Abstract Abstract
Cichlids represent one of the most species-rich families of fishes and have attracted the attention of evolutionary biologists due to the rapid radiation occurring in some groups and the importance of some species in the world aquaculture. Cytogenetic analysis was conducted in 10 cichlid species from the Araguaia River, Amazon Basin, Brazil. The chromosome number was 2n=48 for all analyzed species except for Laetacara araguaiae Ottoni et Costa, 2009 (2n=44). Chromosomal polymorphism was detected only in Geophagus proximus (Castelnau, 1855), which exhibits an extra large submetacentric and and a dot-like chromosomes. Moreover, the C-banding revealed a general pericentromeric heterochromatic pattern and some additional blocks for some species. The heterochromatic blocks corresponding to AgNOR bearing regions were observed in all species and also corresponded to CMA3 positive blocks, which were observed in terminal regions. Besides the general conserved chromosomal and heterochromatin patterns for South American cichlids, the presence of GC-rich heterochromatin was quite different in the species Biotodoma cupido (Heckel, 1840), Geophagus proximus, Retroculus lapidifer (Castelnau, 1855), Crenicichla strigata Günther, 1862 and Heros efasciatus Heckel, 1840. The results suggest that independent events of heterochromatin modification occurred during chromosome evolution in the group, regardless of the conservation of macro-chromosomal structure.
Keywords: chromosome evolution, fish chromosomes, genome, Cichlidae
Introduction
The family Cichlidae includes more than 3000 species comprising one of the most species-rich families of vertebrates (Nelson 2006). Cichlids are distributed mainly in Latin America, Africa and Madagascar, with only few species in South India and the Middle East (Genner et al. 2007). Cichlids found in the great eastern lakes of Africa have served as a model system for the study of evolution (Kornfield and Smitth 2000, Kocher 2004, Genner et al. 2007), and several species have received increasing scientific attention because of their great importance to tropical and subtropical aquaculture (Pullin 1991). This family represents a monophyletic group, and the limits and interrelationships of all four subfamilies (Etroplinae, Ptychochrominae, Cichlinae and Pseudocrenilabrinae) are well supported by molecular and morphological data (Smith et al. 2008).
The African and Neotropical cichlids, Pseudocrenilabrinae and Cichlinae, respectively, are both monophyletic and represent sister groups (Smith et al. 2008). There are 51 genera and 406 species recognized in Neotropical cichlids (Kullander 1998, 2003). The most recent proposed phylogeny of the group denotes the tribes Cichlini, Retroculini, Astronotini, Chaetobranchini, Geophagini, Cichlasomatini and Heroini as members of the Cichlinae clade (Smith et al. 2008).
The chromosome numbers of approximately 135 species of cichlids have been determined. Although more than 60% of the species present karyotypes with 2n=48, the diploid number ranges from 2n=32 to 2n=60 (Poletto et al. 2010, for review). African cichlids have a modal diploid number of 2n=44, whereas the modal number for Neotropical cichlids is 2n=48. Even though chromosomal data are known for several cichlid species, the amount of available data is not representative of the high diversity of species in the group. The chromosomal data already published for the Cichlinae clade focus mostly on the description of chromosome morphology and mapping of 45S rDNA (Poletto et al. 2010), and the heterochromatin patterns of only few species are described (Table 1). The aim of this work was to contribute in the study of the heterochromatin patterns of South American cichlids and their possible involvement in karyotypic diversification in the group.
Table 1.
Synthesis of the cichlid species analyzed with respect to the karyotypic formulae, heterochromatin distribution and CMA3 patterns. m/sm, metacentric and submetacentric chromosomes; st/a, subtelocentric and acrocentric chromosomes; mi, microchromosomes; q, the long arm of a chromosome; p, the short arm of a chromosome; PeriC or C, pericentromeric regions; Prox, proximal portion of a chromosome; Term, Terminal portion of a chromosome; Int, interstitial portion of a chromosome; Adj, adjacent region; NOR, nucleolus organizing region; The numbers in the column “Additional blocks” indicate the number of chromosomes with the described pattern; in some cases, the ranking of these chromosomes are indicated in parentheses.
| Tribes and species | Origin of animals | 2n | Karyotypic formulae | Heterochromatin distribution | CMA3 + blocks | References | |
|---|---|---|---|---|---|---|---|
| General pattern | Additional blocks | ||||||
| Cichlini | |||||||
| Cichla piquiti Kullander et Ferreira, 2006 | Das Mortes river, Araguaia basin, MT State, Brazil | 48 | 48st/a | PeriC | NOR; term 2 | NOR (term) | This work |
| Cichla kelberi Kullander et Ferreira, 2006 | Araguaia river, MT State, Brazil | 48 | 48st/a | C | NOR; int 1 q | absent | Teixeira et al. 2009 |
| Cichla monoculus Spix et Agassiz, 1831 | Uatumã and Solimões rivers, AM State, Brazil | 48 | 48a | PeriC | NOR; int 1 q | absent | Brinn et al. 2004 |
| Cichla temensis Humboldt, 1821 | Uatumã and Jaú rivers, AM State, Brazil | 48 | 48a | PeriC | NOR; int 1 q | absent | Brinn et al. 2004 |
| Retroculini | |||||||
| Retroculus lapidifer lapidifer (Castelnau, 1855) | Das Mortes river, Araguaia basin, MT State, Brazil | 48 | 48st/a | PeriC | NOR; term 1 q | NOR (term) and PeriC | This work |
| Astronotini | |||||||
| Astronotus ocellatus (Agassiz, 1831) | Tietê river, SP State, Brazil | 48 | 16m/sm + 32st/a | C | NOR | absent | Mazzuchelli and Martins 2009 |
| Geophagini | |||||||
| Apistogramma trifasciata (Eigenmann et Kennedy, 1903) | Paraná river, Missiones, Argentina | 46 | 16m/sm + 30st/a | PeriC | absent | absent | Roncati et al. 2007 |
| Biotodoma cupido (Heckel, 1840) | Das Mortes river, Araguaia basin, MT State, Brazil | 48 | 4m/sm + 44st/a | PeriC | NOR; some prox blocks | NOR (int) | This work |
| Crenicichla britskii Kullander, 1982 | Jupiá river, PR State, Brazil | 48 | 8m/sm + 40st/a | PeriC | NOR; 1 p almost completely heterochromatic (1st pair) | absent | Benzaquem et al. 2008 |
| Crenicichla strigata Günther, 1862 | Das Mortes river, Araguaia basin, MT State, Brazil | 48 | 6m/sm + 42st/a | PeriC | NOR; some prox blocks | NOR (term) and PeriC | This work |
| Crenicichla prope johanna Heckel, 1840 | Negro and Solimões rivers, AM State, Brazil | 48 | 8m/sm + 40st/a | PeriC | NOR; term 1 q (19th pair) | absent | Benzaquem et al. 2008 |
| Crenicichla cincta Regan, 1905 | Negro and Solimões rivers, AM State, Brazil | 48 | 8m/sm + 40st/a | PeriC | adj NOR | absent | Benzaquem et al. 2008 |
| Crenicichla iguassuensis Haseman, 1911 | Iguaçu river, PR State, Brazil | 48 | 4m + 4sm + 14st + 26a | PeriC | Some term blocks | NOR | Mizoguchi et al. 2007 |
| Crenicichla inpa Ploeg, 1991 | Negro and Solimões rives, AM State, Brazil | 48 | 6m/sm + 42st/a | PeriC | adj NOR | absent | Benzaquem et al. 2008 |
| Crenicichla lepidota Heckel, 1840 | São Gonçalo stream and Polegar lake, RS State, Brazil | 48 | 4m + 4sm + 40st/a | PeriC | term 1 p and 1 q (1st pair); int 1 q (1st pair) | NOR | Perazzo et al. 2010 |
| Crenicichla lepidota Heckel, 1840 | Porto Rico region, Paraná river basin, PR State, Brazil | 48 | 2m + 4sm + 42st/a | PeriC | int 2 (1st and 5th pairs) | absent | Martins et al. 1995 |
| Crenicichla lugubris Heckel, 1840 | Negro and Solimões rivers, AM State, Brazil | 48 | 8m/sm + 40st/a | PeriC | NOR; int 1 q (2nd pair) | absent | Benzaquem et al. 2008 |
| Crenicichla niederleinii (Holmberg, 1891) | Paraná river, Missiones, Argentina | 48 | 6m/sm + 42st/a | PeriC | absent | absent | Roncati et al. 2007 |
| Crenicichla reticulata (Heckel, 1840) | Negro and Solimões river, AM State, Brazil | 48 | 6m/sm + 42st/a | PeriC | adj NOR; int 1 q (10th pair) | absent | Benzaquem et al. 2008 |
| Crenicichla sp.1 | Iguaçu river, PR State, Brazil | 48 | 4m + 4sm + 14st + 26a | PeriC | Some term blocks | NOR | Mizoguchi et al. 2007 |
| Crenicichla sp. 2 | Iguaçu river, PR State, Brazil | 48 | 4m + 4sm + 14st + 26a | PeriC | Some term blocks | NOR | Mizoguchi et al. 2007 |
| Geophagus brasiliensis (Quoy et Gaimard, 1824) | Socavão and Verde rivers, PR State, Brazil | 48 | 6sm + 42st/a | PeriC/C | absent | NOR | Vicari et al. 2006 |
| Geophagus brasiliensis (Quoy et Gaimard, 1824) | Jaguarriaíva river, PR State, Brazil | 48 | 6sm + 42st/a | PeriC/C | Some int blocks | NOR | Vicari et al. 2006 |
| Geophagus brasiliensis (Quoy et Gaimard, 1824) | Saco da Alemoa, Gasômero, RS State, Brazil | 48 | 4sm + 44st/a | PeriC | NOR | NOR | Pires et al. 2010 |
| Geophagus brasiliensis (Quoy et Gaimard, 1824) | Cambezinho and Três Bocas stream, Tibagi river basin, PR State, Brazil | 48 | 4sm + 44st/a | C | NOR | NOR | Pires et al. 2008 |
| Geophagus brasiliensis (Quoy et Gaimard, 1824) | Pirapo river, Paranapanema basin, PR State, Brazil | 48 | 8sm + 40st/a | PeriC | prox 1 p (10th pair) | absent | Martins et al. 1995 |
| Geophagus proximus (Castelnau, 1855) | Das Mortes river, Araguaia basin, MT State, Brazil | 48 | 4m/sm + 44st/a | PeriC | NOR; 1 p almost completely heterochromatic | NOR (int) | This work |
| Gymnogeophagus balzanii (Perugia, 1891) | Paraná river, Missiones State, Argentina | 48 | 2m/sm + 46st/a | PeriC | absent | absent | Roncati et al. 2007 |
| Geophagus gymnogenys (Hensel, 1870) | Saco da Alemoa, Barra do Ribeiro, Gasômetro, RS State, Brazil | 48 | 4m + 44st/a; 6m + 42st/a | PeriC | NOR | NOR | Pires et al. 2010 |
| Geophagus labiatus (Hensel, 1870) | Saco da Alemoa, Forqueta river, RS State, Brazil | 48 | 4m + 4sm + 40st/a | PeriC | absent | NOR | Pires et al. 2010 |
| Gymnogeophagus sp. | Paraná river, Missiones, Argentina | 48 | 2m/sm + 46st/a | PeriC | absent | absent | Roncati et al. 2007 |
| Satanoperca jurupari (Heckel, 1840) | Das Mortes river, Araguaia basin, MT State, Brazil | 48 | 4m/sm + 44st/a | PeriC | absent | NOR | This work |
| Satanoperca pappaterra (Heckel, 1840) | Porto rico region, Parana river basin, PR State, Brazil | 48 | 6sm + 42st/a | PeriC | absent | absent | Martins et al. 1995 |
| Cichlasomatini | |||||||
| Aequidens tetramerus Heckel, 1840 | Araguaia river, MT State, Brazil | 48 | 12m/sm + 36st/a | PeriC | absent | NOR | This work |
| Australoheros facetus (Jenyns, 1842) | São Gonçalo stream and Polegar lake, RS State, Brazil | 48 | 22sm + 26st/a | PeriC/C | absent | NOR | Perazzo et al. 2010 |
| Bujurquina vittata (Heckel, 1840) | Paraná river, Missiones, Argentina | 44 | 22m/sm + 8st/a + 14 mi | PeriC | NOR; p arm of 5th pair completely heterochromatic | absent | Roncati et al. 2007 |
| Cichlasoma dimerus (Heckel, 1840) | Paraná river, Missiones, Argentina | 48 | 8m/sm + 40st/a | PeriC | absent | absent | Roncati et al. 2007 |
| Cichlasoma facetum (Jenyns, 1842) | Tarumã lake, PR State, Brazil | 48 | 10sm + 38 st/a | PeriC/C | absent | NOR | Vicari et al. 2006 |
| Cichlasoma paranaense Kullander, 1983 | Porto rico region, Parana river basin, PR State, Brazil | 48 | 20sm + 28 st/a | PeriC | prox 2 p (2nd and 9th pairs) | absent | Martins et al. 1995 |
| Laetacara araguaiae Ottoni et Costa, 2009 | Araguaia river, MT State, Brazil | 44 | 4m/sm + 40st/a | PeriC | absent | NOR | This work |
| Laetacara prope dorsigera (Heckel, 1840) | Paraná river, PR State, Brazil | 43 | 5m + 38a | C | NOR | absent | Martins-Santos et al. 2005 |
| 44 | 4m + 40a | ||||||
| 45 | 3m + 42a | ||||||
| 46 | 2m + 44a | ||||||
| Heroini | |||||||
| Heros efasciatus Heckel, 1840 | Araguaia river, MT State, Brazil | 48 | 8m/sm + 40st/a | PeriC | absent | NOR (term) and int 1 p | This work |
| Mesonauta festivus (Heckel, 1840) | Das Mortes river, Araguaia basin, MT State, Brazil | 48 | 14m/sm + 34st/a | PeriC | NOR; term 2 q | NOR (term) | This work |
| Pterophyllum scalare (Schultze, 1823) | Jari river, PA State, Brazil | 48 | 12m/sm + 36st/a | PeriC/C | 1 p almost completely heterochromatic (1st pair) | NOR, some centromeres | Nascimento et al. 2006 |
| Symphysodon aequifasciatus Pellegrin, 1904 | Bauana lake, Tefé river, AM State, Brazil | 60 | 8m/sm + 8st/a +4mi; 50m/sm + 6st/a +4mi | PeriC | Some prox blocks; int 1 q (1st pair) | absent | Mesquita et al. 2008 |
| Symphysodon discus Heckel, 1840 | Boi-boi stream, Negro river, AM State, Brazil | 60 | 50m/sm + 10st/a; 54m/sm + 6st/a | PeriC | Some prox blocks | absent | Mesquita et al. 2008 |
| Symphysodon haraldi Schultz, 1960 | Manacapuru river, AM State, Brazil | 60 | 52m/sm + 4st/a +4mi | PeriC | Some prox blocks | absent | Mesquita et al. 2008 |
Material and methods
Specimens and chromosome preparation
It was analyzed 10 South American cichlid species of the subfamily Cichlinae: Cichla piquiti Kullander et Ferreira, 2006 (4 individuals: sex not identified), Retroculus lapidifer (Castelnau, 1855) (6 individuals: 3 ♀ and 1 ♂, and 2 sex not identified), Biotodoma cupido (Heckel, 1840) (5 individuals: 2 ♀, and 3 ♂), Crenicichla strigata Günther, 1862 (12 individuals: 5 ♀, 5 ♂, and 2 sex not identified), Geophagus proximus (Castelnau, 1855) (9 individuals: 4 ♀, 2 ♂, and 3 sex not identified), Satanoperca jurupari (Heckel, 1840) (15 individuals: 7 ♀, 5 ♂, and 3 sex not identified), Aequidens tetramerus Heckel, 1840 (44 individuals: 21 ♀, 14 ♂, and 9 sex not identified), Laetacara araguaiae Ottoni et Costa, 2009 (5 individuals: 1 ♀, 1 ♂, and 3 sex not identified), Heros efasciatus Heckel, 1840 (5 individuals: 5 females) and Mesonauta festivus (Heckel, 1840) (5 individuals: 2 ♀, 1 ♂, and 2 sex not identified), which belong to the tribes Cichlini, Retroculini, Geophagini, Cichlasomatini and Heroini (Table 1). All individuals analyzed were not juveniles. Wild specimens were collected in several rivers that are part of the Araguaia River system, which is situated in the quadrant bounded by the coordinates 52°24'00"W, 15°30'S (DMS) and 52°05'00"W, 15°58'S (DMS) in the region of Barra do Garças, Mato Grosso State, Brazil. The sampling of wild animals was performed in accordance with Brazilian laws for environmental protection (wild collection permit, SISBIO/15729–1). The animals were maintained for 24 hours in an aired aquarium at a temperature ranging from 25°C to 28°C before collecting tissue samples. The fish were euthanized with a lethal dose of benzocaine followed by spinal section (Protocol 01204 – Committee of Ethical in Animal Experimentation – UNESP – São Paulo State University, Brazil) before removal of the kidneys for chromosome preparation.
Mitotic chromosome preparations were obtained from kidney cells according to Bertollo et al. (1978). The animals were treated with a 0.0125% solution of colchicine, which was injected at a volume of 1mL/100g of body weight at approximately 45–60 min before euthanasia and chromosome preparation. The kidney tissues were dissected, and the cells were dissociated in a hypotonic solution of KCl 0.075 M with a syringe and remained in the solution for 25 min. The cells were fixed in 3:1 methanol-acetic acid solution and used to prepare slides that were stained with 5% Giemsa solution in phosphate buffer at pH 7 for 10 min.
Differential chromosome staining and banding
The chromosome structure was analyzed through silver nitrate staining, Chromomycin A3 (CMA3) staining and C-banding.
To detect nucleolus organizer regions (NORs), the silver staining of the chromosomes was performed according to Howell and Black (1980). The slides were stained with 2% Giemsa for 10 to 15 sec, washed in water and air-dried for later microscopic analysis.
The constitutive heterochromatin was detected using saline solution according to Sumner (1972) with the following adjustments. The slides were initially treated with 0.2 N HCl at 42°C for 5 min, washed in water and rapidly air-dried. The slides were then immersed in 5% barium hydroxide solution that was freshly prepared and filtered at 42°C for 30 sec to 1 min. The treatment was stopped by submerging the slides in 0.2 N HCl and washing them extensively in running water. The slides were immersed in saline solution (2xSSC) at 60°C for 45 min. After completing this step, the slides were air-dried and stained with 5% Giemsa in phosphate buffer at pH 6.8–7.0. Alternatively, the slides were stained with propidium iodide, which also provides excellent results.
The CMA3 staining was conducted according to the method by Schweizer (1976) with minor adjustments. This was done by immersing the slides in 0.2% MgCL2 in McIlvaine buffer, pH 7.0, at 25°C for 10 min. The slides were withdrawn, agitated briefly to remove excess solution, mounted with 150 µL of 0.05% CMA3 in McIlvane buffer under coverslips and then stored in dark boxes for 15 min at 25°C. After this step, the coverslips were removed by washing the slides in McIlvaine buffer. The slides were incubated in a solution of freshly prepared of 0.012% Methyl-green/Hepes for 15 min, rinsed in a solution of Hepes 0.13%/NaCl 0.87% and air-dried. Finally, the slides were mounted with 45–90 µl of glycerol 97.4%/propyl gallate 2.5%. Prior to analysis, the slides were stored in the dark at 4°C for at least one week before analysis by fluorescence microscopy.
Chromosome analysis
The chromosome spreads were analyzed using an Olympus BX 61 microscope, and the images were captured with the Olympus DP71 digital camera with the software Image-Pro MC 6.0. There were analyzed 30 metaphase spreads for all cytogenetic procedures performed for each animal sample. Karyotypes were arranged in the order of decreasing chromosome size, and the chromosomes were classified as either meta/submetacentrics (m/sm) or subtelo/acrocentrics (st/a).
Results
All of the species analyzed have 2n=48 except Laetacara araguaiae, which showed a diploid number of 2n=44 and the karyotype formula of 4m/sm + 40st/a. Moreover, chromosomal polymorphism was found in Geophagus proximus, which presented two karyotype formulae, 4m/sm + 44st/a or 5m/sm + 42st/a + 1 dot-like chromosome (Fig. 1, Table 1).
Figure 1.
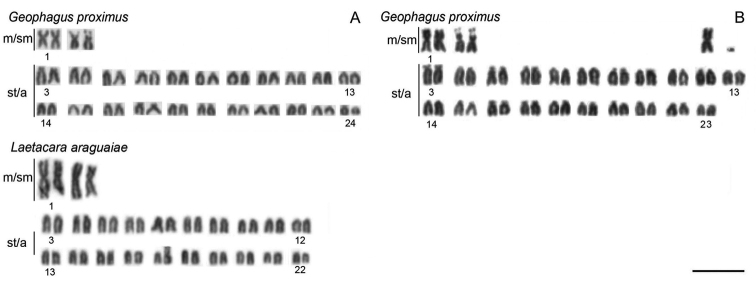
Representative karyotypes of Geophagus proximus and Laetacara araguaiae species. For Geophagus proximus,two karyotypes are presented, a normal (A) and a polymorphic karyotype, showing in the upper right corner one extra large metacentric and one dot-like chromosome (B). Bar = 10 µm.
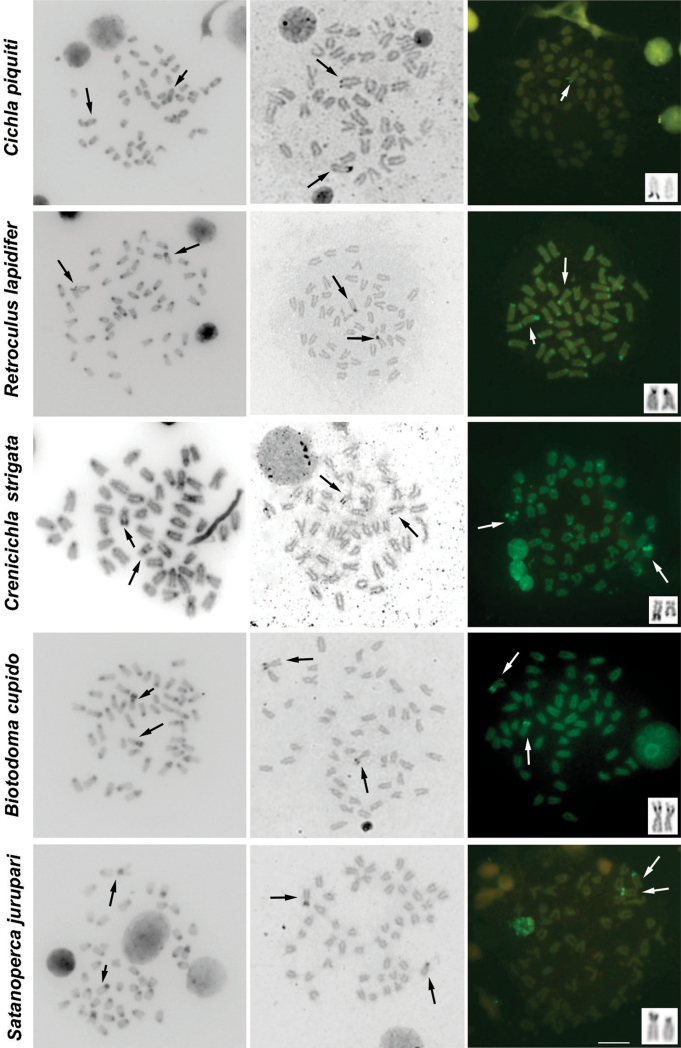
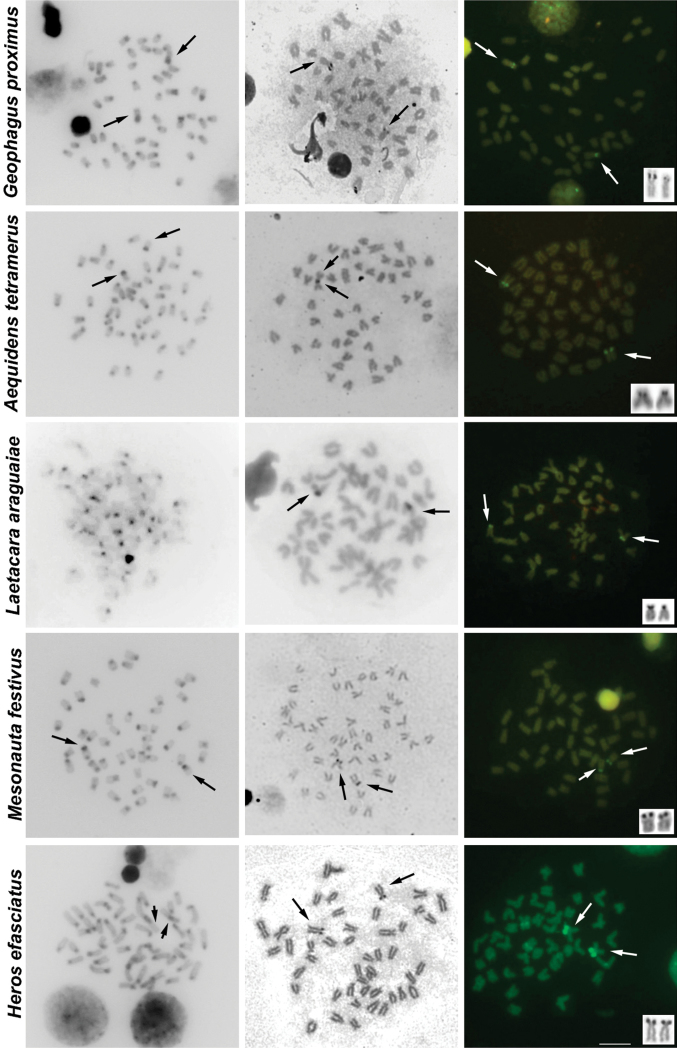
The results of C-banding revealed the heterochromatin generally restricted to pericentromeric regions. Additional blocks of heterochromatin were noticed in Cichla piquiti, Retroculus lapidifer, Biotodoma cupido, Crenicichla strigata, Geophagus proximus and Mesonauta festivus (Figs 2, 3, Table 1).
Figure 2.
Metaphases of several cichlid species under different chromosome treatments. The species are indicated on the left. The first, second and third columns show C-banded, AgNOR- and CMA3- stained metaphases, respectively. The third column shows chromosomes bearing AgNORs in the box. The arrows indicate the NOR-bearing chromosomes. Bar = 10 μm.
Figure 3.
Metaphases of several cichlid species under different chromosome treatments. The species are indicated on the left. The first, second and third columns show C-banded, AgNOR- and CMA3- stained metaphases, respectively. The third column shows chromosomes bearing AgNORs in the box. The arrows indicate the NOR-bearing chromosomes. For some metaphases (without arrows) it was not possible to identify the NOR-carrying chromosomes. Bar = 10 µm.
Characteristic heterochromatic blocks corresponding to AgNOR bearing regions (two blocks, one in each homologue) were observed in all species, and these blocks were consistent with CMA3 positive (CMA3+) blocks (Figs 2, 3, Table 1). These AgNOR/CMA3+ blocks were present in terminal regions; however, positional variation was observed in Biotodoma cupido (Fig. 2) and Geophagus proximus (Fig. 3), which the blocks are present in interstitial regions. Moreover, Retroculus lapidifer (Fig. 2) and Crenicichla strigata (Fig. 2) displayed CMA3+ blocks in pericentromeric regions of almost all chromosomes, and Heros efasciatus (Fig. 3) displayed a positive interstitial signal in one chromosome pair. Size variation was also observed in AgNOR/CMA3+ blocks between homologous chromosomes in Cichla piquiti (Fig. 2), Crenicichla strigata (Fig. 2) and Satanoperca jurupari (Fig. 2). Other chromosomal areas were CMA3 neutral in all of the species analyzed (Figs 2, 3).
Discussion
The diploid number reported for the species in this study, in general are in agreement with the conserved 2n=48 chromosomes commonly found in South American cichlids and in contrast with the presence of 2n=44 chromosomes in African cichlids. All species, except Laetacara araguaiae, had their diploid number already described (Poletto et al. 2010). Moreover, some cichlid species display the occurrence of specific chromosomal rearrangements, such as pericentric inversions, translocations and fission or fusion rearrangements, that occurred during their evolutionary history and deviate their karyotypic formulae from common pattern observed for cichlids (revised by Feldberg et al. 2003, Mesquita et al. 2008, Poletto et al. 2010).
Chromosomal variability was observed in derived lineages, such as the Geophagini and the Cichlasomatini tribes (Feldberg et al. 2003, Poletto et al. 2010). Thus, the diploid number variation observed here in Laetacara araguaiae and the polymorphism observed in Geophagus proximus, which belong to Cichlasomatini and Geophagini tribes, respectively, could reflect the higher chromosomal variation found in these tribes. In fact, another species of Laetacara Kullander, 1986, Laetacara prope dorsigera (Heckel, 1840),generally displayed 2n=44 chromosomes with an intraspecific variation in the diploid number that ranges from 2n=43 to 2n=46, which are thought to have originated from centric chromosomal fusions (Martins-Santos et al. 2005). In Geophagus proximus, the polymorphism is a consequence of a Robertsonian translocation between two st/a chromosomes that results in a large metacentric chromosome and a dot-like element. However, it is inconclusive if this rearrangement occurred between homologous or non-homologous chromosomes due to the great similarities among the st/a chromosomes in Geophagus proximus.
Chromosomal rearrangements such the ones reported here could lead to the karyotypic diversification of the species. In fact, chromosomal rearrangements have contributed to karyotypic evolution in a range of fishes, including the cichlids Symphysodon (Heckel, 1840) (Mesquita et al. 2008, Gross et al. 2009a), salmonids (Allendorf and Thorgaard 1984) and Gobius fallax Sarato, 1889(Thode et al. 1988), among others. Moreover chromosomal rearrangements may result in intraspecific variation as broadly reported in some fish species: in the origin of neo-Y sex chromosomes (Uyeno and Miller 1971, 1972, Bertollo et al. 1983, 1997, Almeida-Toledo et al. 1984, 1988, 2000, Silva and Margarido 2005), in karyotypic diversification of species complex of Gymnotus carapo Linnaeus, 1758 (Milhomem et al. 2008), in Hoplias malabaricus (Bloch, 1794) (Bertollo et al. 1997) and in Erythrinus erythrinus (Bloch et Schneider, 1801) (Bertollo et al. 2004).
Although the cichlid cytogenetics suggests that the ancestral karyotype (2n=48 st/a) could have undergone major changes (pericentric inversions, fusions, fissions and chromosomal translocations) in the macro-structure of the South American species (Feldberg et al. 2003, Poletto et al. 2010), these studies show that this family of fish has a relatively conserved diploid number. Despite of the absence of conclusive data about chromosomal rearrangements rate that occurs in cichlids, it could be suggested that this group has an intermediate level of chromosomal stability compared to birds and mammals, which are more stable and variable, respectively. It is predicted that chromosomal rearrangements can be one of the evolutionary forces that affect the reproductive isolation and speciation processes (Noor et al. 2001, Rieseberg 2001), which create higher levels of species diversity. However, birds and cichlids display greater species richness than what is observed in mammals; this is contrary to the more stable karyotypes of birds and cichlids. Therefore chromosomal rearrangements may be not the most decisive evolutionary process in the cichlids speciation.
C-banding analyses in this study revealed that the conserved pattern of heterochromatin distribution was mostly restricted to the pericentromeric regions of cichlid chromosomes, which has been commonly reported in American and African representatives but with variations in both groups (Kornfield et al. 1979, Majumdar and McAndrew 1986, Feldberg et al. 2003, and others reported in Table 1). Additional heterochromatic blocks were present in almost all species analyzed, and exceptions were observed in Satanoperca jurupari (Geophagini), Aequidens tetramerus (Cichlasomatini), Laetacara araguaiae (Cichlasomatini) and Heros efasciatus (Heroini). For all species, one of these blocks was related to AgNOR regions, which seems to be a common feature in cichlids and other fish (Pendás et al. 1993, Artoni et al. 2008, Souza et al. 2008, Venere et al. 2008, among others cited in Table 1).
Concerning the singular heterochromatic blocks reported here, Cichla piquiti, Crenicichla strigata and Geophagus proximus show variability in the positions, extensions and number of these blocks compared to the other species in each genus. Moreover, the divergent patterns are observed in Crenicichla Heckel, 1840 and Geophagus Heckel, 1840. This variability can be also observed in the Laetacara genus; in this case, Laetacara araguaiae does not have any additional heterochromatic blocks, whereas Laetacara prope dorsigera has heterochromatic NORs as additional blocks (Martins-Santos et al. 2005). Moreover, both of the Satanoperca Günther, 1862 species analyzed do not have any additional heterochromatic blocks. Comparisons within every genera Retroculus Eigenmann et Bray, 1894, Biotodoma Eigenmann et Kennedy, 1903, Aequidens Eigenmann et Bray, 1894, Heros Heckel, 1840 and Mesonauta Günther, 1862 are not possible because this is the first C-banding analysis for these genera. Heterochromatic variations can be observed when comparing the additional heterochromatic blocks patterns within the tribes Geophagini, Cichlasomatini and Heroinitribes. This analysis could support the current idea that these groups display some of the highest chromosomal variability for the Cichlidae family (Feldberg et al. 2003, Poletto et al. 2010). However, they are the most studied group concerning heterochromatin analysis, and it is not clear if this variability reflects higher chromosomal variability or a sampling effort (for all comparisons see Table 1).
The fluorochrome CMA3 showed the presence of GC-rich blocks coinciding with AgNOR sites in all species, which is a common trait in cichlids. The variation in the extension of these blocks also matches the size variation in the AgNOR sites in some species. Additional CMA3+ blocks are uncommon patterns in cichlids species, but they have been reported here for some species. In addition, this trait has only been previously reported in the Heroini species Pterophyllum scalare (Schultze, 1823) (Nascimento et al. 2006). The general pattern of base-pair richness of the heterochromatin indicates some level of compartmentalization of this genomic content at both intragenomic and intraspecific levels. Finally, based on the present and previously reported data, it seems possible that there is a relationship between CMA3+ blocks and AgNOR regions in cichlid species. Furthermore, the variation may be an exception in this group of fish and could suggest that the sequences presented in these regions may possess some dynamism in cichlids genomes.
With respect to AgNOR, length variation between homologous chromosomes could be explained by the duplication or deletion of 45S rDNA repeat units. All AgNOR sites in the species analyzed here are heterochromatic as aforementioned. The length variation detected and extensively observed in other organisms may be caused by the presence of repetitive sequences, errors during the replication process, unequal crossing-over (Ashley and Ward 1993, Pendás et al. 1993, Boron et al. 2006, Gross et al. 2010) and likely non-reciprocal translocation between these regions (revised in Wasko and Galetti 2000).
Conclusion
The heterochromatin, CMA3+ blocks and AgNOR regions are classic cases of enriched repetitive elements regions, such as satellite DNA, transposable elements, and rDNA. Among cichlids, it has been reported that the pericentromeric regions, which are commonly evidenced by C-banding, are repositories for a great amount of repetitive elements, such as transposable elements (Gross et al. 2009b, Mazzuchelli and Martins 2009, Teixeira et al. 2009, Valente et al. 2011). Repetitive sequences are highly dynamic in genome evolution; for example, pericentromeric DNA are rapidly evolving regions in eukaryotic genomes (Haaf and Willard 1997, Csink and Henikoff 1998, Murphy and Karpen 1998) due to the accumulation of repetitive sequences by recombination suppression (Topp and Dawe 2006, Grewal and Jia 2007). In fact, the results reported here and in previous work do not show any phylogenetic relationships in terms of constitutive heterochromatin, NOR and CMA3+ blocks; therefore, the actual number, position and length variation of sites are not related to any homology. All of the variation observed in these regions may be related to the intrinsic dynamism of repeated sequences and independent heterochromatin modifications that do not follow the diversification of taxa.
Acknowledgements
The study was supported by Fundação de Amparo à Pesquisa do Estado de Mato Grosso (FAPEMAT), Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP), Coordenadoria de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq).
Appendix 1
Synthesis of the cichlid species analyzed with respect to the karyotypic formulae, heterochromatin distribution and CMA3 patterns. Table with synthesis of karyotypic traits of cichlids. File format: Microsoft Excel Spreadsheet (xsl).
References
- Allendorf FW, Thorgaard GH. (1984) Tetraploidy and the evolution of Salmonid fishes. In: Turner BJ. (Eds). Evolution of Fishes.Plenum, New York: 1-53
- Almeida-Toledo LF, Foresti F, Daniel MFVZ, Toledo-Filho SA. (2000) Sex chromosome evolution in fish: the formation of the neo-Y chromosome in Eigenmannia (Gymnotiformes). Chromosoma 109 (3): 197-200.10.1007/s004120050428 [DOI] [PubMed] [Google Scholar]
- Almeida-Toledo LF, Foresti F, Toledo-Filho SA. (1984) Complex sex chromosome system in Eigenmannia sp. (Pisces, Gymnotiformes). Genetica 64 (3): 165-169.10.1007/BF00115340 [Google Scholar]
- Almeida-Toledo LF, Viegas-Pequignot E, Foresti F, Toledo-Filho SA, Dutrillaux B. (1988) BrdU replication patterns demonstrating chromosome homeologies in two fish species, genus Eigenmannia. Cytogenetic and Cell Genetic 48 (2): 117-120.10.1159/000132603 [Google Scholar]
- Artoni RF, Gross MC, Schneider CH, Vicari MR, Almeida MC, Matoso DA. (2008) Epifluorescence and light microscopy evidencing structural and functional polymorphism of ribosomal DNA in fish (Teleostei: Astyanax fasciatus). Micron 39 (8): 1156-1159.10.1016/j.micron.2008.05.011 [DOI] [PubMed] [Google Scholar]
- Ashley T, Ward DC. (1993) A “hot spot” of recombination coincides with interstitial telomeric sequence in the Armenian hamster. Cytogenetic and Cell Genetic 62(2–3): 169-171.10.1159/000133464 [DOI] [PubMed] [Google Scholar]
- Bertollo LAC, Fontes MS, Fenocchio AS, Cano J. (1997) The X1X2Y sex chromosome system in the fish Hoplias malabaricus. I. G-, C- and chromosome replication banding. Chromosome Research 5 (7): 493-499.10.1023/A:1018477232354 [DOI] [PubMed] [Google Scholar]
- Bertollo LAC, Oliveira C, Molina WF, Margarido VP, Fontes MS, Pastori MC, Falcão JN, Fenocchio AS. (2004) Chromosome evolution in the erythrinid fish, Erythrinus erythrinus (Teleostei: Characiformes). Heredity 93: 228-233.10.1038/sj.hdy.6800511 [DOI] [PubMed] [Google Scholar]
- Bertollo LAC, Takahashi CS, Moreira-Filho O. (1978) Cytotaxonomic consideration on Hoplias lacerdae (Pisces, Erythrinidae). Brazilian Journal of Genetics 1(2): 103–120 [http://web2.sbg.org.br/GMB/edicoesAnteriores/Indice/indiceVol01N2.html] [Google Scholar]
- Bertollo LAC, Takahashi CS, Moreira-Filho O. (1983) Multiple sex chromosomes in the genus Hoplias (Pisces, Erythrinidae). Cytologia 48(1): 1–12 [http://www.journalarchive.jst.go.jp/english/jnltoc_en.php?cdjournal=cytologia1929&cdvol=48&noissue=1] [Google Scholar]
- Benzaquem DC, Feldberg E, Porto JIR, Gross MC, Zuanon JAS. (2008) Cytotaxonomy and karyoevolution of the genus Crenicichla (perciformes, cichlidae). Genetics and Molecular Biology 31 (1): 250-255.10.1590/S1415-47572008000200016 [Google Scholar]
- Boron A, Ozouf-Costaz C, Coutanceau JP, Woroniecka K. (2006) Gene mapping of 28S and 5S rDNA sites in the spined loach Cobitis taenia (Pisces, Cobitidae) from a diploid population and a diploid-tetraploid population. Genetica 128(1–3): 71-79.10.1007/s10709-005-5536-8 [DOI] [PubMed] [Google Scholar]
- Brinn MNA, Porto JIR, Feldberg E. (2004) Karyological evidence for interspecific hybridization between Cichla monoculus and C. temensis (perciformes, cichlidae) in the amazon. Hereditas 141 (3): 252-257.10.1111/j.1601-5223.2004.01830.x [DOI] [PubMed] [Google Scholar]
- Csink AK, Henikoff S. (1998) Something from nothing: the evolution and utility of satellite repeats. Trends in Genetics 14 (5): 200-204.10.1016/S0168-9525(98)01444-9 [DOI] [PubMed] [Google Scholar]
- Feldberg E, Porto JIR, Bertollo LAC. (2003) Chromosomal changes and adaptation of cichlid fishes during evolution. In: Val AL, Kapoor BG. (Eds). Fish adaptations.Science Publishers INC, New Delhi, New York: 285-308
- Genner MJ, Seehausen O, Lunt DH, Joyce DA, Shaw PW, Carvalho GR, Turner GF. (2007) Age of cichlids: new dates for ancient lake fish radiations. Molecular Biology and Evolution 24 (5): 1269-1282.10.1093/molbev/msm050 [DOI] [PubMed] [Google Scholar]
- Grewal SIS, Jia S. (2007) Heterochromatin revisited. Nature Reviews Genetics 8: 35-46.10.1038/nrg2008 [DOI] [PubMed] [Google Scholar]
- Gross MC, Feldberg F, Cella DM, Schneider MC, Schneider CH, Porto JIR, Martins C. (2009a) Intriguing evidence of translocations in Discus fish (Symphysodon, Cichlidae) and report of the largest meiotic chromosomal chain observed in vertebrates. Heredity 102: 435-441.10.1038/hdy.2009.3 [DOI] [PubMed] [Google Scholar]
- Gross MC, Schneider CH, Valente GT, Porto JIR, Martins C, Feldberg E. (2009b) Comparative Cytogenetic Analysis of the Genus Symphysodon (Discus Fishes, Cichlidae): Chromosomal Characteristics of Retrotransposons and Minor Ribosomal DNA. Cytogenetic and Genome Research 127 (1): 43-53.10.1159/000279443 [DOI] [PubMed] [Google Scholar]
- Gross MC, Schneider CH, Valente GT, Martins C, Feldberg E. (2010) Variability inter and intraspecific of 18S rDNA locus among Symphysodon fishes: chromosomal rearrangements. Journal of Fish Biology 76 (5): 1117-1127.10.1111/j.1095-8649.2010.02550.x [DOI] [PubMed] [Google Scholar]
- Haaf T, Willard HF. (1997) Chromosome-specific alpha-satellite DNA from the centromere of chimpanzee chromosome 4. Chromosoma 106 (4): 226-232.10.1007/s004120050243 [DOI] [PubMed] [Google Scholar]
- Howell WM, Black DA. (1980) Controlled silver-staining of nucleolus organizer regions with a protective colloidal developer: a 1-step method. Cellular and Molecular Life Sciences 36 (8): 1014-1015.10.1007/BF01953855 [DOI] [PubMed] [Google Scholar]
- Kocher TD. (2004) Adaptive evolution and explosive speciation: the cichlid fish model. Nature Reviews Genetics 5 (4): 288-298.10.1038/nrg1316 [DOI] [PubMed] [Google Scholar]
- Kornfield IL, Ritte U, Richler C, Wahrman J. (1979) Biochemical and cytological differentiation among cichlid fishes of the Sea of Galilee. Evolution 33(1): 1–14 [http://www.jstor.org/pss/2407360] [DOI] [PubMed] [Google Scholar]
- Kornfield I, Smith PF. (2000) African cichlid fishes: Model system for evolutionary biology. Annual Review of Ecology and Systematics 31: 163-196.10.1146/annurev.ecolsys.31.1.163 [Google Scholar]
- Kullander SO. (1998) A phylogeny and classification of the South American Cichlidae (Teleostei: Perciformes). In: Malabarba LR, Reis RE, Vari RP, Lucena ZM, Lucena CAS. (Eds). Phylogeny and classification of Neotropical fishes.EDIPUCRS, Porto Alegre: 461-498
- Kullander SO. (2003) Family Cichlidae. In: Reis RE, Kullander SO, Ferraris CJ. (Eds). Check list of the freshwater fishes of South and Central America.EDIPUCRS, Porto Alegre: 605-654
- Majumdar KC, McAndrew BJ. (1986) Relative DNA content of somatic nuclei and chromosomal studies in three genera, Tilapia, Sarotherodon, and Oreochromis of the tribe Tilapiini (Pisces, Cichlidae). Genetica 68 (3): 175-188.10.1007/BF02424441 [Google Scholar]
- Martins IC, Portella-Castro ALB, Julio Jr. HF. (1995) Chromosome analysis of 5 species of the Cichlidae family (Pisces, Perciformes) from the Paraná River. Cytologia 60(3): 223–231 [http://www.journalarchive.jst.go.jp/english/jnltoc_en.php?cdjournal=cytologia1929&cdvol=60&noissue=3] [Google Scholar]
- Martins-Santos IC, Portela-Castro ALB, Julio Jr. HF. (2005) Chromosomal polymorphism and speciation in Laetacara cf. dorsigera (Teleostei, Perciformes, Cichlidae) from the river Paraná PR Brazil. Caryologia 58(2): 95–101 [http://www1.unifi.it/caryologia/past_volumes/58_2/58_2abst.html] [Google Scholar]
- Mazzuchelli J, Martins C. (2009) Genetica 136 (3): 461-469.10.1007/s10709-008-9346-7 [DOI] [PubMed] [Google Scholar]
- Mesquita DR, Porto JIR, Feldberg E. (2008) Chromosomal variability in the wild ornamental fish Symphysodon spp. (Perciformes, Cichlidae) from Amazon. Neotropical Ichthyology 6 (2): 181-190.10.1590/S1679-62252008000200005 [Google Scholar]
- Milhomem SSR, Pieczarka JC, Crampton WGR, Silva DS, Souza ACP, Carvalho Jr. JR, Nagamachi CY. (2008) Chromosomal evidence for a putative cryptic species in the Gymnotus carapo species-complex (Gymnotiformes, Gymnotidae). BMC Genetics 9: 1-75.10.1186/1471-2156-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi SMHK, Portela-Castro ALB, Martins-Santos IC. (2007) Cytogenetic characterization of Crenicichla (Pisces, Perciformes) of the Iguaçu River. Genetics and Molecular Research 6(3): 650–656 [http://www.geneticsmr.com/issue/6/all] [PubMed] [Google Scholar]
- Murphy TD, Karpen GH. (1998) Centromeres take flight: alpha satellite and the quest for the human centromere. Cell 93 (3): 317-320.10.1016/S0092-8674(00)81158-7 [DOI] [PubMed] [Google Scholar]
- Nascimento A, Souza ACP, Feldberg E, Carvalho Jr. JR, Barros RMS, Pieczarka JC, Nagamachi CY. (2006) Cytogenetic analysis on Pterophyllum scalare (perciformes, cichlidae) from Jari river, Pará state. Caryologia 59(3): 138–143 [http://www1.unifi.it/caryologia/past_volumes/59_2/59_2abst.html] [Google Scholar]
- Nelson JS. (2006) Fishes of the world. John Wiley and Sons, New York, 601 pp. [Google Scholar]
- Noor MA, Grams KL, Bertucci LA, Reiland J. (2001) Chromosomal inversions and the reproductive isolation of species. Proceedings of the National Academy of Sciences of the United States of America 98 (21): 12084-12088.10.1073/pnas.221274498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendás AM, Morán P, Garcia-Vasquez E. (1993) Ribosomal RNA genes are interspersed throughout a heterochromatic chromosome arm in Atlantic salmon. Cytogenetic and Cell Genetics 63 (2): 128-130.10.1159/000133517 [DOI] [PubMed] [Google Scholar]
- Perazzo G, Noleto RB, Vicari MR, Machado PC, Gava A, Cestari MM. (2010) Chromosomal studies in Crenicichla lepidota and Australoheros facetus (Cichlidae, Perciformes) from extreme Southern Brazil. Reviews in Fish Biology and Fisheries 21 (3): 509-515.10.1007/s11160-010-9170-x [Google Scholar]
- Pires LB, Giuliano CL, Dias AL. (2008) Karyotype similarities among two populations of Geophagus brasiliensis (Perciformes, Cichlidae) from the Tibagi River basin/PR/Brazil. Caryologia 61(2): 135–138 [http://www1.unifi.it/caryologia/past_volumes/61_2/61_2abst.html] [Google Scholar]
- Pires LB, Giuliano CL, Dias AL. (2010) Cytogenetic characterization of Geophagus brasiliensis and two species of Gymnogeophagus (Cichlidae:Geophaginae) from Guaíba Lake, RS, Brazil. Folia biologica 58(1–2): 29-34.10.3409/fb58_1-2.29-34 [DOI] [PubMed] [Google Scholar]
- Poletto AB, Ferreira IA, Cabral-de-Mello DC, Nakajima RT, Mazzuchelli J, Ribeiro HB, Venere PC, Nirchio M, Kocher TD, Martins C. (2010) Chromosome differentiation patterns during cichlid fish evolution. BMC Genetics 11: 1-50.10.1186/1471-2156-11-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullin RSV. (1991) Cichlids in aquaculture. In: Keenleyside MHA. (Eds). Cichlid fishes: behaviour, ecology and evolution.Chapman & Hall, London: 280-309
- Rieseberg LH. (2001) Chromosomal rearrangements and speciation. Trends in Ecology & Evolution 16 (7): 351-358.10.1016/S0169-5347(01)02187-5 [DOI] [PubMed] [Google Scholar]
- Roncati HA, Pastori MC, Fenocchio AS. (2007) Cytogenetic Studies and Evolutive Considerations on Fishes of the Family Cichlidae (Perciformes) from Parana River (Argentina). Cytologia 72 (4): 379-384.10.1508/cytologia.72.379 [Google Scholar]
- Schweizer D. (1976) Reverse fluorescent chromosome banding with chromomycin and DAPI. Chromosoma 58 (4): 307-324.10.1007/BF00292840 [DOI] [PubMed] [Google Scholar]
- Silva EB, Margarido VP. (2005) An X1X1X2X2/X1X2Y multiple sex chromosome system in a new species of the genus Gymnotus (Pisces, Gymnotiformes). Environmental Biology of Fishes 73 (3): 293-297.10.1007/s10641-005-2144-5 [Google Scholar]
- Smith WL, Chakrabarty P, Sparks JS. (2008) Phylogeny, taxonomy, and evolution of Neotropical cichlids (Teleostei: Cichlidae: Cichlinae). Cladistics 24 (5): 625-641.10.1111/j.1096-0031.2008.00210.x [Google Scholar]
- Souza IL, Santos-Silva LK, Venere PC, Moreira-Filho O. (2008) Molecular cytogenetics of Salminus fish (Characiformes) based on 5S and 18S rRNA genes hybridization, fluorochrome staining and C-banding. Micron 39 (7): 1036-1041.10.1016/j.micron.2007.09.007 [DOI] [PubMed] [Google Scholar]
- Sumner AT. (1972) A simple technique for demonstrating centromeric heterochromatin. Experimental Cell Research 75 (1): 304-306.10.1016/0014-4827(72)90558-7 [DOI] [PubMed] [Google Scholar]
- Teixeira WG, Ferreira IA, Cabral-de-Mello DC, Mazzuchelli J, Valente GT, Pinhal D, Poletto AB, Venere PC, Martins C. (2009) Organization of repeated DNA elements in the genome of the cichlid fish Cichla kelberi and its contributions to the knowledge of fish genomes. Cytogenetic and Genome Research 125 (3): 224-234.10.1159/000230006 [DOI] [PubMed] [Google Scholar]
- Thode G, Martinez G, Ruiz JL, Lopéz JR. (1988) A complex chromosomal polymorphism in Gobius fallax (Gobiidae, Perciformes). Genetica 76 (1): 65-71.10.1007/BF00126011 [Google Scholar]
- Topp CN, Dawe RK. (2006) Reinterpreting pericentromeric heterochromatin. Current Opinion in Plant Biology 9 (6): 647-653.10.1016/j.pbi.2006.09.008 [DOI] [PubMed] [Google Scholar]
- Uyeno T, Miller RR. (1971) Multiple sex chromosomes in a Mexican Cyprinodontidae fish. Nature 231: 452-453.10.1038/231452a0 [DOI] [PubMed] [Google Scholar]
- Uyeno T, Miller RR. (1972) Second discovery of multiple sex chromosome among fishes. Experientia 28 (2): 223-225.10.1007/BF01935773 [DOI] [PubMed] [Google Scholar]
- Valente GT, Mazzuchelli J, Ferreira IA, Poletto AB, Fantinatti BEA, Martins C. (2011) Cytogenetic Mapping of the Retroelements Rex1, Rex3 and Rex6 among Cichlid Fish: New Insights on the Chromosomal Distribution of Transposable Elements. Cytogenetic and Genome Research 133 (1): 34-42.10.1159/000322888 [DOI] [PubMed] [Google Scholar]
- Venere PC, Souza IL, Silva LKS, Dos Anjos MB, De Oliveira RR, Galetti Jr. PM. (2008) Recent chromosome diversification in the evolutionary radiation of the freshwater fish family Curimatidae (Characiformes). Journal of Fish Biology 72 (8): 1976-1989.10.1111/j.1095-8649.2008.01814.x [Google Scholar]
- Vicari MR, Artoni RF, Moreira-Filho O, Bertollo LAC. (2006) Basic and molecular cytogenetics in freshwater Cichlidae (Osteichthyes, Perciformes). Karyotypic conservadorism and divergence. Caryologia 59(3): 260–266 [http://www1.unifi.it/caryologia/past_volumes/59_3/59_3abst.html] [Google Scholar]
- Wasko AP, Galetti Jr. PM. (2000) Mapping 18S ribosomal genes in fish of the genus Brycon (Characidae) by fluorescence in situ hybridization (FISH). Genetics and Molecular Biology 23 (1): 135-138.10.1590/S1415-47572000000100025 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Synthesis of the cichlid species analyzed with respect to the karyotypic formulae, heterochromatin distribution and CMA3 patterns. Table with synthesis of karyotypic traits of cichlids. File format: Microsoft Excel Spreadsheet (xsl).