Abstract
Myelodysplastic syndrome, characterized by ineffective hematopoiesis and cytopenias, remains a lethal disease. Until recently, patients with myelodysplastic syndrome have been managed supportively with blood product transfusions and growth factors, until they succumb to infections, bleeding complications or transformation to acute leukemia. The discovery that epigenetic factors play an important role in cancer, and specifically in myelodysplastic syndrome, has led to the recent approval of several new therapies that will make a significant impact on this disease. Epigenetics refers to a number of biochemical modifications to chromatin that do not alter the primary DNA sequence, but play an important role in genomic regulation at the level of gene transcription. Epigenetic factors can be passed on from a cell to its progeny and can mimic traditional genetic lesions that are implicated in cancer. Unlike genetic abnormalities, however, epigenetic changes, such as DNA methylation or histone deacetylation, can be manipulated pharmacologically. Recently developed hypomethylating agents and histone deacetylase inhibitors have shown significant biological and clinical activity in myelodysplastic syndrome. These drugs have been well-tolerated by patients and have been shown to alter the course of this disease. In order to use these drugs optimally, however, we need to better understand the role of these epigenetic changes: how they contribute to the disease process, how we can use them to better select patients and how we can use combinations to target them more effectively.
Keywords: DNA methylation, histone deacetylase inhibitor, leukemia
Myelodysplastic syndromes (MDS) are a heterogeneous group of malignant clonal disorders characterized by bone marrow dysplasia, ineffective hematopoiesis, resultant cytopenias and the potential to transform into acute myelogenous leukemia (AML). MDS is a disease of older patients, with a median age at diagnosis of approximately 70 years [1–4]. This association of incidence with increasing age may suggest lifelong accumulation of genetic damage from prolonged toxic exposure or genetic predisposition. Genetic instability with detectable cytogenetic abnormalities, clonal evolution and transformation to AML is an important characteristic of the disease. In addition to age, risk factors for MDS include exposure to chemotherapy, ionizing radiation, benzene, other organic solvents, diesel fuel, smoking and immunosuppression [5]. Indeed, treatment-related (chemotherapy, radiotherapy) or ‘secondary’ MDS is a well-recognized entity. The French–American–British (FAB) Cooperative Group first classified this heterogeneous disorder into five subgroups based on morphologic findings in the peripheral blood and bone marrow [6]. Evolution of our understanding of the biological characteristics and prognosis of different forms of MDS has led to the more recent adoption of the WHO classification system [7]. Although the WHO system attempts to incorporate all known, relevant genetic, immunophenotypic, biological and clinical information to define specific disease entities, perhaps our best available tool for determining overall prognosis and guiding treatment in MDS is the International Prognostic Scoring System (IPSS) [8]. Utilizing a scoring system that takes into account the patient’s cytogenetics, percentage blasts and number of cytopenias, the IPSS divides untreated patients with MDS into four categories (low risk, intermediate-1, intermediate-2 and high risk). Although not perfect, the IPSS does a good job at estimating the survival and risk of AML transformation for untreated patients with MDS. One limitation of the IPSS is that it was designed to provide prognostic information for untreated patients at diagnosis and not throughout the course of their disease. The recently proposed WHO classification-based prognostic scoring system (WPSS) offers a ‘time-dependent’ scoring system that allows patients to be continuously re-evaluated as they progress in their disease and through treatment [9]. Using the variables of WHO subgroup, cytogenetics and transfusion requirement, the WPSS offers a scoring system similar to IPSS that divides patients into five prognostic subgroups, ranging from ‘very low’ to ‘very high’ risk. In both a learning and validation cohort, the WPSS was able to separate a heterogeneous group of MDS patients and offer meaningful prognostic information regarding survival and risk of AML transformation. However, even this scoring system has some important limitations: it excludes patients who have secondary MDS and chronic myelomonocytic leukemia (CMML) with leukocytosis; the cytogenetic categories used are similar to the IPSS and are not optimally segregated; the study groups consisted of previously untreated patients; and, finally, the score does not take into account the effect of age, performance status, severity of thrombocytopenia or duration of MDS.
In an effort to broaden the applicability of the IPSS, investigators at the MD Anderson Cancer Center (MDACC; TX, USA) applied the existing IPSS to MDS subtypes not included in the original study, specifically to patients with secondary MDS, patients with prior therapy and patients with CMML and leukocytosis. In addition to showing that the IPSS could be successfully applied to these subsets, they expanded the analysis to develop a new prognostic scoring system. In addition to refining the cytogenetic subsets, they included important features, such as age, performance status, degree of cytopenias and transfusion requirements, in the new scoring system. Using a study set and a validation test set, the investigators showed the new system to be broadly applicable, with improved prognostic separation over the existing IPSS. When the IPSS was tested on the new system, it did not offer any further prognostic power. The new scoring system is much more widely applicable and includes important clinical features that affect the prognosis of MDS. As it becomes more widely used and validated, it will become an important tool in the treatment of MDS.
Myelodysplastic syndrome remains a lethal disease, with allogeneic bone marrow transplant being the only potentially curative therapy – a therapy that is difficult in an elderly population with age-related comorbidities. Until recently, there has been no effective, MDS-specific therapy other than supportive care and growth factor supplementation. The past few years have brought the approval of three important drugs that have proven efficacy in the treatment of MDS and may change its natural history. The first, lenalidomide, is a potent thalidomide analog shown to decrease transfusion requirements and lead to complete cytogenetic remissions in a significant proportion of patients with the 5q- chromosomal abnormality [10]. Patients with MDS and an isolated 5q deletion present with a distinct ‘5q- syndrome’ characterized by a hypoproliferative anemia and dysplastic megakaryocytes. Based on a single-arm, multi-institutional study in transfusion-dependent patients with the 5q- abnormality showing significant transfusion independence and cytogenetic responses [11], lenalidomide was approved for low- and intermediate-1-risk patients with MDS who are transfusion-dependent and have the 5q- chromosomal abnormality. The next two agents, 5-azacytidine (5-aza) and 5-aza-2′-deoxycytidine (decitabine) are both hypomethylating agents that fall into the category of epigenetic therapy. The approval of 5-aza in May 2004 marked not only the first US FDA-approved treatment for MDS in the USA, but also the first epigenetic-based therapy for human disease [12]. In this review, we will discuss the concept of epigenetics, its role in MDS and the therapeutic agents currently available and in development that can modulate the epigenome.
Epigenetics
Although classically considered a disease of genetic alterations, such as mutations and deletions, it is becoming more apparent that the initiation and progression of cancer is driven by both genetic and epigenetic aberrations [13,14]. The epigenome is defined as that heritable cellular information, other than the DNA sequence itself, that is passed on from a cell to its progeny. Although not coded as part of DNA base pairs, epigenetic information has its own code that is precisely inherited and able to exert its own biological effects. The best-known epigenetic mechanisms include DNA methylation and histone protein modification, such as acetylation, methylation and phosphorylation. In general, hypermethylation of CpG islands in the promoter regions of critical tumor suppressor genes or hypoacetylation of histones in a cancer cell lead to changes in the local chromatin structure that signal repression of gene transcription. In this manner, methylation of a gene promoter or deacetylation of regional histones can functionally mimic a catastrophic genetic event such as a deletion, a loss-of-function mutation or chromosomal loss. Acting in concert with existing genetic abnormalities, aberrant epigenetic events can fulfill the requirements of Knudson’s two-hit hypothesis and contribute to the transformation of a normal cell into a malignant one.
DNA methylation
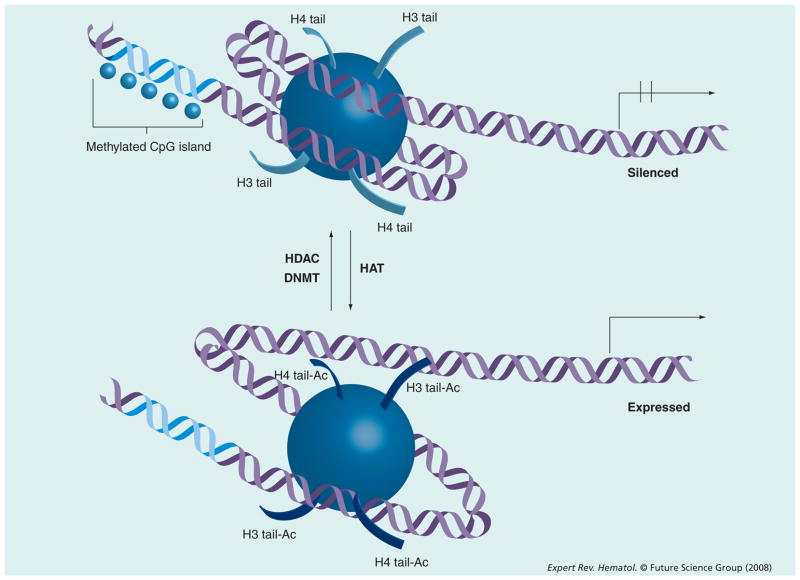
DNA methylation (the addition of a methyl group to a DNA base) in mammals occurs exclusively at the 5-carbon of a cytosine that sequentially precedes a guanine – a so-called CpG dinucleotide. This reaction is catalyzed by enzymes known as DNA methyltransferases (DNMTs). There are three known human DNMTs. DNMT1 is responsible for ‘maintenance’ methylation. That is, it conserves appropriate DNA methylation during mitosis. The exact functions of DNMT 3a and 3b are still being further characterized [15]. Relative to what would be mathematically expected, CpG dinucleotides are relatively under-represented in the human genome and exist either sparsely throughout the genome in noncoding regions or in clusters known as CpG islands [16,17]. These CpG islands are found most often in close association with genes, either in their promoter regions, or within the first exon. Methylation of the CpG islands leads to changes in the local chromatin structure that leads to repression of gene transcription (Figure 1). In effect, methylation of a gene promoter leads to loss of expression of the downstream gene. Acting independently or in concert with existing genetic abnormalities, aberrant gene promoter methylation can potentially lead to tumor suppressor gene inactivation and contribute to the transformation of a normal cell into a malignant one [16]. This has been described in several known human cancers. For example, in various familial cancer syndromes, the first ‘hit’ is the germ-line inherited genetic defect. The second hit has been shown in some instances to be gene promoter methylation of the normal allele – effectively leading to loss of heterozygosity and progression to cancer [18,19].
Figure 1. Epigenetic control of transcription.
Histone acetyl transferase (HAT) and histone deacetylase (HDAC) work in an opposing manner and lead to open or closed (silenced) chromatin states. DNA methylation of CpG islands is catalyzed by DNA methyltransferase (DNMT) and leads to transcriptional silencing.
Ac: Acetylated; H3: Histone.
Although DNA methylation has been recognized for many years as an important mechanism contributing to X chromosome inactivation and imprinting [20,21], more recently its importance in cancer has been recognized. The first reports of methylation and its importance in cancer reported a global decrease in methylation within the genomes of cancer cells compared with normal cells – implying that this was a feature of carcinogenesis [22–25]. However, further investigation uncovered another side to the story; in parallel with global DNA hypomethylation in neoplastic cells relative to normal cells, there was significant hypermethylation at the promoter regions of various genes [24,26]. This hypermethlyation was linked to gene silencing and loss of protein expression. Silencing of important tumor suppressor genes in this manner could contribute the tumorigenesis.
Unlike genetic defects in cancer, which cannot be modified pharmacologically, epigenetic defects are potentially reversible. The development of active compounds that inhibit DNMT and reverse DNA methylation have made it feasible to target the epigenome to therapeutic ends. Although these DNMT inhibitors (DNMTi) are often referred to as ‘hypomethylating agents’, it is important to realize that these agents do not catalyze the active removal of methyl groups from cytosines, but inhibit the addition of methyl groups on a subsequent round of DNA replication. This requires a cycle of cell division after exposure to a DNMTi to lead to hypomethylation. We use these terms interchangeably in this review. In theory, reactivation of aberrantly suppressed tumor suppressor genes using these drugs could restore the cells’ normal signaling pathways and lead to apoptosis or differentiation of abnormal cells. Both preclinical and clinical studies of these agents have demonstrated activity and generated a great deal of excitement regarding the potential of these drugs.
Histone modification
Nuclear histones make up the protein core of a nucleosome, the basic unit of human chromatin packaging. A nucleosome is made up of segment of 146 bp of DNA that is wrapped around a histone octamer containing two copies each of the proteins histone H2A, H2B, H3 and H4. This segment is followed by a small stretch of DNA called the linker, followed by an adjacent nucleosome. Biochemical modifications to these component histones, including acetylation, methylation or phosphorylation, alter the conformation as well as the electrostatic charges that affect protein–protein and protein–DNA interaction [27]. These modifications, therefore, can alter chromatin topology and have structural as well as functional consequences – including regulation of gene transcription (Figure 1). These biochemical modifications of histones are regulated by cellular enzymes that respond to complex cellular physiology, and represent an epigenetic control of gene expression. The countless potential combinations of histone modifications and their effect on chromatin topology is often referred to as the ‘histone code’.
Although advances are being made in identifying the role of histone methylation in cancer [28], the best studied of these modifications thus far in the clinical setting is histone acetylation. Histone acetyltransferases and histone deacetylases (HDACs) work in an opposing manner by acetylating and deacetylating nuclear histones, respectively. Acetylation of lysine residues on histones H3 and H4 leads to an open or permissive chromatin configuration that favors gene transcription. Removal of these acetyl residues by HDACs leads to a closed, compact or repressive configuration that can functionally lead to silencing of gene expression [27]. Phylogenetic analysis reveals HDACs to be highly conserved enzymes that have homologs in animals, plants, fungi and bacteria [29]. Four classes (i–iv) of HDACs have been identified in humans and are grouped according to structural homology (detailed review in [30]). The availability of HDAC inhibitors (HDACi) provides another opportunity to potentially manipulate chromatin configuration and epigenetically affect gene transcription. A number of structurally distinct HDACi are currently being developed for potential applications in cancer. All currently available HDACi mainly inhibit class I and class II HDACs. Although histones represent the bulk of the target for HDACs, it is important to note that there are several other intracellular substrates for HDACs that are regulated by reversible acetylation. Among those important in cancer include transcription factors (SMAD7 and RUNX), p53, HSP90 chaperone protein, STAT3 and Ku70 (involved in sequestering BAX) [30]. The antineoplastic effects of HDACi, therefore, may be broad and not yet completely defined. Demonstration of their safety in humans and clinical activity in hematologic malignancies has led to further investigation into and development of these compounds.
Importance of epigenetics in MDS
The current working hypothesis of the mechanism of activity of epigenetic therapy in leukemia and MDS is that reactivation of aberrantly suppressed genes restores their tumor suppressive ability and mediates clinical responses. Box 1 includes a nonexhaustive list of the most commonly methylated genes in MDS and AML. Although several groups have shown the presence of epigenetically suppressed genes in a wide range of hematologic malignancies [31–34] (reviewed in [35]), treatment responses to hypomethylating agents, HDACi, or their combination have not directly correlated with reactivation of these genes. In fact, most of the knowledge generated about the efficacy of epigenetic therapy in MDS has been empiric in nature. That is to say, initial Phase I trials of hypomethylating agents and HDACi in a broad group of tumor types showed a signal of activity in hematologic malignancies. This has led to larger, disease-specific trials in AML and MDS, which uncovered significant clinical benefit. Correlative studies associated with these clinical trials have indeed shown induction of global hypomethylation or reactivation of selected genes. However, one of the major limitations of this therapy is its relative nonspecificity. Since epigenetic therapy has broad-reaching effects within a cell, it is not clear whether global hypomethylation is important, or whether reactivation of a specific set of genes is important. For that matter, it is not clear which set of genes is important. With our current agents, we cannot ensure that the desired ‘tumor suppressor’ genes are the only ones to be reactivated. No single gene or set of genes has yet been shown to definitively correlate with response. Several groups have investigated the use of methylated genes as biomarkers to predict relapse or prognosis in leukemia [36–39]. More recently, another group has shown an increase in methylation of a set of genes from diagnosis to relapse in patients with AML, alluding to the role of DNA methylation in disease progression and/or chemotherapy resistance [40]. However, the importance of these genes in the pathophysiology of the disease needs to be scrutinized further. Ultimately, the application of gene-expression arrays and other high-throughput techniques may allow us to better define a gene-expression signature that is specific for response in a particular disease, but this work is ongoing.
Box 1. Most commonly methylated genes in myelodysplastic syndrome and acute myelogenous leukemia.
p15
E-cadherin
SOCS-1
p73
DAPK-1
HIC-1
RARβ2
CRBP1
Calcitonin
ER (estrogen receptor)
MGMT
For example, at the 2007 meeting of the American Society of Hematology (ASH), several groups investigated the methylation patterns of normal, AML and MDS cells. Jiang et al. sought to assess the ‘methylome’ of normal CD34+ cells and compare it with that of primary AML blasts and selected CD34+ AML cell lines using chromatin immunoprecipitation coupled with a human gene promoter array [41]. Using this technology, they were able to separate distinct ‘methylation signatures’ that defined primary AML cells, AML cell lines and CD34+ cells from normal volunteers. Overall, they found a relative increase in promoter hypermethylation in AML primary samples and cell lines compared with the normal controls, helping support the hypothesis of using hypomethylating agents in this disease. However, they also found considerable interpatient variability, as well as marked differences between primary AML samples and cell lines. This again highlights the relative heterogeneity of this disease and perhaps some of the shortfalls of our in vitro models. When the investigators examined the methylated promoters in the AML samples, they found a number of important tumor suppressor genes, apoptotic genes and other loci that were previously demonstrated to play a role in AML [41]. This again lends support to the role of aberrant hypermethylation in the pathogenesis of AML/MDS. In another study, Figueroa et al. similarly sought to define the ‘methylome’ of neoplastic cells from patients with MDS and AML and compare them with CD34+ cells from normal controls using a high-throughput method for detecting whole genome promoter methylation [42]. They examined samples from 13 patients with MDS, 16 patients with de novo AML (diploid karyotype) and eight healthy donors. They found a strikingly larger number of hypermethylated genes in patients with MDS when compared with either AML or healthy donors. Analyzing the data from their array, they were able to describe an aberrant methylation signature of 736 genes that differentiated MDS cells from normal CD34+ cells, reflecting extensive epigenetic dysregulation in this disease. When compared with de novo AML, there were also a significantly greater number of methylated genes in the MDS patients, suggesting that these are indeed different diseases. This could explain why the activity of the hypomethylating agents is more prominent in MDS as compared with AML. It also implies that the activity of drugs such as 5-aza and decitabine in MDS owes more to their DNA methyltransferase inhibitory activity and less to their cytotoxic activity. When these investigators further examined the differentially methylated promoters between MDS and AML samples they also found pathophysiologically significant genes, such as RUNX2, DAPK2, CEBPZ, MDM2 and others.
These studies are important for beginning to understand the role of epigenetics in MDS. As these and other data continue to mature, we may be better able to define subsets of patients who can benefit from these therapies and perhaps how we can augment the activity of these therapies. Despite the ambiguity associated with the mechanism of action of epigenetic therapy, one thing cannot be debated; well-conducted clinical trials have shown the safety and striking clinical efficacy of these agents in the treatment of MDS.
Clinical experience with epigenetic therapy
Hypomethylating agents
There are currently two hypomethylating agents that are available and FDA-approved for the treatment of MDS: 5-aza and decitabine. At low doses, both drugs are inhibitors of DNMT and therefore lead to hypomethylation. At higher doses, however, both drugs are cytotoxic.
5-azacytidine
5-aza is an analog of the naturally occurring pyrimidine nucleoside cytidine in which the 5-carbon atom of the pyrimidine ring is replaced with a nitrogen atom. Since it is composed of a ribose sugar, 5-aza is mono-, di- and tri-phosphorylated prior to becoming incorporated into RNA. Here it disrupts RNA metabolism and protein synthesis, presumably leading to cytotoxicity. 5-azacytidine diphosphate is also a substrate of ribonucleotide reductase, which converts the nucleoside to 5-aza-deoxycytidine diphosphate. This is then phosphorylated by nucleoside diphosphate kinases to 5-aza-deoxycytidine, which is incorporated into DNA. Here, at varying doses, it can lead to inhibition of DNA synthesis – resulting in cytotoxicity, or inhibition of DNMT1 – leading to DNA hypomethylation [12].
Initial studies in MDS were conducted by the Cancer and Leukemia Group B (CALGB) and demonstrated the safety and significant clinical activity of intravenous or subcutaneous administration of 5-aza (Table 1). After showing efficacy in two single-arm trials (CALGB 8421, 8921) a randomized, controlled Phase III, multicenter clinical trial was conducted comparing 5-aza treatment to the best supportive care [43,44]. The trial enrolled 191 patients from all five FAB subtypes of MDS who had cytopenias or required transfusions. The patients were randomized to either 5-aza received at a dose of 75mg/m2/day subcutaneously daily for 7 days every 28 days versus the best supportive care, with the primary efficacy end point of overall response (OR). The trial was designed to allow for patients to crossover from the observation arm to treatment with 5-aza if they met prespecified criteria of disease progression. The study demonstrated an OR rate (ORR) of 60% in the treatment arm (including 7% complete response [CR], 16% partial response [PR] and 37% hematological improvement) compared with an ORR of 5% (no CR or PR) in the supportive arm. There was also an improved time to leukemic transformation or death in the treatment arm, as well as a suggestion of possible survival benefit. Given the cross-over design of the trial and lack of power, a statistically significant survival benefit was not shown. This study (CALBG 9221) led to the FDA approval of 5-aza for the treatment of MDS in the USA [43].
Table 1.
Clinical experience with single-agent 5-azacytdine in myelodysplastic syndrome and acute myelogenous leukemia.
| n | Complete response (n [%]) | Partial response (n [%]) | Hematologic improvement (n [%]) | Overall response rate (n [%]) | Ref. | |
|---|---|---|---|---|---|---|
| CALGB 8421 iv. azacitidine | 48 | 7 (15) | 1 (2) | 13 (27) | 21 (44) | [44] |
| CALGB 8921 sc. azacytidine | 70 | 12 (17) | 0 (0) | 16 (23) | 28 (40) | [44] |
| Updated CALGB 9221 sc. azacytidine | 99 | 10 (10) | 1 (1) | 36 (36) | 47 (47) | [43,44] |
| CALGB 9221 sc. azacytidine crossover after observation | 51 | 3 (6) | 2 (4) | 13 (25) | 18 (35) | [43,44] |
CALGB: Cancer and Leukemia Group B; iv.: Intravenous; sc.: Subcutaneous.
Since this trial failed to show a survival benefit, a second randomized trial was performed to study the effect of 5-aza on overall survival. The much anticipated results from this trial were recently reported at the 2007 ASH meeting [45]. This was a Phase III international, multicenter, randomized prospective trial designed to demonstrate the superiority of 5-aza plus best supportive care in prolonging overall survival compared with so-called conventional care regimens (CCRs) plus best supportive care. CCRs were defined as:
Best supportive care
Low-dose ara-C (20 mg/m2/day × 14 days, every 28 days)
Standard chemotherapy including induction/consolidation
Prior to being randomized to either arm, investigators preselected patients with high or int-2 risk into one of the three CCRs. The trial did not allow use of erythropoietin and analysis was by intent-to-treat principle. A total of 358 patients with higher risk MDS were randomized between the two arms. The 5-aza was well-tolerated among the treated patients and its safety profile was consistent with previous reports. Most notably, the study demonstrated a significant survival benefit for patients in the 5-aza arm compared with those in the CCR arm (hazard ratio: 0.58; 95% confidence interval: 0.43, 0.77). The Kaplan–Meier estimated overall survival was 24.4 months for 5-aza compared with 15 months with CCR (p = 0.0001). This trial, which still has not been published in manuscript form, confirms previous observations of 5-aza in MDS and establishes its role as the standard of care in this population.
5-aza-2′-deoxycytidine
Decitabine is an analog of 2′-deoxycytidine. Similar to 5-aza, the 5-carbon of the pyrimidine ring has been replaced by nitrogen. The difference is that decitabine is built on a deoxyribose backbone, allowing it to become incorporated directly into DNA after being acted on by cellular kinases. After rapidly entering into the cell by a nucleotide-specific transport system, decitabine is activated by sequential phosphorylation from 5-aza-dCMP to 5-aza-dCDP, and 5-aza-dCTP by deoxycytidine kinase, dCMP kinase and diphosphokinase, respectively [46]. DNA polymerase catalyzes the incorporation of decitabine’s active metabolite 5-aza-dCTP into DNA, thereby producing DNA lesions. Once incorporated into DNA, decitabine covalently binds to DNA-methyltransferase and traps it, leading to its irreversible inhibition. At high doses, decitabine is cytotoxic owing to DNA cross-linking and DNA synthesis arrest. At lower decitabine doses, the cell survives with depleted levels of DNA-methyltransferase, leading to DNA hypomethylation with subsequent DNA replication [47].
Decitabine was first studied in patients with MDS in a Phase I study of low-dose decitabine by Zagonel et al. [48]. Although previous Phase I studies had shown the maximum tolerated dose (MTD) of decitabine to be higher, this study aimed to examine a lower dose that perhaps took advantage of its hypomethylating, differentiating effects. In total, ten patients with advanced MDS were treated with either:
45 mg/m2/day intravenously divided into three 4-h infusions for 3 days
50 mg/m2 intravenously continuous infusion for 3 days
Half the patients responded, with four out of the ten patients achieving a complete hematologic response. This evidence of a well-tolerated dosing schedule and significant improvement in trilineage hematopoiesis led to further development of this drug in MDS.
Wijermans etal. performed a Phase II study of lower dose decitabine in elderly patients with high-risk MDS [49]. They initially started with a dose of 50mg/m 2/day administered as a continuous infusion for 3 days and repeated every 6 weeks. After the first 21 patients displayed greater than expected myelotoxicity, the dose was reduced to 40 mg/m2/day in the last eight patients. A total of 29 patients were enrolled in the trial. Out of 28 evaluable patients, 15 (54%) had an objective response, including eight CRs (29%), five PRs (18%) and two with hematologic improvement (HI; 7%). The median duration of response was longer than 31 weeks. Including those patients who did not meet criteria for response, but had stable disease, the overall progression-free interval was 33 weeks (range: 6–83 weeks). This encouraging data prompted the same group from The Netherlands to lead a multicenter, Phase II study to confirm their findings [50]. In this trial, decitabine was administered at a dose of 15 mg/m2 intravenously over 4 h every 8 h (total daily dose 45 mg/m2) for 3 consecutive days, every 6 weeks. A total of 66 patients with intermediate-1 to high-risk MDS (by IPSS) from seven centers were enrolled on the trial. The investigators reported an ORR of 49% (32/66) with 20% (13/66) CR, 4.5% (3/66) PR and 24% (16/66) HI. Further analysis of patients enrolled on these two studies as well as a third, compassionate-use trial revealed that decitabine was also able to induce significant cytogenetic responses [51]. Out of 115 successfully karyotyped patients in this cohort, 61 had chromosomal abnormalities prior to treatment. Of those 61 patients, 19 (31%) had a major cytogenetic response after a median of three cycles.
Following this promising data from the European experience, a multicenter randomized Phase III trial of decitabine versus best supportive care was conducted in the USA [52]. In this trial, decitabine was administered at a dose of 15 mg/m2 intravenously over 3 h, every 8 h daily for 3 consecutive days once every 6 weeks. Supportive care, including transfusions, antibiotics and hematopoietic growth factors, was provided according to generally accepted guidelines to patients on both arms. A total of 170 patients representing all FAB subtypes of MDS were enrolled on the trial – 89 randomized to decitabine and 81 to supportive care alone. The ORR of patients on the decitabine arm was 17% (15/89) including eight CRs (9%) and 7 PRs (8%). The supportive care arm had no responses. The time to first response was 3.3 months (2–9.7 months) with a median duration of response of 10.3 months (4.1–13.9 months). The rate of HI on the decitabine arm was 13 versus 7% on the supportive care arm. This translated into an overall improvement rate (CR + PR + HI) of 30 versus 7% in favor of the treatment arm (p < 0.001). There was a trend towards longer time to progression to AML or death for the decitabine arm (12.1 vs 7.8 months; p = 0.16) but it was not statistically significant. Of the patients who were evaluable for cytogenetic response, 35 versus 10% achieved complete cytogenetic remission in favor of the decitabine cohort. Decitabine was relatively well-tolerated in this population, with the expected myelosuppression being the most common side effect. Results from this and previous studies led to the FDA approval of decitabine for patients with MDS.
The next advances in the use of decitabine for MDS involved optimizing the dose and schedule of decitabine. Drawing from clinical and preclinical observations, investigators from the MDACC sought to refine the dosing of decitabine to further exploit its hypomethylating effects, specifically by lowering the dose and prolonging the exposure. In a Phase I study, Issa et al. evaluated low-dose, prolonged exposure schedules of decitabine in patients with relapsed/refractory acute leukemias [53]. Patients were treated in cohorts that received decitabine at doses of 5, 10, 15 or 20 mg/m2 intravenously over 1 h daily for 10 days (5 on, 2 off, 5 on). Two additional cohorts also received doses of 15mg/m 2 daily for 15 or 20 days to better delineate this dose level. A total of 50 patients were treated in this study, 44 with AML/MDS, five with CML and one with ALL. The drug was well-tolerated at all dose levels and schedules, with responses observed at all dose levels. However, a dose of 15 mg/m2 for 10 days appeared to induce the most responses (11/17 or 65%). Responses (CR + PR) were observed in patients with AML (22%), MDS (58%) and CML (80%) [53,54]. Prolonged, lower doses of decitabine appeared to have significant activity in this refractory population.
In an effort to further define the optimal delivery schedule of decitabine based on its mechanism, a Phase II adaptively randomized trial of low-dose decitabine in advanced leukemia was conducted at MDACC. The study by Kantarjian et al. aimed to lower the total dose of decitabine to 100 mg/m2 per cycle compared with the standard 145 mg/m2 as well as increase the dose intensity by administering the drug over a prolonged period (5 or 10 days instead of 3), every 4 weeks (instead of the standard 6 weeks) [55]. Three dosing regimens of decitabine were tested:
10 mg/m2 intravenously over 1 h daily for 10 days
20 mg/m2 intravenously over 1 h daily for 5 days
20 mg/m2 subcutaneously daily for 5 days
The Bayesian trial design initially allowed randomization of patients equally to all three arms until 45 patients were enrolled. The characteristics of the randomization then changed, where the randomization was favored to the arm with the best activity. In this way, more patients would be receiving the more effective regimen, and a lower total number of patients would be required to achieve the study objectives. A total of 95 patients (77 with MDS and 18 with CMML) were enrolled on the trial. A total of 32 patients (34%) achieved a complete remission, with 69 patients (73%) achieving some clinical response. The 5-day intravenous schedule was chosen as the most optimal, demonstrating a CR rate of 32%, compared with 21% in the subcutaneous arm and 24% in the 10-day intravenous arm. Correlative studies on the trial also showed a more pronounced degree of hypomethylation achieved with the 5-day intravenous schedule. The authors concluded that the 5-day intravenous schedule of decitabine was tolerable and significantly active, achieving a higher CR than previous standard schedules. A randomized trial comparing the two is needed to confirm this. Table 2 summarizes the published clinical data with single-agent decitabine.
Table 2.
Clinical experience with single-agent decitabine in myelodysplastic syndromes and acute myelogenous leukemia.
| Study | Phase | Dose and schedule | n | Complete response (n [%]) | Partial response (n [%]) | Hematologic improvement (n [%]) | Overall response rate (n [%]) | Ref. |
|---|---|---|---|---|---|---|---|---|
| Zagonel et al. | I/II | 45–50 mg/m2/day iv. × 3 days | 10 | 4 (40) | 0 | 1 (10) | 5 (50) | [48] |
|
| ||||||||
| Wijerman et al. | II | 40–50 mg/m2/day iv. × 3 days | 28 | 8 (29) | 5 (18) | 2 (7) | 15 (54) | [49] |
|
| ||||||||
| Wijerman et al. | II | 45 mg/m2/day iv. × 3 days | 66 | 13 (20) | 3 (4.5) | 16 (24) | 32 (49) | [50] |
|
| ||||||||
| Kantarjian et al. | III | 45 mg/m2/day iv. × 3 days | 89 | 8 (9) | 7 (8) | 12 (13) | 27 (30) | [52] |
|
| ||||||||
| Issa et al. | I | 5 mg/m2/day iv. × 10 days | 6 | 1 | 0 | 0 | 1 (17) | [53] |
| 10 mg/m2/day iv. × 10 days | 7 | 1 | 0 | 0 | 1 (14) | |||
| 15 mg/m2/day iv. × 10 days | 6 | 4 | 1 | 0 | 5 (83) | |||
| 20 mg/m2/day iv. × 10 days | 8 | 0 | 0 | 1 | 1 (13) | |||
| 15 mg/m2/day iv. × 15 days | 8 | 1 | 0 | 0 | 1 (13) | |||
| 15 mg/m2/day iv. × 20 days | 3 | 0 | 0 | 0 | 0 (0) | |||
| 15 mg/m2/day iv. × 10 days | 10 | 2 | 0 | 3 | 5 (45) | |||
| Total | 50 | 9 (18) | 1 (2) | 4 (8) | 14 (28) | |||
|
| ||||||||
| Kantarjian et al. | II | 10 mg/m2/day iv. × 10 days | 17 | 4 (24) | [55] | |||
| 20 mg/m2/day iv. × 5 days | 64 | 25 (39) | ||||||
| 20 mg/m2/day sc. × 10 days | 14 | 3 (21) | ||||||
| Total | 95 | 32 (34) | 1 (1) | 13 (14) | 46 (48) | |||
iv.: Intravenous; sc.: Subcutaneous.
Histone deacetylase inhibitors
Histone deacetylase inhibitors have emerged as multifunctional cancer agents with some signal of activity in a broad range of malignancies [30,56,57]. Several compounds, from both natural and artificial sources, are currently under development. The compounds differ in chemical structure, specificity and potency with respect to HDAC inhibition. Their safety and efficacy has been shown in multiple clinical studies and has led to FDA approval of the first HDACi, suberoylanilide hydroxamic acid (SAHA, vorinostat), in cutaneous T-cell lymphoma [58]. Unlike the hypomethylating agents, HDACi have had more modest success as single agents in the treatment of patients with MDS/AML. As we will discuss later, although HDACi demonstrate some single-agent activity in these diseases, their optimal use may be in combination with other agents.
A number of different HDACi have been tested in patients with MDS and acute leukemia (Table 3). Agents include phenylbuyrate [59], valproic acid [60,61], depsipeptide [62,63], MGCD0103 [64], LBH589 [65], ms-275 [66] and vorinostat [67]. Each has been shown to have some single-agent clinical benefit, albeit small. The first HDACi to be tested in patients with MDS and demonstrate clinical benefit was sodium phenylbutyrate. In two studies by Gore et al., responses were observed in four out of 27 (15%) and two out of 23 patients (9%), respectively [59,68]. Perhaps the most extensively studied HDACi in MDS is valproic acid (VPA). VPA is a fatty acid chain derivative used clinically as an anticonvulsant and mood stabilizer, which has weak HDACi activity at millimolar serum levels [69]. In an intial study, 18 patients with MDS were treated with single-agent VPA [61]. Eight patients (44%) responded to treatment, including seven patients with hematologic improvement, one with PR and no CRs. Five additional patients on the same study treated with a combination of VPA plus all-trans retinoic acid (ATRA) had no response. In a follow-up report of 75 (43 with MDS), 30% of MDS patients achieved a response, including 13 with HI, one CR and one with PR [70]. When divided by IPSS risk, the response rates by group were: 70% in low-risk, 24% in intermediate-1, 11% in intermediate-2 and 18% in high-risk. These encouraging studies with a relatively weak HDACi prompted investigation of more potent agents. Unfortunately, the increase in potency did not translate into dramatically increased activity in MDS. For example, a Phase I trial of the potent, ‘pan-HDAC’ inhibitor, vorinostat was conducted in patients with advanced leukemia or MDS [67]. The majority of the patients had the diagnosis of AML (76%) or MDS (7%). Out of 41 patients enrolled, there were a total of seven responses (17%), including two CRs, two CRs with incomplete blood count recovery and three HI. Other Phase I trials with HDACis, carried out mainly in patients with AML or high-risk MDS have shown marginal responses and reasonable tolerability.
Table 3.
Clinical experience with histone deacetylase inhibitors and their combinations.
| Study | Phase | n | Major toxicity | Overall response rate (%) | Ref. |
|---|---|---|---|---|---|
| MS-275 | I | 39 | Neuro | 0 | [66] |
| Depsipeptide | I | 10 | Constitutonal | 0 | [62] |
| Vorinostat | I | 41 | GI | 17 | [67] |
| MGCD0103 | I | 29 | GI | 10 | [64] |
| LBH589 | I | 29 | Cardiac | 0 | [65] |
| VPA | II | 75 | Neuro | 24 | [70] |
| AZA + PB | I | 36 | Neuro | 30 | [75] |
| DAC + VPA | I/II | 54 | Neuro | 22 | [76] |
| AZA + VPA | I/II | 53 | Neuro | 42 | [77] |
| AZA + MGCD0103 | I/II | 52 | GI | 36 | [78] |
AZA: 5-azacytidine; DAC: Decitabine; GI: Gastrointestinal; Neuro: Neurotoxicity; PB: Phenylbutyrate; VPA: Valproic acid.
The side-effect profile of HDACi seems to be fairly uniform across the class of agents. Most of the Phase I trials report reasonable tolerability at their respective MTDs. The most common dose-limiting toxicities (DLTs) were fatigue, nausea, vomiting and diarrhea. Overall, the most common reported drug-related adverse events and laboratory abnormalities were diarrhea, nausea, fatigue, anorexia, vomiting, thrombocytopenia and hypokalemia. Among the grade 3/4 toxicities reported were nausea, vomiting, fatigue and thrombocytopenia.
Combination epigenetic therapy
DNA methylation and histone acetylation are dynamic processes that are intimately interrelated and cooperate in the epigenetic regulation of gene expression [71]. Methylated sequences can recruit correpressor complexes that often contain HDAC activity. In this manner, promoter methylation and regional histone de-acetylation by these correpressor complexes trigger a compacted chromatin configuration that is inaccessible by the transcriptional machinery. Targeting both DNA methylation and histone deacetylases, therefore, could theoretically lead to greater reversal of aberrant gene silencing and potentially increased anti-tumor activity. This concept has been tested in vitro and has shown significant synergy [72,73]. The in vitro data prompted several Phase I and II studies to assess the clinical activity of the combination of a hypomethylating agent and an HDACi.
The first HDACi to be studied in this manner was phenyl-butyrate (PB), combined with 5-aza, in patients with MDS or AML [74]. The patients were administered a fixed dose of PB (375 mg/kg/day by continuous infusion for 7 days) that was started after the completion of 5-aza (administered at a dose of 75 mg/m2/day subcutaneously for 5, 10 or 14 days). A total of 32 patients were enrolled on the trial (13 MDS, one CMML, 15 MDS-AML and three relapsed AML), with 29 evaluable for response. Eleven out of 29 (38%) patients responded, including five CRs, one PR and five HIs. Three of the major responders with pretreatment cytogenetic abnormalities also had a complete cytogenetic response. Histone deacetylation and DNA hypomethylation was detected in patients after treatment. The authors concluded that this was a reasonably well-tolerated combination with a suggestion of higher response rates than 5-aza alone. A second smaller study using the combination of 5-aza and PB enrolled a total of 12 patients (10 AML, 2 MDS) [75]. These patients were also treated sequentially with 5-aza (75 mg/m2/day subcutaneously × 7days) followed by PB (200 mg/kg/day intravenously over 1–2 h × 5 days). Out of ten evaluable patients, there were three PRs, no CRs and two patients with stable disease. Again, histone acetylation was observed in patients treated on the trial, but it did not correlate with response.
Valproic acid has also been tested extensively in combination with hypomethylating agents. In a Phase I/II study, Garcia-Manero et al. studied the concomitant administration of decitabine and VPA in patients with AML and MDS [76]. A total of 54 patients were treated (48 AML, six MDS), of which 11 were previously untreated. Patients were treated with a fixed dose of decitabine (15 mg/m2/day intravenously × 10 days) along with escalating doses of VPA (20–50 mg/kg daily × 10 days). The 50-mg/kg dose was found to be safe and was chosen as the Phase II dose. A total of 12 patients (22%) had some objective response, including ten CRs (19%), and two with complete remission in the absence of total platelet recovery (CRp; 3%). Among the ten untreated elderly patients with AML/MDS, five (50%) had a response (four CR, one CRp). Neurotoxicity in the form of transient confusion and somnolence was observed in elderly patients at the 50-mg/kg dose and was potentiated by other neurotropic agents. The authors noted that responders had higher free VPA levels at day 10 when compared with the nonresponders. Both DNA hypomethylation and histone deacetylation were observed, but neither correlated with response. A closely related Phase I/II study combined 5-aza with VPA and ATRA in a population of patients with AML and MDS [77]. A total of 53 patients (49 AML, four MDS) were treated, of which 33 were previously untreated. Patients were given fixed doses of 5-aza (75 mg/m2/day subcutaneously × 7 days) and ATRA (45 mg/m2/day orally × 5 days starting on day 3) along with escalating doses of VPA. The MTD of VPA in this combination was 50 mg/kg/day × 7 days and was used as the Phase II dose. The ORR was 22 out of 53 (42%) including 12 CR, three CRp, and seven HI. In the previously untreated subset, the ORR was 17 out of 33 (52%) with 11 CR, three CRp and three HI. These results were especially encouraging in the treatment of elderly AML, as all the patients in this group were aged over 60 years and there was no induction-related mortality. Reversible neurotoxicity in the form of confusion and somnolence was once again associated with the high doses of VPA in this population. However, once again responders had significantly higher levels of VPA than nonresponders. It was also important to note that, unlike single-agent 5-aza, the time to response was shorter with the combination.
To better define the optimal dose of decitabine and the contribution of VPA in the combination, a Phase I trial was conducted of decitabine with or without VPA in patients with AML [78]. A total of 25 patients were treated, 12 of whom were previously untreated. The treatment plan consisted of a two-step process. Part A treated patients with escalating doses of decitabine alone for 10 days (starting at 15 mg/m2/day intravenously) to determine the optimal biologic dose (OBD) – defined as the dose that led to upregulation of estrogen receptor or p15 mRNA by 100% or greater relative to pretreatment levels. Once the OBD of 20 mg/m2/day was determined, part B of the trial treated patients with that dose of decitabine along with escalating doses of VPA. The MTD for VPA in the combination was found to be 20 mg/kg/day, with the DLT being neurotoxcity, including grade III encephalopathy. Out of 21 evaluable patients, the authors reported a 52% response rate, including four CR (19%), four incomplete CR (19%) and three PR (14%). In the subset of untreated patients, four out of nine (44%) evaluable patients achieved a CR. In this small study, the authors concluded that the impact of adding VPA to decitabine on clinical responses was not clear, but the time to response with the combination was shorter than with decitabine alone.
Building upon these early combination epigenetic trials and following the observation that higher VPA levels were associated with responders, newer, more potent HDACis have been combined with 5-aza or decitabine. Several studies are ongoing, exploring the combinations of MS-275 and 5-aza [79], MGCD0103 and 5-aza [80], as well as vorinostat and decitabine [81,82]. These Phase I/II clinical trials have been presented in their preliminary form and report safety and tolerability of the combinations. Clinical activity is noted with response rates ranging from 20 to 44%. Continued follow-up and mature data from these trials should give us a better idea of the potential of these combinations. To summarize, the role of combination epigenetic therapy in the treatment of MDS is not clear. Our current knowledge from these studies suggests that the combinations are safe and have clinical activity. However, the impact of the addition of the HDACi on clinical efficacy is not exactly clear. Ongoing randomized trials comparing hypomethylating agent alone vesus hypomethylating agent plus HDACi are currently ongoing and should help answer this question. What we have learned from these early studies is that the prolonged time-to-response observed with hypomethylating agents alone is reduced with the addition of an HDACi. Whether this earlier response translates into clinical benefit remains to be seen. Another potentially important role for combination epigenetic therapy is in the treatment of elderly patients with AML. The majority of patients on these trials had the diagnosis of AML rather than MDS with a median age of greater than 60 years. The relatively good response rates to ‘induction’ with these combinations, coupled with low induction-related mortality, make these encouraging regimens to study in this population.
Expert commentary
Until recently, the MDS were considered a group of premalignant bone marrow disorders. Younger patients, a minority, were treated with allogeneic stem cell transplantation, and older ones with palliative and supportive care measures such as transfusion support and hematopoietic growth factor administration. Over the last few years, we have experienced a number of very significant improvements in the understanding, classification and treatment of this heterogeneous group of disorders. One of these has been the introduction of DNA methyltransferase inhibitors, or hypomethylating agents, in MDS. Current available agents, 5-aza and 5-aza-2′-deoxycitidine, have significant activity in MDS and have been shown to alter the natural course of the disease with acceptable toxicities. That said, these therapies have several significant limitations that we need to overcome. First, response rates probably occur in 50% of patients treated and therefore more potent epigenetic interventions are needed. Second, it is expected that at some point most patients will lose response to the therapy, requiring prolonged courses of therapy administration and the need of subsequent interventions for patients that lose a response. Third, there is very little understanding in terms of the molecular mechanisms that mediate response to epigenetic therapy in MDS. At the present time, there are no known epigenetic biomarkers that predict for response or survival, or lack of, in patients treated with these agents. Ongoing extensive genome-wide epigenetic studies may shed light on this problem. New combinations, perhaps with the histone deacetylase inhibitors, may result in increased response rates or longer duration of response. This will need to be tested in randomized studies. Finally, it is possible that other molecular mechanisms involving DNA damage/repair and mitochondria dysfunction could modulate in part the clinical activity of these drugs, a phenomenon that may help in the development of other rationale combinations. Further understanding of resistance mechanisms, dose and schedule optimizations (for lower risk vs higher risk patients) may also contribute towards improving current results with these agents.
Five-year view
In addition to achieving durable responses, complete responses and major cytogenetic responses in our patients with MDS, we need more therapies that are proven to show significant survival benefit and are able to significantly alter the course of the disease. We now have evidence of improved survival with 5-aza, with anticipated results from a decitabine study also in progress. Next, combination epigenetic therapy needs to be scrutinized, to determine whether using hypomethylating agents and HDACi offer similar benefits of improved response rate, decreased rate of transformation to AML and, ultimately, improved survival over the current standard-of-care. The use of low-dose hypomethylating agents as maintenance therapy after induction therapy for AML/MDS [83] or after stem cell transplantation [84] to improve progression-free and overall survival is currently being investigated and is an important area of research.
We also need to investigate the importance of other epigenetic modifications including histone methylation and phosphorylation and whether these are valid biological targets for treatment in MDS. Recent work on histone methylation has shown several different facets of transcriptional regulation that can collaborate or act independently of DNA methylation and histone acetylation. For example, methylation of lysine residues 9 and 27 of histone H3 (H3-K9, H3-K27) are associated with gene silencing, while methylation of lysine 4 (H3-K4) is associated with open chromatin and active transcription [85]. More recently, EZH2-mediated tri-methylation of H3-K27 has been shown to cause tumor suppressor gene silencing independent of DNA methylation [28]. Micro-RNAs are another important area of research that has been shown to be important in the biology of chronic lymphocytic leukemia. miRNAs are noncoding RNAs with important regulatory functions that are encoded by miRNA genes. The noncoding miRNAs are specific for various protein coding mRNAs within a cell. By establishing complementarity to these coding mRNAs, the miRNAs lead to translational repression or outright degradation – thus controlling protein expression at the translational level. Importantly, the regulation of miRNA expression has been shown to be controlled by epigenetic processes and modified by epigenetic therapy [86–89]. Alternatively, miRNAs can also control members of the epigenetic machinery. miRNAs that directly inhibit DNMT or HDAC have also been described [90–93]. This complex interplay between epigenetics and miRNAs and their role in MDS should unlock an exciting area of clinical and translation research. Finally, we still have much to learn about the biology of MDS and why epigenetic therapy is effective in a subset of our patients. Teams of researchers and clinicians need to work towards developing both clinical and molecular biomarkers of response and resistance to help us better select patients who will benefit from epigenetic therapy.
Key issues.
Myelodysplastic syndrome (MDS) is a lethal disease marked by progressive cytopenias, infections, transfusion requirements and progression to acute leukemia. Prognosis of patients with MDS can be estimated by evolving scoring systems that take into account age, performance status, cytogenetics, cytopenias, blast percentage in bone marrow and transfusion dependence.
Epigenetic changes to the chromatin such as DNA methylation and histone acetylation can interact with genetic changes to contribute to the pathogenesis and progression of cancer.
Although there is evidence of aberrant DNA methylation in the neoplastic cells of patients with MDS and acute myelogenous leukemia (AML), investigation is still ongoing to define their exact roles in the pathogenesis and progression of these diseases.
As a result of multiple clinical trials confirming their efficacy and, more recently, suggesting a survival benefit, epigenetic therapy with DNA methyltransferase (DNMT) inhibitors has become part of the standard-of-care for the treatment of patients with MDS.
The two available DNMT inhibitors, 5-azacytidine and decitabine, are well tolerated in most patients, with some myelosuppression. Time to response with these agents can be up to 3–4 months.
Histone deacetylase inhibitors (HDACi) as single agents have limited efficacy in the treatment of MDS and AML and are associated with gastrointestinal toxicity and fatigue.
Combinations of HDACi and DNMT inhibitors are shown to be synergistic in vitro. Preliminary results of early phase clinical trials show these combinations to be safe and active. The time to response with the combinations are noted to be somewhat quicker than single-agent DNMTs.
Further investigation into the combination of HDACi and DNMT inhibitors needs to continue to confirm their efficacy and find a survival benefit.
Studies uncovering the pathogenetic role of epigenetics in MDS and AML are ongoing, and should allow us to better select patients and improve outcome.
Acknowledgments
TMK is supported by NIH grant K12 CA088084. GG-M is supported by a Physician Scientist Program from MD Anderson Cancer Center, the Leukemia and Lymphoma Society of America, and also from NIH grant 5PO1 CA108631.
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
For reprint orders, please contact reprints@expert-reviews.com
Contributor Information
Tapan M Kadia, Email: tkadia@mdanderson.org, Assistant Professor of Medicine, Department of Leukemia, University of Texas, MD Anderson Cancer Center, 1515 Holcombe Blvd, Houston, TX 77030, USA, Tel.: +1 713 563 3534, Fax: +1 713 794 4297.
Guillermo Garcia-Manero, Email: ggarciam@mdanderson.org, Associate Professor of Medicine, Chief, Section of MDS, Department of Leukemia, University of Texas, MD Anderson Cancer Center, 1515 Holcombe Blvd, Houston, TX 77030, USA, Tel.: +1 713 745 3428, Fax: +1 713 794 4297.
References
- 1.Aul C, Giagounidis A, Germing U. Epidemiological features of myelodysplastic syndromes: results from regional cancer surveys and hospital-based statistics. Int J Hematol. 2001;73(4):405–410. doi: 10.1007/BF02994001. [DOI] [PubMed] [Google Scholar]
- 2.Germing U, Strupp C, Kundgen A, et al. No increase in age-specific incidence of myelodysplastic syndromes. Haematologica. 2004;89(8):905–910. [PubMed] [Google Scholar]
- 3.Iglesias Gallego M, Sastre Moral JL, Gayoso Diz P, Garcia Costa A, Ros Forteza S, Mayan Santos JM. Incidence and characteristics of myelodysplastic syndromes in Ourense (Spain) between 1994–1998. Haematologica. 2003;88(10):1197–1199. [PubMed] [Google Scholar]
- 4.Ma X, Does M, Raza A, Mayne ST. Myelodysplastic syndromes: incidence and survival in the United States. Cancer. 2007;109(8):1536–1542. doi: 10.1002/cncr.22570. [DOI] [PubMed] [Google Scholar]
- 5.Corey SJ, Minden MD, Barber DL, Kantarjian H, Wang JC, Schimmer AD. Myelodysplastic syndromes: the complexity of stem-cell diseases. Nat Rev Cancer. 2007;7(2):118–129. doi: 10.1038/nrc2047. [DOI] [PubMed] [Google Scholar]
- 6.Bennett JM, Catovsky D, Daniel MT, et al. Proposals for the classification of the myelodysplastic syndromes. Br J Haematol. 1982;51(2):189–199. [PubMed] [Google Scholar]
- 7.Vardiman JW, Harris NL, Brunning RD. The World Health Organization (WHO) classification of the myeloid neoplasms. Blood. 2002;100(7):2292–2302. doi: 10.1182/blood-2002-04-1199. [DOI] [PubMed] [Google Scholar]
- 8.Greenberg P, Cox C, LeBeau MM, et al. International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood. 1997;89(6):2079–2088. [PubMed] [Google Scholar]
- 9.Malcovati L, Germing U, Kuendgen A, et al. Time-dependent prognostic scoring system for predicting survival and leukemic evolution in myelodysplastic syndromes. J Clin Oncol. 2007;25(23):3503–3510. doi: 10.1200/JCO.2006.08.5696. [DOI] [PubMed] [Google Scholar]
- 10.List A, Kurtin S, Roe DJ, et al. Efficacy of lenalidomide in myelodysplastic syndromes. N Engl J Med. 2005;352(6):549–557. doi: 10.1056/NEJMoa041668. [DOI] [PubMed] [Google Scholar]
- 11.List A, Dewald G, Bennett J, et al. Lenalidomide in the myelodysplastic syndrome with chromosome 5q deletion. N Engl J Med. 2006;355(14):1456–1465. doi: 10.1056/NEJMoa061292. [DOI] [PubMed] [Google Scholar]
- 12.Kaminskas E, Farrell AT, Wang YC, Sridhara R, Pazdur R. FDA drug approval summary: azacitidine (5-azacytidine, Vidaza) for injectable suspension. Oncologist. 2005;10(3):176–182. doi: 10.1634/theoncologist.10-3-176. [DOI] [PubMed] [Google Scholar]
- 13.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer – a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6(2):107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 14.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128(4):683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 16.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349(21):2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 17.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci USA. 2002;99(6):3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Esteller M, Fraga MF, Guo M, et al. DNA methylation patterns in hereditary human cancers mimic sporadic tumorigenesis. Hum Mol Genet. 2001;10(26):3001–3007. doi: 10.1093/hmg/10.26.3001. [DOI] [PubMed] [Google Scholar]
- 19.Herman JG, Latif F, Weng Y, et al. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc Natl Acad Sci USA. 1994;91(21):9700–9704. doi: 10.1073/pnas.91.21.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goto T, Monk M. Regulation of X-chromosome inactivation in development in mice and humans. Microbiol Mol Biol Rev. 1998;62(2):362–378. doi: 10.1128/mmbr.62.2.362-378.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 22.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301(5895):89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 23.Gaudet F, Hodgson JG, Eden A, et al. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300(5618):489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 24.Issa JP. DNA methylation as a therapeutic target in cancer. Clin Cancer Res. 2007;13(6):1634–1637. doi: 10.1158/1078-0432.CCR-06-2076. [DOI] [PubMed] [Google Scholar]
- 25.Lapeyre JN, Becker FF. 5-Methylcytosine content of nuclear DNA during chemical hepatocarcinogenesis and in carcinomas which result. Biochem Biophys Res Commun. 1979;87(3):698–705. doi: 10.1016/0006-291x(79)92015-1. [DOI] [PubMed] [Google Scholar]
- 26.Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- 27.Garcia-Manero G, Issa JP. Histone deacetylase inhibitors: a review of their clinical status as antineoplastic agents. Cancer Invest. 2005;23(7):635–642. doi: 10.1080/07357900500283119. [DOI] [PubMed] [Google Scholar]
- 28.Kondo Y, Shen L, Cheng AS, et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet. 2008;40(6):741–750. doi: 10.1038/ng.159. [DOI] [PubMed] [Google Scholar]
- 29.Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol. 2004;338(1):17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 30.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;6(1):38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 31.Esteller M. Profiling aberrant DNA methylation in hematologic neoplasms: a view from the tip of the iceberg. Clin Immunol. 2003;109(1):80–88. doi: 10.1016/s1521-6616(03)00208-0. [DOI] [PubMed] [Google Scholar]
- 32.Guo SX, Taki T, Ohnishi H, et al. Hypermethylation of p16 and p15 genes and RB protein expression in acute leukemia. Leuk Res. 2000;24(1):39–46. doi: 10.1016/s0145-2126(99)00158-7. [DOI] [PubMed] [Google Scholar]
- 33.Melki JR, Vincent PC, Clark SJ. Concurrent DNA hypermethylation of multiple genes in acute myeloid leukemia. Cancer Res. 1999;59(15):3730–3740. [PubMed] [Google Scholar]
- 34.Takahashi T, Shivapurkar N, Reddy J, et al. DNA methylation profiles of lymphoid and hematopoietic malignancies. Clin Cancer Res. 2004;10(9):2928–2935. doi: 10.1158/1078-0432.ccr-03-0716. [DOI] [PubMed] [Google Scholar]
- 35.Galm O, Herman JG, Baylin SB. The fundamental role of epigenetics in hematopoietic malignancies. Blood Rev. 2006;20(1):1–13. doi: 10.1016/j.blre.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 36.Agrawal S, Unterberg M, Koschmieder S, et al. DNA methylation of tumor suppressor genes in clinical remission predicts the relapse risk in acute myeloid leukemia. Cancer Res. 2007;67(3):1370–1377. doi: 10.1158/0008-5472.CAN-06-1681. [DOI] [PubMed] [Google Scholar]
- 37.Garcia-Manero G, Daniel J, Smith TL, et al. DNA methylation of multiple promoter-associated CpG islands in adult acute lymphocytic leukemia. Clin Cancer Res. 2002;8(7):2217–2224. [PubMed] [Google Scholar]
- 38.Shen L, Toyota M, Kondo Y, et al. Aberrant DNA methylation of p57KIP2 identifies a cell-cycle regulatory pathway with prognostic impact in adult acute lymphocytic leukemia. Blood. 2003;101(10):4131–4136. doi: 10.1182/blood-2002-08-2466. [DOI] [PubMed] [Google Scholar]
- 39.Wong IH, Ng MH, Huang DP, Lee JC. Aberrant p15 promoter methylation in adult and childhood acute leukemias of nearly all morphologic subtypes: potential prognostic implications. Blood. 2000;95(6):1942–1949. [PubMed] [Google Scholar]
- 40.Kroeger H, Jelinek J, Estecio MR, et al. Aberrant CpG island methylation in acute myeloid leukemia is accentuated at relapse. Blood. 2008;112(4):1366–1373. doi: 10.1182/blood-2007-11-126227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang Y, Falk J, Jin M, Liu D, Maciejewski JP. Global methylome of normal and malignant hematopoietic stem cells. ASH Annual Meeting Abstracts. 2007;110(11):2118. [Google Scholar]
- 42.Figueroa ME, Fandy T, McConnell MJ, et al. Myelodysplastic syndrome (MDS) displays profound and functionally significant epigenetic deregulation compared to acute myeloid leukemia (AML) and normal bone marrow cells. ASH Annual Meeting Abstracts. 2007;110(11):345. [Google Scholar]
- 43.Silverman LR, Demakos EP, Peterson BL, et al. Randomized controlled trial of azacitidine in patients with the myelodysplastic syndrome: a study of the cancer and leukemia group B. J Clin Oncol. 2002;20(10):2429–2440. doi: 10.1200/JCO.2002.04.117. [DOI] [PubMed] [Google Scholar]
- 44.Silverman LR, McKenzie DR, Peterson BL, et al. Further analysis of trials with azacitidine in patients with myelodysplastic syndrome: studies 8421, 8921, and 9221 by the Cancer and Leukemia Group B. J Clin Oncol. 2006;24(24):3895–3903. doi: 10.1200/JCO.2005.05.4346. [DOI] [PubMed] [Google Scholar]
- 45.Fenaux P, Mufti GJ, Santini V, et al. Azacitidine (AZA) treatment prolongs overall survival (OS) in higher-risk MDS patients compared with conventional care regimens (CCR): results of the AZA-001 Phase III Study. ASH Annual Meeting Abstracts. 2007;110(11):817. [Google Scholar]
- 46.Momparler RL. Pharmacology of 5-aza-2′-deoxycytidine (decitabine) Semin Hematol. 2005;42(3 Suppl 2):S9–S16. doi: 10.1053/j.seminhematol.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 47.Santini V, Kantarjian HM, Issa JP. Changes in DNA methylation in neoplasia: pathophysiology and therapeutic implications. Ann Intern Med. 2001;134(7):573–586. doi: 10.7326/0003-4819-134-7-200104030-00011. [DOI] [PubMed] [Google Scholar]
- 48.Zagonel V, Lo Re G, Marotta G, et al. 5-aza-2′-deoxycytidine (decitabine) induces trilineage response in unfavourable myelodysplastic syndromes. Leukemia. 1993;7(Suppl 1):30–35. [PubMed] [Google Scholar]
- 49.Wijermans PW, Krulder JW, Huijgens PC, Neve P. Continuous infusion of low-dose 5-aza-2′-deoxycytidine in elderly patients with high-risk myelodysplastic syndrome. Leukemia. 1997;11(1):1–5. doi: 10.1038/sj.leu.2400526. [DOI] [PubMed] [Google Scholar]
- 50.Wijermans P, Lubbert M, Verhoef G, et al. Low-dose 5-aza-2′-deoxycytidine, a DNA hypomethylating agent, for the treatment of high-risk myelodysplastic syndrome: a multicenter Phase II study in elderly patients. J Clin Oncol. 2000;18(5):956–962. doi: 10.1200/JCO.2000.18.5.956. [DOI] [PubMed] [Google Scholar]
- 51.Lubbert M, Wijermans P, Kunzmann R, et al. Cytogenetic responses in high-risk myelodysplastic syndrome following low-dose treatment with the DNA methylation inhibitor 5-aza-2′-deoxycytidine. Br J Haematol. 2001;114(2):349–357. doi: 10.1046/j.1365-2141.2001.02933.x. [DOI] [PubMed] [Google Scholar]
- 52.Kantarjian H, Issa JP, Rosenfeld CS, et al. Decitabine improves patient outcomes in myelodysplastic syndromes: results of a Phase III randomized study. Cancer. 2006;106(8):1794–1803. doi: 10.1002/cncr.21792. [DOI] [PubMed] [Google Scholar]
- 53.Issa JP, Garcia-Manero G, Giles FJ, et al. Phase 1 study of low-dose prolonged exposure schedules of the hypomethylating agent 5-aza-2′-deoxycytidine (decitabine) in hematopoietic malignancies. Blood. 2004;103(5):1635–1640. doi: 10.1182/blood-2003-03-0687. [DOI] [PubMed] [Google Scholar]
- 54.Atallah E, Kantarjian H, Garcia-Manero G. The role of decitabine in the treatment of myelodysplastic syndromes. Expert Opin Pharmacother. 2007;8(1):65–73. doi: 10.1517/14656566.8.1.65. [DOI] [PubMed] [Google Scholar]
- 55.Kantarjian H, Oki Y, Garcia-Manero G, et al. Results of a randomized study of 3 schedules of low-dose decitabine in higher-risk myelodysplastic syndrome and chronic myelomonocytic leukemia. Blood. 2007;109(1):52–57. doi: 10.1182/blood-2006-05-021162. [DOI] [PubMed] [Google Scholar]
- 56.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5(9):769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 57.Liu T, Kuljaca S, Tee A, Marshall GM. Histone deacetylase inhibitors: multifunctional anticancer agents. Cancer Treat Rev. 2006;32(3):157–165. doi: 10.1016/j.ctrv.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 58.Mann BS, Johnson JR, Cohen MH, Justice R, Pazdur R. FDA approval summary: vorinostat for treatment of advanced primary cutaneous T-cell lymphoma. Oncologist. 2007;12(10):1247–1252. doi: 10.1634/theoncologist.12-10-1247. [DOI] [PubMed] [Google Scholar]
- 59.Gore SD, Weng LJ, Figg WD, et al. Impact of prolonged infusions of the putative differentiating agent sodium phenylbutyrate on myelodysplastic syndromes and acute myeloid leukemia. Clin Cancer Res. 2002;8(4):963–970. [PubMed] [Google Scholar]
- 60.Kuendgen A, Schmid M, Knipp S, et al. Valproic acid (VPA) achieves high response rates in patients with low-risk myelodysplastic syndromes. ASH Annual Meeting Abstracts. 2005;106(11):789. [Google Scholar]
- 61.Kuendgen A, Strupp C, Aivado M, et al. Treatment of myelodysplastic syndromes with valproic acid alone or in combination with all-trans retinoic acid. Blood. 2004;104(5):1266–1269. doi: 10.1182/blood-2003-12-4333. [DOI] [PubMed] [Google Scholar]
- 62.Byrd JC, Marcucci G, Parthun MR, et al. A Phase 1 and pharmacodynamic study of depsipeptide (FK228) in chronic lymphocytic leukemia and acute myeloid leukemia. Blood. 2005;105(3):959–967. doi: 10.1182/blood-2004-05-1693. [DOI] [PubMed] [Google Scholar]
- 63.Odenike OM, Alkan S, Sher D, et al. The histone deacetylase inhibitor (HDI) depsipeptide has differential activity in core binding factor AML. ASH Annual Meeting Abstracts. 2006;108(11):1956. [Google Scholar]
- 64.Garcia-Manero G, Assouline S, Cortes J, et al. Phase 1 study of the oral isotype specific histone deacetylase inhibitor MGCD0103 in leukemia. Blood. 2008;112(4):981–989. doi: 10.1182/blood-2007-10-115873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Giles F, Fischer T, Cortes J, et al. A Phase I study of intravenous LBH589, a novel cinnamic hydroxamic acid analogue histone deacetylase inhibitor, in patients with refractory hematologic malignancies. Clin Cancer Res. 2006;12(15):4628–4635. doi: 10.1158/1078-0432.CCR-06-0511. [DOI] [PubMed] [Google Scholar]
- 66.Gojo I, Jiemjit A, Trepel JB, et al. Phase 1 and pharmacologic study of MS-275, a histone deacetylase inhibitor, in adults with refractory and relapsed acute leukemias. Blood. 2007;109(7):2781–2790. doi: 10.1182/blood-2006-05-021873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garcia-Manero G, Yang H, Bueso-Ramos C, et al. Phase 1 study of the histone deacetylase inhibitor vorinostat (suberoylanilide hydroxamic acid [SAHA]) in patients with advanced leukemias and myelodysplastic syndromes. Blood. 2008;111(3):1060–1066. doi: 10.1182/blood-2007-06-098061. [DOI] [PubMed] [Google Scholar]
- 68.Gore SD, Weng LJ, Zhai S, et al. Impact of the putative differentiating agent sodium phenylbutyrate on myelodysplastic syndromes and acute myeloid leukemia. Clin Cancer Res. 2001;7(8):2330–2339. [PubMed] [Google Scholar]
- 69.Gottlicher M, Minucci S, Zhu P, et al. Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J. 2001;20(24):6969–6978. doi: 10.1093/emboj/20.24.6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kuendgen A, Knipp S, Fox F, et al. Results of a Phase 2 study of valproic acid alone or in combination with all-trans retinoic acid in 75 patients with myelodysplastic syndrome and relapsed or refractory acute myeloid leukemia. Ann Hematol. 2005;84(Suppl 1):61–66. doi: 10.1007/s00277-005-0026-8. [DOI] [PubMed] [Google Scholar]
- 71.Jones PL, Wolffe AP. Relationships between chromatin organization and DNA methylation in determining gene expression. Semin Cancer Biol. 1999;9(5):339–347. doi: 10.1006/scbi.1999.0134. [DOI] [PubMed] [Google Scholar]
- 72.Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999;21(1):103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 73.Yang H, Hoshino K, Sanchez-Gonzalez B, Kantarjian H, Garcia-Manero G. Antileukemia activity of the combination of 5-aza-2′-deoxycytidine with valproic acid. Leuk Res. 2005;29(7):739–748. doi: 10.1016/j.leukres.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 74.Gore SD, Baylin S, Sugar E, et al. Combined DNA methyltransferase and histone deacetylase inhibition in the treatment of myeloid neoplasms. Cancer Res. 2006;66(12):6361–6369. doi: 10.1158/0008-5472.CAN-06-0080. [DOI] [PubMed] [Google Scholar]
- 75.Maslak P, Chanel S, Camacho LH, et al. Pilot study of combination transcriptional modulation therapy with sodium phenylbutyrate and 5-azacytidine in patients with acute myeloid leukemia or myelodysplastic syndrome. Leukemia. 2006;20(2):212–217. doi: 10.1038/sj.leu.2404050. [DOI] [PubMed] [Google Scholar]
- 76.Garcia-Manero G, Kantarjian HM, Sanchez-Gonzalez B, et al. Phase 1/2 study of the combination of 5-aza-2′-deoxycytidine with valproic acid in patients with leukemia. Blood. 2006;108(10):3271–3279. doi: 10.1182/blood-2006-03-009142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Soriano AO, Yang H, Faderl S, et al. Safety and clinical activity of the combination of 5-azacytidine, valproic acid, and all-trans retinoic acid in acute myeloid leukemia and myelodysplastic syndrome. Blood. 2007;110(7):2302–2308. doi: 10.1182/blood-2007-03-078576. [DOI] [PubMed] [Google Scholar]
- 78.Blum W, Klisovic RB, Hackanson B, et al. Phase I study of decitabine alone or in combination with valproic acid in acute myeloid leukemia. J Clin Oncol. 2007;25(25):3884–3891. doi: 10.1200/JCO.2006.09.4169. [DOI] [PubMed] [Google Scholar]
- 79.Gore SD, Jiemjit A, Silverman LB, et al. Combined methyltransferase/histone deacetylase inhibition with 5-azacitidine and MS-275 in patients with MDS, CMMoL and AML: clinical response, histone acetylation and DNA damage. ASH Annual Meeting Abstracts. 2006;108(11):517. [Google Scholar]
- 80.Garcia-Manero G, Yang AS, Klimek V, et al. Phase I/II study of MGCD0103, an oral isotype-selective histone deacetylase (HDAC) inhibitor, in combination with 5-azacitidine in higher-risk myelodysplastic syndrome (MDS) and acute myelogenous leukemia (AML) ASH Annual Meeting Abstracts. 2007;110(11):444. [Google Scholar]
- 81.Ravandi F, Faderl S, Thomas D, et al. Phase I study of suberoylanilide hydroxamic acid (SAHA) and decitabine in patients with relapsed, refractory or poor prognosis leukemia. ASH Annual Meeting Abstracts. 2007;110(11):897. [Google Scholar]
- 82.Yee KWL, Minden MD, Brandwein J, et al. A Phase I trial of two sequence-specific schedules of decitabine and vorinostat in patients with acute myeloid leukemia (AML) ASH Annual Meeting Abstracts. 2007;110(11):908. doi: 10.3109/10428194.2015.1018248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Grovdal M, Khan R, Aggerholm A, et al. Maintenance treatment with azacytidine for patients with high risk myelodysplastic syndromes or acute myeloid leukaemia in complete remission after intensive chemotherapy. ASH Annual Meeting Abstracts. 2007;110(11):818. [Google Scholar]
- 84.Jabbour E, Giralt S, Kantarjian H, et al. Efficacy of azacytidine (5-AC) given as maintenance or salvage therapy for patients (pts) with acute leukemia post allogeneic stem cell transplantation (HSCT) ASH Annual Meeting Abstracts. 2007;110(11):3013. [Google Scholar]
- 85.Lachner M, Jenuwein T. The many faces of histone lysine methylation. Curr Opin Cell Biol. 2002;14(3):286–298. doi: 10.1016/s0955-0674(02)00335-6. [DOI] [PubMed] [Google Scholar]
- 86.Fabbri M. MicroRNAs and cancer epigenetics. Curr Opin Investig Drugs. 2008;9(6):583–590. [PubMed] [Google Scholar]
- 87.Lehmann U, Hasemeier B, Christgen M, et al. Epigenetic inactivation of microRNA gene hsa-mir-9–1 in human breast cancer. J Pathol. 2008;214(1):17–24. doi: 10.1002/path.2251. [DOI] [PubMed] [Google Scholar]
- 88.Lujambio A, Ropero S, Ballestar E, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67(4):1424–1429. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- 89.Saito Y, Liang G, Egger G, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9(6):435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 90.Chen JF, Mandel EM, Thomson JM, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38(2):228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Duursma AM, Kedde M, Schrier M, le Sage C, Agami R. miR-148 targets human DNMT3b protein coding region. RNA. 2008;14(5):872–877. doi: 10.1261/rna.972008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fabbri M, Garzon R, Cimmino A, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA. 2007;104(40):15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tuddenham L, Wheeler G, Ntounia-Fousara S, et al. The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett. 2006;580(17):4214–4217. doi: 10.1016/j.febslet.2006.06.080. [DOI] [PubMed] [Google Scholar]