times for the paternal allele on linkage group 2 which originates from C. perifretum.
A general pattern of viability and fertility of F1 crosses between C. rhenanus and C. perifretum was observed for replicated pairs of populations and directions of crosses. Likewise, our experiments did not suggest that F2 crosses were strongly impaired, but showed allele frequency distortions that suggest partial incompatibilities that were associated with varying genomic regions. χ2-tests for deviation from the expected 1:2:1 mendelian segregation indicated that groups of linked markers deviated significantly in the same manner within mapping families (Table 5). However, none of these effects was common or even fixed for the parental species or populations. Of the 27 candidate regions detected among all families, 19 regions were not shared and four were shared by maximally two out of seven mapping families. Moreover, of the 27 regions, 15 regions showed an excess of C. perifretum alleles and 12 regions showed an excess of C. rhenanus alleles (χ2-test, P=0.5637) that is not significantly biased towards one of the parental species. Markers that were assigned map positions based on synteny relations with the stickleback added evidence that confirms patterns of segregation distortion as observed for markers that were placed directly on the Cottus map. We detected four instances where strong segregation distortion (Supplementary Table S2) coincides with patterns at regularly mapped loci in Cottus (Table 5). The remaining patterns did not correspond with signals in directly mapped markers. Importantly, none of the regions for which evidence for transmission distortion was detected corresponded physically with the candidate genomic regions that affect differentiation of sex (Table 3), and we did not observe an increased mortality of the heterogametic sex (Table 1). As single exception, the marker ctg05522 is located in the region on LG2 that was found to be involved in sex determination in the cross WN(♂) × Broel(♀) (ZG40, ZG46). This locus shows signs of distortion in a single family ZG46, but the sex ratio in this family is skewed in favor of males (Table 1). Accordingly, there are no shifts in genotype frequencies in F2 offspring that would suggest that the presence of the hemizygous allele determining sex affects survival.
Table 5. Genotypic segregation distortion in individual mapping families.
| Cross type | Mapping families | LG | Linked distorted markers |
|---|---|---|---|
| Broel(♂) × WN(♀) | ZG18 | 10 | ctg00684, ctg14120, ctg07255 |
| 12 | ctg05216, ctg11936, ctg00327, ctg03228 | ||
| 13 | ctg04943, ctg00150, ctg25684, ctg02495 | ||
| 20 | ctg00952, ctg02681 | ||
| 24 | ctg03529, ctg06588, ctg03740 | ||
| ZG33 | 11 | ctg12662, Co539-M13, ctg02955 | |
| 12 | ctg01809, ctg13097 | ||
| 18 | ctg07784, ctg03025 | ||
| 20 | ctg02681, Co303-SP6 | ||
| WN(♂) × Broel(♀) | ZG40 | 3 | ctg16139, ctg09944, ctg01723, ctg14942 |
| 3 | ctg07551, ctg02192, ctg03212 | ||
| 5 | ctg05707, ctg03002 | ||
| 22 | ctg08405, ctg04802 | ||
| ZG46 | 9 | Co552, ctg10777 | |
| 9 | ctg03758, ctg01301 | ||
| 9 | ctg25562, ctg02595 | ||
| 19 | ctg01510, ctg02938, ctg03059 | ||
| Naaf(♂) × LB(♀) | ZG39 | 1 | ctg17346, ctg11104 |
| 3 | ctg03212, ctg04800 | ||
| 5 | ctg03280, ctg08226 | ||
| 15 | ctg02216, ctg03596 | ||
| 15 | ctg02768, ctg07422, ctg04258, ctg16543 | ||
| LB(♂) × Naaf(♀) | ZG32 | 6 | ctg03214, ctg11972 |
| 6 | ctg05890, ctg04444 | ||
| ZG38 | 10 | ctg08331, ctg18049, ctg05417, ctg03090, ctg01402 | |
| 13 | ctg00150, ctg25684 | ||
| 22 | ctg02839, ctg08405, ctg04802, ctg09067, ctg00565 |
Genomic regions (linkage groups and marker names given) spanning two or more linked markers with a congruent and significant (P<0.05) deviation from mendelian inheritance are reported. Distorted regions were shared only for pairs of families (marker names bold). Most regions were not shared, and no region had a general effect in all crosses.
To determine whether the candidate regions in Cottus have similarities with sex-determining regions in other fish species, we used Oxford grids (Figure 2 and Supplementary Figure S2) to determine the homology of the Cottus sex QTL with stickleback, medaka or fugu chromosomes. This analysis was complemented with published records (Kikuchi et al., 2007; Takehana et al., 2008; Kondo et al., 2009; Ross et al., 2009) accessible through the reference species (Supplementary Table S3). The region represented by CottE1 on Cottus—LG01 is homologous with parts of stickleback chromosome 18 and medaka chromosome 24, whereas the region represented by ctg06004 on Cottus—LG02 is homologous with parts of stickleback chromosome 20 and medaka chromosome 16. None of these regions are associated with the sex-determining region in stickleback (chromosome 19) and medaka (chromosome 1). The same holds for additional closely related species that are accessible through the reference genomes studied here and for which the sex determination system has been mapped (Supplementary Table S3). As an exception, Cottus—LG02 is homologous with the sex chromosome in Oryzias javanicus (medaka chromosome 16) (Takehana et al., 2008), although with the important difference that the latter species has a female heterogametic sex.
Discussion
A genetic map can have a central role to integrate results on the genetics of speciation (Bernatchez et al., 2010) and facilitate interspecies comparisons of genomes to understand patterns of genome and chromosomal evolution (Backstroem et al., 2008; Sarropoulou et al., 2008). This study has followed these paths for the exploration of a non-model system. We have generated a Cottus linkage map that agrees with the haploid chromosome number of 24 as described for the closely related Cottus gobio (Vitturi and Rasotto, 1990). The map contains an evenly dispersed set of markers that has an appropriate density for QTL mapping (Mauricio, 2001), and the conserved synteny and collinearity with sequenced reference genomes can serve to bridge considerable phylogenetic distances. Cottus shares the highest collinearity and nucleotide sequence similarity with stickleback (Gasterosteus). This result agrees with recent phylogenetic studies that have placed the Gasterosteidae into a clade that contains Cottus among other scorpaeniform fishes (Kawahara et al., 2008; Li et al., 2009). Therefore, the stickleback genome could serve as an excellent reference for the entire Scorpaeniformes clade. In this study, finding approximate genome positions of EST sequences from Cottus based on the Gasterosteus genome was highly successful, and, within collinear regions, the physical distance in the stickleback genome is a reasonable estimator of map distances in Cottus.
It was a central goal of this study to test whether genetic factors exist that may contribute to intrinsic barriers to reproduction between hybridizing species of Cottus. Diverse genetic factors that cause incompatibilities were identified in F2 crosses. Moreover, we have identified the genomic regions that determine sex in the ancestral species and did not find evidence that incompatibility loci reduce the viability of the heterogametic sex in hybrids. Here we discuss how genomic information from reference genomes contributes to understanding the results and discuss the role that the candidate regions identified here may have in the evolutionary processes following hybridization of Cottus in the River Rhine system.
Sex determination differs between species of Cottus and other fishes
A single genetic region was significantly associated with the differentiation of sexes in all crosses, and the sex ratios were generally not imbalanced, which suggests that sex is heritable and has a simple genetic basis (Devlin and Nagahama, 2002). When a male from C. perifretum was mated with a C. rhenanus female to generate F2 crosses, a region on LG02 was recovered, whereas the opposite cross direction (male C. rhenanus crossed to C. perifretum female) revealed a region on LG01. This finding, together with patterns of segregation of microsatellite alleles, suggests that sex in our F2 mapping families is determined by different male heterogametic regions from the two ancestral species. No consistent sex-specific reduction of recombination rates at QTL regions was detected, which suggests that the heterogametic region should be small. A lack of large heterogametic region or distinct Y chromosome agrees with Vitturi and Rasotto's (1990) observation that the closely related C. gobio has no discernible sex chromosome in its karyotype, although heterogametic XY systems have been reported for related species (Devlin and Nagahama, 2002).
Conserved synteny among species represents a bridge to complement initial QTL experiments with results from homologous chromosomal locations. A lack of homology of Cottus LG01 and LG02 with the sex chromosomes of medaka (Chr01) and stickleback (Chr19) suggests that the genetic basis of sex in Cottus differs from the reference genomes and also from patterns observed in closely related species (Kikuchi et al., 2007; Takehana et al., 2008; Kondo et al., 2009; Ross et al., 2009). Interestingly, LG02 corresponds to medaka chromosome 16, which is the sex chromosome in Oryzias javanicus (Takehana et al., 2008) albeit with the key difference that, unlike Cottus, O. javanimuc has a ZW sex determination system. Moreover, the XY sex determination of the African fish Nothobranchius furzeri is also located in a region that is homologous to medaka chromosome 16 (Valenzano et al., 2009), suggesting the possibility that sex in C. perifretum and N. furzeri could be determined by a common system. However, this association has to be reassessed once the candidate regions have been studied in detail. It is not likely that this system is shared because of common ancestry as the species are phylogenetically distant (Supplementary Table S3). Sex is an important evolutionary component, and some key elements of sex-determining pathways are conserved across the animal kingdom (Herpin and Schartl, 2011). However, the mechanism by which the sex differentiation pathway is triggered varies widely (Marin and Baker, 1998; Herpin and Schartl, 2011). The fact that two different regions determine sex in closely related species of Cottus indicates that factors controlling a conserved sex determination pathway evolve rapidly and have a fast turnover, which is not astonishing given the results from other fish species (Mank and Avise, 2009). Detailed genetic studies will have to reveal whether the same genes are repeatedly recruited in different lineages. If this occurs, then similarity of sex-determining regions across large phylogenetic distances as found for Cottus LG02 in this study could be explained through convergent evolution.
Implications for the genetics of speciation and hybridization in Cottus
The presence of genetic incompatibilities may be relevant to understand the persistence of the ancestral C. perifretum and C. rhenanus throughout glacial cycles (Englbrecht et al., 2000) and could affect more recent processes related to hybrid speciation (Nolte and Tautz, 2010) or gene flow between the parental species and invasive sculpins (Stemshorn et al., 2011). No effects on F1 hybrid viability were observed, which is in line with the inference that inviability in fish hybrids was estimated to take 2.2 to 10 times longer to evolve than the assumed divergence up to 2 million years between C. rhenanus and C. perifretum (Englbrecht et al., 2000; Russell, 2003; Bolnick and Near, 2005; Stelkens et al., 2009). On the other hand, the time to evolve effects that manifest in the F2 generation is expected to be shorter (Edmands, 1999; Maheshwari and Barbash, 2011; White et al., 2011). In agreement with this, the screen for patterns of segregation distortion in F2 crosses of Cottus revealed a number of candidate regions that are associated with offspring survival in our mapping families (Table 5). However, the absence of signs for segregation distortion at the sex-determining regions and the absence of signs for a reduced viability of the males suggest that none of the incompatibilities manifest specifically in the heterogametic sex. Hence, effects of the incompatibility factors we have identified between C. perifretum and C. rhenanus apparently differ from organisms like birds (Sætre et al., 2003) or mice (White et al. 2011; Janousek et al., 2012), in which species divergence and the dynamics of hybridization are strongly determined by effects related to Haldane's rule. According to Qvarnström and Bailey (2009), the situation in Cottus is representative of the first steps of speciation where differentiation is primarily driven by ecologically driven selection that targets autosomal genes. We add that this situation may also be very general in speciation processes of species where the heterogametic regions in the genome are very limited. Genetically differentiated sex chromosomes are expected to contribute to intrinsic barriers to reproduction (Wolf et al., 2010), but the evolution of incompatibilities associated with heteromorphic sex chromosomes requires some evolutionary stability (Devlin and Nagahama, 2002; Bachtrog et al., 2008). The comparison with other fish genomes suggests that heterogametic regions in Cottus are subject to rapid evolutionary turnover—that is, that loci that trigger the differentiation of sexes repeatedly evolved at different and very confined chromosomal regions. This evolutionary instability makes it unlikely that the respective regions contribute to barriers to reproduction between C. rhenanus and C. perifretum.
Incompatibilities in our experiments were hardly consistent between crosses with a strong representation of effects that were encountered in single families. This strongly suggests that incompatibilities vary at the level of differentiated populations and individuals. It is possible that unknown environmental factors differed among our laboratory tanks or that intrinsically caused transmission distortion (Koide et al., 2008) had a role that warrants further studies. However, with respect to the evolutionary processes triggered by natural hybridization in Cottus, our results suggest that these are neither affected by species specific nor by very strong intrinsic genetic factors. Invasive sculpins have emerged from a complex pattern of admixture from diverse sources (Nolte et al., 2005a; Stemshorn et al., 2011). Hence, a diverse set of genetic incompatibility factors will have entered the admixed gene pool, but most likely none at very high frequencies. According to this study, such factors would cause fluctuating and genotype-specific patterns of selection in natural hybrids. The incompatibility loci identified here may thus constitute a fitness burden for admixed individuals, affect hybrid zone structure and slow down secondary gene flow into the invasive gene pool (Nolte et al., 2006, 2009; Stemshorn et al., 2011). On the other hand, they are unlikely to be purged or fixed rapidly in the admixed gene pool because of the family-specific nature of the effects and because balanced proportions of loci cause transmission distortion toward either one or the other one of the ancestral species. Likewise, natural hybrids of Cottus may carry alternative suites of genetic factors that determine sex depending on their paternal origin. The genetic basis for sex would be subject to strong balancing selection when a dominant factor biases sex ratios. However, six out of seven of the crosses analyzed here suggest that both sex-determining regions cause a balanced sex ratio in a hybrid genetic background. Moreover, effects on the viability of the heterogametic sex appear to be absent. Accordingly, it would be possible that a mosaic genetic architecture of sex determination prevails in the admixed lineage or in hybrid zones for some time without inducing strong selection.
The results from this screen have not found evidence for prevalent genetic factors that trace back to the ancestral species and determine general evolutionary trends in invasive sculpins. This result confirms our previous inference that patterns of introgression vary among individual loci for different hybrid zones (Nolte et al., 2009), which is in line with genetic incompatibilities that are unique to local demes. On the other hand, invasive Cottus remain differentiated from all ancestral populations studied to date. We have previously described strong genotype–environment associations for Cottus in the River Rhine basin. Particularly, general patterns of adaptive divergence are replicated in nature, indicating that they are independent from local genotypic variance. This is true for the parallel molding of hybrid zone clines across habitat borders (Nolte et al., 2006) and the evolution of multiple lineages of invasive sculpins in ecologically perturbed habitats (Stemshorn et al., 2011). This implies that local genetic variance must interact with more general ecological factors to determine evolutionary processes after hybridization in Cottus within the lower River Rhine basin.
Data archiving
Loci included in the consensus map for Cottus with locus name, segregating alleles that distinguish C. perifretum and C. rhenanus populations and the sequence are summarized in Supplementary Table S1. Data for QTL mapping of sex determination regions in Cottus are summarized in Supplementary Table S4. Both supplementary tables are deposited as excel files at the Dryad repository: doi:10.5061/dryad.7s393.
Acknowledgments
We thank K de Gelas, A Kobler, L Bervoets and F Volckaert for their benevolent help in obtaining breeding stocks of Cottus perifretum. K Lessenich, A Mellin (Berirksregierung Köln) and T Heilbronner from the Siegfischereigenossenschaft have given permits to conduct the research. Cottus were kept and bred in the laboratory of MPI in Plön with permission from S Hauschildt (Veterinaeramt in Plön). E Bustorf, H Buhtz, S Dembeck, H Harre, G Augustin, D Mertens and D Lemke have contributed to the laboratory work and fish care. We thank K Konrad, F. Chan, B Haubold and D Tautz for comments and discussions and DT and the Max-Planck Society for generous support. The project was funded by a DFG grant to AN.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Abbott R, Albach D, Ansell S, Arntzen JW, Baird SJE, Bierne N, et al. Hybridization and speciation. J Evol Biol. 2013;26:229–246. doi: 10.1111/j.1420-9101.2012.02599.x. [DOI] [PubMed] [Google Scholar]
- Bachtrog D, Hom E, Wong KM, Maside X, Jong P. Genomic degradation of a young Y chromosome in Drosophila miranda. Genome Biol. 2008;9 (2:R30. doi: 10.1186/gb-2008-9-2-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backstroem N, Karaiskou N, Leder EH, Gustafsson L, Primmer CR, Qvarnstroem A, et al. A gene-based genetic linkage map of the collared flycatcher (Ficedula albicollis) reveals extensive synteny and gene-order conservation during 100 million years of avian evolution. Genetics. 2008;179:1479–1495. doi: 10.1534/genetics.108.088195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton NH, Gale KS.1993Genetic analysis of hybrid zones. Pp. 13-45in Hybrid zones and the evolutionary process (ed). Harrison R G, Oxford University Press: Oxford [Google Scholar]
- Bernatchez L, Renaut S, Whiteley AR, Derome N, Jeukens J, Landry L, et al. On the origin of species: insights from the ecological genomics of lake whitefish. Phil Trans R Soc B. 2010;365:1783–1800. doi: 10.1098/rstb.2009.0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne N, Welch J, Loire E, Bonhomme F, David P. The coupling hypothesis: why genome scans may fail to map local adaptation genes. Mol Ecol. 2011;20:2044–2072. doi: 10.1111/j.1365-294X.2011.05080.x. [DOI] [PubMed] [Google Scholar]
- Bolnick DI, Near TJ. Tempo of hybrid inviability in centrarchid fishes (Teleostei: Centrarchidae) Evolution. 2005;59:1754–1767. [PubMed] [Google Scholar]
- Czypionka T, Cheng J, Pozhitkov A, Nolte AW. Transcriptome changes after genome wide admixture in invasive sculpins (Cottus) Mol Ecol. 2012;21:4797–4810. doi: 10.1111/j.1365-294X.2012.05645.x. [DOI] [PubMed] [Google Scholar]
- Devlin RH, Nagahama Y. Sex determination and sex differentiation in fish: an overview of genetic, physiological, and environmental influences. Aquaculture. 2002;208:191–364. [Google Scholar]
- Edmands S. Heterosis and outbreeding depression in interpopulation crosses spanning a wide range of divergence. Evolution. 1999;53:1757–1768. doi: 10.1111/j.1558-5646.1999.tb04560.x. [DOI] [PubMed] [Google Scholar]
- Englbrecht CC, Freyhof J, Nolte AW, Rassmann K, Schliewen U, Tautz D. Phylogeography of the bullhead Cottus gobio (Pisces: Teleostei: Cottidae) suggests a pre-pleistocene origin of the major central European populations. Mol Ecol. 2000;9:709–722. doi: 10.1046/j.1365-294x.2000.00912.x. [DOI] [PubMed] [Google Scholar]
- Gaut BS. Patterns of chromosomal duplication in Maize and their implications for comparative maps of the grasses. Genome Res. 2001;11:55–66. doi: 10.1101/gr.160601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herpin A, Schartl M. Dmrt1 genes at the crossroads: a widespread and central class of sexual development factors in fish. FEBS J. 2011;278:1010–1019. doi: 10.1111/j.1742-4658.2011.08030.x. [DOI] [PubMed] [Google Scholar]
- Hillier LW, Marth GT, Quinlan AR, Dooling D, Fewell G, Barnett D, et al. Whole—genome sequencing and variant discovery in C. elegans. Nature Methods. 2008;5:183–188. doi: 10.1038/nmeth.1179. [DOI] [PubMed] [Google Scholar]
- Janousek V, Wang L, Luzynski K, Dufkova P, Vyskocilova MM, Nachman MW, et al. Genome-wide architecture of reproductive isolation in a naturally occurring hybrid zone between Mus musculus musculus and M. m. domesticus. Mol Ecol. 2012;21:3032–3047. doi: 10.1111/j.1365-294X.2012.05583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara R, Miya M, Mabuchi K, Lavoue S, Inoue JG, Satoh TP, et al. Interrelationships of the 11 gasterosteiform families (sticklebacks, pipefishes, and their relatives): A new perspective based on whole mitogenome sequences from 75 higher teleosts. Mol Phyl Evol. 2008;46:224–236. doi: 10.1016/j.ympev.2007.07.009. [DOI] [PubMed] [Google Scholar]
- Kikuchi K, Kai W, Hosokawa A, Mizuno N, Suetake H, Asahina K, et al. The sex-determining locus in the tiger pufferfish, Takifugu rubripes. Genetics. 2007;175:2039–2042. doi: 10.1534/genetics.106.069278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapen D, Knaepkens G, Bervoets L, Taylor MI, Eens M, Verheyen E. Conservation units based on mitochondrial and nuclear DNA variation among European bullhead populations (Cottus gobio L., 1758) from Flanders, Belgium. Conserv Genet. 2003;4:129–140. [Google Scholar]
- Koide Y, Ikenaga M, Sawamura N, Nishimoto D, Matsubara K, Onishi K, et al. The evolution of sex-independent transmission ratio distortion involving multiple allelic interactions at a single locus in rice. Genetics. 2008;180:409–420. doi: 10.1534/genetics.108.090126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M, Nanda I, Schmid M, Schartl M. Sex determination and sex chromosome evolution: insights from medaka. Sex Dev. 2009;3:88–98. doi: 10.1159/000223074. [DOI] [PubMed] [Google Scholar]
- Li B, Dettaï A, Cruaud C, Couloux A, Desoutter-Meniger M, Lecointre G. RNF213, a new nuclear marker for acanthomorph phylogeny. Mol Phyl Evol. 2009;50:345–363. doi: 10.1016/j.ympev.2008.11.013. [DOI] [PubMed] [Google Scholar]
- Lukens L, Zou F, Lydiate D, Parkin I, Osborn T. Comparison of a Brassica oleracea genetic map with the genome of Arabidopsis thaliana. Genetics. 2003;164:359–372. doi: 10.1093/genetics/164.1.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari S, Barbash DA. The Genetics of Hybrid Incompatibilities. Annu Rev Genet. 2011;45:331–355. doi: 10.1146/annurev-genet-110410-132514. [DOI] [PubMed] [Google Scholar]
- Mallet J. Hybrid speciation. Nature. 2007;446:279–283. doi: 10.1038/nature05706. [DOI] [PubMed] [Google Scholar]
- Mank JE, Avise JC. Evolutionary diversity and turn-over of sex determination in Teleost Fishes. Sex Dev. 2009;3:60–67. doi: 10.1159/000223071. [DOI] [PubMed] [Google Scholar]
- Marin I, Baker BS. The evolutionary dynamics of sex determination. Science. 1998;281:1990–1994. doi: 10.1126/science.281.5385.1990. [DOI] [PubMed] [Google Scholar]
- Mauricio R. Mapping quantitative trait loci in plants: uses and caveats for evolutionary biology. Nat Rev Genet. 2001;2:370–381. doi: 10.1038/35072085. [DOI] [PubMed] [Google Scholar]
- Nolte AW, Freyhof J, Stemshorn KC, Tautz D. An invasive lineage of sculpins, Cottus sp. (Pisces,Teleostei) in the Rhine with new habitat adaptations has originated form hybridization between old phylogeographic groups. Proc R Soc B. 2005a;272:2379–2387. doi: 10.1038/rspb.2005.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte AW, Stemshorn KC, Tautz D. Direct cloning of microsatellite loci from Cottus gobio through a simplified enrichment procedure. Mol Ecol Notes. 2005b;5:628–636. [Google Scholar]
- Nolte AW, Freyhof J, Tautz D. When invaders meet locally adapted types: rapid moulding of hybrid zones between two species of sculpins (Cottus, Pisces) in the Rhine system. Mol Ecol. 2006;15:1983–1993. doi: 10.1111/j.1365-294X.2006.02906.x. [DOI] [PubMed] [Google Scholar]
- Nolte AW, Gompert Z, Buerkle CA. Variable patterns of introgression in two sculpin hybrid zones suggest that genomic isolation differs among populations. Mol Ecol. 2009;18:2615–2627. doi: 10.1111/j.1365-294X.2009.04208.x. [DOI] [PubMed] [Google Scholar]
- Nolte AW, Tautz D. Understanding the onset of hybrid speciation. Trends Genet. 2010;26:54–58. doi: 10.1016/j.tig.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Qvarnström A, Bailey RI. Speciation through evolution of sex-linked genes. Heredity. 2009;102:4–15. doi: 10.1038/hdy.2008.93. [DOI] [PubMed] [Google Scholar]
- Ross JA, Urton JR, Boland J, Shapiro MD, Peichel CL. Turnover of sex chromosomes in the stickleback fishes (Gasterosteidad) PLoS Gentics. 2009;5 (2:e1000391. doi: 10.1371/journal.pgen.1000391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell S. Evolution of intrinsic post-zygotic reproductive isolation in fish. Ann Zool Fennici. 2003;40:321–329. [Google Scholar]
- Sætre GP, Borge T, Lindroos K, Haavie J, Sheldon BC, Primmer C, et al. Sex chromosome evolution and speciation in Ficedula flycatchers. Proc R Soc Lond B. 2003;270:53–59. doi: 10.1098/rspb.2002.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarropoulou E, Nousdili D, Magoulas A, Kotoulas G. Linking the genomes of nonmodel teleosts through comparative genomics. Marine Biotech. 2008;10:227–233. doi: 10.1007/s10126-007-9066-5. [DOI] [PubMed] [Google Scholar]
- Stelkens R, Young KA, Seehausen O. The accumulation of reproductive incompatibilities in African cichlid fish. Evolution. 2009;64 (3:617–633. doi: 10.1111/j.1558-5646.2009.00849.x. [DOI] [PubMed] [Google Scholar]
- Stemshorn KC, Nolte AW, Tautz D. A genetic map of Cottus gobio (Pisces,Teleostei) based on microsatellites can be linked to the physical map of Tetraodon nigroviridis. J Evol Biol. 2005;18:1619–1624. doi: 10.1111/j.1420-9101.2005.00929.x. [DOI] [PubMed] [Google Scholar]
- Stemshorn KC, Reed FA, Nolte AW, Tautz D. Rapid formation of distinct hybrid lineages after secondary contact of two fish species (Cottus sp.) Mol Ecol. 2011;20:1475–1491. doi: 10.1111/j.1365-294X.2010.04997.x. [DOI] [PubMed] [Google Scholar]
- Takehana Y, Hamaguchi S, Sakaizumi M. Different origins of ZZ/ZW sex chromosomes in closely related medaka fishes, Oryzias javanicus and O. hubbsi Chromosomes Res. 2008;16:801–811. doi: 10.1007/s10577-008-1227-5. [DOI] [PubMed] [Google Scholar]
- Taylor SJ, Rojas LD, Ho SW, Martin NH. Genomic collinearity and the genetic architecture of floral differences between the homoploid hybrid species Iris nelsonii and one of its progenitors, Iris hexagona. Heredity. 2013;110:63–70. doi: 10.1038/hdy.2012.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzano DR, Kirschner J, Kamber RA, Zhang E, Weber D, Cellerino A, et al. Mapping loci associated with tail color and sex determination in the short-lived fish Nothobranchius furzeri. Genetics. 2009;183:1385–1395. doi: 10.1534/genetics.109.108670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitturi R, Rasotto MB. Karyotype analysis of Cottus gobio L. Pisces Cottidae. Cytobios. 1990;62:81–86. [Google Scholar]
- Volckaert FAM, Hanfling B, Hellemans B, Carvalho GR. Timing of population dynamics of bullhead Cottus gobio (Teleostei: Cottidae) during the Pleistocene. J Evol Biol. 2002;15:930–944. [Google Scholar]
- White MA, Steffy B, Wiltshire T, Payseur BA. Genetic dissection of a key reproductive barrier between nascent subspecies of house mice, Mus musculus domesticus and Mus musculus musculus. Genetics. 2011;169:289–304. doi: 10.1534/genetics.111.129171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf JBW, Lindell J, Backstrom N. Speciation genetics: current status and evolving approaches. Phil Trans R Soc B. 2010;365:1717–1733. doi: 10.1098/rstb.2010.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.