Abstract
C burnetii is a gram-negative bacterium that causes acute and chronic Q fever in humans. Creation of a safe and effective new generation vaccine to prevent Q fever remains an important public health goal. Previous studies suggested that antibody (Ab)-mediated immunity to C. burnetii phase I lipopolysaccharide (PI-LPS) is protective. To identify the potential peptides that can mimic the protective epitopes on PI-LPS, a PI-LPS specific monoclonal antibody (mAb) 1E4 was generated, characterized and used to screen a phage display library. Interestingly, our results indicate that 1E4 was able to inhibit C. burnetii infection in vivo, suggesting that 1E4 is a protective mAb. After three rounds of biopanning by 1E4 from the phage display library, a mimetic peptide, m1E41920 was identified, chemically synthesized and conjugated to KLH for examining its immunogenicity. The results indicate that the synthetic peptide m1E41920 was able to inhibit the binding of 1E4 to PI antigen, suggesting m1E41920 shares the same binding site of 1E4 with the epitopes of PI antigen. In addition, m1E41920-KLH elicited a specific IgG response to PI antigen and immune sera from m1E41920-KLH-immunized mice was able to inhibit C. burnetii infection in vivo, suggesting that m1E41920 may specifically mimic the protective epitope of PI-LPS. Furthermore, m1E41920-KLH was able to confer significant protection against C. burnetii challenge. Thus, m1E41920-KLH is a protective antigen and may be useful for developing a safe and effective vaccine against Q fever. This study demonstrates the feasibility of developing a peptide mimic vaccine against Q fever.
Keywords: Coxiella burnetii, monoclonal antibody, lipopolysaccharide, peptide mimic, vaccine
Introduction
C. burnetii, the etiological agent of Q fever, is an obligate intracellular bacterium that causes acute and chronic infections in humans (1). The highly infectious nature of C. burnetii and its hardiness in adverse environmental conditions make this organism an important zoonotic pathogen. Although formalin-inactivated C. burnetii phase I whole cell vaccine provides near complete protection in animal models as well as human vaccinees, it can induce severe local or systemic adverse reactions when administered to individuals with prior immunity to the agent (2, 3). A formalin-inactivated whole cell vaccine, Q-vax, has been developed and widely used in high-risk individuals in Australia since 1989. Safe use of this vaccine requires screening of potential vaccinees by skin test, serological tests, or in vitro lymphocyte proliferation assay (2–4). Currently, there is no licensed vaccine for preventing Q fever in the US. Creation of a safe and effective vaccine to prevent Q fever remains an important public health goal.
C. burnetii undergoes a lipopolysaccharide (LPS) phase variation in which its virulent smooth LPS phase I (PI) converts to an avirulent rough LPS phase II (PII) upon serial passage in eggs and tissue cultures (5). Similar to other gram-negative bacteria, PI-LPS is a major outer surface component of C. burnetii, which is comprised of three structural domains: the hydrophobic lipid A, the core oligosaccharide (inner and outer core) and the polysaccharide O-antigen. However, PII-LPS lacks the outer-core and the O-antigen (6, 7). It has been shown that PI vaccine (PIV) was more protective than PII vaccine (PIIV) in guinea pig and mouse models (8, 9). Hackstadt et al. demonstrated that PI-and PII-LPS were structurally and antigenically different, but the protein components were indistinguishable between PI and PII (10). The difference between PI- and PII-LPS in their O-antigen polysaccharide expression suggests that the O-antigen of PI-LPS may be the key protective antigen and responsible for PIV-induced protection. An earlier study has shown that PI-LPS was able to elicit Ab responses to PI and PII antigens and to confer protection against virulent C. burnetii challenge in a mouse model (11). One recent study also demonstrated that PI-LPS induced a level of protection similar to PIV, but PII-LPS did not provide measurable protection (9). These studies demonstrated that PI-LPS maybe the key protective antigen. Since the endotoxicity of PI-LPS is very low compared to typical enterobacterial LPS, it is highly innovative to develop a PI-LPS-based vaccine against Q fever. However, since cultivation of C. burnetii is difficult, hazardous, and requires the use of a BL3 facility, it is very difficult to generate large quantities of purified bacteria for isolation of LPS, which, consequently limits the use of C. burnetii LPS to produce vaccines. Therefore, identification of the protective epitopes on PI-LPS and demonstration of the ability of chemically synthesized protective epitopes to induce protective immunity are critical steps toward developing a safe and effective LPS-based vaccine against Q fever.
Peptide mimics have been proposed as potential surrogate antigens of carbohydrates for vaccine development against several microorganisms (12–18). In addition, phage display libraries are commonly used to identify peptide mimics of various surface carbohydrate structures of pathogenic bacteria (13, 18, 19). Several synthetic peptides have been shown to mimic the LPS of bacterial pathogens in either a structural or functional manner and are potentially useful as vaccine candidates and therapeutics targets (19).
In this study, we developed a novel protective monoclonal antibody (mAb) which recognizes a PI specific epitope on PI-LPS and identified a protective peptide mimic of PI-LPS by screening a phage display library with the protective mAb. This report provides the first evidence to demonstrate that there is a protective epitope on PI-LPS and demonstrates the feasibility of development of a PI-LPS-based peptide mimic vaccine against Q fever.
Materials and Methods
C. burnetii strain
Nine Mile phase I (NMI) clone 7 (RSA493) and Nine Mile phase II (NMII) clone 4 (RSA439) organisms were propagated in L929 cells and purified as described previously (20). Purified NMI and NMII organisms were inactivated by 1% formaldehyde solution as described elsewhere (21) and used as whole cell antigen for vaccination and ELISA. The protein concentrations of inactivated NMI and NMII antigens were measured by a Micro BCA™ Protein Assay Kit (Pierce, Rockford, IL).
Animals
Specific-pathogen-free (SPF) 6 week old female BALB/c mice were purchased from the Jackson Laboratories (Bar Harbor, Maine). All mice were housed in sterile microisolator cages under SPF conditions at the University of Missouri laboratory animal facility according to the Guide for the Care and Use of Laboratory Animals. The research protocols described in this report were approved by the Institutional Biosafety Committee and the Animal Care and Use Committee of the MU. All C. burnetii infection experiments were conducted in an Animal Biohazard Safety Level 3 (ABL3) facility at the MU Laboratory of Infectious Disease Research (LIDR).
Generation of mAbs
To generate mAbs against PI-LPS, 6 week old BALB/c mice were immunized with 10 μg of formalin-inactivated C. burnetii Nine Mile PI antigen four times at 3-week intervals and used to isolate splenocytes. The hybridomas were obtained from the fusion of splenocytes from PI-immunized BALB/c mice with SP2/0 myeloma cells according the standard protocol for generation of mAbs (22, 23). Hybridoma supernatants were screened by ELISA for their ability to react with PI antigen. The positive hybridomas were cloned by limiting dilution and isotyped by ELISA. Cloned hybridomas were also analyzed by immunoblotting with proteinase K-treated and untreated PI and PII antigens.
ELISA
ELISA was performed as modified from the method described previously (24) and used to screen hybridoma supernatants for their reactivity with PI and PII antigens and isotype cloned hybridomas. One hundred microliters of inactivated NMI or NMII antigen at 0.5 μg/ml in 0.05 M carbonate/bicarbonate coating buffer (pH 9.6) was added to each well of a 96-well microtiter plate and coated at 4°C for 24 h. The plates were blocked with 1% BSA in PBST buffer (0.05% Tween 20 in PBS) and then incubated with 100μl of hybridoma supernatants or diluted purified mAb at 37°C for 2 h. After washing five times with PBST buffer, the plates were incubated with 100μl of horseradish peroxidase (HRP)-conjugated goat anti-mouse IgM, IgG, IgG1, IgG2a, IgG2b or IgG3 (1:1,000 dilution) at 37°C for 2h. After washing five times with PBST buffer, the Sigma Fast O-Phenylenediamine Dihydrochloride Tablet Sets (Sigma-Aldrich) were used as substrates and the absorbance was measured at 490 nm by the SPECTRA MAX M2 system using SoftMax program (Molecular Devices Corporation, Sunnyvale, CA).
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting
NMI and NMII whole cell antigens were separated by SDS-PAGE and transferred electrophoretically to nitrocellulose membranes in Tris-glycine buffer. The membranes were blocked for 1h at room temperature (RT) in TBS buffer with 0.05% Tween 20 (TBST) and 5% nonfat dry milk, and then incubated with diluted mAb or immune serum at 4°C overnight. After washing five times (5 min each wash) with TBST buffer, the membranes were incubated with HRP-conjugated goat anti-mouse IgG (1:10,000 dilution) for 1 h at RT. The reactions were detected by using an ECL Western blot detection kit (Amersham Pharmacia, Piscataway, N.J.). To determine whether purified 1E4 or immune sera recognize protein or LPS, the reactivity of 1E4 and immune sera to proteinase K-treated and untreated PI and PII antigens were also tested by Western blotting. Digestion of PI and PII whole cell antigens by proteinase K were performed as described previously (25).
Purification of mAb
One hybridoma, 1E4, which recognizes PI-LPS, was cultured in Hybridoma Serum Free Medium (Invitrogen) and purified from the supernatants by using HiTrap protein G HP columns (GE Healthcare) according to the protocol from manufacture. Purified 1E4 was desalted and concentrated by using an Amicon Ultra-15 centrifugal filter device with a 30 kDa molecular-weight cutoff (Millipore). The purity of the purified 1E4 was analyzed by Coomassie blue-staining of SDS-PAGE gel and the protein concentration of 1E4 was measured by the Micro BCA™ Protein Assay Kit.
C. burnetii inhibition assay in vivo
To know if 1E4 is protective, we examined whether 1E4 can inhibit C. burnetii infection in BALB/c mice. Inhibition of C. burnetii was performed by incubation of 1×107 virulent C. burnetii NMI with 1, 10, 100 and 300 μg of purified 1E4, or mouse IgG2a isotype control at 4 °C overnight. Six week-old BALB/c mice were infected by i.p. injection with 1×107 of 1E4 or control IgG2a-treated C. burnetii NMI. In addition, mice infected with 1×107 of PBS-treated C. burnetii NMI were used as negative controls. Splenomegaly and bacterial burden in the spleen were measured at 14 days post infection and used as indicators to evaluate the ability of 1E4 to inhibit C. burnetii infection in BALB/c mice.
Quantitative PCR assay
A High Pure PCR Template Preparation Kit (Roche Molecular Biochemicals, Indianapolis, IN) with modifications was used for extraction of DNA templates from spleen samples as described previously (9). Real time-PCR was performed as described previously (26) with modifications using Applied Biosystems 7300/7500 Real Time PCR System. The recombinant plasmid DNA (com1 gene ligated into pET23a vector) (27) was used as standard DNA to quantify com1 gene copy numbers in spleens.
Sequence Analysis of Variable Region of mAb 1E4
To determine the cDNA-derived amino acid sequences of variable heavy (VH) and light (VL) chains of 1E4, total RNA was extracted from 105 of 1E4 producing hybridoma cells using QIAgen RNeasy Mini Kit (Qiagen, Valencia, CA). The cDNA was synthesized using QuantiTect Rev. Transcription Kit (Qiagen) according to the manufacturer’s instructions. The VH and VL genes of 1E4 from the synthesized cDNA were amplified by PCR using previously published (28–30) and newly designed primers. Since the mouse immunoglobulin gene sequences are highly diverse in variant region, the VH and VL genes of 1E4 were amplified from the synthesized cDNA by PCR using eight forward primers as described previously (28–30) and a newly designed heavy chain reverse primer RCHg2a based on the first domain of the constant region of VH gene encoding mouse IgG2a. The sequences of primers used in this study are shown in Table 1. The PCR products of VH and VL genes were purified from an agarose gel by using QIAquick gel extraction kit (Qiagen) and sequenced using the constant region reverse PCR primers at the MU’s DNA core facility. The sequences were compared against the mouse IgG database using IMGT/V-Quest (www.imgt.cines.fr/home.html) and the mouse IgG germline V-gene database using IgBlast (www.ncbi.nlm.nih.gov/igblast).
Table 1.
Primer sequences.
| Primer | Sequence |
|---|---|
| FVLk | 5′-gac att gag ctc acc cag tct cca-3′ |
| RCLk | 5′-ggc tcg agg aag atg gat aca gtt ggt gca-3′ |
| FVH1 | 5′-ag gtg cag ctc gag gag tca gga cc-3′ |
| FVH2 | 5′-gag gtc cag ctc gag cag tct gga cc-3′ |
| FVH3 | 5′-cag gtc caa ctc gag cag cct ggg gc-3′ |
| FVH4 | 5′-gag gtt cag ctc gag cag tct ggg gc-3′ |
| FVH5 | 5′-gag gtg aag ctc gag gaa tct gga gg-3′ |
| FVH6 | 5′-gag gta aag ctc gag gag tct gga gg-3′ |
| FVH7 | 5′-gaa tgt cag ctc gag gag tct ggg gg-3′ |
| FVH8 | 5′-gag gtt cag ctc gag cag tct gga gc-3′ |
| RCHg2a | 5′-gtt ctg act agt ggg cac tct ggg ctc-3′ |
Screening a phage display peptide library
To identify the peptides that can mimic the protective epitopes of PI-LPS, the Ph.D.-12 Phage Display Peptide Library (New England Biolabs Inc., Beverly, MA) was screened with 1E4 in a solid-phase support system. Three subsequent biopanning rounds were carried out according to the manufacturer’s instructions. Briefly, 100, 10, and 1 μg /ml of 1E4 were used in the first, second and third round panning, respectively. Amplified phage clones were purified by precipitating the phage particles with PEG/NaCl (20% polyethylene glycol 8000 and 2.5 M NaCl). The selected phage DNAs were extracted by using iodide buffer (10 mM Tris-HCl [pH 8], 1 mM EDTA, 4 M NaI) according to the manufacturer’s instructions. Sequence analysis of the peptide inserts was performed by automated dideoxyoligonucleotide sequencing using the −96 M13 primer (New England Biolabs Inc.) at the MU’s DNA core facility. The inserted peptide sequences were deduced from the DNA sequences.
Competitive inhibition
Selected phage clones were analyzed by ELISA to determine the degree of binding to 1E4. In addition, to determine whether identified peptide mimics can specifically mimic the protective epitopes of PI antigen, the selected phages were further tested for their binding ability with 1E4 to compete against binding with PI antigen by a competitive inhibition ELISA. Approximately 1010 of purified phage particles in 0.05 M carbonate/bicarbonate coating buffer (pH 9.6) were coated in a 96-well microtiter plate at 4°C overnight. One hundred microliters of 5μg/ml 1E4 with or without 2μg/ml of PI antigen was added into the wells of the phages coated plate. The binding abilities of 1E4 with or without PI antigen were detected with horseradish peroxidase-conjugated anti-mouse IgG. The inhibition index (A490 without PI-A490 with PI/A490 without PI) was used to evaluate the ability of PI antigen to inhibit 1E4 binding with selected phage particles.
1E4 binding to peptide-conjugate
The amino acid sequence analysis of phagotopes and competitive inhibition results indicated that a mimetic peptide, m1E41920, was identical between two selected phase clones (m1E419 and m1E420) and resulted in highly competitive binding with 1E4 against PI antigen. The m1E41920 amino acid sequences (SLTWHKHELHRK) were chemically synthesized and conjugated to KLH by NeoBioSci™ (Cambridge, MA). In addition, a dodecapeptide (ETQHLTRDSTTR) was also chemically synthesized, conjugated and used as control. ELISA was used to test if synthetic m1E41920 and m1E41920-KLH can specifically bind to 1E4. One hundred microliters of 1 μg/ml m1E41920, m1E41920-KLH, control peptide, control peptide-KLH or KLH were coated in a 96-well microtiter plate at 4°C overnight. One hundred microliters of 5μg/ml 1E4 was added into the synthetic peptide coated plate. The binding ability of 1E4 with synthetic peptides was detected with horseradish peroxidase-conjugated anti-mouse IgG. In addition, the synthetic m1E41920 was further tested for its binding ability with 1E4 to compete against binding with PI antigen by a competitive inhibition ELISA. Five μg/ml of 1E4 was mixed with different concentrations of m1E41920 or control peptide and added into a 96-well microtiter plate which was coated with 0.5μg/ml of PI antigen. The ability of m1E41920 to inhibit 1E4 binding with PI antigen was detected with horseradish peroxidase-conjugated anti-mouse IgG.
Molecular modeling and computational docking
The automatic modeling of 1E4 variable domains was established by canonical structure method from RosettaAntibody (www.antibody.graylab.jhu.edu/). The structure of mimetic peptide m1E41920 was modeled using SAM-T08 (www.compbio.soe.ucsc.edu/SAM_T08/T08-query). The two predicted PDB files were loaded onto the RosettaDock (www.rosettadock.graylab.jhu.edu/) web server to obtain the 1E4-m1E41920 complex model. Structure analysis and graphical renderings of the top one docking prediction were performed by using PyMOL (Delano Scientific, San Carlos, CA).
Immunization of mice with m1E41920-KLH
To examine the immunogenicity of m1E41920-KLH, 6 week old female BALB/c mice were immunized with 5 μg of m1E41920-KLH plus adjuvant four times at 3-week intervals. At each immunization, one mouse was subcutaneously injected with a mixture of 50 μl of antigen (containing 5 μg of m1E41920-KLH) in PBS and 50 μl of Aluminum hydroxide (Sigma). In addition, a group of BALB/c mice were vaccinated with 5 μg of KLH plus adjuvant and used as controls. Serum samples were collected from each immunized group of mice at one week after final-immunization and stored −80 °C until used. Immune sera from m1E41920-KLH-immunized mice was tested for IgG responses to PI antigen by ELISA. The reactivity of immune sera from m1E41920-KLH-immunized mice with PI and proteinase K-digested PI antigen was also analyzed by Western blotting as described above. In addition, the binding ability of immune sera from m1E41920-KLH-immunized mice with PI antigen was also evaluated by a competitive inhibition ELISA, which used m1E41920 peptide to inhibit the binding of the immune sera to PI antigen.
Inhibition of C. burnetii infection by immune sera from peptide-KLH-immunized mice
To know whether immune sera from m1E41920-KLH-immunized mice is protective, we examined if treatment of virulent C. burnetii with immune sera could inhibit C. burnetii infection in BALB/c mice. 1×107 virulent C. burnetii NMI was incubated with 30 μl of normal mouse sera or immune sera from m1E41920-KLH-immunized mice at 4 °C overnight. In addition, 1×107 virulent C. burnetii NMI was treated with 30 μl of immune sera from PIV-vaccinated BALB/c mice or 300 μg of purified 1E4 in the same manner and used as positive controls. Six week-old BALB/c mice were infected by i.p. injection with 1×107 of normal mouse sera, immune sera and 1E4-treated C. burnetii NMI, respectively. Splenomegaly and bacterial burden in the spleen were measured at 14 days post infection and used as indicators to evaluate the ability of immune serum from m1E41920-KLH-immunized mice to inhibit C. burnetii infection in BALB/c mice with negative and positive controls.
Protective efficacy of m1E41920-KLH against C. burnetii infection
To determine whether m1E41920-KLH can confer protection against C. burnetii challenge, BALB/c mice were immunized with 5 μg of m1E41920-KLH plus adjuvant four times at 3-week intervals. At each vaccination, one mouse was subcutaneously injected with a mixture of 50 μl of antigen (containing 5 μg of m1E41920-KLH) in PBS and 50 μl of Aluminum hydroxide. BALB/c mice were vaccinated with KLH plus adjuvant or PIV and used as negative and positive control, respectively. All mice were sacrificed at 14 days post challenge. Serum samples were collected from each immunization group of mice at pre-challenge and stored at −80 °C until used. Immune sera from m1E41920-KLH-immunized mice were tested for IgG responses to PI antigen by ELISA as described above. Immunized and control mice were challenged at 28 days post final-immunization by i.p. injection with 1×107 C. burnetii as described previously (27). The protective efficacy of m1E41920-KLH was evaluated by comparing both splenomegaly and C. burnetii bacterial burden in the spleen with negative and positive controls.
Histopathology
Spleens were collected from mice at 14 days post challenge with C. burnetii, fixed in 10% formalin PBS, prepared as 5-μm paraffin-embedded sections by standard methods, and sliced. Slides were stained with hematoxylin and eosin and examined in a blinded fashion for evaluation of histopathology.
Statistical analysis
Statistical comparisons were performed with Prism 5.0 (GraphPad Software Inc., San Diego, CA.). Results expressed as means ± standard deviations were compared with Student’s t test and ANOVA test. Differences were considered significant at a P value of <0.05.
RESULTS
mAb 1E4 specifically recognizes PI-LPS
Analysis of hybridoma supernatants by ELISA with PI and PII antigens identified one cloned hybridoma 1E4, which produces PI-specific mAb (data not shown). Figure 1A shows the isotyping of 1E4 by ELISA with PI and PII antigens. The results indicate that in the PI antigen coated wells, 1E4 reacted with anti-IgG and -IgG2a secondary antibodies but did not react with anti-IgM, -IgG1, -IgG2b and -IgG3 secondary antibodies, suggesting that the isotype of 1E4 is IgG2a. In addition, in the PII antigen coated wells, 1E4 did not react with any subclass secondary antibodies. Figure 1B shows the reactivity of 1E4 with PI and PII antigens, and proteinase K digested PI and PII antigens in Western blotting. 1E4 specifically reacted with a 14 kDa band in both proteinase K undigested and digested PI antigens but not react with either proteinase K undigested nor digested PII antigens. In addition, the binding of 1E4 with living C. burnetii PI organisms was also confirmed by indirect immunofluorescence assay with purified 1E4 and NMI infected L929 cells (data not shown). These results suggest that 1E4 specifically recognizes epitopes of PI-LPS.
Figure 1.
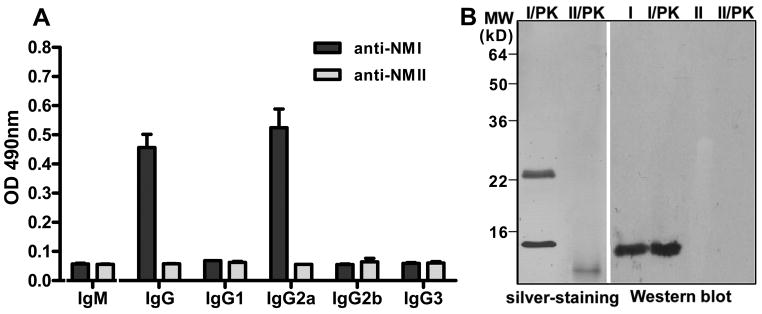
Characterization of 1E4 by ELISA and Western blotting. Panel A, isotyping of 1E4 by ELISA with PI antigen. A 96-well microtiter plate was coated with inactivated PI or PII antigen and incubated with the hybridoma supernatant from cloned hybridoma 1E4. The reactivity of 1E4 with anti-IgM, -IgG, -IgG1, -IgG2a -IgG2b and -IgG3 was measured by OD 490. The values represent the average absorbance at 490 nm of duplicate wells. Panel B, the reactivity of 1E4 with PI and PII antigens, and proteinase K digested PI and PII antigens in Western blotting. I: PI antigen; I/PK, proteinase K-treated PI antigen; II: PII antigen; II/PK, proteinase K-treated PII antigen.
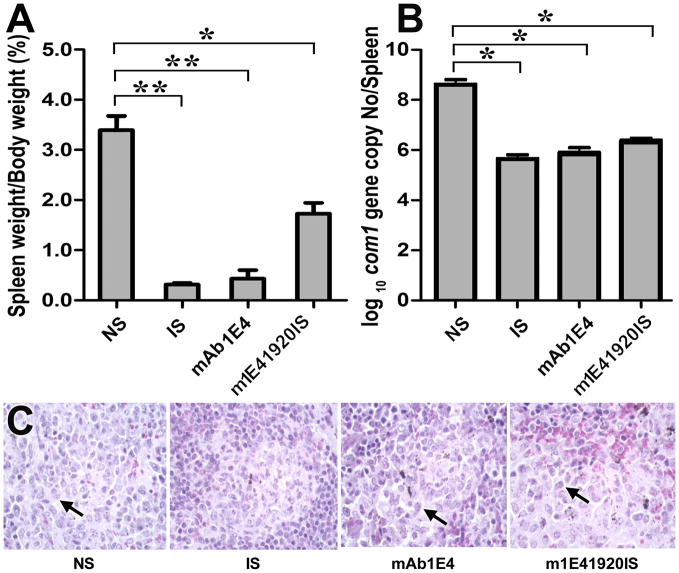
1E4 treatment of virulent C. burnetii can inhibit infection in vivo
To determine whether 1E4 is protective, we examined if treatment of virulent C. burnetii with 1E4 can inhibit C. burnetii infection in BALB/c mice. As shown in Figure 2A, compared to mice infected with PBS or mouse IgG2a isotype control-treated C. burnetii, splenomegaly was significantly reduced in mice infected with 100 or 300 μg of 1E4-treated C. burnetii (P<0.01), while mice infected with 1 or 10 μg of 1E4-treated C. burnetii developed severe splenomegaly. In support of splenomegaly results, compared to mice infected with PBS or mouse IgG2a isotype control-treated C. burnetii, significantly lower C. burnetii genome copies were detected in spleens from mice infected with 100 or 300 μg of 1E4-treated C. burnetii, while similar numbers of C. burnetii were detected in spleens from mice infected with 1 or 10 μg of 1E4-treated C. burnetii (Fig. 2B). In addition, compared to mice infected with 100 μg of 1E4-treated C. burnetii, splenomegaly and C. burnetii numbers in spleens were significantly lower in mice infected with 300 μg of 1E4-treated C. burnetii. The histopathological differences were observed in the spleen between mice infected with PBS, IgG2a isotype control, or lower doses (1 or 10 μg) of 1E4-treated C. burnetii and mice infected with 100 or 300 μg of 1E4-treated C. burnetii. Figure 2C showed pathological changes in the spleen of mice infected with IgG2a isotype control, 10, 100 or 300 μg of 1E4-treated C. burnetii. Robust extramedullary hematopoesis and increased macrophages with more aggregates occurred in mice infected with PBS, IgG2a isotype control, or lower doses (1 or 10 μg) of 1E4-treated C. burnetii, while there were decreased extramedullary hematopoesis and few to no macrophages in mice infected with 100 or 300 μg of 1E4-treated C. burnetii. In addition, multifocal moderate to large accumulations of macrophages (arrow) were present throughout red pulp of spleens of mice infected with PBS, IgG2a isotype control, or lower doses (1 or 10 μg) of 1E4-treated C. burnetii. In contrast, multifocal accumulations of macrophages (arrow) were fewer and smaller in red pulp of spleens of mice infected with 100 of 1E4-treated C. burnetii. In the spleen of mice infected with 300 μg of 1E4-treated C. burnetii, there were only two small accumulations of macrophages were present in red pulp of spleen of 1 out of 4 mice. Thus, pathological changes in the spleen correlated to splenomegaly and bacterial burden in the spleen. These results demonstrate that 1E4 was able to inhibit C. burnetii infection in vivo in a dose-dependent manner, suggesting that 1E4 may directly neutralize or become bactericidal towards C. burnetii to block C. burnetii infection.
Figure 2.
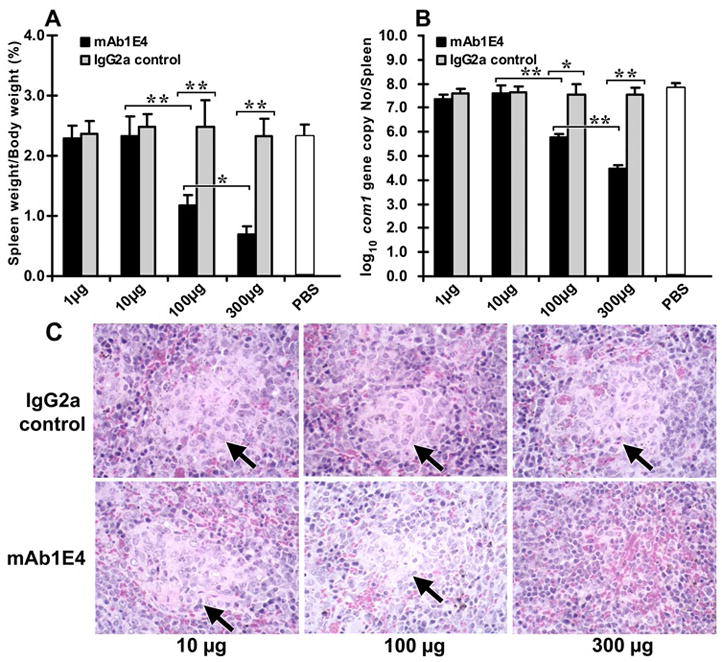
Evaluation of the ability of 1E4 to inhibit C. burnetii infection in vivo. The inhibition of C. burnetii was performed by incubation of 1×107 virulent C. burnetii NMI with 1, 10, 100 and 300 μg of purified 1E4, or mouse IgG2a isotype control at 4 °C overnight. Six week-old BALB/c mice were infected by i.p. injection with 1×107 of 1E4 or control IgG2a-treated C. burnetii NMI. Mice infected with 1×107 of PBS-treated C. burnetii NMI were used as negative controls. Splenomegaly and bacterial burden in the spleen were measured at 14 days post infection and used as indicators to evaluate the ability of 1E4 to inhibit C. burnetii infection in BALB/c mice. Panel A, splenomegaly was measured by spleen weight as a percentage of body weight. Panel B, bacterial burden in the spleen was determined by real time-PCR and reported as log10 of C. burnetii com1 gene copy numbers. Panel C, Pathological changes in the spleen at 14 days post challenge. The data presented in each group is the average with standard deviation of four mice. *, P<0.05; **, P<0.01.
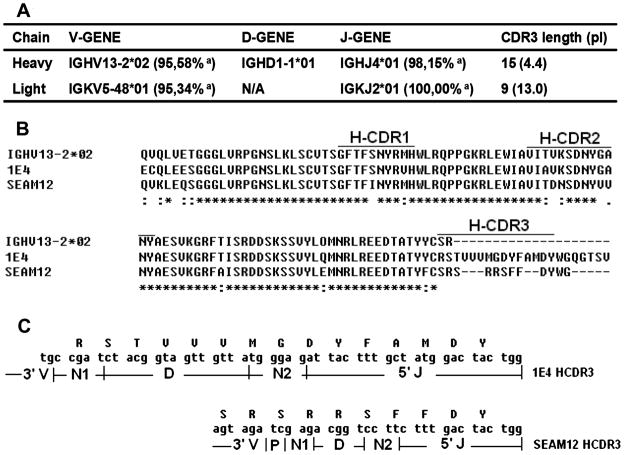
1E4 has unique VH and VL gene usage and complementarity determining region (CDR) sequence
The 1E4 VH gene was only amplified by using FVH7-RCHg2a primer pairs (data not shown). As shown in Figure 3A, the IMGT/V-QUEST analysis of 1E4 nucleotide sequences {GenBank accession numbers: JX139949 (VH gene) (http://www.ncbi.nlm.nih.gov/nuccore/JX139949) and JX139950 (VL gene) (http://www.ncbi.nlm.nih.gov/nuccore/JX139950)} and indicate that the 1E4 VH gene is encoded by a rarely reported murine germline family IGHV13-2. BLAST search of the 1E4 VH gene nucleotide and amino acid sequences in the GenBank non-redundant sequence databases found that only one reported bacterial carbohydrate antigen specific antibody, the anti-Neisseria meningitidis group B polysaccharide mAb SEAM12 (GenBank: DQ113493.1), was derived from IGHV13-2 (31). Figure 3B shows the alignment analysis of respective amino acid sequences of 1E4, IGHV13-2 and SEAM12 VH gene. Compared to the germline gene IGHV13-2, several mutations were identified in HCDR1 and HCDR2 regions of 1E4 and SEAM12 VH genes. Interestingly, the HCDR3 region differed in length, pI and hydrophobicity between 1E4 and SEAM12 (Figure 3B and C). The HCDR3 of 1E4 has 15 amino acids but there are only 10 amino acids found in the HCDR3 of SEAM12. The HCDR3 of 1E4 has a much lower pI (4.4) than the HCDR3 of SEAM12 (pI 10.75). 1E4 contains three highly hydrophobic valine amino acids and the average normalized Kyte-Doolittle hydrophobicity of HCDR3 loop is greater than 1.12 (32). In contrast, the SEAM12 HCDR3 contains three highly charged arginine amino acids and the average hydrophobicity is just -1.29.
Figure 3.
Analysis of VH and VL gene usage and CDR sequence of 1E4. Panel A, analysis of 1E4 VH and VL gene usage by IMGT/V-QUEST. a: identity to germline genes. Panel B, comparison of amino acid sequence of 1E4 VH gene with the anti-Neisseria meningitidis group B polysaccharide mAb SEAM12 and murine germline family IGHV13-2 by ClustalW program. A star (*) indicates the same amino acid residue identity in a position, while a dot (:) indicates a different residue, and a dash (-) indicates inserted spaces placed in the sequence to provide maximum identity. Panel C, comparison of the HCDR3 conjunction between 1E4 and SEAM12. N: nucleotide addition mutation; P: P deletion mutation.
Selection of 1E4-specific phage clones
The Ph.D.-12 Phage Display Peptide Library was bio-panned against 1E4. After three rounds of enrichment, the specific phage titers from round to round of biopanning was increased (data not shown) and 20 plaques were randomly selected, amplified and sequenced. The amino acid sequences of the inserted regions were deduced and analyzed. After being scanned by the web service SAROTUP which is an acronym for “Scanner And Reporter Of Target-Unrelated Peptides” (www.immunet.cn/sarotup/), nonspecific sequences were excluded from the selected phage clones. As shown in Figure 4A, 16 mimetic peptides (with unique sequences) that could bind to 1E4 were identified after three rounds of biopanning against 1E4. Interestingly, the mimetic peptide, m1E41920, SLTWHKHELHRK, was identified twice (m1E419, m1E420) and shared several consensus motifs with the mimetic peptides, m1E42, m1E43, m1E44, m1E49 and miE16. In addition, m1E41920 also shared similar discontinued amino acids with the most identified mimetic peptides.
Figure 4.
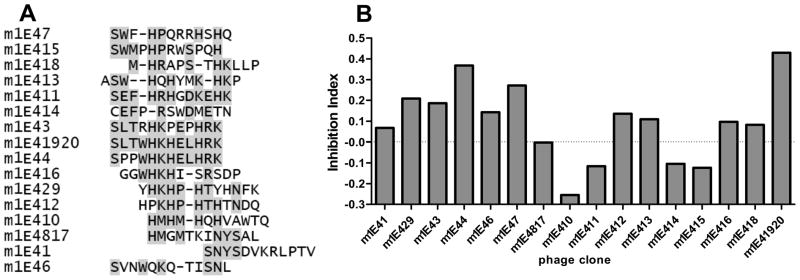
Identification of 1E4-specific phage clones. Panel A, amino acid sequence alignment of nineteen 1E4 recognized phagotopes. The consensus residues are shadowed. Panel B, the reactivity of recombinant phage clones with 1E4 was analyzed by ELISA. A 96-well microtiter plate was coated with individual 1010 of purified phage particles and incubated with 5μg/ml of 1E4 with or without 2μg/ml of PI antigen. The Inhibition of 1E4 binding by PI antigen was measured by the inhibition index {(A490 without inhibitor-A490 with inhibitor)/A490 without inhibitor}.
Binding properties of 1E4-specific phage clones
Each of the sequenced unique phage clones was analyzed by ELISA to determine their ability to bind to 1E4. The results indicate that all identified 1E4-specific phage clones were able to bind to 1E4, while several phage clones including m1E44, m1E46, m1E47, m1E411 and m1E41920 showed higher affinity with 1E4 than other phage clones (data not shown). To determine whether identified 1E4-specific phage clones can specifically mimic the protective epitopes of PI antigen, the selected phage clones were further tested for their binding ability with 1E4 to compete against binding with PI antigen by a competitive inhibition ELISA. Figure 4B shows the ability of PI antigen to inhibit 1E4 binding with selected phage clones as measured by inhibition index (A490 without PI-A490 with PI/A490 without PI). The results indicate that the phage clones, m1E44 and m1E41920 showed the highest inhibition index, suggesting these two phage clones may contain mimic epitopes that bind to the same 1E4 binding site as PI-LPS. Collectively, these results suggest that the mimetic peptide, m1E41920, SLTWHKHELHRK, may mimic the protective epitope of PI-LPS.
1E4 binding to synthetic mimic peptide m1E41920 and m1E41920-KLH conjugate
The m1E41920 amino acid sequences were chemically synthesized and conjugated to KLH to further characterize their properties. ELISA was used to test if synthetic m1E41920 and m1E41920-KLH could specifically bind to 1E4. As shown in Figure 5A, 1E4 bound to both synthetic peptide m1E41920 and m1E41920-KLH conjugate but did not bind to the control peptide, the control peptide KLH conjugate or KLH alone. These results indicate that both synthetic peptide m1E41920 and m1E41920-KLH conjugate retain their binding ability to 1E4 as in M13 phage particles. In addition, the synthetic peptide m1E41920 was further tested for its binding ability with 1E4 to compete against binding with PI antigen through the competitive inhibition ELISA. As shown in Figure 5B, the synthetic peptide m1E41920 was able to inhibit the binding of 1E4 to PI antigen in a dose dependent manner while the control peptide did not inhibit the binding of 1E4 to PI antigen even at the highest concentration. These results indicate that the synthetic peptide m1E41920 shares the same binding site of 1E4 with the epitopes of PI antigen. Since 1E4 may recognize protective epitopes of PI-LPS, our results suggest that the synthetic peptide m1E41920 may mimic the protective epitopes of PI-LPS.
Figure 5.
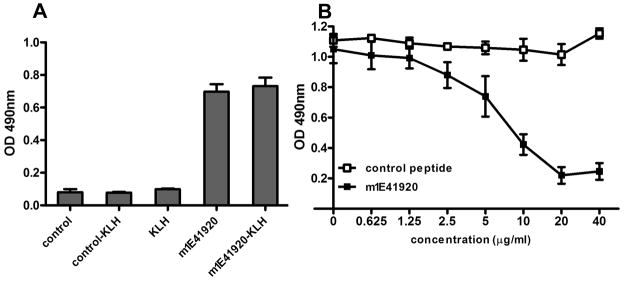
Evaluation of the binding ability of the synthetic mimetic peptide m1E41920 and m1E41920-KLH conjugate with 1E4 by ELISA and competitive ELISA. Panel A, binding activity of m1E41920 and m1E41920-KLH conjugate with 1E4 was measured by ELISA. A 96-well microtiter plate was coated with m1E41920, m1E41920-KLH, control peptide, control-KLH or KLH and incubated with 5μg/ml of 1E4. The values represent the average absorbance at 490 nm of duplicate wells. Panel B, competitive inhibition ELISA analysis of the ability of synthetic peptide m1E41920 to inhibit 1E4 binding with PI antigen. A 96-well microtiter plate was coated with inactivated PI antigen and incubated with 5μg/ml of 1E4 mixed with different concentrations of m1E41920 or control peptide. Points plotted represent the average absorbance at 490 nm of duplicate wells.
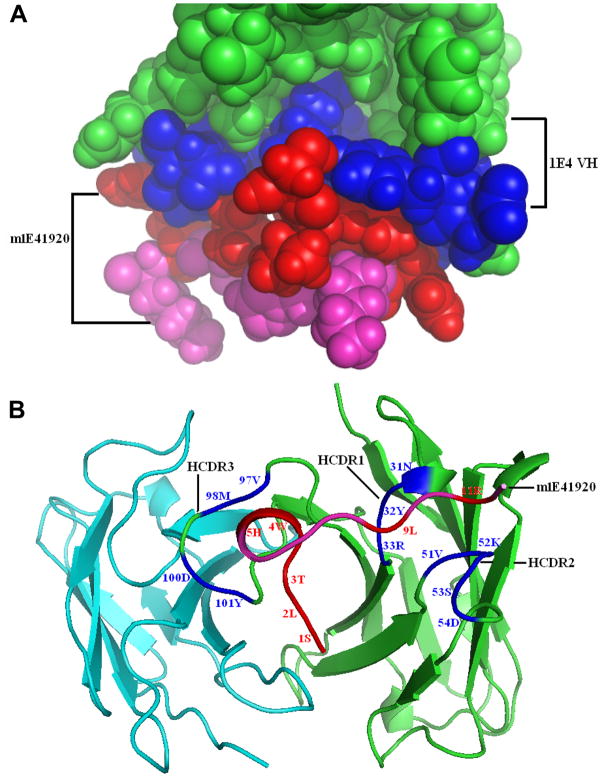
m1E41920 fits well into the 1E4 groove in the docking model
As shown in Figure 6A and B, the mimetic peptide m1E41920 fits well into the binding site of the parent mAb 1E4 VH chain. The directly contacted m1E41920 and 1E4 VH residues were identified and displayed as red and blue spacefill graphs, respectively (Figure 6A), while there was no direct contacted residue in the 1E4 VL chain (not shown). The model docking structure of the 1E4-m1E41920 complex was displayed as a cartoon graph in Figure 6B. The mimetic peptide m1E41920 fits into the heavy chain groove with the middle four residue domain (WHKH) in a helix turn whereas it directly contacts HCDR1, HCDR2 and HCDR3 in an extended conformation through the N terminal domain SLTWH and C terminal domain -L-R-, respectively. This suggests that the m1E41920 central domain is necessary to maintain the conformation and the two terminal domains are involved in binding to HCDRs. It may also help to explain why m1E41920 is a stronger inhibitor as only m1E41920 contains the complete motif SLTWH, which may be very important for the 1E4 binding. In addition, although 1E4 HCDR3 has 15 residues (Figure 3B), only the four HCDR3 central residues (VM-DY), which are de novo generated by D-J conjugation and N insertion, directly contact five N terminal residues (SLTWH) of m1E41920 (Figure 3C and 6B). This suggests that m1E41920 can bind to the LPS binding site of 1E4. These data provide theoretical evidences to support the m1E41920 structural mimicry of PI-LPS based on its ability to bind to the same HCDR3 residues of 1E4.
Figure 6.
Models of 1E4-m1E41920 complex established by the molecular modeling and docking procedures. Panel A, the images were generated using Pymol and the 1E4-m1E41920 interaction model was showed as a spacefill graph. The directly contacted m1E41920 and 1E4 VH residues in direct contact were identified and displayed as red and blue spacefill graph, respectively. The contacted residues of the 1E4 VH paratopes in complex with m1E41920 are shown in blue and the remainders are shown in green. The peptide mimic m1E41920 fits well into the binding site of the parent mAb 1E4 VH chain. Panel B, the model docking structure of the 1E4-m1E41920 complex was displayed as a cartoon graph. The contacted residues and amino acid abbreviations are shown in the corresponding color described in panel A. The peptide mimic m1E41920 fits into the heavy chain groove with the middle four residue domain (WHKH) in a helix turn whereas it directly contacts HCDR1, HCDR2 and HCDR3 in an extended conformation through the N terminal domain SLTWH and C terminal domain -L-R-, respectively.
PI specific Abs elicited by mimetic peptide-KLH conjugate
To determine whether m1E41920-KLH can induce a specific Ab response against PI antigen, immune sera from m1E41920-KLH-immunized mice was tested by ELISA for the presence of anti-PI specific IgG. As shown in Figure 7A, anti-PI specific IgG was detected in immune sera from m1E41920-KLH-immunized mice but not detected in immune sera from mice immunized with KLH alone. These results indicate that m1E41920-KLH was able to elicit specific IgG response against PI antigen. To determine whether anti-m1E41920 specific Ab can recognize specific epitopes on PI-LPS, the reactivity of immune sera from m1E41920-KLH-immunized mice was analyzed by Western blotting with PI and proteinase K-digested PI antigen. Interestingly, immune sera from m1E41920-KLH-immunized mice reacted with a 14 kDa proteinase K resistant band, which was identical to 1E4 recognized antigen in proteinase K-digested PI (Fig. 7B). In addition, the binding ability of immune sera from m1E41920-KLH-immunized mice with PI antigen was evaluated by a competitive inhibition ELISA. As shown in Figure 7C, the synthetic peptide m1E41920 was able to inhibit the binding of immune sera from m1E41920-KLH-immunized mice to PI antigen in a dose dependent manner, while the synthetic control peptide did not affect the binding of immune sera from m1E41920-KLH-immunized mice to PI antigen. These results suggest that m1E41920 mimics epitopes on PI-LPS.
Figure 7.
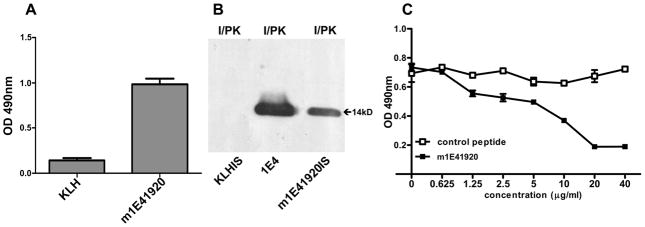
Analysis of m1E41920-KLH elicited antibody response to PI antigen. Panel A, m1E41920-KLH elicited IgG response to PI antigen. Immune sera from m1E41920-KLH-immunized mice were tested for IgG response to PI antigen by ELISA. A 96-well microtiter plate was coated with inactivated PI antigen and incubated with immune sera from KLH-or m1E41920-KLH-immunized mice at 1:400 dilutions. The reactivity of immune sera from KLH- or m1E41920-KLH-immunized mice with anti-IgG was measured by OD 490. The values represent the average absorbance at 490 nm of duplicate wells. Panel B, Western blotting analysis of the reactivity of 1E4 and immune sera from KLH- or m1E41920-KLH-immunized mice with proteinase K digested PI antigens. Panel C, evaluation of the binding ability of immune sera from m1E41920-KLH-immunized mice with PI antigen by the competitive inhibition ELISA. A 96-well microtiter plate was coated with inactivated PI antigen and incubated with 1:400 diluted pooled immune sera from m1E41920-KLH-immunized mice mixed with different concentrations of synthetic peptide m1E41920 or control peptide. Points plotted represent the average absorbance at 490 nm of duplicate wells.
Immune sera from peptide-KLH-immunized mice was able to inhibit C. burnetii infection in vivo
To determine if immune sera from m1E41920-KLH-immunized mice is protective, we examined if treatment of virulent C. burnetii with the immune sera can inhibit C. burnetii infection in BALB/c mice. As shown in Figure 8A, compared to mice infected with normal mouse sera-treated C. burnetii, splenomegaly was significantly reduced in mice infected with immune sera from PIV-vaccinated mice (P<0.01), 1E4 (P<0.01) or immune sera from m1E41920-KLH-immunized mice (P<0.05)-treated C. burnetii. In addition, compared to mice infected with C. burnetii-treated by immune sera from PIV-vaccinated mice or 1E4, splenomegaly was significantly higher in mice infected with C. burnetii-treated by immune sera from m1E41920-KLH-immunized mice (P<0.05). In support of splenomegaly results, compared to mice infected with normal mouse sera-treated C. burnetii, significantly lower C. burnetii genome copies were detected in spleens from mice infected with immune sera from PIV-vaccinated mice, 1E4 or immune sera from m1E41920-KLH-immunized mice-treated C. burnetii (P<0.05) (Fig. 8B). As shown in Figure 8C, histopathological differences were observed in the spleen between mice infected with normal mouse sera-treated C. burnetii and mice infected with immune sera-treated C. burnetii There were more and larger multifocal accumulations of macrophages (arrow) in red pulp of spleens of mice infected with normal mouse sera-treated C. burnetii than mice infected with immune sera from PIV-vaccinated mice, 1E4 or immune sera from m1E41920-KLH-immunized mice-treated C. burnetii. This indicates that immune sera from PIV-vaccinated mice, 1E4 or immune sera from m1E41920-KLH-immunized mice provided similar levels of protection against C. burnetii challenge-induced inflammatory responses. These results indicate that immune sera from m1E41920-KLH-immunized mice was able to inhibit C. burnetii infection in vivo, while immune sera from PIV-vaccinated mice or 300 μg of 1E4 had a stronger ability than immune sera from m1E41920-KLH-immunized mice to inhibit C. burnetii infection. These results suggest that m1E41920 specifically may mimic the protective epitope of PI-LPS.
Figure 8.
Evaluation of the ability of immune sera from m1E41920-KLH-immunized mice to inhibit C. burnetii infection in BALB/c mice. The inhibition of C. burnetii was performed by incubation of 1×107 virulent C. burnetii with 30 μl of normal mouse sera or immune sera from m1E41920-KLH-immunized mice at 4 °C overnight. In addition, 1×107 virulent C. burnetii NMI was treated with 30 μl of immune sera from PIV-vaccinated BALB/c mice or 300 μg of purified 1E4 in the same manner and used as positive controls. Six week-old BALB/c mice were infected by i.p. injection with 1×107 of normal mouse sera, immune sera and 1E4-treated C. burnetii, respectively. Splenomegaly and bacterial burden in the spleen were measured at 14 days post infection and used as indicators to evaluate the ability of immune sera from m1E41920-KLH-immunized mice to inhibit C. burnetii infection in BALB/c mice with negative and positive controls. Panel A, splenomegaly was measured by spleen weight as percentage of body weight. Panel B, bacterial burden in the spleen was determined by real time-PCR and reported as log10 of C. burnetii com1 gene copy numbers. Panel C, Pathological changes in the spleen at 14 days post challenge. The data presented in each group is the average with standard deviation of four mice. *, P<0.05; **, P<0.01.
m1E41920-KLH was able to confer protection against C. burnetii challenge
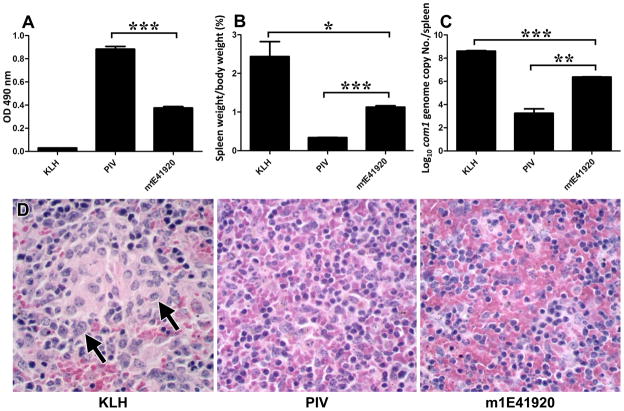
Figure 9A shows m1E41920-KLH induced a specific IgG response to PI antigen as measured by ELISA. Compared to KLH-immunized control mice, m1E41920-KLH-immunized mice elicited significant IgG response to PI antigen but the IgG level was significantly lower than PIV-immunized mice. Immunization of BALB/c mice with 5 μg of m1E41920-KLH plus adjuvant four times at 3-week intervals was able to induce a specific IgG response to PI antigen. Figure 9B&C shows the protective efficacy of m1E41920-KLH in BALB/c mice as measured by comparing splenomegaly and the bacterial burden in the spleens with negative and positive controls. As shown in Figure 9B, compared to KLH-immunized mice, splenomegaly was significantly reduced in m1E41920-KLH-immunized mice (P<0.05), while m1E41920-KLH-immunized mice developed more severe splenomegaly than PIV-immunized mice (P<0.001). In support of the splenomegaly results, compared to KLH-vaccinated mice, significantly lower C. burnetii genome copies were detected in m1E41920-KLH- immunized mice (P<0.001). However, compared to PIV- immunized mice, significantly higher C. burnetii genome copies were detected in m1E41920-KLH- immunized mice (P<0.01) (Fig. 9C). In addition, multifocal accumulations of large macrophages (arrow) are present in red pulp of spleens of KLH-immunized mice but not in PIV or m1E41920-KLH-immunized mice (Fig. 9D), which indicates that PIV- or m1E41920-KLH-immunized mice provided similar levels of protection against C. burnetii chanllege-induced inflammatory responses. These results suggest that m1E41920-KLH was able to confer significant protection against C. burnetii challenge but the levels of protection were lower than PIV.
Figure 9.
Evaluation of the protective efficacy of m1E41920-KLH against C. burnetii infection in BALB/c mice. Panel A, m1E41920-KLH induced specific IgG response to PI antigen as measured by ELISA. A 96-well microtiter plate was coated with inactivated PI antigen and incubated with immune sera from KLH-, PIV- or m1E41920-KLH-immunized mice at 1:400 dilutions. The reactivity of immune sera with anti-IgG was measured by OD 490. The values represent the average absorbance at 490 nm of duplicate wells. Panel B, splenomegaly was measured by spleen weight as percentage of body weight. Panel C, bacterial burden in the spleen was determined by real time-PCR and reported as log10 of C. burnetii com1 gene copy numbers. Panel D, Pathological changes in the spleen at 14 days post challenge. The data presented in each group is the average with standard deviation of four mice. *, P<0.05; **, P<0.01; ***, P<0.001.
Discussion
Although it has been shown that PI-LPS was able to confer significant protection against C. burnetii infection, the feasibility of using PI-LPS to produce vaccines is limited by the difficulties of purification of large quantities of PI- LPS. To identify the protective epitopes on PI-LPS and prove the concept that peptide mimics of PI-LPS can confer protective immunity against C. burnetii infection, we developed a novel protective mAb that recognizes a PI specific epitope on PI-LPS and identified a protective peptide mimic of PI-LPS by screening a phage display library with the protective mAb. This study provides the first evidence to demonstrate that there is a protective epitope on PI-LPS and prove the feasibility of development of a peptide mimic vaccine against Q fever.
Several PI-LPS specific mAbs have been developed for analysis of antigenicity of immunogenic components of C. burnetii LPS and detection of the virulent form of C. burnetii (33–36). Hotta et al (34) reported generation of 19 PI-LPS specific mAbs and demonstrated that these mAbs were able to be separated into three groups based on their reactivity with O-polysaccharide chains (27 kDa of PI-LPS), O-polysaccharide chains (15 to 27 kDa of PI-LPS) and outer-core oligosaccharides (14 kDa of PI-LPS). Palkovicova et al (36) developed a virenose targeted mAb (IgG2b subclass) that recognized O-polysaccharide chains and suggested that this virenose unique mAb may be a useful biomarker for detection of virulent C. burnetii. However, it still remains unknown whether PI-LPS recognized mAbs are protective. In this study, we generated a novel PI specific mAb 1E4. Western blotting analysis of 1E4 demonstrated that 1E4 specifically reacted with a 14 kDa band in both proteinase K digested and undigested PI antigens, suggesting 1E4 recognizes epitopes on PI-LPS. Since previous study demonstrated that the 14 kDa of PI-LPS are outer-core oligosaccharides of PI-LPS (34), our results suggest that 1E4 may recognize outer-core oligosaccharides of PI-LPS. Comparison of the respective amino acid sequence of 1E4 with IGHV13-2 and SEAM12 demonstrated that the HCDR3 region of 1E4 showed unique features in its length, pI and hydrophobicity (Figure 3B and C). Although the reasons why 1E4 has a longer special 15 amino acids, a lower pI (4.4) and higher hydrophobic HCDR3 are unclear, such unique features of 1E4 might be associated with the special chemical composition of its targeted antigen, PI-LPS. C. burnetii infection in mice does not cause death and clear clinical signs. However, infection can induce significant splenomegaly. Splenomegaly has been used as an indicator to monitor severity of C. burnetii infection in mice (27). In addition, because C. burnetii is difficult to grow on a plate and does not form clear plaques in cell culture, we are unable to use traditional methods to measure the C. burnetii burden in animal tissues. Recently, a quantitative real time-PCR procedure has been developed and used to accurately measure the number of C. burnetii in the spleen (37). One recent study demonstrated (9) that splenomegaly was correlated with infection dose and also correlated with bacterial loading in the spleen as measured by Real Time PCR. This suggests that splenomegaly can be a useful indicator to monitor severity of infection and may be useful to evaluate the protective efficacy of vaccine. In this study, we used splenomegaly, and bacterial burden and pathological changes in the spleen to evaluate the ability of 1E4 in inhibiting C. burnetii infection and the protective efficacy of mimic peptide vaccine-induced protection against C. burnetii challenge in BALB/c mice. The results provided additional evidence to support that mouse splenomegaly sub-lethal challenge model can be used to measure the ability of Ab-mediated protection and the protective efficacy of vaccine candidates against C. burnetii infection. In addition, our results suggest that pathological change in the spleen can be used as an additional indicator to evaluate the ability of Ab-mediated protection and the protective efficacy of vaccines against C. burnetii infection.
Our previous published work (9) and unpublished data have demonstrated that both passive transfer of immune sera or Abs and pre-mixed immune sera or Abs with C. burnetii were able to transfer significant protection to naive recipient mice. Although the mechanisms of Ab-mediated protection against C. burnetii infection remain unclear, the Ab binding with bacteria is necessary for direct bactericide, neutralization or opsonization. To reduce the influence of unpredictable factors in vivo variables, such as immunoglobulin biodistribution and catabolism on the Ab-pathogen complex formation, immune sera, mAb were pre-mixed with C. burnetii for evaluating its ability to inhibit C. burnetii infection in this study. In addition, pre-mixing immune sera or mAb with C. burnetii may be able to provide quantitative measurement of their ability to inhibit C. burnetii infection. To determine whether 1E4 is a protective mAb, we mixed different concentrations of 1E4 with 107 of virulent C. burnetii overnight to evaluate its ability to inhibit C. burnetii infection in BALB/c mice. Interestingly, compared to IgG2a isotype control, treatment of virulent C. burnetii with 1E4 was able to inhibit C. burnetii infection in vivo in a dose-dependent manner. This result demonstrates that pre-mixing mAb with C. burnetii can be used to measure its ability to inhibit C. burnetii infection in vivo. Although the mechanisms of 1E4-mediated inhibition of C. burnetii infection remain undefined, this result suggests that 1E4 is a protective mAb. To our knowledge, this is the first evidence demonstrating that there may be a protective epitope on PI-LPS. This study extended the previous studies and supports the hypothesis that PI-LPS is the key protective antigen and responsible for PIV-induced protection.
To identify the potential peptides that can mimic the protective epitopes on PI-LPS, the protective mAb 1E4 was used to screen the Ph.D.-12 phage display library. Based on sequence analysis, 1E4-phage binding assay and PI-antigen competitive inhibition ELISA, a mimetic peptide, m1E41920 was identified. To further characterize the properties and immunogenicity of m1E41920, the amino acid sequences of m1E441920 were chemically synthesized and conjugated to KLH. Our results indicate that the synthetic peptide m1E41920 was able to inhibit the binding of 1E4 to PI antigen, suggesting m1E41920 shares the same binding site of 1E4 with the epitopes of PI antigen. In addition, m1E41920-KLH elicited a specific IgG response to PI antigen and immune sera from m1E41920-KLH-immunized mice was able to inhibit C. burnetii infection in vivo, suggesting that m1E41920 may specifically mimic the protective epitope of PI-LPS. Although there is no obvious motif for carbohydrate mimicking mimotopes, the most identified carbohydrate mimotopes are usually rich in aromatic amino acids which resemble sugar moieties in their size and cyclic shape (38). For example, W/YXY was found in mimotopes of GXM (39), YPY was proposed as a mannose mimic and YRY was initially implicated in the mimicry of the major C polysaccharide of Neisseria meningitides (40–42). In addition, a recently reported mimotope of N. meningitidis group C polysaccharide (LIPFHKHPHHRG) also contained the consensus HKH motif (43). Interestingly, a unique motif, W/HXH, was found in 1E4 recognized mimotopes. This may be due to the imidazole ring of histidine becoming aromatic at all pH values and mimicing the sugar cyclic structures. These observations provide further evidence to support that a particular peptide structure is required for carbohydrate mimicry.
In general, to be considered as carbohydrate mimicry, a peptide mimic must meet at least two criteria. First, it should not only be able to bind to the carbohydrate targeting mAb but also inhibit the parent mAb from binding the target carbohydrate. Second, it should be capable of eliciting antibodies that recognize the carbohydrate being mimicked (44). In this study, our results demonstrated that the synthetic mimetic peptide m1E41920 was able to bind to the parent mAb 1E4 and inhibit the binding of 1E4 to PI antigen in a dose dependent manner. In addition, immunogenicity analysis indicated that m1E41920-KLH was able to elicit specific Abs against PI antigen and that immune sera from m1E41920-KLH-immunized mice reacted with a 14 kDa band in both PI and proteinase K digested PI antigens, which was identical with 1E4 recognized PI antigen. These results suggest that m1E41920 may mimic both antigenicity and immunogenicity of the epitope on PI-LPS. Furthermore, our docking modeling analysis provided theoretical evidence to support that the m1E41920 structural mimics the epitope on PI-LPS. Thus, our results suggest that m1E41920 may be a carbohydrate mimicking mimotope. Future study using purified PI-LPS will help to determine whether m1E41920 mimic a specific carbohydrate epitope on PI-LPS.
The ability of m1E41920-KLH-immunized mouse sera to inhibit C. burnetii infection in BALB/c mice was also examined. Interestingly, compared to normal mouse sera-treated C. burnetii, treatment of virulent C. burnetii with immune sera from m1E41920-KLH-immunized mice was able to significantly reduce the severity of C. burnetii infection in BALB/c mice as measured by splenomegaly and bacterial burden in the spleens. However, the inhibition activity of m1E41920-KLH-immunized mouse sera was significantly lower than immune sera from PIV-vaccinated mice. This result suggests that unidentified epitopes of PI-LPS or other antigenic components of PIV recognized Abs might be also involved in PIV-immunized mouse sera induced protection. In despite of m1E41920-KLH-immunized mouse sera had lower ability than immune sera from PIV-vaccinated mice in inhibiting C. burnetii infection, our results demonstrate that m1E41920-KLH-immunized mouse sera significantly inhibited C. burnetii infection in vivo, suggesting that m1E41920 mimics the protective epitopes on PI-LPS. In addition, the observation in Figures 8A and 8B that the effect of each treatment (IS, 1E4 and 1E41020IS) similar on bacterial burden but not on spleen weight can be explained by the following possibilities: 1) immune sera from IE41920-KLH immunized mice might be able inhibit bacterial replication but were unable to protect from pathological changes; and 2) since the genomic copy numbers were calculated by log10 function, this result might be due to some scientific error.
We also examined whether vaccination with m1E41920-KLH can confer protection against C. burnetii challenge in BALB/c mice. Interestingly, compared to KLH-immunized control mice, m1E41920-KLH was able to confer significant protection against C. burnetii challenge as measured by splenomegaly, and the bacterial burden and pathological changes in the spleens. However, compared to PIV-immunized mice, m1E41920-KLH-immunized mice developed more severe splenomegaly and significantly higher C. burnetii genome copies were detected in their spleens. These results indicate that m1E41920-KLH can provide significant protection against C. burnetii challenge but the levels of protection are lower than PIV, suggesting unidentified protective epitopes on PI-LPS or other antigenic components in PIV are also contributed to PIV-mediated protection. On the other hand, since the mouse splenomegaly model relies on IP infection with high dose of C. burnetii, m1E41920-KLH alone may not be able to provide complete protection against high dose challenge but might be provide enough protection against natural acquired C. burnetii infection. Nonetheless, our results suggest m1E41920-KLH is a protective antigen and may be useful for developing a safe and effective peptide mimic vaccine against Q fever.
Although the peptide mimics have been considered as potential surrogate antigens of carbohydrates for vaccine development against several microorganisms (19), only few studies have achieved protective efficacy testing of mimetic peptides. Previous studies suggested that the most widely used parameters for selecting peptide mimics, such as high-affinity binding to a relevant anti-carbohydrate antibody, competition with native carbohydrate antigen for antibody binding, and induction of a robust anti-peptide response, maybe insufficient and not predictive of whether a mimetic peptide is capable of eliciting an protective carbohydrate cross-reactive immune response (14). The mimotope vaccine strategies are based on the concept of reverse vaccinology, in which protective Abs can be used as probes to identify mimetic peptides that can re-induce protective Ab responses in vivo. This is understandable since only mimotopes identified by protective Abs have a greater chance of being recognized by the receptor on the monoclonal B cell which coded the parental protective Abs. There was a successful example that a mimetic peptide was identified by using the protective mAb 2E9 to Cryptococcus neoformans capsular polysaccharide, which demonstrated its ability to prolong the survival of mice infected by C. neoformans (14). Similarly, our results demonstrate that the identification of protective mAb 1E4 led to the identification of protective mimic peptide m1E41920. On the contrary, in studies using carbohydrate-specific antibodies in which the protectivity has not been well defined, it is always more difficult to obtain the ideal mimotope vaccine candidates. Thus, this study suggests that using protective Abs as probes is critical to identify protective mimetic peptides. In addition, several studies have confirmed that a mimotope peptide should not only mimic salient features, but also bind to the same Ab residue groups as the carbohydrate epitopes (13, 45). As shown in the present study, sequencing analysis and computational docking results provide theoretical evidence to suggest that m1E41920 can not only simulate the sugar cyclic structures through the imidazole ring of histidine in W/HXH motif, but also can simulate the mAb/LPS recognition through binding to the same HCDR3 residues. These characteristics might contribute more to the successful identification of the protective mimic peptide in this location. Although it is still too early to make the conclusion that a single mimetic peptide of PI-LPS can be an ideal Q fever vaccine, further testing whether m1E41920-KLH can confer significant protection against multiple strains of C. burnetti aerosol challenge in mouse and guinea pig models will help determine whether m1E41920 can be a safe and effective vaccine for preventing human Q fever.
Acknowledgments
This study was supported by Public Health Service grants R21AI75175 (GZ) and RO1AI083364 (GZ) from the National Institute of Allergy and Infection Diseases. We are grateful to Dr. Robert Heinzen at the Rocky Mountain Laboratories, NIAID, NIH and Dr. James Samuel at the Texas A&M Health Science Center for providing C. burnetii Nine Mile phase I and Nine Mile phase II strains. We thank Dr. Jinfen Zhang for help in bioinformatics analysis and the staff at the MU Laboratory of Infectious Disease Research for their assistant. We also thank Dr. Alexandra Elliott and Laura Schoenlaub for critical reading and editing of the manuscript.
Abbreviations used in this paper
- LPS
lipopolysaccharide
- mAb
monoclonal antibody
- PI
C. burentii phase I
- PIV
formalin-inactivated C. burentii Nine Mile phase I vaccine
- KLH
keyhole limpet haemocyanin
- BL3
biosafety level 3
- VH
variable heavy chain
- VL
variable light chain
- CDR
complementarity determining region
- HCDR3
heavy chain complementarity determining region 3
References
- 1.Angelakis E, Raoult D. Q Fever. Vet Microbiol. 2010;140:297–309. doi: 10.1016/j.vetmic.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 2.Bell JF, Lackman DB, Meis A, Hadlow WJ. Recurrent Reaction of Site of Q Fever Vaccination in a Sensitized Person. Mil Med. 1964;129:591–5. [PubMed] [Google Scholar]
- 3.Marmion BP, Ormsbee RA, Kyrkou M, et al. Vaccine prophylaxis of abattoir-associated Q fever. Lancet. 1984;2:1411–4. doi: 10.1016/s0140-6736(84)91617-9. [DOI] [PubMed] [Google Scholar]
- 4.Ackland JR, Worswick DA, Marmion BP. Vaccine prophylaxis of Q fever. A follow-up study of the efficacy of Q-Vax (CSL) 1985–1990. Med J Aust. 1994;160:704–8. [PubMed] [Google Scholar]
- 5.Stoker MG, Fiset P. Phase variation of the Nine Mile and other strains of Rickettsia burnetii. Can J Microbiol. 1956;2:310–21. doi: 10.1139/m56-036. [DOI] [PubMed] [Google Scholar]
- 6.Amano K, Williams JC. Chemical and immunological characterization of lipopolysaccharides from phase I and phase II Coxiella burnetii. J Bacteriol. 1984;160:994–1002. doi: 10.1128/jb.160.3.994-1002.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amano K, Williams JC, Missler SR, Reinhold VN. Structure and biological relationships of Coxiella burnetii lipopolysaccharides. J Biol Chem. 1987;262:4740–7. [PubMed] [Google Scholar]
- 8.Ormsbee RA, Bell EJ, Lackman DB, Tallent G. The Influence of Phase on the Protective Potency of Q Fever Vaccine. J Immunol. 1964;92:404–12. [PubMed] [Google Scholar]
- 9.Zhang G, Russell-Lodrigue KE, Andoh M, Zhang Y, Hendrix LR, Samuel JE. Mechanisms of vaccine-induced protective immunity against Coxiella burnetii infection in BALB/c mice. J Immunol. 2007;179:8372–80. doi: 10.4049/jimmunol.179.12.8372. [DOI] [PubMed] [Google Scholar]
- 10.Hackstadt T, Peacock MG, Hitchcock PJ, Cole RL. Lipopolysaccharide variation in Coxiella burnetti: intrastrain heterogeneity in structure and antigenicity. Infect Immun. 1985;48:359–65. doi: 10.1128/iai.48.2.359-365.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams JC, Hoover TA, Waag DM, Banerjee-Bhatnagar N, Bolt CR, Scott GH. Antigenic structure of Coxiella burnetii. A comparison of lipopolysaccharide and protein antigens as vaccines against Q fever. Ann N Y Acad Sci. 1990;590:370–80. doi: 10.1111/j.1749-6632.1990.tb42243.x. [DOI] [PubMed] [Google Scholar]
- 12.De Bolle X, Laurent T, Tibor A, Godfroid F, Weynants V, Letesson JJ, Mertens P. Antigenic properties of peptidic mimics for epitopes of the lipopolysaccharide from Brucella. J Mol Biol. 1999;294:181–91. doi: 10.1006/jmbi.1999.3248. [DOI] [PubMed] [Google Scholar]
- 13.Dharmasena MN, Jewell DA, Taylor RK. Development of peptide mimics of a protective epitope of Vibrio cholerae Ogawa O-antigen and investigation of the structural basis of peptide mimicry. J Biol Chem. 2007;282:33805–16. doi: 10.1074/jbc.M707314200. [DOI] [PubMed] [Google Scholar]
- 14.Fleuridor R, Lees A, Pirofski L. A cryptococcal capsular polysaccharide mimotope prolongs the survival of mice with Cryptococcus neoformans infection. J Immunol. 2001;166:1087–96. doi: 10.4049/jimmunol.166.2.1087. [DOI] [PubMed] [Google Scholar]
- 15.Harris SL, Craig L, Mehroke JS, et al. Exploring the basis of peptide-carbohydrate crossreactivity: evidence for discrimination by peptides between closely related anti-carbohydrate antibodies. Proc Natl Acad Sci U S A. 1997;94:2454–9. doi: 10.1073/pnas.94.6.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hou Y, Gu XX. Development of peptide mimotopes of lipooligosaccharide from nontypeable Haemophilus influenzae as vaccine candidates. J Immunol. 2003;170:4373–9. doi: 10.4049/jimmunol.170.8.4373. [DOI] [PubMed] [Google Scholar]
- 17.Lesinski GB, Smithson SL, Srivastava N, Chen D, Widera G, Westerink MA. A DNA vaccine encoding a peptide mimic of Streptococcus pneumoniae serotype 4 capsular polysaccharide induces specific anti-carbohydrate antibodies in Balb/c mice. Vaccine. 2001;19:1717–26. doi: 10.1016/s0264-410x(00)00397-2. [DOI] [PubMed] [Google Scholar]
- 18.Phalipon A, Folgori A, Arondel J, Sgaramella G, Fortugno P, Cortese R, Sansonetti PJ, Felici F. Induction of anti-carbohydrate antibodies by phage library-selected peptide mimics. Eur J Immunol. 1997;27:2620–5. doi: 10.1002/eji.1830271022. [DOI] [PubMed] [Google Scholar]
- 19.Monzavi-Karbassi B, Cunto-Amesty G, Luo P, Kieber-Emmons T. Peptide mimotopes as surrogate antigens of carbohydrates in vaccine discovery. Trends Biotechnol. 2002;20:207–14. doi: 10.1016/s0167-7799(02)01940-6. [DOI] [PubMed] [Google Scholar]
- 20.Ho T, Htwe KK, Yamasaki N, Zhang GQ, Ogawa M, Yamaguchi T, Fukushi H, Hirai K. Isolation of Coxiella burnetii from dairy cattle and ticks, and some characteristics of the isolates in Japan. Microbiol Immunol. 1995;39:663–71. doi: 10.1111/j.1348-0421.1995.tb03254.x. [DOI] [PubMed] [Google Scholar]
- 21.Williams JC, Cantrell JL. Biological and immunological properties of Coxiella burnetii vaccines in C57BL/10ScN endotoxin-nonresponder mice. Infect Immun. 1982;35:1091–102. doi: 10.1128/iai.35.3.1091-1102.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reading CL. Theory and methods for immunization in culture and monoclonal antibody production. J Immunol Methods. 1982;53:261–91. doi: 10.1016/0022-1759(82)90175-2. [DOI] [PubMed] [Google Scholar]
- 23.Savitt AG, Mena-Taboada P, Monsalve G, Benach JL. Francisella tularensis infection-derived monoclonal antibodies provide detection, protection, and therapy. Clin Vaccine Immunol. 2009;16:414–22. doi: 10.1128/CVI.00362-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang GQ, Hotta A, Ho T, Yamaguchi T, Fukushi H, Hirai K. Evaluation of a recombinant 27-kDa outer membrane protein of Coxiella burnetii as an immunodiagnostic reagent. Microbiol Immunol. 1998;42:423–8. doi: 10.1111/j.1348-0421.1998.tb02305.x. [DOI] [PubMed] [Google Scholar]
- 25.Zhang G, Samuel JE. Vaccines against Coxiella infection. Expert Rev Vaccines. 2004;3:577–84. doi: 10.1586/14760584.3.5.577. [DOI] [PubMed] [Google Scholar]
- 26.Brennan RE, Samuel JE. Evaluation of Coxiella burnetii antibiotic susceptibilities by real-time PCR assay. J Clin Microbiol. 2003;41:1869–74. doi: 10.1128/JCM.41.5.1869-1874.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang GQ, Samuel JE. Identification and cloning potentially protective antigens of Coxiella burnetii using sera from mice experimentally infected with Nine Mile phase I. Ann N Y Acad Sci. 2003;990:510–20. doi: 10.1111/j.1749-6632.2003.tb07420.x. [DOI] [PubMed] [Google Scholar]
- 28.Orlandi R, Gussow DH, Jones PT, Winter G. Cloning immunoglobulin variable domains for expression by the polymerase chain reaction. Proc Natl Acad Sci U S A. 1989;86:3833–7. doi: 10.1073/pnas.86.10.3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phalipon A, Costachel C, Grandjean C, et al. Characterization of functional oligosaccharide mimics of the Shigella flexneri serotype 2a O-antigen: implications for the development of a chemically defined glycoconjugate vaccine. J Immunol. 2006;176:1686–94. doi: 10.4049/jimmunol.176.3.1686. [DOI] [PubMed] [Google Scholar]
- 30.Wang Z, Raifu M, Howard M, Smith L, Hansen D, Goldsby R, Ratner D. Universal PCR amplification of mouse immunoglobulin gene variable regions: the design of degenerate primers and an assessment of the effect of DNA polymerase 3′ to 5′ exonuclease activity. J Immunol Methods. 2000;233:167–77. doi: 10.1016/s0022-1759(99)00184-2. [DOI] [PubMed] [Google Scholar]
- 31.Moe GR, Dave A, Granoff DM. Molecular analysis of anti-N-propionyl Neisseria meningitidis group B polysaccharide monoclonal antibodies. Mol Immunol. 2006;43:1424–31. doi: 10.1016/j.molimm.2005.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–32. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 33.Hotta A, Kawamura M, To H, Andoh M, Yamaguchi T, Fukushi H, Amano K, Hirai K. Use of monoclonal antibodies to lipopolysaccharide for antigenic analysis of Coxiella burnetii. J Clin Microbiol. 2003;41:1747–9. doi: 10.1128/JCM.41.4.1747-1749.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hotta A, Kawamura M, To H, Andoh M, Yamaguchi T, Fukushi H, Hirai K. Phase variation analysis of Coxiella burnetii during serial passage in cell culture by use of monoclonal antibodies. Infect Immun. 2002;70:4747–9. doi: 10.1128/IAI.70.8.4747-4749.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hotta A, Zhang GQ, Andoh M, Yamaguchi T, Fukushi H, Hirai K. Use of monoclonal antibodies for analyses of Coxiella burnetii major antigens. J Vet Med Sci. 2004;66:1289–91. doi: 10.1292/jvms.66.1289. [DOI] [PubMed] [Google Scholar]
- 36.Palkovicova K, Ihnatko R, Vadovic P, Betinova E, Skultety L, Frangoulidis D, Toman R. A monoclonal antibody specific for a unique biomarker, virenose, in a lipopolysaccharide of Coxiella burnetii. Clin Microbiol Infect. 2009;15(Suppl 2):183–4. doi: 10.1111/j.1469-0691.2008.02218.x. [DOI] [PubMed] [Google Scholar]
- 37.Andoh M, Zhang G, Russell-Lodrigue KE, Shive HR, Weeks BR, Samuel JE. T cells are essential for bacterial clearance, and gamma interferon, tumor necrosis factor alpha, and B cells are crucial for disease development in Coxiella burnetii infection in mice. Infect Immun. 2007;75:3245–55. doi: 10.1128/IAI.01767-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deroo S, Muller CP. Antigenic and immunogenic phage displayed mimotopes as substitute antigens: applications and limitations. Comb Chem High Throughput Screen. 2001;4:75–110. doi: 10.2174/1386207013331309. [DOI] [PubMed] [Google Scholar]
- 39.Valadon P, Nussbaum G, Boyd LF, Margulies DH, Scharff MD. Peptide libraries define the fine specificity of anti-polysaccharide antibodies to Cryptococcus neoformans. J Mol Biol. 1996;261:11–22. doi: 10.1006/jmbi.1996.0438. [DOI] [PubMed] [Google Scholar]
- 40.Oldenburg KR, Loganathan D, Goldstein IJ, Schultz PG, Gallop MA. Peptide ligands for a sugar-binding protein isolated from a random peptide library. Proc Natl Acad Sci U S A. 1992;89:5393–7. doi: 10.1073/pnas.89.12.5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scott JK, Loganathan D, Easley RB, Gong X, Goldstein IJ. A family of concanavalin A-binding peptides from a hexapeptide epitope library. Proc Natl Acad Sci U S A. 1992;89:5398–402. doi: 10.1073/pnas.89.12.5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Westerink MA, Campagnari AA, Giardina P, Apicella MA. Antiidiotype antibodies as surrogates for polysaccharide vaccines. Ann N Y Acad Sci. 1994;730:209–16. doi: 10.1111/j.1749-6632.1994.tb44250.x. [DOI] [PubMed] [Google Scholar]
- 43.Wu Y, Zhang Q, Sales D, Bianco AE, Craig A. Vaccination with peptide mimotopes produces antibodies recognizing bacterial capsular polysaccharides. Vaccine. 2010;28:6425–35. doi: 10.1016/j.vaccine.2010.07.049. [DOI] [PubMed] [Google Scholar]
- 44.Van Regenmortel MH. What is a B-cell epitope? Methods Mol Biol. 2009;524:3–20. doi: 10.1007/978-1-59745-450-6_1. [DOI] [PubMed] [Google Scholar]
- 45.Luo P, Canziani G, Cunto-Amesty G, Kieber-Emmons T. A molecular basis for functional peptide mimicry of a carbohydrate antigen. J Biol Chem. 2000;275:16146–54. doi: 10.1074/jbc.M909121199. [DOI] [PubMed] [Google Scholar]