Abstract
The emerging tick-borne pathogen Anaplasma phagocytophilum infects humans, domestic animals, and wildlife throughout the Holarctic. In the western U.S., the ecology of A. phagocytophilum is particularly complex, with multiple pathogen strains, tick vectors, and reservoir hosts. A recent phylogenetic analysis of A. phagocytophilum strains isolated from various small mammal hosts in California documented distinct clustering of woodrat strains separate from sciurid (chipmunk and squirrel) strains. Here, we identified strains of A. phagocytophilum in various Ixodes tick species in California and related these genotypes to those found among reservoir and clinical hosts from the same areas. The sequences from all of the nidicolous (nest-dwelling) Ixodes ticks grouped within a clade that also contained all of the woodrat-origin A. phagocytophilum strains. Two of the I. pacificus sequences were also grouped within this woodrat clade, while the remaining five belonged to a less genetically diverse clade that included several sciurid-origin strains as well as a dog, a horse, and a human strain. By comparing A. phagocytophilum strains from multiple sources concurrently, we were able to gain a clearer picture of how A. phagocytophilum strains in the western U.S. are partitioned, which hosts and vectors are most likely to be infected with a particular strain, and which tick species and reservoir hosts pose the greatest health risk to humans and domestic animals.
Keywords: Anaplasma phagocytophilum, Ixodes spp., Nidicolous ticks, Ixodes pacificus, ank gene, Phylogeny
Introduction
Anaplasma phagocytophilum, the causative agent of granulocytic anaplasmosis (GA) is a tick-transmitted, intra-leukocytic rickettsial parasite of humans and other animals. Reservoir hosts include small mammals, such as white-footed mice (Peromyscus leucopus) in the eastern U.S. (Telford et al., 1996) and woodrats (Neotoma fuscipes), squirrels (Sciurus spp.), and chipmunks (Tamias spp.) in the western U.S. (Nicholson et al., 1999; Nieto and Foley, 2008, 2009). The tick vectors for A. phagocytophilum are most frequently reported to be the questing ticks of the Ixodes ricinus group, including I. pacificus in the western U.S. and I. scapularis in the eastern U.S. (Foley et al., 2004). While these tick species likely serve as the primary bridge vectors transmitting GA to humans and domestic animals, other nidicolous (i.e. primarily nest dwelling) Ixodes ticks including I. spinipalpis, I. ochotonae, and I. trianguliceps harbor A. phagocytophilum and likely help maintain enzootic cycles of GA (Zeidner et al., 2000; Bown et al., 2003; Foley et al., 2011).
Although A. phagocytophilum was originally classified as 3 distinct organisms – Ehrlichia equi, Ehrlichia phagocytophila, and the agent of human granulocytic ehrlichiosis – morphological, phenotypic, and genetic evidence led to the reclassification of these 3 organisms as the modern A. phagocytophilum in 2001 (Dumler et al., 2001). Despite this reorganization, phenotypic and preliminary genetic data strongly support the presence of multiple distinct strains. For example, strains in Europe commonly cause clinical disease in small hoofstock, but North American strains are neither particularly infectious nor virulent based on data from experimental infections of sheep or cattle (Pusterla et al., 1997; Stuen, 2007; Gorman et al., 2012). Similarly, in England, 2 genetically distinct subpopulations of A. phagocytophilum coexist in separate enzootic cycles, one involving deer and I. ricinus ticks and the other involving field voles and I. trianguliceps (Bown et al., 2009). The North American strain designated Ap-Variant 1 occurs in ticks and deer and is infectious to goats, but not rodents (Massung et al., 2003, 2007). The California strain DU1, originating from a woodrat, can infect rodent species, but not horses (Nieto et al., 2010). Strain MRK, which was isolated from a horse, reproducibly induces severe clinical disease in horses indistinguishable from that induced when human-origin A. phagocytophilum is inoculated into horses and is also infectious to small mammals (Pusterla et al., 1999; Foley et al., 2009b). These clinical and epidemiological distinctions are associated with genotypic segregation into specific clades based on analyses of the 16S rRNA, msp4, msp2, and ank genes (Massung et al., 2003; de la Fuente et al., 2005; Bown et al., 2009; Rejmanek et al., 2011).
In the western U.S., the ecology of A. phagocytophilum is particularly complex, with multiple pathogen strains, tick vectors, and reservoir hosts (Foley et al., 2004). A recent phylogenetic analysis of ank, groESL, and the 23S-5S rRNA genes from 28 Californian rodent strains, the MRK horse strain, and a California dog strain documented distinct clustering of woodrat strains separate from a genetically uniform group consisting of sciurid (chipmunk and squirrel), horse, and dog strains (Rejmanek et al., 2011).
One question that has not been addressed in previous studies of A. phagocytophilum is whether particular A. phagocytophilum genotypes are associated with certain host-specialist ticks. California is home to numerous nidicolous Ixodes spp., many of which infest some of the same small mammal species as I. pacificus (Furman and Loomis, 1984). A recent survey of ticks on small mammals from numerous sites across California revealed that 66% of tick-infested small mammals hosted nidicolous Ixodes species, while the remaining 34% were infested with I. pacificus (Foley et al., 2011). In the current study, our goal was to identify strains of A. phagocytophilum in various Ixodes tick species in California and relate these genotypes to those found among reservoir and clinical hosts from the same areas. For our analysis, we focused on a single gene (ank), which was chosen because it exhibited a high level of polymorphism even among closely related A. phagocytophilum strains and was largely concordant with phylogenetic results from analysis of other gene regions (Rejmanek et al., 2011).
Materials and methods
Sample collection
DNA samples for molecular characterization were obtained from I. pacificus, I. spinipalpis, I. angustus, I. ochotonae, and I. woodi from April 2006 to September 2011. Ticks were collected in central and northern California at the following sites: Soquel Demonstration Forest (SD, Santa Cruz County; 37° 03.25’, 121° 50.68’), Henry Cowell State Park (HC, Santa Cruz County; 37° 08.705’, 122° 11.12’), Samuel P. Taylor State Park (SPT, Marin County; 38° 01.232’, 122° 40.774’), Hendy Woods State Park (HW, Mendocino County; 39° 04.25’, 123° 28.238’), Humboldt Redwoods State Park (HR, Humboldt County; 40° 17.770’, 123° 59.178’), and Archer Taylor Preserve (ATP, Napa County; 38° 20.54’, 122° 25.34’). Two ticks were obtained from the Green Diamond Resource Company (GD), found on a dusky-footed woodrat in an unspecified site of Humboldt County.
Ticks were collected from small mammals trapped and sampled as described previously (Foley et al., 2011). Ticks were found on dusky-footed woodrats, deer mice, a chipmunk (Tamias ochrogenys), an eastern grey squirrel (Sciurus carolinensis), and a human. Trapping and sample collection were carried out as previously described (Foley et al., 2011).To obtain questing ticks, flagging was performed over herbaceous and shrubby vegetation as well as duff and leaf litter using a 1-m2 white cotton flag.
Nucleic acid extraction, PCR, and sequencing
DNA was extracted from ticks following a modified protocol described previously (Humair et al., 2007). Briefly, individual ticks were placed in microcentrifuge tubes, cooled in liquid nitrogen for 3 min, and crushed using a microcentrifuge pestle. Next, 100 µl of 0.7 M NH4OH was added, and the tubes were placed on a 100°C heat block for 15 min. Tubes were then cooled on ice for 30 s followed by an additional 15 min of heating at 100°C with open lids in order to evaporate the ammonia.
All DNA samples were initially screened for the presence of A. phagocytophilum DNA using a highly sensitive real-time TaqMan PCR assay targeting the msp2/p44 gene (Drazenovich et al., 2006). Results of real-time PCR were considered positive if they had a cycle threshold (CT) value <40 and a characteristic amplification curve. Nested conventional PCR assays targeting a section of the ank gene were then performed on all positive samples. Amplification of the ank gene was performed as described by Massung et al. (2007) using external primers ANK-F1 (5’-GAAGAAATTACAACTCCTGAAG-3’) and ANK-R1 (5’-CAGCCAGATGCAGTAACGTG-3’), followed by internal primers ANK-F2 (5’-TTGACCGCTGAAGCACTAAC-3’) and ANK-R2 (5’-ACCATTTGCTTCTTGAGGAG-3’), yielding an approximately 600-bp amplicon. Amplified DNA was visualized on a 1% agarose gel stained with GelStar nucleic acid stain (Lonza, Rockland, ME). Bands of the expected size were excised and cleaned with a Qiagen (Valencia, CA) gel extraction kit. Gel-extracted amplicons were cloned into the pGEM-T Easy vector (Promega, Madison, WI), transformed into Escherichia coli DH5α cells, and plated onto LB agar containing 100 µg/ml ampicillin. Individual colonies were grown overnight in LB broth containing 100 ug/ml ampicillin, and plasmids were purified using a Quantum Prep plasmid miniprep kit (BioRad, Hercules, CA). Following EcoRI digestion, plasmids were assessed for appropriate insert size. Gel-extracted or cloned PCR products were sequenced in both forward and reverse directions on an ABI 3730 sequencer (Davis Sequencing). Consensus sequences were initially aligned using the CLUSTAL_X sequence alignment program (Larkin et al., 2007) and trimmed to a final length of 567 bp. All unique sequences were deposited in GenBank and issued the following accession numbers: KC249918–KC249925.
Phylogenetic analysis
In addition to the ank sequences acquired in the current study, 31 previously reported A. phagocytophilum ank sequences were used in the phylogenetic analysis. These included sequences from 22 A. phagocytophilum strains isolated from small mammal hosts (woodrats, chipmunks, and grey squirrels) sampled in the same areas and during the same time frame as most of the ticks (GenBank accession numbers JF303732–JF303741 and JF776828) (Rejmanek et al., 2011). Additional strains included HZ_NY (the fully sequenced human-origin strain from New York State) (Dunning Hotopp et al., 2006), 2 other human isolates from New York (NY_2 and NY_3), Webster_WI and WI_2 (human isolates from Wisconsin), Mn_Dog (a dog isolate from Minnesota), RI_1 (isolated from an I. scapularis tick in Rhode Island), MRK_CA (originally isolated from an infected horse in Shasta County, CA), Dog_CA, (a strain isolated from an infected dog from Tuolumne County, CA), and CAHU_HGE2, a strain isolated from an infected human from southern Humboldt County, CA (GenBank accession numbers: CP000235 and AF172153, AF100884, AF100885, GU236811, AF100890, AF100894, DQ320648, AF153716, JF303732, and AF172153, respectively).
All DNA sequences were aligned using MUSCLE v3.8 (Edgar, 2004), then translated into amino acid codons, and adjusted by eye to ensure that the sequences were in frame and did not contain stop codons. Identical sequences were excluded from the analysis, then reincluded as polytomies for the phylogeny figures. To examine the effect of missing data, 2 separate data matrices were used: one in which all sequence data in hand were included for each accession, and one in which all sequences were trimmed to the length of the shortest sequence read (the Californian human strain, CAHU_HGE2). The longer alignment consisted of 567 aligned sites and the shorter alignment of 222 aligned sites. The analysis pipeline described below was carried out for both alignments, but, because both the final models selected and the majority-rule consensus trees for each alignment were effectively the same, we will only present results from the longer alignment below.
Preliminary analyses were performed using the GTR model of nucleotide substitution, with a discrete gamma distribution used to accommodate among-site variation in the rate of nucleotide substitution (i.e., GTR+G). We accommodated process heterogeneity by partitioning the alignment by codon position: a separate GTR+G model was applied to each subset of nucleotides comprising the 3 codon positions.
We approximated the joint posterior probability distribution of trees and other parameters of the GTR+G substitution model using the Metropolis Coupled Markov Chain Monte Carlo (MC3) samplers implemented in MrBayes version 3.2 (Ronquist and Huelsenbeck, 2003). Preliminary analysis consisted of 4 independent MC3 analyses run for 1×107 generations, each with 4 incrementally heated chains (with the temp parameter set to 0.2), where the cold chain was thinned by sampling every 1×103 cycles. For the purposes of diagnosing MCMC performance (see below), we also estimated the joint prior probability density by running the MCMC samplers in MrBayes on an ‘empty’ alignment consisting of entirely missing data (e.g., a matrix of sequences where each sequence is a string of question marks equal in length to the corresponding sequence in the original data matrix).
We assessed performance of the MCMC analyses by scrutinizing the samples from each of the 4 independent chains using Tracer 1.4 (http://beast.bio.ed.ac.uk/Tracer). We assessed convergence by comparing the marginal posterior probability density from each chain for each of the model parameters to ensure that estimates from each of the 4 independent chains were effectively identical. We assessed mixing of the chains over each parameter using 3 diagnostics: (1) the rate at which proposed updates to each parameter were accepted (with a target window of 20–70%); (2) by examining the form of the marginal posterior probability densities (to ensure that the density for each parameter was smooth and unimodal); and (3) by comparing the marginal posterior probability density for each parameter to its corresponding prior probability density (to assess the degree of information in the data for each parameter).
We identified weak parameters as those with low ESS values (i.e., <200) and for which the inferred marginal posterior and prior probability densities were similar. We eliminated any weak parameters by collapsing to submodels of the GTR family as necessary, and then running new analyses under these simplified models (using the same details as described above). Specifically, this process resulted in the selection of the following substitution model for final analysis: a separate GTR model was applied to each of 3 partitions (by codon position) with unlinked stationary frequencies, instantaneous rate matrices, and rate priors.
For the phylogenies presented below, all final MrBayes analyses were carried out with 4 independent runs and 4 chains per run. Each analysis ran for 5×107 generations, sampled every 1×103 generations, with a burn-in of 1×104 sampled generations. To summarize the posterior distribution of trees, we present only the majority-rule consensus tree (Holder et al., 2008). Alignments, trace files, and tree posteriors are available upon request.
Results
Six-hundred and sixty-two ticks including 99 I. angustus, 1 I. auritulus, 17 I. ochotonae, 492 I. pacificus, 1 I. sculptus, 31 I. spinipalpis, and 21 I. woodi, were collected at 7 different study sites throughout central and northern California from April 2005 to September 2011. Twenty-five (3.8%) were positive for A. phagocytophilum DNA by real-time PCR, and partial A. phagocytophilum ank gene sequences were successfully amplified and cloned from 13 of them including 6 nidicolous ticks (1 I. angustus, 1 I. ochotonae, 2 I. spinipalpis, and 2 I. woodi) and 7 I. pacificus ticks. Additional information about each A. phagocytophilum strain including the tick life stage it was isolated from, the study site, the source of each tick (i.e. flag or animal host), and the A. phagocytophilum infection status of the animal host each tick was removed from, are presented in Table 1. Interestingly, 2 of the hosts (a woodrat and chipmunk) from which positive ticks were removed did not have detectable A. phagocytophilum infections, suggesting that those ticks were infected during a previous blood meal.
Table 1.
Description of tick A. phagocytophilum strains analyzed in this study.
| Strain ID | Tick species | Life stage | Study Site | Tick source | Host infection status* |
|---|---|---|---|---|---|
| Ia_1013_HW | Ixodes angustus | Nymph | Hendy Woods | Neotoma fuscipes | Positive |
| Io_504_HW | Ixodes ochotonae | Adult | Hendy Woods | Peromyscus sp. | NA |
| Ip_252_SPT | Ixodes pacificus | Nymph | Samuel P. Taylor | flag | NA |
| Ip_728_HW | Ixodes pacificus | Adult | Hendy Woods | flag | NA |
| Ip_755_SD | Ixodes pacificus | Adult | Soquel D. Forest | Homo sapiens | NA |
| Ip_819_ATP | Ixodes pacificus | Adult | Archer Taylor Preserve | flag | NA |
| Ip_902_HW | Ixodes pacificus | Adult | Hendy Woods | flag | NA |
| Ip_954_HW | Ixodes pacificus | Adult | Hendy Woods | Tamias ochrogenys | Negative |
| Ip_1042_HC | Ixodes pacificus | Nymph | Henry Cowell | Neotoma fuscipes | Negative |
| Is_895_HW | Ixodes spinipalpis | Adult | Hendy Woods | Neotoma fuscipes | NA |
| Is_973_HR | Ixodes spinipalpis | Adult | Humboldt Redwoods | Neotoma fuscipes | Positive |
| Iw_957_GD | Ixodes woodi | Adult | Green Diamond | Neotoma fuscipes | Positive |
| Iw_1006_GD | Ixodes woodi | Adult | Green Diamond | Neotoma fuscipes | Positive |
Based on real-time PCR detection of A. phagocytophilum in blood of hosts that ticks were removed from.
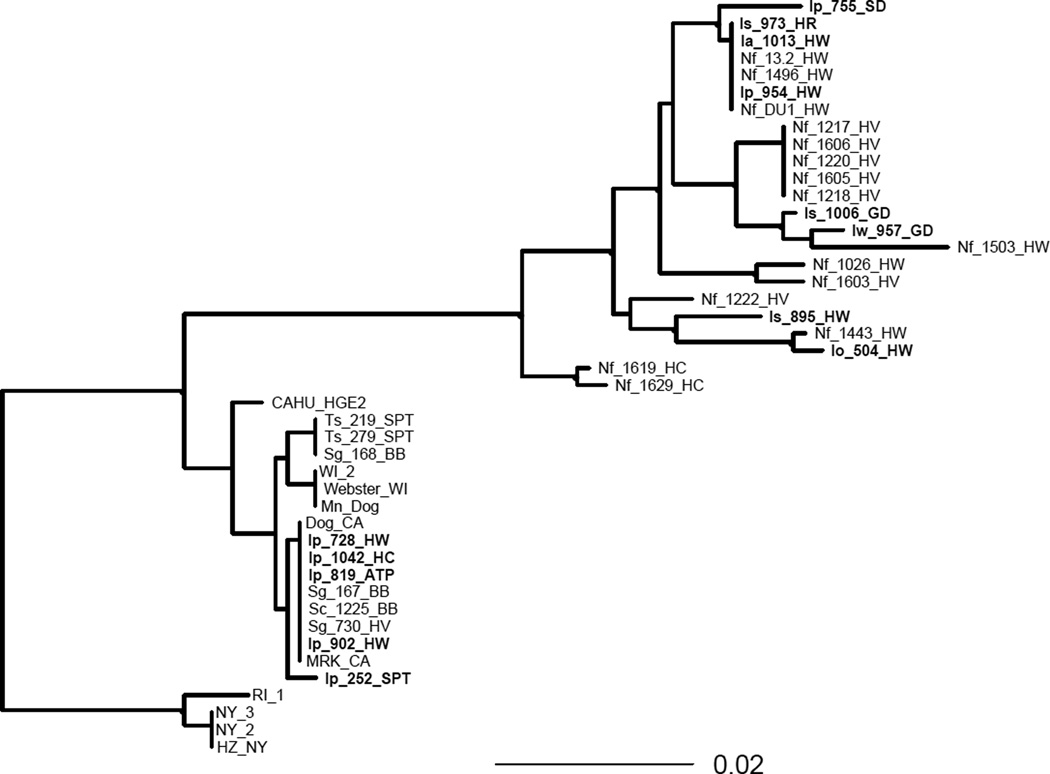
The phylogenetic relationships among all of the tick-derived A. phagocytophilum ank sequences along with sequences from small mammals and clinical hosts from many of the same locations in the current study as well as several sequences from the upper Midwest and East Coast are presented in Fig. 1. The sequences from all of the nidicolous ticks grouped within a clade that also contained all of the woodrat-origin A. phagocytophilum strains. Two of the I. pacificus sequences were also grouped within this woodrat clade, while the remaining 5 belonged to a clade that included several squirrel- and chipmunk-origin strains as well as dog, horse, and human strains. This sciurid/large mammal clade showed very little genetic divergence, with only 3 single nucleotide polymorphisms (SNPs) among all 17 sequences. In contrast, within the woodrat clade, there was a much higher level of polymorphism. For the most part, all of the tick and woodrat strains within this clade were grouped by geographic location. The exception was a strain isolated from an I. pacificus tick (Ip_755_SD) from Soquel Demonstration forest (SD), one of the southernmost sampling sites. Its ank sequence phylogenetically grouped most closely with tick and woodrat strains from Hendy Woods (HW) and Humboldt Redwoods (HR). The 3 New York-origin human strains (HZ_NY, NY_2, and NY_3) along with the Rhode Island tick strain (RI_1) formed their own separate clade.
Fig. 1.
Majority-rule consensus phylogram inferred from an alignment of Anaplasma phagocytophilum ank sequences. Bayesian analyses of phylogeny was performed using MrBayes v. 3.1.2. Identical sequences were excluded from analysis and reincluded for this figure. Black circles indicate >90% nodal support based on Bayesian posterior probabilities. Branch lengths are in units of expected number of nucleotide substitutions per site. Ank sequences derived from ticks in this study are presented in bold text. HZ_NY, NY_2, and NY_3 = human strains from New York State; RI_1 = tick strain from Rhode Island; Webster_WI and WI2 = human strains from Wisconsin; Mn_Dog = dog strain from Minnesota; CAHU_HGE2 = human strain from California; MRK_CA = horse strain from California; Dog_CA = dog strain from California; Ip = Ixodes pacificus; Ia = Ixodes angustus; Io = Ixodes ochotonae; Is = Ixodes spinipalpis; Iw = Ixodes woodi; Sg = Sciurus griseus; Sc = Sciurus carolinensis; Ts = Tamias sonomae; Nf = Neotoma fuscipes; HV = Hoopa Valley; BB = Big Basin; SPT = Samuel P. Taylor; HW = Hendy Woods; HC = Henry Cowell; APT = Archer Taylor preserve; GD = Green Diamond Resource Company.
Discussion
The complex ecology of A. phagocytophilum in the far-western U.S. features multiple reservoir host species, multiple bacterial strains (Foley et al., 2009b; Rejmanek et al., 2011), and differences in dynamics of hosts and vectors across diverse landscapes (Foley et al., 2009a, 2011). There is also a large diversity of Ixodes tick species. Data in the present study revealed that, in addition to I. pacificus, at least 4 nidicolous tick species (I. angustus, I. ochotonae, I. spinipalpis, and I. woodi) may serve as vectors for A. phagocytophilum in California. Importantly, all of the nidicolous ticks in this study were infected with A. phagocytophilum strains characteristic of woodrat infections (Rejmanek et al., 2011). In contrast, all but 2 I. pacificus ticks were infected with a phylogenetically distinct strain previously detected in squirrel and chipmunk reservoir hosts, humans, dogs, and horses.
The detection of both human/sciurid strains and woodrat strains in ticks and small mammals in many of the same locations indicates the presence of at least 2 coexisting, yet discrete, enzootic cycles of A. phagocytophilum in California. A similar scenario has been reported by Bown et al. (2009) in the United Kingdom. In their system, a unique strain of A. phagocytophilum was detected only in field voles (Microtus agrestis) and their associated nidicolous tick, I. trianguliceps, whereas in the same habitats, a genetically divergent strain of A. phagocytophilum was detected exclusively in roe deer (Capreolus capreolus) and the questing tick I. ricinus. Interestingly, none of the I. ricinus ticks tested in their study were infected with the vole-associated A. phagocytophilum strain even though blood meal analysis showed that over half of the questing I. ricinus nymphs had fed on voles as larvae. This led the authors to conclude that I. ricinus was not functioning as a bridge vector of the vole-associated A. phagocytophilum strain to other animals including deer, livestock, and humans. By contrast, 2 out of 7 I. pacificus ticks tested in our study were infected with woodrat strains, demonstrating that I. pacificus in the western U.S. may function as a bridge vector of multiple A. phagocytophilum strains. And while at least one woodrat strain appeared to be minimally infectious to horses and likely humans as well (Foley et al., 2009b), I. pacificus is likely able to vector multiple strains of A. phagocytophilum in the wild, and some woodrat strains may have broader host tropism than DU1. Intriguingly, the one A. phagocytophilum-positive I. pacificus in this study that was found on a person was infected with a woodrat-like strain.
The phylogenetic tree indicates that the clade comprising woodrat strains is genetically much more variable than the human/sciurid clade, even though the strains comprising both clades come from ticks and animals sampled at numerous sites spanning a similarly large geographic range across central and northern California (approximately 500 km north to south). One possible explanation for the extensive genetic variability within the woodrat clade is that nidicolous ticks tend to be closely associated with their small mammal hosts and are therefore limited in their ability to disperse over long distances. This would allow geographically distant strains to accumulate mutations and diverge overtime. The observation of geographically clustered groups within the woodrat clade lends support to this hypothesis. In contrast, I. pacificus is a questing tick that often feeds on larger mammals such as deer, coyotes, and bobcats, which can carry the tick across much greater distances (Furman and Loomis, 1984). I. pacificus has also been found on numerous domestic and migratory bird species allowing even greater dispersal potential (Furman and Loomis, 1984; Wright et al., 2011). It is conceivable that an A. phagocytophilum strain readily vectored by I. pacificus could spread across a large geographical area in a short enough time span that only limited genetic variation would be detected among distantly sampled isolates. Interestingly, sequences from the 3 Midwestern human and dog-origin strains included in our analysis were almost identical to the other strains within the human/sciurid clade suggesting a fairly recent evolutionary history between the California and Midwestern strains. In contrast, sequences comprising the 4 East Coast human and tick-origin strains were markedly different from all the other strains tested.
While our data support the conclusion that there are at least 2 discrete and coexisting enzootic cycles of A. phagocytophilum in the western U.S., there are still many unanswered questions regarding the different strains involved in these cycles. For one, only a few woodrat strains (including DU1) have ever been experimentally tested in the laboratory (Nieto et al., 2010; Rejmanek et al., 2012). Given the considerable genetic diversity observed among these strains, it may be premature to assign similar phenotypic properties to all of them. Another important question is whether any of these strains can provide cross-immune protection against each other. Although experimental work in sheep suggests that superinfection with multiple A. phagocytophilum strains is possible, it has yet to be demonstrated in other species (Stuen et al., 2009). In the current study, we found no evidence of superinfection. In fact, our data suggest a high level of host tropism among reservoir hosts, that is, woodrat A. phagocytophilum strains were only detected in woodrats, while sciurid strains were only detected in sciurids. To some extent, host tropism was also evident among nidicolous ticks, which were only infected with woodrat A. phagocytophilum strains. This is interesting because except for I. woodi, which almost exclusively feeds on woodrats, the other nidicolous ticks targeted in this study have a fairly broad host range, which includes among other hosts, squirrels and chipmunks (Furman and Loomis, 1984; Foley et al., 2011). However, this finding should be interpreted cautiously since all of the A. phagocytophilum-positive nidicolous ticks except one, were removed from woodrats. The one nidicolous tick not removed from a woodrat (an adult I. ochotonae) was found on a deer mouse, a poor host for A. phagocytophilum in California (Foley et al., 2008; Rejmanek et al., 2011), indicating that the tick was most likely infected during a previous blood meal, presumably on a woodrat. Determining the extent of host tropism among the reservoir hosts and their associated nidicolous ticks in our system and testing whether different strains can provide cross-immune protection will require future laboratory work as well as additional testing of nidicolous ticks from other hosts.
In order to most effectively understand the ecology and genetic variability of A. phagocytophilum in the western U.S., we strategically focused on strains from multiple sources, including reservoir hosts, clinical hosts, and numerous tick vectors throughout central and northern California. Only by comparing strains from all of these different sources concurrently were we able to gain a clearer picture of how A. phagocytophilum strains are partitioned, which hosts and vectors are most likely to be infected with a particular strain, and which tick species and reservoir hosts pose the greatest health risk to humans and domestic animals. Furthermore, based on these findings, we would urge vector control agencies charged with the task of A. phagocytophilum detection to start genotyping PCR-positive ticks whenever possible in order to more accurately assess the potential risk to humans.
Acknowledgments
We acknowledge contributions to collection of samples from the field by Katryna Fleer, Mourad Gabriel, Greta Wengert, and Nate Nieto. Laboratory support was provided by Joy Worth, Jen Truong, and Joey Tse. We thank the following for logistical support: Pat Freeling, Scott Struckman, and Rene Pasquinelli at Hendy Woods State Park, Jay Harris at Humboldt Redwoods State Park, Thomas Sutfin and Edgar Orre at Soquel Demonstration Forest, Keith Hamm, Brendan Lynch, and Lowell Diller at Green Diamond Resource Company, and the staffs of Henry Cowell State Park and Samuel P. Taylor State Park. Funding was provided by the National Institutes of Health Allergy and Infectious Disease Evolution of Infectious Disease program #RO1 GM081714.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bown KJ, Begon M, Bennett M, Woldehiwet Z, Ogden NH. Seasonal dynamics of Anaplasma phagocytophila in a rodent-tick ( Ixodes trianguliceps) system, United Kingdom. Emerg. Infect. Dis. 2003;9:63–70. doi: 10.3201/eid0901.020169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bown KJ, Lambin X, Ogden NH, Begon M, Telford G, Woldehiwet Z, Birtles RJ. Delineating Anaplasma phagocytophilum ecotypes in coexisting, discrete enzootic cycles. Emerg. Infect. Dis. 2009;15:1948–1954. doi: 10.3201/eid1512.090178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente J, Massung RF, Wong SJ, Chu FK, Lutz H, Meli M, von Loewenich FD, Grzeszczuk A, Torina A, Caracappa S, Mangold AJ, Naranjo V, Stuen S, Kocan KM. Sequence analysis of the msp4 gene of Anaplasma phagocytophilum strains. J. Clin. Microbiol. 2005;43:1309–1317. doi: 10.1128/JCM.43.3.1309-1317.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drazenovich N, Foley J, Brown RN. Use of real-time quantitative PCR targeting the msp2 protein gene to identify cryptic Anaplasma phagocytophilum infections in wildlife and domestic animals. Vector Borne Zoonotic Dis. 2006;6:83–90. doi: 10.1089/vbz.2006.6.83. [DOI] [PubMed] [Google Scholar]
- Dumler JS, Barbet AF, Bekker CP, Dasch GA, Palmer GH, Ray SC, Rikihisa Y, Rurangirwa FR. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia descriptions of six new species combinations and designation of Ehrlichia equi and ‘HGE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. Evol. Microbiol. 2001;51:2145–2165. doi: 10.1099/00207713-51-6-2145. [DOI] [PubMed] [Google Scholar]
- Dunning Hotopp JC, Lin M, Madupu R, Crabtree J, Angiuoli SV, Eisen JA, Seshadri R, Ren Q, Wu M, Utterback TR, Smith S, Lewis M, Khouri H, Zhang C, Niu H, Lin Q, Ohashi N, Zhi N, Nelson W, Brinkac LM, Dodson RJ, Rosovitz MJ, Sundaram J, Daugherty SC, Davidsen T, Durkin AS, Gwinn M, Haft DH, Selengut JD, Sullivan SA, Zafar N, Zhou L, Benahmed F, Forberger H, Halpin R, Mulligan S, Robinson J, White O, Rikihisa Y, Tettelin H. Comparative genomics of emerging human ehrlichiosis agents. PLoS. Genet. 2006;2:e21. doi: 10.1371/journal.pgen.0020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley J, Rejmanek D, Fleer K, Nieto N. Nidicolous ticks of small mammals in Anaplasma phagocytophilum -enzootic sites in northern California. Ticks Tick Borne. Dis. 2011;2:75–80. doi: 10.1016/j.ttbdis.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley JE, Nieto NC, Foley P. Emergence of tick-borne granulocytic anaplasmosis associated with habitat type and forest change in northern California. Am. J. Trop. Med. Hyg. 2009a;81:1132–1140. doi: 10.4269/ajtmh.2009.09-0372. [DOI] [PubMed] [Google Scholar]
- Foley JE, Foley P, Brown RN, Lane RS, Dumlers JS, Madigan JE. Ecology of Anaplasma phagocytophilum and Borrelia burgdorferi in the western United States. J. Vector Ecol. 2004;29:41–50. [PubMed] [Google Scholar]
- Foley JE, Nieto NC, Massung R, Barbet A, Madigan J, Brown RN. Distinct ecologically relevant strains of Anaplasma phagocytophilum. Emerg. Infect. Dis. 2009b;15:842–843. doi: 10.3201/eid1505.081502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley JE, Nieto NC, Adjemian J, Dabritz H, Brown R. Anaplasma phagocytophilum infection in small mammal hosts of Ixodes ticks, Western United States. Emerg. Infect. Dis. 2008;14:1147–1150. doi: 10.3201/eid1407.071599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furman D, Loomis E. The Ticks of California (Acari: Ixodida) Bulletin of the California Insect Survey. 1984;25:247. [Google Scholar]
- Gorman JK, Hoar BR, Nieto NC, Foley JE. Evaluation of Anaplasma phagocytophilum infection in experimentally inoculated sheep and determination of Anaplasma spp seroprevalence in 8 free-ranging sheep flocks in California and Oregon. Am. J. Vet. Res. 2012;73:1029–1034. doi: 10.2460/ajvr.73.7.1029. [DOI] [PubMed] [Google Scholar]
- Holder MT, Sukumaran J, Lewis PO. A justification for reporting the majority-rule consensus tree in Bayesian phylogenetics. Syst. Biol. 2008;57:814–821. doi: 10.1080/10635150802422308. [DOI] [PubMed] [Google Scholar]
- Humair PF, Douet V, Moran Cadenas F, Schouls LM, Van De Pol I, Gern L. Molecular identification of bloodmeal source in Ixodes ricinus ticks using 12S rDNA as a genetic marker. J. Med. Entomol. 2007;44:869–880. doi: 10.1603/0022-2585(2007)44[869:miobsi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Massung RF, Priestley RA, Miller NJ, Mather TN, Levin ML. Inability of a variant strain of Anaplasma phagocytophilum to infect mice. J. Infect. Dis. 2003;188:1757–1763. doi: 10.1086/379725. [DOI] [PubMed] [Google Scholar]
- Massung RF, Levin ML, Munderloh UG, Silverman DJ, Lynch MJ, Gaywee JK, Kurtti TJ. Isolation and propagation of the Ap-Variant 1 strain of Anaplasma phagocytophilum in a tick cell line. J. Clin. Microbiol. 2007;45:2138–2143. doi: 10.1128/JCM.00478-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson WL, Castro MB, Kramer VL, Sumner JW, Childs JE. Dusky-footed woodrats ( Neotoma fuscipes) as reservoirs of granulocytic Ehrlichiae (Rickettsiales: Ehrlichieae) in northern California. J. Clin. Microbiol. 1999;37:3323–3327. doi: 10.1128/jcm.37.10.3323-3327.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto N, Foley J. Evaluation of squirrels (Rodentia: Sciuridae) as ecologically significant hosts for Anaplasma phagocytophilum in California. J. Med. Entomol. 2008;45:763–769. doi: 10.1603/0022-2585(2008)45[763:eosrsa]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Nieto NC, Foley JE. Reservoir competence of the redwood chipmunk ( Tamias ochrogenys) for Anaplasma phagocytophilum. Vector Borne Zoonotic Dis. 2009;9:573–577. doi: 10.1089/vbz.2008.0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto NC, Madigan JE, Foley JE. The dusky-footed woodrat ( Neotoma fuscipes) is susceptible to infection by Anaplasma phagocytophilum originating from woodrats, horses, and dogs. Journal of Wildlife Diseases. 2010;46:810–817. doi: 10.7589/0090-3558-46.3.810. [DOI] [PubMed] [Google Scholar]
- Pusterla N, Wolfensberger C, Lutz H, Braun U. [Serologic studies on the occurrence of bovine ehrlichiosis in the cantons Zurich, Schaffhausen, Thurgau, St. Gallen and Obwalden] Schweiz. Arch. Tierheilkd. 1997;139:543–549. [PubMed] [Google Scholar]
- Pusterla N, Pusterla JB, Braun U, Lutz H. Experimental cross-infections with Ehrlichia phagocytophila and human granulocytic ehrlichia-like agent in cows and horses. Vet. Rec. 1999;145:311–314. doi: 10.1136/vr.145.11.311. [DOI] [PubMed] [Google Scholar]
- Rejmanek D, Nieto NC, Barash N, Foley JE. Temporal patterns of tick-borne granulocytic anaplasmosis in California. Ticks Tick Borne Dis. 2011;2:81–87. doi: 10.1016/j.ttbdis.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Rejmanek D, Bradburd G, Foley J. Molecular characterization reveals distinct genospecies of Anaplasma phagocytophilum from diverse North American hosts. J. Med. Microbiol. 2011;61:204–212. doi: 10.1099/jmm.0.034702-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rejmanek D, Foley P, Barbet A, Foley J. Antigen variability in Anaplasma phagocytophilum during chronic infection of a reservoir host. Microbiology. 2012;158:2632–2641. doi: 10.1099/mic.0.059808-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Stuen S. Anaplasma phagocytophilum – the most widespread tick-borne infection in animals in Europe. Vet. Res. Comm. 2007;31(Suppl 1):79–84. doi: 10.1007/s11259-007-0071-y. [DOI] [PubMed] [Google Scholar]
- Stuen S, Torsteinbo WO, Bergström K, Bardsen K. Superinfection occurs in Anaplasma phagocytophilum infected sheep irrespective of infection phase and protection status. Acta. Vet. Scand. 2009;51:41. doi: 10.1186/1751-0147-51-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telford SR, 3rd, Dawson JE, Katavolos P, Warner CK, Kolbert CP, Persing DH. Perpetuation of the agent of human granulocytic ehrlichiosis in a deer tick-rodent cycle. PLOS USA. 1996;93:6209–6214. doi: 10.1073/pnas.93.12.6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SA, Tucker JR, Donohue AM, Castro MB, Kelley KL, Novak MG, Macedo PA. Avian hosts of Ixodes pacificus (Acari: Ixodidae) and the detection of Borrelia burgdorferi in larvae feeding on the Oregon junco. J. Med. Entomol. 2011;48:852–859. doi: 10.1603/me11001. [DOI] [PubMed] [Google Scholar]
- Zeidner NS, Burkot TR, Massung R, Nicholson WL, Dolan MC, Rutherford JS, Biggerstaff BJ, Maupin GO. Transmission of the agent of human granulocytic ehrlichiosis by Ixodes spinipalpis ticks: evidence of an enzootic cycle of dual infection with Borrelia burgdorferi in Northern Colorado. J. Infect. Dis. 2000;182:616–619. doi: 10.1086/315715. [DOI] [PubMed] [Google Scholar]