Abstract
Background
Esophageal adenocarcinoma (EA) is an increasingly common cancer with poor survival. Barrett’s esophagus (BE) is the main precursor to EA, and every year 0.12% to 0.5% of BE patients progress to EA. BE typically arises on a background of chronic gastroesophageal reflux (GERD), one of the risk factors for EA.
Methods
We used genome-wide association data to investigate the genetic architecture underlying GERD, BE, and EA. We applied a method to estimate the variance explained (array heritability, h2 g) and the genetic correlation (rg) between GERD, BE, and EA by considering all single nucleotide polymorphisms (SNPs) simultaneously. We also estimated the polygenic overlap between GERD, BE, and EA using a prediction approach. All tests were two-sided, except in the case of variance-explained estimation where one-sided tests were used.
Results
We estimated a statistically significant genetic variance explained for BE (h2 g = 35%; standard error [SE] = 6%; one-sided P = 1 × 10−9) and for EA (h2 g = 25 %; SE = 5%; one-sided P = 2 × 10−7). The genetic correlation between BE and EA was found to be high (rg = 1.0; SE = 0.37). We also estimated a statistically significant polygenic overlap between BE and EA (one-sided P = 1 × 10−6), which suggests, together with the high genetic correlation, that shared genes underlie the development of BE and EA. Conversely, no statistically significant results were obtained for GERD.
Conclusions
We have demonstrated that risk to BE and EA is influenced by many germline genetic variants of small effect and that shared polygenic effects contribute to risk of these two diseases.
Esophageal adenocarcinoma (EA) is a fatal cancer with a lifetime risk of approximately 0.25% (1,2) and a rapidly increasing incidence in much of the developed world. The mortality rate for EA is high, with less than 20% of patients surviving 5 years (3). The main precursor to EA is Barrett’s esophagus (BE), which is defined as a metaplastic change of the normal stratified squamous epithelium of the esophagus to columnar epithelium-containing goblet cells (4). Recent estimates suggest that 0.12% to 0.5 % of BE cases progress to EA per year (5–7), and clinical guidelines recommend surveillance for persons with BE, with the goal of identifying cancers at an early stage when they have a higher probability of cure (7). BE typically arises in patients with chronic gastroesophageal reflux (GERD), one of the risk factors for EA (8).
Observational studies indicate that a number of factors, including GERD, cigarette smoking, obesity, and Heliobacter pylori CagA seronegativity account for the majority of EA cases, estimated at 76% to 79% (9,10). However, there is emerging evidence for an important contribution of genetic factors as well to the etiology of EA and its precursors. Case reports, familial clusters, and clinical series all have suggested that variations in GERD, BE, and EA risk are underpinned at least in part by genetic factors (11,12). Some studies have suggested that the three conditions may share a genetic background, because risk is increased for GERD, BE, and EA when a relative is affected with any one of the three (13–15). Larger studies of EA and its precursors have also suggested a genetic underlying component (16,17), and previous twin studies estimated a moderate heritability of GERD (30%–40%) (18,19).
A recently published genome-wide association study (GWAS) of BE found common variants at the MHC locus and at chromosome 16q24.1 (20), although as in other complex diseases (21), the variants only explain a small proportion of the genetic variance. No GWASs on GERD and EA have been published to date. It is important to consider, however, that even if the individual effect size at any given single nucleotide polymorphism (SNP) is small, collectively these SNPs could still account for a substantial proportion of variation in risk (22,23). Therefore, studies of combined risk alleles throughout the genome might identify individuals at higher risk for disease (24).
One way to investigate the extent of genetic contribution to disease is to estimate the heritability (h2), usually defined as the proportion of total phenotypic variation that is due to additive genetic factors, from family history (pedigree data). With this traditional pedigree-based method, phenotypic similarity is related to expected allele sharing across the genome among family members. A novel way of measuring the genetic variance explained in “unrelated” individuals (h2 g), which is the case for most human population studies, is to use information from genetic markers as an alternative to “expected sharing” among family members (25). Estimating h2 g in unrelated individuals by including all SNPs with even a small association with disease can help us understand the genetic architecture behind disease. Yang et al. have shown that it is possible to estimate the realized genetic relationship between unrelated individuals (in the conventional sense) from dense SNP data (25).
To estimate the proportion of variation explained—h2 g by all markers in case–control studies for GERD, BE, and EA—we used a recently developed software program, Genome-wide Complex Trait Analysis (GCTA) (26). We also investigated whether GERD, BE, and EA share a similar genetic background using two methods. The first method estimates genetic correlations from unrelated individuals (rather than families), thereby avoiding confounding by shared environment. The second method uses a large number of autosomal SNPs associated with one trait to predict the risk of developing the other trait (22).
Methods
The datasets used are summarized in Table 1, and a detailed summary of the BE and EA data can be found in Supplementary Table 1 (available online).
Table 1.
Summary of datasets used for chronic gastroesophageal reflux (GERD) (Swedish Twin Registry + UK Twin Registry), Barrett’s esophagus (BE), and esophageal adenocarcinoma (EA)*
| Trait | Case subjects | Control subjects | SNPs | Array |
|---|---|---|---|---|
| GERD | ||||
| Sweden | 994 | 1342 | 643 277 | Illumina Omni Express |
| UK | 451 | 1007 | 551 049 | HumanHapmap 660 |
| GERD total | 1445 | 2349 | 547 971 | |
| BE | 2383 | 2167 | 797 518 | Omni1-Quad |
| EA | 1509 | 2170 | 797 518 | Omni1-Quad |
* A detailed summary of raw data is given in Supplementary Table 1 (available online). SNP = single nucleotide polymorphism.
For EA and BE case subjects (and associated control subjects), we used data and samples collected by investigators in the Barrett’s and Esophageal Adenocarcinoma Consortium (BEACON). We selected a subset of individuals with white-European ancestry from 14 epidemiological studies from three continents. After exclusions, 1509 EA case subjects, 2383 BE case subjects, and 2170 control subjects, typed for 797518 SNPs, were used for analysis.
GERD analyses used a subset of unrelated white-European individuals from two twin studies, the Screening across the Lifespan Twin Study in the nationwide Swedish Twin Registry (27) and the UK St Thomas Adult Twin Registry (28). The Swedish data were used for estimating the variance explained as well as the genetic correlation, and a combined dataset (Sweden + United Kingdom) was used for the polygenic overlap estimates. We only had summary data for the UK samples, and these could therefore only be used for the polygenic overlap analysis. Swedish samples were genotyped on the Illumina Omni Express (Illumina Inc, San Diego, CA). After exclusions, 994 GERD case subjects and 1342 control subjects, typed for 643 277 SNPs were included from this study. The UK samples were genotyped on the Illumina 660 array. After exclusions, 451 GERD cases and 1007 controls, typed for 551049 SNPs were included from this study. After exclusions, the combined GERD dataset (Sweden + United Kingdom) included 1445 GERD case subjects and 2349 control subjects, typed for 547971 SNPs.
Statistical Methods
Individuals were excluded to ensure that no pairs had an estimated genetic relationship greater than 0.025 (approximately a second-cousin relationship). Such individuals were excluded to avoid the possibility that the phenotypic resemblance between close relatives could be because of nongenetic effects (for example, shared environment). Estimates of variance explained by all SNPs can be biased by genotyping errors, and we therefore applied a stricter quality control than for typical GWAS analyses (details in Supplementary Methods, available online). Variance explained for the X chromosome was estimated separately from the autosomes, under both full dosage (correcting for one X chromosome turned off in females) and no dosage compensation models. Twenty eigenvectors (principal components) were calculated using GCTA (26) and included as covariables to capture variance due to population stratification. Sex was included as a covariable for all disease traits, and continent of sample origin (Europe, North America, and Australia) was included as a covariable for BE and EA.
We used GCTA to calculate one genetic relationship matrix for the autosomal chromosomes (1–22) and another for the X chromosome. All three diseases were coded as binary traits (case–control status). The estimated variance explained was transformed from the observed scale to the unobserved continuous “liability” scale using a probit transformation. The continuous scale is independent of the incidence of each category and can therefore be compared across traits or populations (26). Phenotypes were modeled as a linear function of the sum of the additive effects due to all SNPs associated with trait-associated variants and residual effects. Variance components were estimated by residual maximum likelihood (26). For tests for whether a variance component is zero or not, the test is one sided, and under the null hypothesis the test statistic follows a 50:50 mixture of a point mass at zero and the χ1 distribution (29). In our GCTA analysis, one-sided tests were performed for the significance of the autosomal and the sex chromosome–specific variance-explained estimates.
Case–control studies usually have a much larger proportion of case subjects than do general populations, and we therefore needed to correct for disease prevalence/lifetime risk. Prevalence is suitable for conditions that are not fatal, but for cancer, lifetime risk is a more suitable measure of how the case–control sets differed from a random sample of subjects. For lifetime risk estimates, we used 1.6% for BE (30), 0.25% for EA (1,2) and 18% for GERD (31). Because of the uncertainty around these estimates, sensitivity analyses were performed to assess the robustness of the estimates of variance explained (Supplementary Methods, available online).
Genetic correlation measures the proportion of genetic variance that two traits share because of genetic causes. Traditionally, genetic correlations are estimated between quantitative traits in pedigree studies, which can be confounded by shared environmental factors. For disease traits with low prevalence, even if the true genetic correlation is high between diseases, coaggregation of disorders in families might not occur or be distinguishable from chance. Here, we estimated the genetic correlation (rg) between all three traits using a bivariable mixed-effects linear model implemented in GCTA (32). The genetic correlation is the estimated additive genetic covariance between traits normalized by the geometric mean of the individual trait genetic variances (possible values of −1 to +1). The additive genetic covariance was estimated by relating trait covariances between unrelated individuals to genetic relationship estimates from marker data. That is, information comes from the covariance between individuals from different studies (eg, BE and EA). A higher covariance between traits with higher genetic relationship values implies a higher genetic correlation between traits. Case–control status was transformed to the underlying liability scale (33), as described previously for variance explained. To ensure no bias due to shared BE and EA control subjects in the per-disease analysis, control subjects were divided between BE and EA and matched with case subjects from the same continent.
Trait prevalence is higher in males than females for both BE (34) and EA (35). To investigate differences between sexes in variance of liability captured by SNPs, we also fit a bivariable model for sex where male case subjects and male control subjects were used as the first trait and female case subjects and female control subjects were used as the second trait. A high genetic correlation between the trait in males and the trait in females implies little or no sex-specific genetic effects.
We also investigated whether GERD, BE, and EA share a similar genetic background by using a large number of autosomal SNPs in one trait to predict the risk of developing the other trait (22). This method of prediction analysis has previously been used and published for other complex diseases such as schizophrenia and bipolar disorder (22) and endometriosis (36). Systematic evaluation of the overlap between the traits was based on profile scores, for which we estimated the relative risk for each SNP of interest based on a discovery set (eg, BE), with this profile and then tested in a target set of interest (eg, EA). Profiles were constructed using varying proportions of SNPs (top 10% P values, top 20%, and so on). Continent of sample origin, sex, and the first 20 principal components were used as covariables in the logistic regression for both the discovery set analysis and the target set analysis. Control subjects were divided between BE and EA and matched with case subjects from the same continent.
In the target sample logistic regression analysis, variance explained in disease status by the profile score was quantified using the difference in the Nagelkerke’s pseudo R 2 for the model including the score vs a model without the score (37). We refer to this variance explained as R 2.
Previous studies have shown that smoking, a high body mass index (BMI) and a large waist-to-hip ratio (WHR) are risk factors for GERD, BE, and EA (38–44). We therefore stratified BE and EA cases by smoking history (never smoked or smokers), BMI (lean/normal BMI ≤ 25 kg/m2, overweight BMI > 25 and ≤ 30 kg/m2, obese BMI > 30 kg/m2) and WHR (lean WHR ≤ 0.9 and obese WHR > 0.9). Males are over-represented in our data, and a WHR threshold for males was therefore used. We did not have the covariable data to allow us to stratify the GERD twin samples.
Results
We found that both BE and EA have a statistically significant polygenic component underlying disease risk. Autosomal h2 g estimates were 35% (standard error [SE] = 6%; one-sided P = 1 × 10−9) for BE and were 25% (SE = 5%; one-sided P = 2 × 10−7) for EA (Table 2). We also estimated a small but statistically significant h2 g for the X chromosome for EA (1.6%; SE = 1%; one-sided P = .04) (Table 3). Statistically significant results were not obtained for GERD, where wide 95% confidence intervals (0% to 42%; SE = 21%) only allowed us to exclude a heritable component greater than 42% (Table 2) in this dataset.
Table 2.
Estimates of proportion of variation due to common genetic variants for chronic gastroesophageal reflux (GERD), Barrett’s esophagus (BE), and esophageal adenocarcinoma (EA)*
| Phenotype | Case subjects | Control subjects | Variance explained h2 g (SE) | P |
|---|---|---|---|---|
| GERD† | 994 | 1342 | 0.0 (0.21)‡ | .50 |
| BE | 2255 | 1958 | 0.35 (0.06) | 1×10−9 |
| EA | 1397 | 1947 | 0.25 (0.05) | 2×10−7 |
* Proportion of variation and associated P values for the likelihood ratio test were estimated using a linear mixed model incorporating 797 518 single nucleotide polymorphisms for BE and EA and 643 277 single nucleotide polymorphisms for GERD from the genome-wide association studies panel after additional quality control. Case and control subject numbers are lower than for the original genome-wide association study because of stricter quality control (removal of related individuals and individuals with missing genotypes greater than 1%). SE = standard error.
† GERD included only Swedish data.
‡ The large standard error for GERD relative to, for instance, EA is because of the variance explained being scaled by a different life time risk (0.18 vs 0.0025). One from each pair of individuals with a relationship greater than 0.025 (between and within control and case subjects) were excluded, which is why the number of individuals for BE (removed n = 128), EA (removed n = 112), and controls (removed nBE = 209, nEA = 220) differ compared with Table 1.
Table 3.
Estimates of proportion of variation due to common genetic variants by the X chromosome for chronic gastroesophageal reflux (GERD), Barrett’s esophagus (BE), and esophageal adenocarcinoma (EA)*
| Phenotype | Case subjects | Control subjects | Proportion of variance (SE)† | |
|---|---|---|---|---|
| dc0 | dc1 | |||
| GERD‡ | 994 | 1342 | 0.0 (0.04) | 0.0 (0.02) |
| BE | 2255 | 1958 | 0.01 (0.01) | 0.007 (0.006) |
| EA | 1397 | 1947 | 0.016 (0.01) | 0.013 (0.008) |
* One from each pair of individuals with a relationship greater than 0.025 (between and within control and case subjects) were excluded, reducing the number of participants as follows: BE (removed n = 128), EA (removed n = 112) and controls (removed nBE = 209, nEA = 220). SE = standard error.
† For proportion of variance, dc1 is the estimate corrected for full dosage score and dc0 is not corrected. Bold results are statistically significantly different from zero.
‡ GERD included only Swedish data.
The bivariable analysis showed a strong genetic correlation between BE and EA (rg = 1.0; SE. = 0.37) (Table 4), which means that BE and EA share a large proportion of common genes. We also estimated a strong genetic correlation (rg = 0.96; SE = 0.52) between males and females for BE, suggesting that the majority of genes underlying risk in males also influence risk in females, even though male cases were over-represented (35,45) (Table 5). The male–female correlation was also close to one for EA, but with a high standard error because of limited sample size (Table 5). Again, these analyses did not provide any statistically significant results for GERD, with a predicted null genetic variance (Table 2) precluding a reliable estimate of genetic correlation between GERD and BE and GERD and EA (Table 4).
Table 4.
Bivariable analysis for gastroesophageal reflux (GERD), Barrett’s esophagus (BE), and esophageal adenocarcinoma (EA) datasets*
| Subset (1/2) | Cases (trait 1/2) | Controls (trait 1/2) | rg (SE) |
|---|---|---|---|
| BE/EA | 1986/1403 | 789/1175 | 1 (0.37) |
| BE/GERD† | 1982/934 | 787/1282 | NA‡ |
| EA/GERD† | 1400/928 | 1167/1283 | NA‡ |
* Results shown in bold are statistically significantly different from zero. One from each pair of individuals with a relationship greater than 0.025 (between and within controls and cases for both traits) were excluded (ie, within BE/EA (removed nBE = 397, nEA = 105), BE/GERD (removed nBE = 401, nGERD = 60) and EA/GERD (removed nEA = 109, nGERD = 66). rg = estimated genetic correlation; SE = standard error.
† GERD included only Swedish samples.
‡ Because the additive genetic variance was estimated to be zero for GERD, we could not estimate the genetic correlation for BE/GERD or EA/GERD.
Table 5.
Bivariable analysis of sex difference for gastroesophageal reflux (GERD), Barrett’s esophagus (BE), and esophageal adenocarcinoma (EA) datasets*
| Disease | Males, case/control | Females, case/control | h2 g (SE), males only | h2 g (SE), females only | rg (SE), males/females |
|---|---|---|---|---|---|
| GERD† | 429/584 | 565/758 | 0.17 (0.47) | 0.0 (0.36) | NA‡ |
| BE | 1495/1542 | 486/422 | 0.37 (0.08) | 0.30 (0.26) | 0.96 (0.52) |
| EA | 1239/1541 | 165/415 | 0.26 (0.06) | 0.30 (0.36) | 1 (0.85) |
* One from each pair of individuals with a relationship greater than 0.025 (between and within controls and case subjects in both sexes) were excluded, which is why the number of individuals for BE, EA, and controls differ compared with Table 1. h2g = estimated genetic variance explained; rg = estimated genetic correlation; SE = standard error.
† GERD included only Swedish samples.
‡ Because the additive genetic variance was estimated to be zero for GERD in females we could not estimate the genetic correlation for GERD males/females.
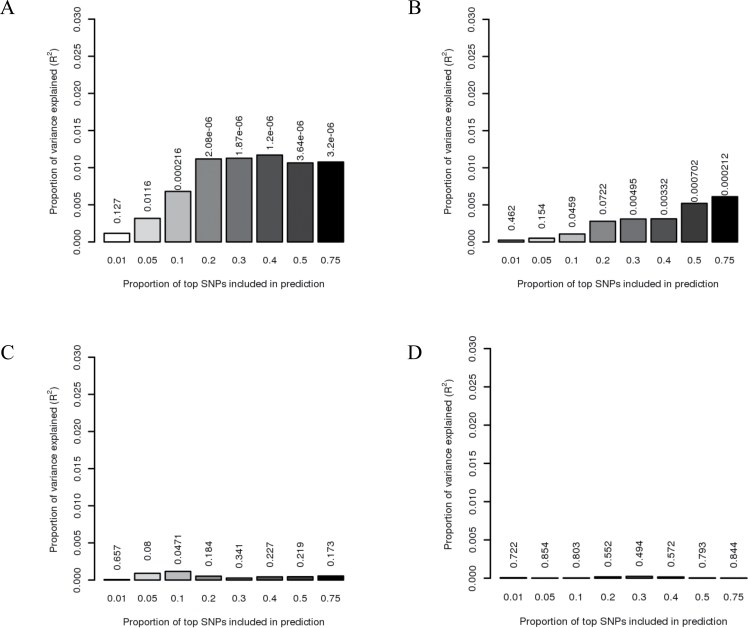
We estimated a statistically significant polygenic overlap between BE and EA (one-sided P = 1 × 10−6). The polygenic profile score estimated from a large number of SNP effects in BE (>20% of all SNPs) explains 1.2% of the variance in EA (Figure 1A). Predicting whether the reverse is true, where SNPs associated with EA are used to predict risk of BE, suggests a lower genetic overlap of approximately 0.5% (Figure 1B). This is primarily because estimates for each SNP from the association study when using EA as a discovery set were less accurate because the EA dataset is smaller. The prediction analysis showed no overlap between BE and GERD or EA and GERD (Figure 1, C and D). Although the statistical power decreased using smaller subsets of the data, we observed an increase in the polygenic overlap among cases with low BMI (≤25 kg/m2) when using EA to predict BE (2%) (Table 6).
Figure 1.
Allele specific score prediction. A) Barrett’s esophagus (BE) predicts esophageal adenocarcinoma (EA). BE as discovery set and EA as target set. B) EA predicts BE. EA as discovery set and BE as the target set. C) Gastroesophageal reflux (GERD) (Sweden + UK datasets) predicts BE. D) GERD (Sweden + UK datasets) predicts EA. C and D) GERD as discovery set where (C) uses BE as target set and (D) uses EA as target set. Variance explained in the target dataset on the basis of allele-specific scores derived in the discovery dataset for eight statistical significance thresholds (x-axis). The y-axis indicates variance explained, based on Nagelkerke’s pseudo R 2. The P value for the target dataset analysis is found above each bar. (A) and (B) show that the results are not driven by a few highly associated regions, indicating a substantial number of common variants underlying disease. SNP = single nucleotide polymorphism.
Table 6.
Polygenic overlap for smaller subsets, stratified for body mass index, waist-to-hip ratio, and smoking status*
| Prediction discovery/target | Subset | No. of cases target set | No. of controls target set | R 2 | P |
|---|---|---|---|---|---|
| BE/EA | 1498 | 1199 | 0.012 | 1×10−6 | |
| EA/BE | 2092 | 957 | 0.006 | 2×10−4 | |
| BE/EA | BMI1 | 215 | 1199 | 0.01 | 2×10−6 |
| EA/BE | BMI1 | 377 | 957 | 0.02 | 1×10−5 |
| BE/EA | BMI2 | 429 | 1199 | 0.017 | 7×10−6 |
| EA/BE | BMI2 | 728 | 957 | 0.012 | 6×10−5 |
| BE/EA | BMI3 | 262 | 1199 | 0.014 | 3×10−4 |
| EA/BE | BMI3 | 598 | 957 | 0.010 | 4×10−4 |
| BE/EA | WHR > 0.9 | 43 | 1199 | 0.005 | 0.14 |
| EA/BE | WHR > 0.9 | 321 | 957 | 0.007 | 9×10−3 |
| BE/EA | WHR ≤ 0.9 | 5 | 1199 | 0.014 | 0.35 |
| EA/BE | WHR ≤ 0.9 | 138 | 957 | 0.005 | 8×10−2 |
| BE/EA | Never smoked | 314 | 1199 | 0.013 | 3×10−4 |
| EA/BE | Never smoked | 652 | 957 | 0.010 | 7×10−4 |
| BE/EA | Smoked | 974 | 1199 | 0.009 | 8×10−4 |
| EA/BE | Smoked | 1306 | 957 | 0.0050 | 4×10–3 |
* Sex, 20 principal components, and continent of participant origin are included as covariaBles. Body mass index (BMI) is a proxy for estimating human body fat based on an individual’s weight and height (kg/m2). BMI1 includes individuals with a BMI less than or equal to 25kg/m2 (lean and normal people), BMI2 includes overweight individuals with a BMI between 25 and 30kg/m2, and BMI3 is obese people with a BMI greater than 30kg/m2. Waist-to-hip ratio (WHR) is the ratio of the circumference of the waist to that of the hips. A WHR greater than 0.9 defines abdominal obesity for males, and a WHR greater than 0.85 defines abdominal obesity for females. In this study, 0.9 was used as a threshold because of overrepresentation of males in the data. One from each pair of individuals with a relationship greater than 0.025 (between and within control and case subjects for both traits) were excluded, which is why the numbers of individuals differ compared with Table 1. BE = Barrett’s esophagus; EA = esophageal adenocarcinoma.
Discussion
Rates of EA have been rising rapidly during the past three decades in most Western countries, suggesting strong environmental influences on incidence. However, changes in trait means do not preclude there being substantial variance due to genetic factors [one simple quantitative trait example would be human height, where despite increases in mean height throughout the 20th century, the mean difference between countries remained stable and trait variance and heritability estimates are broadly similar across countries (46)]. A likely explanation, compatible with our data, is that susceptibility to BE/EA is not distributed in the population uniformly. Rather, some people have higher susceptibility than others because of their genetic load, which in the context of changing environmental conditions increases their absolute risk of cancer.
Traditional heritability estimates from twin studies differ from these analyses in two ways. First, in the case where expected sharing is used among (close) family members, all of the genome contributes to the estimate of h2. In contrast, estimating the genetic relationship between unrelated individuals uses information on only the portion of the genome tagged by SNPs on the array used. This makes h2 g a lower bound for h2 (33). Second, twin studies typically sample from the general population and hence provide heritability estimates for the condition up to the age at which the twins were ascertained (47). Twin studies generally, but not always, ascertain individuals who are aged less than 85 years. In contrast, we assumed that lifetime risk (up to age 85 years) was of primary interest for our h2 g calculations.
The method implemented in GCTA depends upon specification of prevalence of the trait in the population—essentially one has to parameterize how different the case–control sample would be from a randomly drawn sample from the general population. The general population has a lifetime risk of esophageal cancer approximating 0.5% (1,2,48,49). This estimate is inclusive of both major histological types of cancer of the esophagus: esophageal adenocarcinoma and esophageal squamous cell carcinoma, which differ in epidemiology, risk factors, and presumed pathogenesis. The adenocarcinoma subtype accounts for 50% or more of esophageal cancer cases in North America and Western Europe (1–3), whereas esophageal squamous cell carcinoma is the predominant form worldwide and accounts for 90% of all esophageal cancers (50,51). In this study, we used a lifetime risk estimate of 0.25% for esophageal adenocarcinoma, which could be an underestimate because our samples were obtained from different parts of the developed world; however, although estimates of variance explained require specification of the lifetime risk, they are relatively robust to mis-specification of this parameter (26).
The main aim of the polygenic overlap analysis is not to predict risk but to evaluate the influence of many common variants of small effects on risk of disease and to assess the overlap between diseases. A polygenic trait is defined as a trait influenced by many genes. The polygenic overlap prediction estimates of variance explained (R 2) are low because of each individual SNP being estimated with substantial error from the association study. These estimates are also constrained to be not more than the total genetic variance (heritability) for the trait. However, the estimated overlap is highly statistically significant for BE/EA and clearly shows that both traits have a polygenic architecture (because the variance explained continues to rise as one goes from the top 1% of SNPs to the top 20% of SNPs etc, implying that many SNPs contribute) as well as a shared genetic background. A new method with greater flexibility than GCTA in terms of modeling large effect loci (in addition to small effect loci under the infinitesimal model) was recently published (52). However, for this study, we assumed an infinitesimal model for our traits of interest (where there was no evidence for a strong departure from the infinitesimal model; eg, through there being variants of large effect). Figure 1A shows that the R 2 estimate decreases when adding more than 40% of the top SNPs. This is because of the inclusion of less statistically significant P values, which represent noise.
Our GERD dataset was not informative enough to allow us to reach a conclusion on the array heritability for GERD—the 95% confidence interval (0% to 42%) overlapped with the traditional heritability estimates from twin studies (30%–40%) (18,19). Array heritability may represent an underestimation of heritability as compared with that calculated from twin studies because the latter also takes into account rare variants that are usually absent from genome-wide genotyping arrays. In other diseases, array heritability has been reported to be between a quarter and half of the heritability estimated from twin studies (26). Our GERD study was less informative for estimating array heritability because, although the number of case subjects was similar to EA, the ascertainment was less extreme than EA (lifetime risk for EA = 0.0025 vs lifetime risk for reflux = 0.18). This is apparent in the standard error for GERD, which is approximately four times greater than the EA standard error, despite roughly similar sample sizes (0.21 and 0.05, respectively). There is also a limitation of identifying GERD using a questionnaire, compared with clinically identifying BE and EA patients.
Given, the null estimate of array heritability, we were unable to reliably estimate a genetic correlation between GERD and EA or BE. As an alternative to examining possible polygenic overlap between GERD and EA and BE, we applied a prediction approach. Although this prediction approach showed a clear relationship between EA and BE (P =1 × 10−6), there was no statistically significant prediction of GERD with EA or BE.
A longitudinal study for patients developing BE from GERD, EA from GERD, and EA from BE would be an ideal approach to estimating a genetic overlap between all three diseases. However, in the absence of this kind of data, we have shown that we can make useful inferences from the cross-sectional data collected here.
We have shown that the risks of BE and EA are influenced by genetic variants that are common in the population and that a large proportion of the genes associated with these two diseases are shared between them. These results emphasize the important role of genetic variants in BE and EA risk and suggest that future GWASs should consider combining these two disease types to create larger sample sizes that may identify further genetic associations.
Funding
This work was supported by the National Cancer Institute (R01CA136725 to DCW and TLV); funds from the Swedish Research Council (VR) to MD; the Swedish Ministry of Higher Education; the Swedish Research Council (M-2005-1112); GenomEUtwin (EU/QLRT-2001-01254; QLG2-CT-2002-01254); NIH DK (U01-066134); and the Swedish Foundation for Strategic Research (SSF), the Heart and Lung foundation (no. 20070481). The Kaiser Permanente Study was supported by a grant from US National Institutes of Health (RO1 DK63616). SM is supported by an Australian NHMRC Career Development Award. LB was supported by the US National Cancer Institute (CA59636). AHW was supported by the California Tobacco Related Research Program (3RT-0122 and 10RT-0251). The Romero Registry Consortium is supported in part by the American Digestive Health Foundation Endoscopic Research Award, the American College of Gastroenterology Junior Faculty Development Award, the Glaxo Wellcome Inc Institute for Digestive Health Clinical Research Award, and the Miles and Shirley Fiterman Center for Digestive Diseases at Mayo Clinic, Rochester, Minnesota. The Romero Registry also has charitable gifts from five industry partners (Affymetrix, AstraZeneca, Santarus, Takeda and Wyeth). AGC was supported in part by the Nova Scotia Research Foundation (N419); YRwas supported in part by a grant from the National Institutes of Health (NIDDK 02956), the Robert Wood Johnson Foundation Harold Amos Medical Faculty Development Program, and the Fraternal Order of the Eagles. The US Multicenter Study was funded by U01-CA57949 to TLV, U01-CA57983 to MDG, and U01- CA57923 to HAR.
DCW is supported by a Future Fellowship from the Australian Research Council. The Study of Digestive Health was funded by a grant from the US National Cancer Institute (RO1 CA 001833). The Australian Cancer Study was funded by a grant from the National Health & Medical Research Council of Australia (199600).
Supplementary Material
Data acquisition was approved by the institutional review board or research ethics committee of each participating institution providing data for the study. Permission to participate in the BEACON consortium was also provided by these boards if required by a study’s home institution.
References
- 1. Howlader N, Noone A, Krapcho M, et al. SEER Cancer Statistics Review, 1975–2009 (Vintage 2009 Populations). http://seer.cancer.gov/csr/1975_2009_pops09/ Accessed September 27, 2013 [Google Scholar]
- 2. Powell J, McConkey CC, Gillison EW, Spychal RT. Continuing rising trend in oesophageal adenocarcinoma. Int J Cancer. 2002;102(4):422–427 [DOI] [PubMed] [Google Scholar]
- 3. Holmes RJ, Vaughan TL. Epidemiology and pathogenesis of esophageal cancer. Semin Rad Oncol. 2007;17(1):2–9 [DOI] [PubMed] [Google Scholar]
- 4. Wong D, Paulson T, Prevo L, et al. p16INK4a Lesions are common, early abnormalities that undergo clonal expansion in barrett’s metaplastic epithelium. Cancer Res. 2001;61(22):8284–8289 [PubMed] [Google Scholar]
- 5. Maley C, Rustgi A. Barrett’s esophagus and its progression to adenocarcinoma. J Natl Compr Canc Netw. 2006;4(4):367–374 [DOI] [PubMed] [Google Scholar]
- 6. Hvid-Jensen F, Pedersen L, Drewes A, Sørensen H, Funch-Jensen P. Incidence of adenocarcinoma among patients with Barrett’s esophagus. N Engl J Med. 2011(365):1375–1383 [DOI] [PubMed] [Google Scholar]
- 7. American Gastroenterological Association. Spechler S, Sharma P, Souza R, Inadomi J, Shaheen N. American Gastroenterological Association medical position statement on the management of Barrett’s esophagus. Gastroenterology. 2011;140(3):1084–1091 [DOI] [PubMed] [Google Scholar]
- 8. Reid B, Li X, Galipeau P, Vaughan T. Barrett’s oesophagus and oesophageal adenocarcinoma: time for a new synthesis. Nat Rev Cancer. 2010;10(2):87–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Olsen C, Pandeya N, Green A, Webb P, Whiteman D, Australian Cancer Study Population attributable fractions of adenocarcinoma of the esophagus and gastroesophageal junction. Am J Epidemiol. 2011;1(174):582–590 [DOI] [PubMed] [Google Scholar]
- 10. Engel L, Chow W, Vaughan T, et al. Population attributable risks of esophageal and gastric cancers. J Natl Cancer Inst. 2003;95(18):1404–1413 [DOI] [PubMed] [Google Scholar]
- 11. Poynton A, Walsh T, O’Sullivan G, Hennessy T. Carcinoma arising in familial Barrett’s esophagus. Am J Gastroenterol. 1996;91(9):1855–1856 [PubMed] [Google Scholar]
- 12. Romero Y, Cameron A, Locke G, et al. Familial aggregation of gastroesophageal reflux in patients with Barrett’s esophagus and esophageal adenocarcinoma. Gastroenterol. 1997;113(5):1449–1456 [DOI] [PubMed] [Google Scholar]
- 13. Chak A, Lee T, Kinnard M, et al. Familial aggregation of Barrett’s oesophagus, oesophageal adenocarcinoma, and oesophagogastric junctional adenocarcinoma in Caucasian adults. Gut. 2002;51(3):323–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Munitiz V, Parrilla P, Ortiz A, Martinez-de-Haro L, Yelamos J, Molina J. High risk of malignancy in familial Barrett’s esophagus: presentation of one family. J Clin Gastroenterol. 2008;42(7):806–809 [DOI] [PubMed] [Google Scholar]
- 15. Groves C, Jankowski J, Barker F, Holdstock G. A family history of Barrett’s oesophagus: another risk factor? Scand J Gastroenterol. 2005;40(9):1127–1128 [DOI] [PubMed] [Google Scholar]
- 16. Chak A, Faulx A, Kinnard M, et al. Identification of Barrett’s esophagus in relatives by endoscopic screening. Am J Gastroenterol. 2004;99(11):2107–2114 [DOI] [PubMed] [Google Scholar]
- 17. Fitzgerald R. Complex diseases in gastroenterology and hepatology: GERD, Barrett’s, and esophageal adenocarcinoma. Clin Gastroenterol Hepatol. 2005;3(6):529–537 [DOI] [PubMed] [Google Scholar]
- 18. Mohammed I, Cherkas L, Riley S, Spector T, Trudgill N. Genetic influences in gastro-oesophageal reflux disease: a twin study. Gut. 2003;52(8):1085–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cameron A, Lagergren J, Henriksson C, Nyren O, Locke GR, 3rd, Pedersen NL. Gastroesophageal reflux disease in monozygotic and dizygotic twins. Gastroenterology. 2002;122(1):55–59 [DOI] [PubMed] [Google Scholar]
- 20. The Esophageal Adenocarcinoma Genetics Consortium, The Wellcome Trust Case Control Consortium. Su Z, et al. Common variants at the MHC locus and at chromosome 16q24.1 predispose to Barrett’s esophagus. Nat Genet. 2012;44(10):1131–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stranger B, Stahl E, Raj T. Progress and promise of genome-wide association studies for human complex trait genetics. Genetics 2011;187(2):367–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. International Schizophrenia Consortium. Purcell S, Wray N, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460(7256):748–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gibson G. Rare and common variants: twenty arguments. Nat Rev Genet. 2012;13(2):135–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wray N, Goddard M, Visscher P. Prediction of individual genetic risk to disease from genome-wide association studies. Genome Res. 2007;17(10):1520–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang J, Lee S, Goddard M, Visscher P. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88(1):76–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee S, Wray N, Goddard M, Visscher P. Estimating missing heritability for disease from genome-wide association studies. Am J Hum Genet. 2011;88(3):294–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lichtenstein P, Holm N, Verkasalo P, et al. Environmental and heritable factors in the causation of cancer—analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343(2):78–85 [DOI] [PubMed] [Google Scholar]
- 28. Spector TD, Williams F MK. The UK Adult Twin Registry (TwinUK). Twin Res Hum Genet. 2006;9(6):899–906 [DOI] [PubMed] [Google Scholar]
- 29. Self GS, Liang K-Y. Asymptotic properties of maximum likelihood estimators and likelihood ratio tests under nonstandard conditions. J Am stat Assoc. 1987;82(398):605–610 [Google Scholar]
- 30. Ronkainen J, Aro P, Storskrubb T, et al. Prevalence of Barrett’s esophagus in the general population: an endoscopic study. Gastroenterology. 2005;129(6):1825–1831 [DOI] [PubMed] [Google Scholar]
- 31. Dent J, El-Serag H, Wallander M, Johansson S. Epidemiology of gastro-oesophageal reflux disease: a systematic review. Gut. 2005;54(5):710–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee S, Yang J, Goddard M, Visscher P, Wray N. Estimation of pleiotropy between complex diseases using single-nucleotide polymorphism-derived genomic relationships and restricted maximum likelihood. Bioinformatics. 2012;28(19):2540–2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vattikuti S, Guo J, Chow C. Heritability and genetic correlations explained by common SNPs for metabolic syndrome traits. PLoS Genet. 2012;8(6)e1002637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Blankenstein MV, Looman C, Johnston B, Caygill C. Age and sex distribution of the prevalence of Barrett’s esophagus found in a primary referral endoscopy center. Am J Gastroenterol. 2005;100(3):568–576 [DOI] [PubMed] [Google Scholar]
- 35. Derakhshan M, Liptrot S, Paul J, Brown I, Morrison D, McColl K. Oesophageal and gastric intestinal-type adenocarcinomas show the same male predominance due to a 17 year delayed development in females. Gut. 2009;58(1):16–23 [DOI] [PubMed] [Google Scholar]
- 36. Painter J, Anderson C, Nyholt D, et al. Genome-wide association study identifies a locus at 7p15.2 associated with endometriosis. Nat Genet. 2011;43(1):51–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nagelkerke NJD. A note on a general definition of the coefficient of determination. Biometrika. 1991;78(3):691–692 [Google Scholar]
- 38. Corley D, Kubo A, Levin T, et al. Abdominal obesity and body mass index as risk factors for Barrett’s esophagus. Gastroenterology. 2007;133(1):34–41 [DOI] [PubMed] [Google Scholar]
- 39. Corley D, Kubo A, Zhao W. Abdominal obesity and the risk of esophageal and gastric cardia carcinomas. Cancer Epidemiol Biomarkers Prev. 2008;17(2):352–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cook M, Shaheen N, Anderson L, et al. Cigarette smoking increases risk of Barrett’s esophagus: an analysis of the Barrett’s and Esophageal Adenocarcinoma Consortium. Gastroenterology. 2012;142(4):744–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Edelstein Z, Farrow D, Bronner M, Rosen S, Vaughan T. Central adiposity and risk of Barrett’s esophagus. Gastroenterology. 2007;133(2):403–411 [DOI] [PubMed] [Google Scholar]
- 42. El-Serag H, Graham D, Satia J, Rabeneck L. Obesity is an independent risk factor for GERD symptoms and erosive esophagitis. Am J Gastroenterol. 2005;100(6):1243–1250 [DOI] [PubMed] [Google Scholar]
- 43. Nilsson M, Johnsen R, Ye W, Hveem K, Lagergren J. Lifestyle related risk factors in the aetiology of gastro-oesophageal reflux. Gut. 2004;53(12):1730–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wu P, Ma L, Dai G, et al. The association of metabolic syndrome with reflux esophagitis: a case–control study. Neurogastroenterol Motil. 2011;23(11):989–994 [DOI] [PubMed] [Google Scholar]
- 45. Kulig M, Nocon M, Vieth M, et al. Risk factors of gastroesophageal reflux disease: methodology and first epidemiological results of the ProGERD study. J Clin Epidemiol. 2004;57(6):580–589 [DOI] [PubMed] [Google Scholar]
- 46. Silventoinen K, Sammalisto S, Perols M, et al. Heritability of adult body height: a comparative study of twin cohorts in eight countries. Twin Res. 2003;6(5):399–408 [DOI] [PubMed] [Google Scholar]
- 47. So HC, Gui AH, Cherny SS, Sham PC. Evaluating the heritability explained by known susceptibility variants: a survey of ten complex diseases. Genet Epidemiol. 2011;35(5):310–317 [DOI] [PubMed] [Google Scholar]
- 48. Freedman ND, Abnet CC, Leitzmann MF, et al. A prospective study of tobacco, alcohol, and the risk of esophageal and gastric cancer subtypes. Am J Epidemiol. 2007;165(12):1424–1433 [DOI] [PubMed] [Google Scholar]
- 49. Brown LM, Devesa SS. Epidemiologic trends in esophageal and gastric cancer in the United States. Surg Oncol Clin N Am. Apr 2002;11(2):235–256 [DOI] [PubMed] [Google Scholar]
- 50. Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol. 2006;24(14):2137–2150 [DOI] [PubMed] [Google Scholar]
- 51. Corley D, Buffler P. Oesophageal and gastric cardia adenocarcinomas: analysis of regional variation using the Cancer Incidence in Five Continents database. Int J Epidemiol. 2001;30(6):1415–1425 [DOI] [PubMed] [Google Scholar]
- 52. Zhou X, Carbonetto P, Stephens M. Polygenic modeling with Bayesian sparse linear mixed models. PLos Gen. 2013;9(2):e1003264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.