Abstract
PURPOSE
New prognostic markers to guide treatment decisions in early stage non-small cell lung cancer are necessary to improve patient outcomes. In this report, we assess the utility of a pre-defined mRNA expression signature of cell cycle progression genes (CCP score) to define 5-year risk of lung cancer related death in patients with early stage lung adenocarcinoma.
EXPERIMENTAL DESIGN
A CCP score was calculated from the mRNA expression levels of 31 proliferation genes in stage I and II tumor samples from two public microarray data sets (Director’s Consortium (DC) and GSE31210). The same gene set was tested by quantitative PCR in 381 formalin-fixed paraffin-embedded (FFPE) primary tumors. Association of the CCP score with outcome was assessed by Cox proportional hazards analysis.
RESULTS
In univariate analysis the CCP score was a strong predictor of cancer-specific survival in both the DC cohort (p=0.00014, HR 2.08, 95%CI 1.43–3.02) and GSE31210 (p=0.0010, HR 2.25, 95%CI 1.42–3.56). In multivariate analysis the CCP score remained the dominant prognostic marker in the presence of clinical variables (p=0.0022, HR 2.02, 95%CI 1.29–3.17 in DC, p=0.0026, HR 2.16, 95%CI 1.32–3.53 in GSE31210). On a quantitative PCR platform the CCP score maintained highly significant prognostic value in FFPE derived mRNA from clinical samples in both univariate (p=0.00033, HR 2.10, 95%CI 1.39–3.17) and multivariate analyses (p=0.0071, HR 1.92, 95%CI 1.18–3.10).
CONCLUSIONS
The CCP score is a significant predictor of lung cancer death in early stage lung adenocarcinoma treated with surgery and may be a valuable tool in selecting patients for adjuvant treatment.
Keywords: non-small cell lung cancer, adenocarcinoma, prognosis, expression signature
Introduction
Despite the introduction of adjuvant chemotherapy, survival rates in early stage non-small cell lung cancer remain considerably lower than in early stages of any other major cancer type. Surgical resection is the standard treatment of patients with early stage NSCLC. Adjuvant chemotherapy has been recommended as standard of care for stage II and III patients based on results of several randomized studies and most recently summarized in the Lung Adjuvant Cisplatin Evaluation (LACE) meta-analysis of multiple cisplatin-based trials (1–3). In contrast, no significant treatment benefit could be demonstrated for patients with stage I disease (3,4) leaving surgery alone as the only curative treatment option. Nevertheless, five-year survival rates in stage I NSCLC patients are as low as 50% in stage IB and reach only 70% in IA patients (5,6), indicating a considerable high-risk subpopulation that could benefit from more aggressive therapy. Advances in surgical techniques, for example video-assisted thoracic surgery (VATS) are expected to increase the number of patients with improved performance status and the ability to undergo additional post-surgical treatment options (7–9). In addition, low-dose computed tomographic screening for high-risk populations will lead to increased numbers of patients with early stage diagnoses (10).
Many clinical and molecular factors have been evaluated as possible markers to improve staging and treatment decisions in stage I patients. This includes the use of pathological parameters, such as tumor size, to guide adjuvant treatment in stage IB (4). Histologic (11), immunohistochemistry (12) and genetic markers (13) have been suggested as possible factors for patient stratification. Microarray studies have produced a large number of tumor RNA expression signatures associated with survival and outcome in NSCLC (reviewed in 14,15). However, most of these expression profiles, developed from fresh frozen samples, have never been rigorously tested in formalin-fixed and paraffin-embedded (FFPE) tissue samples and their performance on clinical samples remains unknown. Recently, a 14-gene profile was developed on a quantitative PCR platform and tested in two large cohorts of non-squamous, resectable non-small cell lung cancer showing prognostic separation of low and high risk patients (16, 17). However, neither the 14-gene signature nor any other expression profile has gained sufficient acceptance to become standard of care.
In contrast, tumor RNA signatures have been highly successful as prognostic tools in breast cancer. A careful evaluation and comparison of prognostic breast RNA profiles revealed a common component of cell cycle regulated mRNAs which contains the major prognostic power of each expression profile (18–21). The expression levels of cell cycle progression (CCP) genes measure tumor growth irrespective of the underlying genetic aberrations. The prognostic power of proliferation in lung cancer was first shown in early studies with the mitotic index (22) as well as the examination of single gene expression data (23, 24, 25). More recently, proliferation has been the focus of one of fourteen methods applied to the microarray data in the Director’s Consortium (26). In addition, a proliferation signature derived from meta-analysis of breast cancer data was successfully applied to lung cancer (20) and the prognostic signature derived by Park et al (27) has a strong cell cycle related component. We therefore decided to test an independently developed signature of cell cycle genes in lung adenocarcinoma. The cell cycle proliferation (CCP) score was first applied to prostate cancer where it is a strong predictor of death from prostate cancer in biopsies (28) and of biochemical recurrence in post-prostatectomy samples (29). The CCP signature was established as a quantitative PCR assay to accommodate FFPE clinical specimens and implemented in a Clinical Laboratory Improvements Amendments (CLIA)-certified laboratory.
This study investigates the utility of the CCP score as a predictor of survival in patients with early stage lung adenocarcinoma. To focus on the group of patients with the most possible clinical utility, we selected stage I and II patients with the goal of identifying low-risk patients who may forego adjuvant treatment and high-risk patients with increased need for more aggressive treatment options. The CCP score was tested retrospectively in three large, independent patient sets for its ability to predict death from lung cancer.
Patients & Methods
Microarray data
Clinical and pathological characteristics of the different patient sets with microarray data tested are detailed in Table 1. A patient set of 442 resectable lung adenocarcinomas was collected at four US institutions through the Director’s Consortium (DC) for the development and validation of gene expression signatures in lung adenocarcinoma (26). Expression values, clinical and pathology data for the data set were retrieved from the NCI website https://caarraydb.nci.nih.gov/caarray/publicExperimentDetailAction.do?expId1/41015945236141280. Patient inclusion criteria, sample processing and clinical data collection have been described in detail (26). For the complete data set, pathological stage of each patient according to IASLC 6th edition criteria was determined from available TNM staging variables resulting in 371 stage I and II patients. From this subset 92 patients were excluded due to incomplete clinical data. 19 patients were omitted for unknown progression status, two patients for incomplete resection and a further two patients for death within 30 days from surgery. The remaining 256 patients were used for analysis (Supplementary Figure S1A).
Table 1.
Patient clinical and pathological characteristics
| DC N=256 |
GSE31210 N=204 |
IEO N=174 |
MDACC N=207 |
|
|---|---|---|---|---|
|
| ||||
| N (%) | N (%) | N (%) | N (%) | |
| Age at Diagnosis | ||||
| Median | 64 | 60 | 64 | 66 |
| SD | 10 | 8 | 8 | 11 |
| Sex | ||||
| Male | 111 (43) | 95 (47) | 122 (70) | 94 (45) |
| Female | 145 (57) | 109 (53) | 52 (30) | 113 (55) |
| Smoking Status | ||||
| Never | 34 (13) | 105 (51) | 26 (15) | 34 (16) |
| Former* | 200 (78) | 99* (49) | 47 (27) | 93 (45) |
| Current* | 22 (9) | 101 (58) | 80 (39) | |
| T stage | ||||
| T1a† | na | 64 (37) | 42 (20) | |
| T1b† | 90† (35) | na | 56 (32) | 32 (15) |
| T2a†† | na | 54 (31) | 105 (51) | |
| T2b†† | 157†† (61) | na | 0 (0) | 17 (8) |
| T3 | 9 (4) | na | 0 (0) | 11 (5) |
| N status | ||||
| N0 | 199 (78) | na | 174 (100) | 186 (90) |
| N1 | 57 (22) | na | 0 (0) | 21 (10) |
| Tumor Size < 3 cm | ||||
| Yes | na | na | 137 (78) | 103 (50) |
| No | na | na | 37 (21) | 104 (50) |
| Stage | ||||
| IA | 77 (30) | 190 (53) | 120 (69) | 64 (31) |
| IB | 113 (44) | 53 (26) | 54 (31) | 99 (48) |
| IIA# | 13 (5) | 0 (0) | 27 (13) | |
| IIB# | 53 (21) | 42# (21) | 0 (0) | 17 (8) |
| Adjuvant Treatment | ||||
| Yes | 77 (30) | 0 (0) | 19 (11) | 46 (22) |
| No | 179 (70) | 204 (100) | 155 (89) | 161 (78) |
| Pleural Invasion | ||||
| Yes | na | na | 24 (14) | 80 (39) |
| No | na | na | 150 (89) | 127 (61) |
| Recurrence < 5 years | ||||
| Yes | 111 (43) | 53 (26) | 36 (21) | 55 (27) |
| No | 136 (53) | 151 (74) | 138 (79) | 152 (73) |
| Disease related death at 5y | ||||
| Yes | 71 (28) | 25 (12) | 28 (16) | 34 (16) |
| No | 185 (72) | 179 (88) | 146 (84) | 173 (84) |
Refers to “ever” smokers.
Refers to 6th edition T1.
Refers to 6th edition T2.
Refers to stage II.
CEL files and patient data of an adenocarcinoma cohort collected at the University of Tokyo are available under GSE31210 from the GEO database. Patient selection, sampling and processing have been described previously (30). All patients in the GSE31210 set were chemo-naive. From the total data set of 226 stage I and II samples, 22 with incomplete resection were excluded, leaving 204 patients for analysis (Supplementary Figure S1A).
FFPE Sample Cohorts
Archived FFPE samples from surgically resected lung adenocarcinomas were obtained from 2 institutions: The University of Texas MD Anderson Cancer Center (MDACC) and European Institute of Oncology (IEO). Clinical and pathological characteristics of the different patient sets with FFPE tumors examined are detailed in Table 1. The MDACC patients had been ascertained between 1997 and 2008, were surgically treated and had complete resection. Initial patient selection criteria were based on staging according to IASLC 6th edition guidelines. Subsequently, tumors were re-staged using the current IASLC 7th edition criteria. The total cohort of 294 samples yielded 234 stage I and II lung adenocarcinoma patients. Fifteen samples were excluded for reasons related to clinical data: nine patients were excluded for missing data related to tumor size, adjuvant treatment and/or pleural invasion, three patients were excluded for having less than one month of follow-up, and three patients were excluded for having synchronous non-lung tumors. Of the remaining 219 samples, 207 produced CCP scores and were included in the analysis. The IEO provided 186 archived FFPE samples from 185 patients diagnosed with stage I lung adenocarcinoma ascertained between 1999 and 2005. All patients had complete surgical resection. Tumor staging followed the 7th edition of IASLC guidelines. One patient was excluded due to death 12 days after surgery and two patients were omitted for missing clinical variables. Of the remaining set, 174 samples produced expression scores and were included in the analysis. (Supplementary Figure S1B).
Sample Processing
Microarray data
For each data set, microarrays were subjected to robust multi-array analysis (RMA) for normalization of gene expression. CCP scores were calculated as the average RMA normalized signal of 31 cell cycle genes. Where multiple probes per gene were present, all probes per gene were averaged first, followed by averaging across 31 genes.
Formalin Fixed Samples
Clinical specimens were analyzed for the expression of 31 cell cycle and 15 house keeping genes by quantitative PCR according to a previously published protocol (29) summarized in Supplementary Materials. The performance of housekeeper genes is used to monitor and ensure adequate RNA quality. A list of genes can be found in Supplementary Tables S1 and S2. The success rate for obtaining CCP scores from formalin-fixed paraffin-embedded samples was 96% in the IEO cohort and 95% in the MDACC patient set.
Expression Signature
The CCP score is the un-weighted average of 31 cell cycle genes normalized by the average of 15 housekeeping genes. Each unit change is equivalent to a 2-fold change in expression levels. Details of the CCP score calculation as well as failure criteria for housekeeping genes and cell cycle genes were described previously (29) and have been summarized in the Supplementary Materials. Calculation for the lung cancer CCP score was identical except centering according to a set of 98 commercial lung adenocarcinomas (Proteogenex, Culver City, CA, USA, Asterand, Detroit, MI, USA).
Statistical Analysis
All analyses were carried out with the R (version 2.15.1; The R Foundation for Statistical Computing, Vienna, Austria, http://R-project.org).
Clinical Data
In each cohort, clinical data were required to include age of patient at surgery, gender, smoking status, TNM staging and adjuvant treatment status. Adjuvant treatment was defined as chemotherapy and/or radiation. FFPE cohorts had additional pathological data on tumor size and pleural invasion that were incorporated into modeling. To account for different effects of adjuvant treatment by stage, we included an interaction term for treatment and stage, where applicable. Details on modeling of clinical variables by cohort are provided in Supplementary Materials.
Survival Analysis
We evaluated the prognostic value of the CCP score using univariate and multivariate Cox proportional hazards models. The primary endpoint was disease related death within five years of surgery. Disease related death was defined as death following recurrence. Patients who were lost to follow-up or died of other causes were censored at the last observation.
Univariate p-values were based on the partial likelihood ratio. Multivariate p-values were based on the partial likelihood ratio for the change in deviance from a full model (which included all relevant covariates) versus a reduced model (which included all covariates except for the covariate being evaluated, and any interaction terms involving the covariate being evaluated). All reported p-values are two-sided. Hazard ratios for the CCP score are per interquartile range of the CCP distribution.
A proportional hazards violation related to processing site was detected in the DC cohort, and a proportional hazards violation related to stage was detected in the GSE31210 cohort. To address these violations, univariate and multivariate Cox proportional hazards models were stratified by processing site for the DC cohort and by stage for the GSE31210 cohort (Supplementary Materials).
Absolute treatment benefit
To assess the association of the CCP score with absolute benefit from adjuvant treatment, we employed the Zhang and Klein (31) method to construct confidence bands for the difference of two survival curves under the Cox proportional hazards model. Additionally, we constructed Cox proportional hazards models with contrast coding to test for increased benefit from adjuvant treatment among patients with high CCP scores relative to patients with low CCP scores. CCP scores were categorized as high or low using the median as the cutoff point (Supplementary Methods).
Results
Test of the CCP signature in lung adenocarcinoma microarray datasets
We tested the ability of the CCP score to predict five-year death from disease after resection with curative intent in public expression microarray data. Since the CCP score had been defined previously, no further gene selection or training in lung cancer was necessary. 256 stage I-II lung adenocarcinoma patients with complete clinical data from the DC cohort (26) and 204 stage I-II patients from a Japanese study on lung adenocarcinoma (30) were analyzed independently. In both cohorts patients had undergone complete resections and RNA from frozen surgery specimens had been subjected to gene expression analysis on Affymetrix expression microarrays. Twenty-eight percent of the DC cohort had died at 5 years, providing 71 events for analysis. Within five years from surgery 25 deaths were observed in the GSE31210.
In univariate Cox proportional hazards analysis, the CCP score was significantly predictive of five-year outcome in both the DC cohort (p=0.00014) and the Japanese patient set (p=0.0010). The hazard ratio per interquartile range of the CCP score was 2.08 (95% CI 1.43–3.02) in the DC cohort and 2.25 (95% CI 1.42–3.56) in the Japanese data set. As expected, stage was highly prognostic (p<0.0001) in the DC patients. As shown in Table 2, among clinical variables, only adjuvant treatment in the DC cohort was significantly associated with outcome. However, adjuvant treatment did not reduce the risk of death (HR 2.78, 95% CI (1.69–4.58)). In the Japanese cohort, EGFR and KRAS mutation and EML4-ALK translocation status were available. Neither of the mutation classes nor a combination of mutations contributed prognostic information in this cohort.
Table 2.
The CCP score is an independent predictor of lung cancer related death in stage I and II lung adenocarcinoma
| Director’s Consortium (DC)* Events/N: 71/256 | GSE31210** Events/N: 25/204 | |||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Univariate | Multivariate | Univariate | Multivariate | |||||
|
| ||||||||
| HR (95%CI) | P | HR (95%CI) | P | HR (95%NCI) | P | HR (95%NCI) | P | |
| CCP# | 2.08 (1.43–3.02) | 0.00014 | 2.02 (1.29–3.17) | 0.0022 | 2.25 (1.42–3.56) | 0.0010 | 2.16(1.32–3.53) | 0.0026 |
| Age | 1.02 (0.99–1.04) | 0.23 | 1.03 (1.01–1.06) | 0.01 | 1.03 (0.97–1.09) | 0.28 | 1.05 (0.99–1.11) | 0.11 |
| Gender | 0.83 | 0.81 | 0.08 | 0.68 | ||||
| Male | 1 | 1 | 1 | 1 | ||||
| Female | 1.05 (0.66–1.69) | 0.94 (0.56–1.56) | 0.49 (0.21–1.11) | 0.79 (0.26–2.45) | ||||
| Smoking | 0.81 | 0.97 | 0.04 | 0.46 | ||||
| Never | 1 | 1 | 1 | 1 | ||||
| Ever | 1.09 (0.54–2.2) | 1.02 (0.47–2.18) | 2.37 (1.01–5.55) | 1.56 (0.48–5.09) | ||||
| Stage## | <0.0001 | 0.02 | -** | ** | ||||
| IA | 1 | 1 | ||||||
| IB | 2.98 (1.27–7.01) | 1.89 (0.71–5.04) | ||||||
| IIA | 4.63 (1.43–14.93) | 4.51 (0.88–23.14) | ||||||
| IIB | 7.50 (3.14–17.94) | 5.71 (2.00–16.29) | ||||||
| Adj.Treatment§ | 2.78 (1.69–4.58) | <0.0001 | 2.66 (0.32–22.43) | 0.14 | - | - | ||
| Stage: Adj.T. | 0.73 | |||||||
Results from univariate and multivariate Cox proportional hazards analysis
Analysis is stratified by site (see supplementary methods).
Analysis is stratified by stage (see supplementary methods).
Hazard ratio is reported per interquartile range of the CCP score.
Hazard ratio for untreated patients
Hazard ratio for stage IA
The prognostic utility of the CCP score after adjustment for clinical parameters was evaluated in multivariate Cox proportional hazards regression. The CCP score remained the most significant predictor of 5-year disease survival in both microarray data sets (p=0.0022 for the DC cohort, p=0.0026 for GSE31210) with hazard ratios per interquartile range of 2.02 (95% CI 1.29–3.17) in the DC data and 2.16 (95% CI 1.32–3.53) in GSE31210. Results from univariate and multivariate Cox proportional hazards analysis are summarized in Table 2.
We tested for an interaction between the CCP score and any of the clinical variables by introducing an interaction term into the model. None of these interaction terms reached significance at the 5% level. Scaled Schoenfeld residuals versus untransformed time were used to evaluate the appropriateness of the proportional hazards assumption. No evidence was found supporting time dependence for the hazard ratio of the CCP score. To evaluate the possibility that CCP score might have a non-linear effect, second- and third-order polynomials for CCP score were tested in Cox proportional hazards models but were not significant at the 5% level.
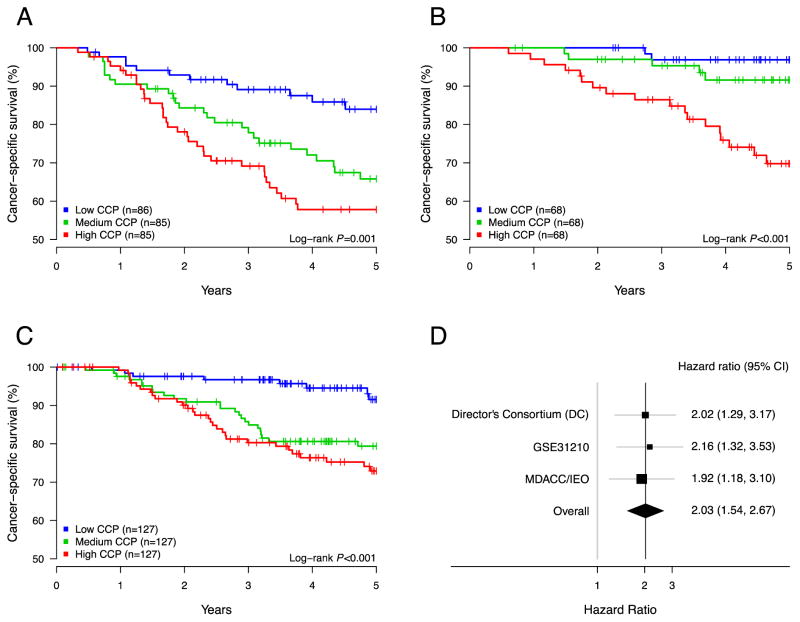
Kaplan-Meier curves visualize the separation of low and high risk patients according to CCP score (Figure 1A–B). For illustration purposes, the patient sets were divided into equally sized groups based on terciles of the CCP score. For these low, intermediate and high CCP score patient groups five-year survival rates were 84%, 68% and 56%, respectively, in the DC cohort and 97%, 92% and 70% in the Japanese data set. Thus, the lowest tercile of CCP score consistently identifies a low risk subgroup of early stage adenocarcinoma.
Figure 1.
Association between CCP scores and lung cancer death in several cohorts: Director’s consortium (A), GSE31210 (B) and MDACC/IEO (C). Each patient set was separated into three equally sized groups based on CCP scores. The lowest tercile of CCP scores defines a subpopulation with higher survival. (D) Forest plot of hazard ratios for the interquartile CCP range observed in the three study sets.
Prognostic utility of the CCP score in formalin-fixed samples
To validate the prognostic use of the CCP score in FFPE specimens, surgically resected tumors were obtained from MDACC and IEO (Table 1). The MDACC cohort comprised 204 stage I and II specimens with a median follow-up time for patients alive at the date of last follow-up of 132 months. The IEO cohort consisted of 174 stage I patients with a median follow-up time for patients alive at the date of last follow-up of 80 months. At five years from surgery, 34 (16%) of patients in the MDACC cohort had died of disease and 28 (16%) deaths had occurred in the IEO patient set. A statistical comparison of the two cohorts found no significant differences in the distribution of pathological or clinical parameters (Supplementary Methods). Thus, to improve statistical power, the cohorts were combined for survival analysis.
Each unit of CCP score represents a two-fold change in mRNA expression. The median CCP score was 0.020, and the interquartile range extended from −0.87 to 0.91. A significant variation in CCP scores was observed in all stages, in particular stage IA and IB, with only a minor shift towards higher CCP scores in stage II.
Results from univariate and multivariate Cox proportional hazards analysis are summarized in Table 3. In univariate analysis, the CCP score was the most significant predictor of outcome (p=0.00033). An increase in CCP score resulted in increased risk of dying from lung cancer with a hazard ratio of 2.10 (95% CI 1.39–3.17). Several clinical parameters also were prognostic, including stage (p=0.0043), gender (p=0.0019), tumor size (p=0.0069), pleural invasion (p=0.011) and age (p=0.031). When added to clinical variables in multivariate analysis, the CCP score remained the dominant prognostic factor (p=0.0071) with a hazard ratio of 1.92 (95% CI 1.18–3.10). Among the clinical factors, only pleural invasion (p=0.0090) and gender (p=0.012) retained significance in multivariate analysis. None of the clinical variables showed a significant interaction with the CCP score. No time dependence for the hazard ratio of the CCP score or non-linear effects of the CCP score was observed.
Table 3.
The CCP score predicts outcome in stage I and II FFPE samples
| MDACC/IEO Events/N: 62/381
| ||||
|---|---|---|---|---|
| Univariate | Multivariate | |||
|
| ||||
| HR (95%CI) | P | HR (95%CI) | P | |
| CCP* | 2.10 (1.39–3.17) | 0.00033 | 1.92 (1.18–3.10) | 0.0071 |
| Age | 1.03 (1.00–1.06) | 0.031 | 1.02 (0.99–1.05) | 0.12 |
| Gender | 0.0019 | 0.012 | ||
| Male | 1 | 1 | ||
| Female | 0.43 (0.24–0.75) | 0.46 (0.24–0.86) | ||
| Smoking | 0.32 | 0.99 | ||
| Never | 1 | 1 | ||
| Former | 1.89 (0.78–4.58) | 1.05 (0.40–2.72) | ||
| Current | 1.73 (0.72–4.16) | 1.06 (0.39–2.89) | ||
| Stage** | 0.0043 | 0.15 | ||
| IA | 1 | 1 | ||
| IB | 1.82 (1.03–3.22) | 1.03 (0.44–2.45) | ||
| IIA | 2.97 (1.26–7.05) | 1.41 (0.39–5.13) | ||
| IIB | 4.91 (1.97–12.28) | 2.02 (0.48–8.43) | ||
| Adjuvant T§ | 0.79 (0.38–1.66) | 0.52 | 2.38 (0.79–7.19) | 0.13 |
| Tumor Size# | 1.25 (1.07–1.44) | 0.0069 | 1.10 (0.89–1.37) | 0.39 |
| Pleural Invasion | 2.00 (1.2–3.33) | 0.01 | 2.65 (1.27–5.52) | 0.0090 |
| Cohort | 1.22 (0.74–2.02) | 0.43 | 1.18 (0.62–2.26) | 0.61 |
| Stage: Treatment | - | 0.088 | ||
Results from univariate and multivariate Cox proportional hazards analysis
Hazard ratio is reported per interquartile range of the CCP score
Hazard ratio for untreated patients
Hazard ratio is reported per cm, rounded to the nearest mm.
Hazard ratio for stage I
To illustrate the results, a Kaplan-Meier curve using the same terciles of the CCP score as used for the analysis of the microarray data is presented in Figure 1C. Five-year lung cancer specific survival for the low CCP patient group was 92%, decreasing to 79% and 73% in the intermediate and high CCP patients. Heterogeneity analysis between the three data sets (DC, GSE31210 and MDACC/IEO) did not identify any significant cohort differences. This is supported by the consistent hazard ratios for the CCP score observed the three data sets (Figure 1D).
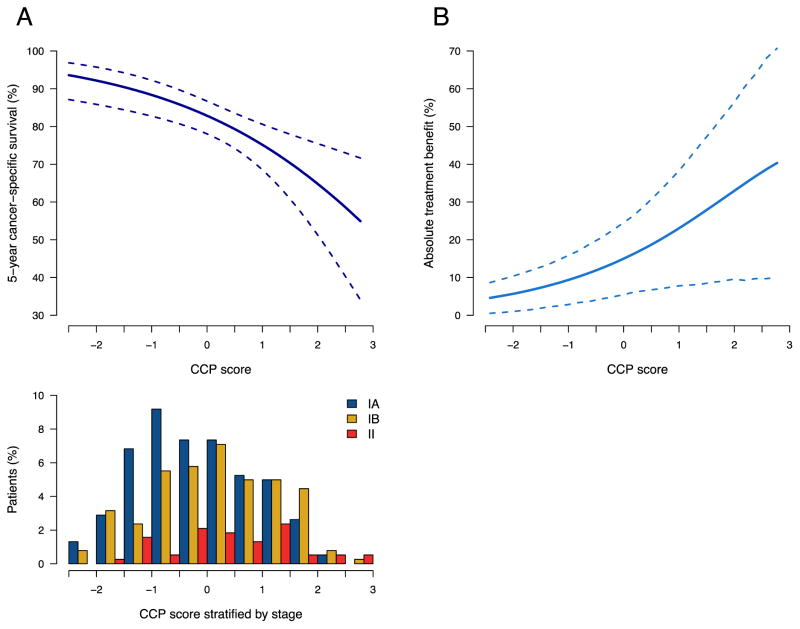
Low CCP scores are clearly associated with improved outcome and the predicted probability of five-year death from lung cancer increases as a function of the CCP score. Figure 2A shows the relationship between the risk of lung cancer related death and the CCP score measured by quantitative PCR in the FFPE cohort. The CCP score provides additional prognostic information within each stage group. This is visualized in Figure 2A by the wide distribution of CCP scores within each stage and supported by the hazard ratio in multivariate Cox proportional hazards regression.
Figure 2.
(A) Five year predicted risk of lung cancer related death as a function of CCP score analyzed by quantitative PCR in FFPE samples (top panel) and distribution of CCP scores within stage IA, IB and II patient samples (bottom panel).
(B) Absolute benefit from adjuvant treatment depending on CCP score. Association of the expression score with treatment benefit was derived from the difference in survival ratios between the treated and untreated patients in the MDACC cohort.
Risk stratification in stage I patients
To verify the utility of the CCP score in patients with stage IA and IB tumors, we conducted a sub-analysis in stage I samples. This subset consisted of 163 patients from the MDACC set and 174 patients from the IEO cohort. Each patient set was divided into two equally sized groups based on the median CCP score. A statistically significant separation was observed in Kaplan-Meier survival curves of five-year lung cancer related survival between the low and high CCP groups (Supplemental Figure S2). Cancer-specific survival rates at five years were 90% for patients with CCP scores below the median and 77% for patients with scores higher than the median CCP. The effect was similar in the MDACC cohort (low CCP 90%, high CCP 79%) and the IEO patients (low CCP 90%, high CCP 75%).
Prediction of adjuvant treatment benefit
To explore the possibility of using the CCP score as a predictive tool, we examined the relationship between CCP score and absolute benefit from adjuvant treatment in the MDACC cohort. The 207 patients included 46 patients who had received adjuvant therapy. Most importantly, the treated patient set from the MDACC cohort showed significant improvement (p=0.030, HR=0.32) in 5 year survival (Kaplan-Meier estimate 92.25%, 95% CI 77.70%–97.46%) compared to patients not receiving adjuvant treatment (Kaplan-Meier estimate 77.56%, 95% CI 69.46%–83.76%). The number of treated patients in the IEO data set was insufficient to provide statistical power and adjuvant therapy provided no measurable survival benefit. Therefore, association with treatment benefit was restricted to the MDACC cohort. To evaluate the relationship between CCP score and absolute treatment benefit, we categorized CCP scores as being high or low using the median CCP value as the cutoff point. Cox proportional hazards analysis with contrast coding indicated significantly greater absolute treatment benefit for patients with high CCP scores compared to patients with low CCP scores (p=0.0060) (Figure 2B). The association between CCP score and absolute treatment benefit maintained significance after adjusting for clinical variables (p=0.024).
Discussion
A major goal of biomarker development is directed at improving patient-specific treatment decisions. Tools for clinicians to utilize to help make final determination of treatment, especially in “cured” patients have been utilized in diseases such as breast cancer (Oncotype DX or Mammaprint) (32,33). Treatment of early stage lung cancer, specifically stage I, has been challenging for physicians as these patients are “cured” and have not demonstrated significant benefit from adjuvant cytotoxic chemotherapy, which carries sometimes significant toxicity. In this paper, we tested a previously developed and validated 31-gene expression signature in three independent data sets as a prognostic tool in early stage lung adenocarcinoma, Multiple aspects distinguish this study from previously published lung signatures: i) the testing of a single, predefined gene profile, ii) the use of cohorts with adequate statistical power, iii) inclusion of all relevant and available clinical variables in the survival analysis, iv) the presentation of solid evidence for the utility of the expression score in addition to standard clinical parameters and v) the validation of the expression score on a quantitative PCR-based platform suitable for the analysis of routine clinical FFPE specimens.
The data presented here establish the CCP score as a reliable prognostic marker in early stage lung adenocarcinoma. The CCP score effectively identified patients groups of reduced or increased risk of death after surgical resection and added significant prognostic information not contained in clinical variables. Importantly, none of the data sets described in this study had been used in the development of the gene profile. The composition of the signature centers on a characteristic trait of tumor cells. In contrast, other prognostic profiles aggregate the highest ranked genes in survival analysis from a single training data set into a prognostic score. Inevitably, these lists contain genes that appear correlated with outcome due to statistical noise rather than true biological association. Not surprisingly, many of these signatures lack robustness when applied to additional data sets. In contrast, the CCP signature was previously shown to be a superior prognostic tool in prostate cancer (28,29).
Expression of CCP genes is elevated in dividing cells and the CCP score likely reflects the proportion of actively dividing cells in the tumor. The CCP score thus provides a measure of the inherent aggressiveness of the tumor which is related to outcome. Proliferation associated genes have been correlated with outcome in lung cancer by individual gene expression (23, 24, 25), or immunohistochemistry (22,34). However, single genes lack robustness as expression markers in clinical specimens where nucleic acid quality is compromised by formalin-fixation and may lead to the loss of individual signals. We addressed this issue by creating a signature that uses redundancy to minimize technical variability. In addition, signal averaging reduces biological variability from copy number variations.
Outcomes after resection in stage I and II NSCLC remain considerably worse compared to other solid tumor types (35). Multiple randomized trials have shown limited improvement in survival from adding adjuvant platinum-containing systemic therapy after surgical treatment and benefit of adjuvant therapy is largely restricted to stage II and IIIA patients (3). No improvement in survival could be shown for stage I patients in a large meta-analysis of five trials (3), nor in a randomized controlled trial of stage IB disease treated with adjuvant paclitaxel and carboplatin (4). Thus, clinical TNM staging remains the main determinant of post-surgery treatment (eg chemotherapy) for early stage non-small cell lung cancer and stage I patients remain largely untreated despite five-year survival rates of less that 50% (1,2). Most of these deaths are likely due to occult disease at the time of surgery and those cases would be expected to benefit from systemic therapy. Heterogeneity of patients for whom surgery was curative likely contributes to the failure to show a treatment benefit in stage I patients. This highlights the need for prognostic markers to identify patients with an increased risk of death not captured by current clinical evaluation tools. The development of such markers has been the goal of many microarray studies in the lung cancer field and several signatures have been described as prognostic tools in early stage NSCLC. As highlighted in a recent review (14), many of these signatures are not independently validated, failed to show independence from clinical variables or were not established on platforms suitable for clinical use. A notable exception is the recently published Lung RS score (16), which uses quantitative PCR to examine the expression levels of 14 genes in formalin fixed tissue. The score was shown to predict overall survival in non-squamous, non-small cell lung cancer, separating patients into groups with low, intermediate and high risk of five-year mortality in cohorts of mixed stages and in a sub-analysis of tumors <2cm (17). The Lung RS study supports the idea that expression signatures can be established for use in formalin fixed lung tissue and that they may have similar importance in modifying treatment decisions in lung adenocarcinoma as pioneered in estrogen receptor positive breast cancer. The present study distinguishes itself from the 14 gene score by focusing on a gene set that is directly related to a well-established feature of tumor cells. In addition, the choice of outcome measure is more relevant to the intended clinical application. For the purpose of informing treatment decisions in resectable lung cancer, association with overall survival is less informative than prediction of lung-cancer specific survival. In a patient population that is often of advanced age and frequently burdened with co-morbidities, potential treatment benefits need to be weighed against therapy side effects in the context of both non-cancer related and cancer-specific mortality.
Adjuvant chemotherapy in NSCLC has limited effectiveness and the estimated absolute benefit in a large meta-analysis was a rather modest at approximately 5% (1,3). Therefore, it is desirable not only to identify patients at high risk of lung cancer-related death, but also those patients with the greatest likelihood of therapeutic benefit. Patients not likely to benefit could be directed to clinical trials or avoid the treatment and its toxicities altogether. Showing the association of a biomarker with treatment benefit requires a cohort with one treated and one untreated arm and a measurable treatment benefit between both arms. In many cohorts treatment fails to improve outcome. For example, in the DC data set, those patients treated fare worse than untreated patients (14). The data set from MD Anderson Cancer Center included patients who had received adjuvant therapy and, more importantly, the treated patients showed a significant treatment benefit. We observed a statistically significant association between treatment benefit and CCP score. This observation requires further confirmation in other data sets, but it suggests that the CCP score may provide predictive information about therapy benefit in addition to its prognostic utility. We recognize that for clinical purposes a risk threshold that guides treatment decisions is useful. However, in this first study in lung adenocarcinoma, we avoided setting predefined thresholds since treatment decisions in resectable NSCLC are subject to other important considerations, most notably stage, but also age, performance status and preexisting conditions. All of these factors influence the balance between risk of cancer related death, risk of non-cancer related death and expected therapy benefit. A clinically meaningful risk threshold for recommending treatment should be drawn when these factors are appropriately integrated. We will address this issue with a combined score in further prospectively collected studies
In summary, our study shows convincingly that the CCP score is a significant prognostic marker in lung adenocarcinoma that stratifies early stage patients more effectively than conventional clinical parameters. Its CLIA-certified quantitative PCR platform is compatible with clinical practice patterns and may help differentiate patient populations with early stage disease to improve treatment selection.
Supplementary Material
Statement of Translational Relevance.
Adjuvant treatment recommendations for early stage non-small cell lung cancer are based only on pathological staging. Patients with stage I disease generally do not receive adjuvant chemotherapy despite frequent recurrences and those that do are based arbitrarily on tumor size. Conversely, all stage II cases are treated, yet few patients receive significant benefit. Hence, additional information from biomarkers could improve treatment decisions. In this report, we employ a previously described expression signature and show its utility in defining lung adenocarcinoma patient subsets with low and high risk of death from lung cancer. This molecular marker has been validated on a platform suitable for the analysis of formalin-fixed and paraffin-embedded tumor specimens and may lead to more refined treatment selection algorithms in patients with early stage lung adenocarcinoma.
Acknowledgments
Myriad Genetics paid the costs of shipping the paraffin-embedded samples and performing all qPCR assays. In addition, this study was supported in part by the UT Lung Specialized Programs of Research Excellence grant (P50CA70907; to I.I.W.), and the MD Anderson Cancer Center Support Grant CA016672.
Footnotes
Author contributions:
Ignacio I. Wistuba: study design, study plan, data interpretation, report writing
Carmen Behrens: study design, clinical data collection
Francesca Lombardi: clinical data collection
Susanne Wagner: study plan, experimental design, report writing
Junya Fujimoto: sample processing, pathological data collection
Gabriela Raso: sample processing, pathological data collection
Lorenzo Spaggiari: patient and data collection
Domenico Galetta: patient and data collection
Robyn Riley: sample preparation and assay
Elisha Hughes: statistical analysis
Julia Reid: statistical analysis
Zaina Sangale: pathology review and sample processing
Steven G. Swisher: clinical data collection
Neda Kalhor: pathological data collection and review
Cesar A. Moran: pathological data collection and review
Alexander Gutin: study design and data interpretation
J.S. Lanchbury: study design, data interpretation, report writing
Massimo Barberis: study design, data interpretation, report writing
Edward S. Kim: study design, data interpretation, report writing
Conflicts of Interest: AG, EH, JSL, JR, RR, ZS and SW are employees of Myriad Genetics.
References
- 1.Pisters KM, Evans WK, Azzoli CG, Kris MG, Smith CA, Desch CE, et al. Cancer Care Ontario and American Society of Clinical Oncology adjuvant chemotherapy and adjuvant radiation therapy for stages I–IIIA resectable non-small cell lung cancer guideline. J Clin Oncol. 2007;25:5506–18. doi: 10.1200/JCO.2007.14.1226. [DOI] [PubMed] [Google Scholar]
- 2.Crino L, Weder W, van Meerbeeck J, Felip E ESMO Guidelines Working Group. Early stage and locally advanced (non-metastatic) non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Annals of Oncology. 2010;21 (Suppl 5):v103–15. doi: 10.1093/annonc/mdq207. [DOI] [PubMed] [Google Scholar]
- 3.Pignon JP, Tribodet H, Scagliotti GV, Douillard JY, Shepherd FA, Stephens RJ, et al. Lung adjuvant cisplatin evaluation: a pooled analysis by the LACE Collaborative Group. J Clin Oncol. 2008;26:3552–9. doi: 10.1200/JCO.2007.13.9030. [DOI] [PubMed] [Google Scholar]
- 4.Strauss GM, Herndon JE, 2nd, Maddaus MA, Johnstone DW, Johnson EA, Harpole DH, et al. Adjuvant paclitaxel plus carboplatin compared with observation in stage IB non-small cell lung cancer: CALGB 9633 with the Cancer and Leukemia Group B, Radiation Therapy Oncology Group, and North Central Cancer Treatment Group Study Groups. J Clin Oncol. 2008;26:5043–51. doi: 10.1200/JCO.2008.16.4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsuboi M, Ohira T, Saji H, Miyajima K, Kajiwara N, Uchida O, et al. The present status of postoperative adjuvant chemotherapy for completely resected non-small cell lung cancer. Ann Thorac Cardiovasc Surg. 2007;13:73–7. [PubMed] [Google Scholar]
- 6.Chansky K, Sculier JP, Crowley JJ, Giroux D, Van Meerbeeck J, Goldstraw P. The International Association for the Study of Lung Cancer Staging Project: prognostic factors and pathologic TNM stage in surgically managed non-small cell lung cancer. J Thorac Oncol. 2009;4:792–801. doi: 10.1097/JTO.0b013e3181a7716e. [DOI] [PubMed] [Google Scholar]
- 7.Li Z, Liu H, Li L. Video-assisted thoracoscopic surgery versus open lobectomy for stage I lung cancer: A meta-analysis of long-term outcomes. Exp Therapeutic Med. 2012;3:886–92. doi: 10.3892/etm.2012.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cajipe MD, Chu D, Bakaeen FG, Casal RF, LeMaire SA, Coselli JS, et al. Video-assisted thoracoscopic lobectomy is associated with better perioperative outcomes than open lobectomy in a veteran population. Am J Surg. 2012;204:607–12. doi: 10.1016/j.amjsurg.2012.07.022. [DOI] [PubMed] [Google Scholar]
- 9.Scott WJ, Allen MS, Darling G, Meyers B, Decker PA, Putnam JB, et al. Video-assisted thoracic surgery versus open lobectomy for lung cancer: a secondary analysis of data from the American College of Surgeons Oncology Group Z0030 randomized clinical trial. J Thorac Cardiovasc Surg. 2010;139:976–81. doi: 10.1016/j.jtcvs.2009.11.059. [DOI] [PubMed] [Google Scholar]
- 10.The National Lung Screening Trial Research Team. Reduced Lung-Cancer Mortality with Low-Dose Computed Tomographic Screening. N Engl J Med. 2011;365:395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsubura D, Morikawa T, Goto A, Nakajima J, Fukayama M, Niki T. Subepithelial myofibroblast in lung adenocarcinoma: a histological indicator of excellent prognosis. Mod Path. 2009;22:776–85. doi: 10.1038/modpathol.2009.27. [DOI] [PubMed] [Google Scholar]
- 12.Kadara H, Behrens C, Yuan P, Solis L, Liu D, Gu X, et al. A five-gene and corresponding protein signature for stage-I lung adenocarcinoma prognosis. Clin Cancer Res. 2010;17:1490–501. doi: 10.1158/1078-0432.CCR-10-2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graziano SL, Gamble GP, Newman NB, Abbott LZ, Rooney M, Mookherjee S, et al. Prognostic significance of K-ras codon 12 mutations in patients with resected stage I and II non-small-cell lung cancer. J Clin Oncol. 1999;17:668–75. doi: 10.1200/JCO.1999.17.2.668. [DOI] [PubMed] [Google Scholar]
- 14.Subramanian J, Simon R. Gene expression-based prognostic signatures in lung cancer: ready for clinical use? J Natl Cancer Inst. 2010;102:464–74. doi: 10.1093/jnci/djq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kratz JR, Jablons DM. Genomic prognostic models in early-stage lung cancer. Clin Lung Cancer. 2009;10:151–7. doi: 10.3816/CLC.2009.n.021. [DOI] [PubMed] [Google Scholar]
- 16.Kratz JR, He J, Van Den Eeden SK, Zhu ZH, Gao W, Pham PT, et al. A practical molecular assay to predict survival in resected non-squamous, non-small cell lung cancer: development and international validation studies. Lancet. 2012;379:823–32. doi: 10.1016/S0140-6736(11)61941-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kratz JR, Van Den Eeden SK, He J, Jablons DM, Mann MJ. A prognostic assay to identify patients at high risk of mortality despite small, node-negative lung tumors. JAMA. 2012;308:1629–31. doi: 10.1001/jama.2012.13551. [DOI] [PubMed] [Google Scholar]
- 18.Beresford MJ, Wilson GD, Makris A. Measuring proliferation in breast cancer: practicalities and applications. Breast Cancer Res. 2006;8:216–27. doi: 10.1186/bcr1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mosley JD, Keri RA. Cell cycle correlated genes dictate the prognostic power of breast cancer gene lists. BMC Medical Genomics. 2008;1:11–25. doi: 10.1186/1755-8794-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Starmans MHW, Krishnapuram B, Steck H, Horlings H, Nuyten DS, van de Vijver MJ, et al. Robust prognostic value of a knowledge-based proliferation signature across large patient microarray studies spanning different cancer types. Br J Cancer. 2008;99:1884–90. doi: 10.1038/sj.bjc.6604746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wirapati P, Sotiriou C, Kunkel S, Farmer P, Pradervand S, Haibe-Kains B, et al. Meta-analysis of gene expression profiles in breast cancer: toward a unified understanding of breast cancer subtyping and prognosis signatures. Breast Cancer Res. 2008;10:R65–76. doi: 10.1186/bcr2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pence JC, Kerns BJ, Dodge RK, Iglehart JD. Prognostic significance of the proliferation index in surgically resected non-small-cell lung cancer. Arch Surg. 1993;128:1382–90. doi: 10.1001/archsurg.1993.01420240090017. [DOI] [PubMed] [Google Scholar]
- 23.Hashimoto H, Ozeki Y, Sato M, Obara K, Matsutani N, Nakagishi Y, et al. Significance of thymidylate synthase gene expression level in patients with adenocarcinoma of the lung. Cancer. 2006;106:1595–601. doi: 10.1002/cncr.21777. [DOI] [PubMed] [Google Scholar]
- 24.Corson TW, Zhu QC, Lau SK, Shepherd FA, Tsao MS, Gallie BL. KIF14 messenger RNA expression is independently prognostic for outcome in lung cancer. Clin Cancer Res. 2007;13:3229–34. doi: 10.1158/1078-0432.CCR-07-0393. [DOI] [PubMed] [Google Scholar]
- 25.Rosell R, Skrzypski M, Jassem E, Taron M, Bartolucci R, Sanchez JJ, et al. BRCA1: A novel prognostic factor in resectable non-small-cell lung cancer. PLoS One. 2007;2:e1129. doi: 10.1371/journal.pone.0001129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shedden K, Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, Gerald WL, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008;14:822–7. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park YY, Park ES, Kim SB, Kim SC, Sohn BH, Chu IS, et al. Development and validation of a prognostic gene-expression signature in lung adenocarcinoma. PLoS One. 2012;7:e44225. doi: 10.1371/journal.pone.0044225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cuzick J, Berney DM, Fisher G, Mesher D, Møller H, Reid JE, et al. Prognostic value of a cell cycle progression signature for prostate cancer death in a conservatively managed needle biopsy cohort. Br J Cancer. 2012;106:1095–9. doi: 10.1038/bjc.2012.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cuzick J, Swanson GP, Fisher G, Brothman AR, Berney DM, Reid JE, et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes for recurrence and death from prostate cancer: A retrospective study in two cohorts. Lancet Oncol. 2011;12:245–55. doi: 10.1016/S1470-2045(10)70295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okayama H, Kohno T, Ishii Y, Shimada Y, Shiraishi K, Iwakawa R, et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinoma. Cancer Res. 2012;72:100–11. doi: 10.1158/0008-5472.CAN-11-1403. [DOI] [PubMed] [Google Scholar]
- 31.Zhang MJ, Klein JP. Confidence bands for the difference of two survival curves under the proportional hazards model. Lifetime Data Anal. 2001;7:243–54. doi: 10.1023/a:1011636407958. [DOI] [PubMed] [Google Scholar]
- 32.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–26. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 33.Buyse M, Loi S, van’t Veer L, Viale G, Delorenzi M, Glas AM, et al. Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J Natl Cancer Inst. 2006;98:1183–92. doi: 10.1093/jnci/djj329. [DOI] [PubMed] [Google Scholar]
- 34.Burke L, Flieder DB, Guinee DG, Brambilla E, Freedman AN, Bennett WP, et al. Prognostic implications of molecular and immunohistochemical profiles of the Rb and p53 cell cycle regulatory pathways in primary non-small cell lung carcinoma. Clin Cancer Research. 2005;11:232–41. [PubMed] [Google Scholar]
- 35.SEER Cancer Statistics – Review. National Cancer Institute; Bethesda, MD: 1995–2008. http://seer.cancer.gov/csr/1975-2008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.